Abstract
Background
Streptococcus suis is an emerging zoonotic pathogen and is the leading cause of bacterial meningitis in adults in Vietnam. Systematic data on the antimicrobial susceptibility profiles of S. suis strains isolated from human cases are lacking. We studied antimicrobial resistance and associated resistance determinants in S. suis isolated from patients with meningitis in southern Vietnam.
Methods
S. suis strains isolated between 1997 and 2008 were investigated for their susceptibility to six antimicrobial agents. Strains were screened for the presence and expression of tetracycline and erythromycin resistance determinants and the association of tet(M) genes with Tn916- like transposons. The localization of tetracycline resistance gene tet(L) was determined by pulse field gel electrophoresis and Southern blotting.
Results
We observed a significant increase in resistance to tetracycline and chloramphenicol, which was concurrent with an increase in multi-drug resistance. In tetracycline resistance strains, we identified tet(M), tet(O), tet(W) and tet(L) and confirmed their expression. All tet(M) genes were associated with a Tn916-like transposon. The co-expression of tet(L) and other tetracycline resistance gene(s) encoding for ribosomal protection protein(s) was only detected in strains with a minimum inhibitory concentration (MIC) of tetracycline of ≥ 64 mg/L
Conclusions
We demonstrated that multi-drug resistance in S. suis causing disease in humans in southern Vietnam has increased over the 11-year period studied. We report the presence and expression of tet(L) in S. suis strains and our data suggest that co-expression of multiple genes encoding distinct mechanism is required for an MIC ≥ 64 mg/L to tetracycline.
Background
Streptococcus suis is an emerging zoonotic pathogen associated with pigs and can cause severe systemic infections in humans. Up to date approximately 800 human S. suis infections have been reported from over twenty countries [1]. The most noticeable incident was a single outbreak in China affecting 215 individuals, of whom 38 died [2]. S. suis serotype 2 is the most common serotype associated with human disease [3] and is the most common cause of acute bacterial meningitis in adults in Vietnam [1,4,5].
Antimicrobial susceptibility profiles and the corresponding resistance determinants of S. suis have been reported in strains isolated from pigs, but there are only limited data from strains isolated from human patients [6-12]. Resistance to tetracycline and macrolide-lincosamide- streptogramin B (MLSB) has been widely reported in S. suis strains isolated from pigs in Asia, Europe and North America [8-10,12-15]. In contrast, resistance to chloramphenicol is uncommon [8,14].
The gene erm(B), which encodes erythromycin ribosomal methylase, is associated with resistance to MLSB, and has been identified in a large number of S. suis strains from Hongkong and Belgium [7,9]. Tetracycline resistance in S. suis was mainly associated with tet(M), tet(O), tet(W) and tet(O/W/32/O); yet, efflux proteins encoded by tet(L) and tet(K), which also determine tetracyline resistance, have not been reported [7,8,10,11,15-19]. The co-existence of tet(M) and tet(O) genes in S. suis serotype 2 has been detected in limited number of strains isolated from humans and pigs in China and Italy [7,10,11].
We have previously shown that 79/95 (83.2%) S. suis serotype 2 strains isolated from adults with meningitis in Vietnam were resistant to tetracycline, and 19/94 (20.2%) were resistant to erythromycin [4]. We have also described the concurrent presence of tet(L), tet(M), tet(O) and erm(B) genes in one S. suis serotype 2 strain isolated from a Vietnamese patient [20]. However, longitudinal data of antimicrobial resistance over many years and the associated resistance determinants of a large number of S. suis strains isolated from human patients have not been reported. Here we describe the antimicrobial resistance pattern, trends and the genetic determinants of tetracycline and erythromycin resistance in S. suis strains isolated from adult patients with meningitis in 12 consecutive years (1997-2008), in southern Vietnam.
Methods
Bacterial strains
All strains used in this study were obtained from samples which were collected for diagnostic purposes as part of standard care, at The Hospital for Tropical Diseases (HTD) in Ho Chi Minh City, Vietnam. The study protocol was approved by the ethical committee of the HTD (CS/NĐ/09/13). A total of 175 non-duplicate S. suis strains, isolated from the cerebral spinal fluid (CSF) of adult patients with meningitis admitted to the HTD between March 1997 and November 2008, were investigated. Bacterial culture, identification and serotyping of S. suis was performed as previously described [4]. All strains, with the exception of four strains belong to serotype 14 (n = 3) and serotype 16 (n = 1) [21], were of serotype 2.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by assessing the minimum inhibitory concentration (MIC) for all isolates using E-test (AB Biodisk, Sweden) according to the manufacturer recommendations. The antimicrobials tested were penicillin, ceftriaxone, vancomycin, chloramphenicol, erythromycin and tetracycline. Antimicrobial resistance was assessed using breakpoints recommended by the Clinical and Laboratory Standard Institute (CLSI) guidelines 2008 (M100-S18). There are currently no breakpoints recommended for S. suis, therefore, breakpoints for viridans group Streptococci were used (Table 1). Streptococcus pneumoniae strain ATCC 49619 was used for quality control purpose. Erythromycin resistance phenotypes were identified using the triple disk diffusion test as described [22].
Table 1.
Distribution of minimal inhibitory concentrations of antimicrobial agents tested in Streptococcus suis strains isolated from patients with meningitis
| Antimicrobial Agent | Minimal Inhibitory Concentrations | ||||
|---|---|---|---|---|---|
| Breakpoint* (mg/L) | Range (mg/L) | MIC50# (mg/L) | MIC90# (mg/L) | Resistant strains (%) | |
| Tetracycline | S ≤ 2; R ≥ 8 | 0.125-->256 | 24 | 32 | 88.6 |
| Erythromycin | S ≤ 0.25; R ≥ 1 | 0.016-- >256 | 0.064 | >256 | 22.2 |
| Chloramphenicol | S ≤ 4; R ≥ 16 | 0.5-96 | 3 | 4 | 8.6@ |
| Penicillin | S ≤ 0.12; R ≥ 4 | 0.012-0.064 | 0.032 | 0.047 | 0 |
| Ceftriaxone | S ≤ 0.5 | 0.025-0.38 | 0.094 | 0.125 | 0 |
| Vancomycin | S ≤ 1 | 0.25-0.5 | 0.38 | 0.5 | 0 |
*: Breakpoints are based on equivalent CLSI breakpoints for Streptococcus spp. viridans group
#: MIC50, MIC90 are the MIC values inhibiting growth of 50% and 90% of tested isolates, respectively.
@: Including the intermediate resistant isolates
Detection of erythromycin and tetracycline resistance determinants
Genomic DNA was extracted using DNEasy tissue kit (Qiagen, United Kingdom). The erm(A), erm(B), mef(A), tet(M), tet(O), tet(L) and tet(K) genes were detected by multiplex PCR as previously described [23]. The presence of tet(W) and mosaic gene tet(O/W/32/O) was screened using primers as described in Table 2. DNA sequencing of PCR amplicons was performed to confirm the presence of the full length tet(W) gene using primers tet(W)-F, tetO-2-F, tetO-2-R and tet(W)-R (Table 2).
Table 2.
Primers used to amplify the tetracycline and macrolide resistance determinants and for sequencing purpose
| Primer | Sequence (5' - 3') | Product size | Position in coding sequence | Reference |
|---|---|---|---|---|
| erm(A)-F | CCCGAAAAATACGCAAAATTTCAT | 590 | 15-38 | [23] |
| erm(A)-R | CCCTGTTTACCCATTTATAAACG | 604-582 | ||
| erm(B)-F | TGGTATTCCAAATGCGTAATG | 745 | 203-183 | |
| erm(B)-R | CTGTGGTATGGCGGGTAAGT | 541-522 | ||
| mef(A)-F* | CAATATGGGCAGGGCAAG | 317 | 38-55 | |
| mef(A)-R* | AAGCTGTTCCAATGCTACGG | 352-333 | ||
| tet(M)-F | GTGGACAAAGGTACAACGAG | 406 | 106-125 | |
| tet(M)-R | CGGTAAAGTTCGTCACACAC | 511-492 | ||
| tet(O)- F | AACTTAGGCATTCTGGCTCAC | 515 | 13-43 | |
| tet(O)- R | TCCCACTGTTCCATATCGTCA | 527-507 | ||
| tet(K)-F | GATCAATTGTAGCTTTAGGTGAAGG | 155 | 344-368 | |
| tet(K)-R | TTTTGTTGATTTACCAGGTACCATT | 498-474 | ||
| tet(L)-F | TGGTGGAATGATAGCCCATT | 229 | 384-403 | |
| tet(L)-R | CAGGAATGACAGCACGCTAA | 612-593 | ||
| 16S-rDNA-F | GAGTACGACCGCAAGGTTGA | 100 | 886-905 | |
| 16S-rDNA -R | CTGGTAAGGTTCTTCGCGTTG | 985-964 | ||
| ss-Tn916-1 | GCCATGACCTATCTTATA | 476 | 16083-16100 | [24] |
| ss-Tn916-2 | CTAGATTGCGTCCAA | 16559-16545 | ||
| tet(32)For | GAACCAGATGCTGCTCTT | 620 | 619-637 | [28] |
| Tet(32)Rev | CATAGCCACGCCCACATGAT | 1239-1220 | ||
| SScps2J-F | CAAACGCAAGGAATTACGGTATC | 236 | 209-231 | [29,30] |
| SScps2J-R | CATTTCCTAAGTCTCGCACC | 445-426 | ||
| tet(L)Ng-F | TCGTTAGCGTGCTGTCATTC | 698 | 680-700 | [31] |
| tet(L)-R-pDG364 | CTTAGAAATCCCTTTGAGAAT | 1378-1358 | This study | |
| tet(W)- F | TTGGAATTCTTGCCCATGTAGACGC | 1872 | 18-42 | This study |
| tet(W)- R | TTGTCCAGGCGGTTGTTTGGAC | 1889-1868 | ||
| tet(W)-F-HN | GGTGCAGTTGGAGGTTGTTT | 410 | 1179-1198 | |
| tet(W)-R-HN | CCTTCAATGCCTGTTCCAAT | 1569-1588 | ||
| tet(O)-F-pDG364 | ATGAAAATAATTAACTTAGG | 1920 | 1-19 | |
| tet(O)-R-pDG364 | TTAAGCTAACTTGTGGAACA | 1920-1901 | ||
| ss-tet(M)-whole-F | ACAGACAAAGAACTATCCTTAATG | 2500 | 419-396 | |
| ss-tet(M)-whole-R | GTACCCAGTTTAAGAATACCTTTATC | 161-136 | ||
| Additional primers used for sequencing | ||||
| tet(O)-2-F | TTCAAGACGCCTCCCTGTTC | 679-698 | This study | |
| tet(O)-2-R | ATTTGGCGGGACTTCTATGTGG | 1318-1296 | ||
| ss-tet(M)-Fseq1 | GTTAAATCACTACGATAT | 1763-1745 | ||
| ss-tet(M)-Rseq1 | ATAGTGTTCTTGGAGATA | 906-929 | ||
| ss-tet(M)-Fseq2 | GTATAATTTCATGTGTCG | 1162-1144 | ||
| ss-tet(M)-Rseq2 | AGATGGCGTACAAGCACA | 305-323 | ||
| ss-PAI-P8-tet(M)-R | GCCCTTTTGGGTTTTTGAAT | -33- -14 | ||
| ss-PAItetM-P9over F | GGGAATCCCCATTTTCCTAA | 366-347 | ||
*: These primers were named mef(A/E) in the referenced publication.
S. suis strain BM407, which contains the genes tet(M), tet(O), tet(L) and erm(B), and S. suis strain BM331 containing the full length tet(W) gene, were used as positive control strains [20]. The presence of Tn916-like conjugative transposons was screened for in all tet(M) positive strains by amplification of the xis-Tn gene of Tn916, as described elsewhere [24]. The association of the tet(M) gene with Tn916 was demonstrated in xis-Tn and tet(M) positive strains by an additional PCR using primers Tn916-2 and P8-tet(M)-R (Table 2).
Detection of expression of tetracycline resistance genes
The expression of tetracycline resistance genes was determined in all S. suis strains possessing multiple tetracycline resistance determinants according to techniques described previously with modification [17]. A tetracycline disk (30 ug) was placed on a Mueller-Hinton agar plate inoculated with a 0.5 McFarland suspension of the test S. suis strain. Colonies surrounding the zone of inhibition were harvested for RNA extraction. cDNA was synthesized using random hexamer primers and the subsequent products were used as templates for multiplex PCR for detection of tetracycline resistance genes tet(M), tet(O), tet(L), and tet(K) (Table 1). RT-PCR of cps2J or 16SrRNA gene products was used as internal control.
Pulse Field Gel Electrophoresis (PFGE) and Southern blot to detect the location of tet(L) in S. suis isolates
Agarose plugs were prepared and solidified plugs were processed as described [25]. SmaI digested genomic DNA was electrophoresed by PFGE in a CHEF mapper (BioRad, Vietnam). The separation of plasmid DNA and chromosomal DNA was achieved using the conditions suitable for separation of DNA fragments between 1.2 Kb and 100 Kb. S. suis strain BM407, which was shown previously to contain one plasmid and the tet(L) gene, located on the chromosome, was used as positive control [20]. Ten nanograms (10 ng) of PCR amplicon tet(L) (698 bp) was included as a positive control for Southern blotting in each gel. DNA was transfered to the nitrocellulose membranes using a vacuum blotter (BioRad, Vietnam).
The probe for detection of the tet(L) gene after Southern blotting was prepared using the same PCR amplicon, which was labeled using the ECL direct nucleic acid labeling and detection kit (GE Healthcare, HCMC, Vietnam).
Statistical analysis
We tested for a linear time trend in differential antimicrobial resistance rates using logistic regression with the antimicrobial susceptibility of the strain as the outcome and the calendar year of the strain collection as the covariate. As only one strain was collected in 1997, it was excluded from all statistical analyses. Strains with intermediate resistance were considered resistant for the purpose of these analyses. The MIC of tetracycline was compared between tetracycline resistant S. suis groups. Four groups of tetracycline resistant strains were defined as follows: strains carrying tet(M); strains carrying tet(O); strains carrying tet(M) and tet(O); and strains carrying tet(M) and tet(L) or tet(M), tet(O) and tet(L). The sole strain which contained tet(M) and tet(L) was added to the latter group prior to the analysis. We used a Kruskal-Wallis test for the overall comparison of the four groups and then performed pair-wise comparisons using Wilcoxon rank-sum tests with Holm correction for multiple testing. All reported p-values correspond to two-sided tests and analyses were performed with R 2.8.1 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Antimicrobial susceptibility profiles and macrolide resistance phenotype
All 175 S. suis strains were fully susceptible to penicillin, ceftriaxone and vancomycin (Table 1). Fourteen strains were sensitive to all 6 antibiotics tested. A total of 159/175 (90.9%) strains were resistant to tetracycline and 39/175 (22.2%) strains were resistant to erythromycin. Fifteen (8.6%) strains were resistant to chloramphenicol, including those demonstrating intermediate resistance (Figure 1). The macrolide resistance phenotype was identified using triple-disk tests in 38/39 erythromycin resistant strains. Of these, 37 (97.4%) strains demonstrated a cMLSB phenotype and the remaining strain expressed the iMLSB -A phenotype.
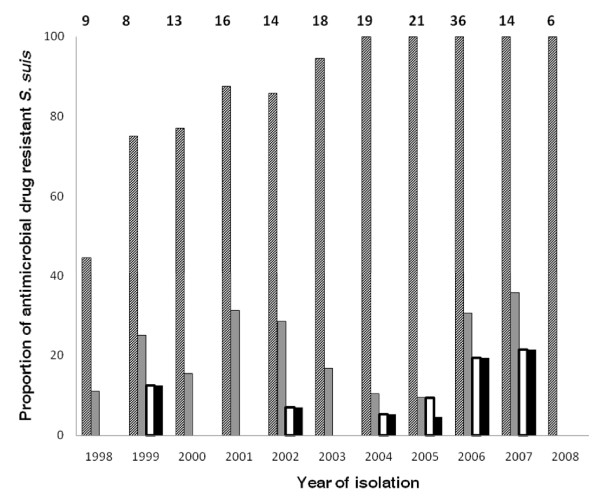
Figure 1.
Proportional distribution of antimicrobial resistant S. suis strains between 1998 and 2008. Streptoccoccus suis strains were isolated from patients with acute bacterial meningitis admitted to the Hospital for Tropical Diseases between 1998 and 2008. The numbers above each column correspond to the total number of strains isolated in each year (the lone isolate from 1997 is not included. Diagonal, grey and white bars represent the proportion of strains resistant to tetracycline, erythromycin and chloramphenicol, respectively. Black bars represent the proportion of MDR strains (resistant to tetracycline, erythromycin and chloramphenicol).
Resistance to tetracycline significantly increased over time (OR = 1.77 [95% CI 1.36-2.30] per year, p < 0.001). The proportion of S. suis strains demonstrating intermediate or full resistance to tetracycline was 80.8% (61/78 strains) in the period 1998-2003 and 99% (95/96 strains) in the period 2004-2008 (Figure 1). A significant increase was also observed in chloramphenicol resistance (OR = 1.36 [95% CI 1.05-1.76] per year, p = 0.02) which rose from 2.5% (2/78 strains) in 1998-2003 to 13% (12/96 strains) in 2004-2008. In contrast, there was no significant increase in erythromycin resistance observed over 11 years (OR = 1.04 (95% CI 0.91-1.19) per year; p = 0.56) and the proportion of erythromycin resistant strains was the same (22%) in both periods of time (Figure 1). Multi-drug resistance (MDR), defined as resistance to tetracycline, erythromycin and chloramphenicol, significantly increased over time (OR = 1.36 [95% CI = 1.04-1.77] per year, p = 0.02). Two (2.5%) and 12 MDR strains (12.5%) were isolated in 1998-2003 and in 2004-2008, respectively (Figure 1).
Tetracycline and erythromycin resistance determinants
We screened for the presence of tetracycline and erythromycin resistance determinants in 169 isolates, including two S. suis serotype 14 and one S. suis serotype 16 strains. The tetracycline resistance determinant tet(M) was successfully amplified in 129/153 (84.3%) tetracycline resistant isolates. The presence of the xis-Tn gene of Tn916-like transposons and its association with tet(M) was further screened by PCRs in strains that contained tet(M). All tet(M) positive S. suis strains produced amplicons for both the xis-Tn gene and the DNA fragments linking xis-Tn and tet(M) genes. The tet(O) gene was found in 33 (21.6%) strains and tet(L) was detected in 5 (3.3%) strains. The three genes tet(M), tet(O) and tet(L) were concomitantly amplified in 4 (2.6%) strains. We detected several different combinations of tetracycline resistance genes in individual strains. The genes tet(M) and tet(L) or tet(M) and tet(O) were found in 6 (3.9%) isolates, whilst single tetracycline resistance gene tet(M) or tet(O) were present in 139 (90.8%) strains. We identified 2/92 tested strains containing the full-length tet(W) gene.
Resistance determinant erm(B) was detected in 36/38 (94.7%) erythromycin resistant strains. All possible combinations of the erm(B) gene with tetracycline resistance genes tet(M), tet (O) and tet(L) were found. We were unable to amplify amplicons of the genes tet(K), tet(O/W/32/O), erm(A) and mef(A) in any of the strains studied. Statistical analysis showed no association between tetracycline and erythromycin resistance of S. suis strains (p = 0.53).
Defining the genomic location of tet(L) in S. suis
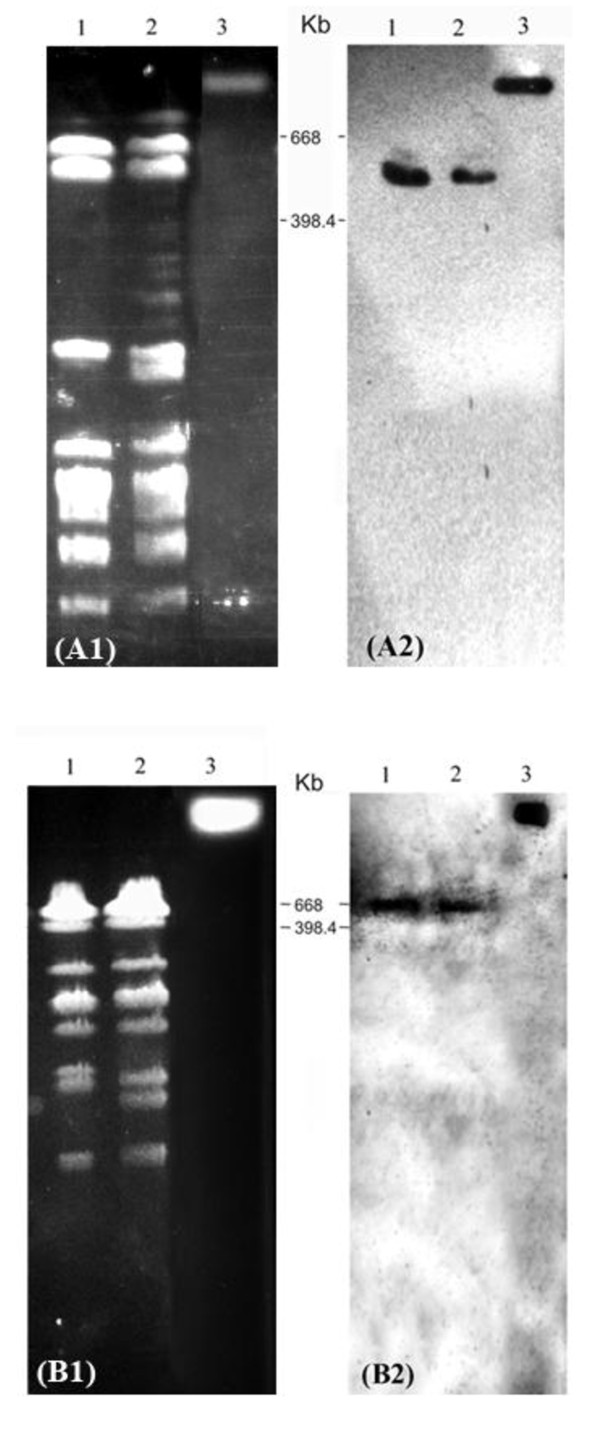
The gene tet(L) was previously found on a conjugative element integrated into the chromosome of S. suis strain BM407 [20]. However, tet(L) is often found on small transmissible plasmids [26]. Using PFGE and Southern blotting we investigated whether tet(L) genes of the other tet(L) PCR positive S. suis strains were also located on the chromosomes or on replicons additional to chromosomes. Figure 2 shows hybridization patterns for the presence of tet(L) after Southern blotting of SmaI digested DNA of four S. suis strains BM308, BM407, EN031 and EN241. Only strain BM407 was found to contain plasmid DNA (data not shown), which confirms a previous observation that this strain contains a 24,579 bp-pBM407 plasmid with unknown function [20]. Southern hybridization confirmed the location of tet(L) on fragments between 398 and 668 Kb in size on the chromosomes of studied strains.
Figure 2.
Chromosomal location of tet(L) genes in S. suis isolates. (A1, B1) PFGE patterns of SmaI digested genomic DNA of four S. suis serotype 2 strains, (A2, B2): Southern blot hybridization of SmaI digested genomic DNA of four strains with 698 bp- tet(L) probe. (A) Lane 1: S. suis BM308; lane 2: S. suis BM407; (B) lane 1: S. suis EN031; lane 2: S. suis EN241. (A, B) Lane 3: 698 bp-tet(L) PCR amplicons, which was added to the well of PFGE gel one hour prior to completion of the PFGE running programme. Molecular weights of marker bands, consisting of XbaI digested genomic DNA of Salmonella serotype Braenderup strain (H9812), spanning the region containing the positive signals of tet(L), are indicated.
Expression of tetracycline resistance determinants
The MICs of tetracycline differed significantly between the 4 groups of strains containing tetracycline resistance genes (p < 0.001, Kruskal-Wallis test). Pair-wise comparisons revealed that the MICs to tetracycline for strains containing tet(M) and tet(L) or tet(M), tet(O) and tet(L) genes was higher than for strains possessing both tet(M) and tet(O) genes, and for strains containing only tet(M) or tet(O) (Table 3).
Table 3.
Groups of S. suis strains with different combinations of tetracycline genes and respective minimal inhibitory concentrations.
| Number of isolates | Group | Tetracycline resistance genes | MIC50* (mg/L) | Range of MIC (mg/L) | p-values# |
|---|---|---|---|---|---|
| 5 | 1 | tet(M), tet(O) and tet(L) or tet(M) and tet(L) | 64 | 64 - 256 | 0.01 (group 1 vs. 2) |
| <0.001 (group 1 vs. 3) | |||||
| 0.002 (group 1 vs. 4) | |||||
| 0.002 (group 2 vs. 3) | |||||
| 0.03 (group 2 vs. 4) | |||||
| 0.08 (group 3 vs. 4) | |||||
| 8 | 2 | tet(M) and tet(O) | 32 | 24 - 48 | |
| 116 | 3 | tet(M) | 24 | 3 - 48 | |
| 22 | 4 | tet(O) | 24 | 16 - 32 | |
* MIC50 is the MIC value inhibiting growth of 50% of tested isolates
# Pair-wise comparisons based on Wilcoxon rank-sum tests with Holm correction for multiple testing
Among the four S. suis strains (BM308, BM407, EN031 and EN241) that possessed the three tetracycline resistance genes tet(M), tet(O), and tet(L), only strain BM407 had a MIC to tetracycline ≥256 mg/L, whilst the MIC was 64 mg/L for the other three strains. We attempted to confirm the co-transcription of the three tetracycline resistance determinants in these four strains. The transcription of all 3 genes simultaneously was only detected in strain BM407. We could not detect the expression of tet(M) in one strain and mRNA from tet(O) was not detected in the other two strains (data not shown). However, we detected the presence of mRNAs from tet(M) and tet(L) in the single strain which contained both these determinants, which had a MIC of 64 mg/L to tetracycline.
Discussion
Invasive human infections by S. suis are becoming increasingly reported in Asia. We have observed an increase in the number of infections caused by S. suis serotype 2 at the Hospital for Tropical Diseases in HCMC since 1998. This increase could be due to increased awareness and/or a true increase in the incidence of disease. Here we have shown an increase in a tetracycline and chloramphenicol resistance over an eleven- year period. This suggests that continuous surveillance for antimicrobial resistance in S. suis is important to guide current and future treatment of human and porcine disease caused by S. suis.
Our data show that all S. suis strains isolated from adult patients over a twelve year period in Vietnam were sensitive to penicillin. Whilst S. suis resistant to penicillin has been reported in isolates from pigs in Denmark, Poland and Portugal [8], it does not appear to be emerging in Vietnam.
We did, however, demonstrate a high proportion of tetracycline resistance (91%). Tetracycline resistance has been reported previously in S. suis isolated from pig in various location in Asia and Europe [7,10-12]. Since S. suis infection in human is associated with exposure to pigs or contaminated pork [1], the increase in tetracycline resistance in strains isolated from humans may be related to tetracycline usage in animal production for prophylaxis or therapy leading to positive selection of resistant strains. Although data on antimicrobial drug use in animal husbandry in Vietnam is currently unavailable, we observed a tetracycline resistance rate of 100% in 45 S. suis serotype 2 strains isolated from a representative sample of healthy slaughterhouse pigs in southern Vietnam in 2006 and 2007, suggesting that current tetracycline resistance rates in pig carriage strains are extremely high (Hoa et al, in preparation).
Resistance to tetracycline was encoded by multiple determinants including tet(L) in this strain collection. We were able to demonstrate that the tet(L) gene was located on the chromosomes of four strains and the tet(L) transcription was detected in all five S. suis strains with tet(L) amplicons positive. The presence of the gene tet(L) has, until now, only been described in the genome sequence of S. suis serotype 2 strain BM407, which was isolated from a patient with meningitis in Vietnam [20]. The most frequently identified tetracycline resistance gene was tet(M) and its association with the presence of Tn916 like elements was confirmed in all tet(M) positive strains. Mobile genetic elements, including transposons and integrative elements, are common in Streptococci such as S. suis [11]. The colonization of S. suis in the upper respiratory and gastrointestinal tracts of healthy pigs is well known, and the transfer of mobile DNA in such locations is common [20].
In our study, strains carrying multiple tetracycline resistance genes were likely to associate with higher MIC to tetracycline than those carrying single or two resistance genes. The difference in MIC to tetracycline in four strains, all harbouring tet(M), tet(O), and tet(L), was likely due to the simultaneous expression of these genes. The proteins encoded by tet(M) and tet(O) prevent tetracycline binding to its target on the bacterial ribosome, whilst tet(L) encodes a membrane-associated protein, which facilitates tetracycline export from the cell [27]. We surmise that the simultaneous expression of tetracycline resistance genes encoding for efflux protein and ribosomal protection proteins in the same S. suis strain results in an elevated MIC to tetracycline.
Erythromycin resistance has been reported in S. suis strains isolated from human infections in Hong Kong, with a proportion of resistant strains similar to that observed in our study (21.2%) [7]. The proportions of resistance to erythromycin of S. suis strains isolated from pig are higher in other countries including Denmark (29.1%), the United Kingdom (36%), the Netherlands (35%) and Poland (30.6%), France (64.6%), China (67.2%), Portugal (75%) and Italy (78%) [8,10,12]. We were unable to amplify target sequences for the genes erm(A), erm(B) and mef(A) in two erythromycin resistant strains. These data suggest that other erythromycin resistance determinants may be present in S. suis and requires additional investigations.
The usage of chloramphenicol in agriculture has been banned in Vietnam since 2003. However, other amphenicols (such as florfenicol) are still allowed to use in agriculture and animal husbandry in Vietnam. The increasing proportion of chloramphenicol resistant S. suis strains isolated from humans in Vietnam may be associated with other amphenicols use. It is also possible that the chloramphenicol resistance determinants are under co-selection in strains that are resistant to other antimicrobials currently approved for veterinary use in the prevention and treatment of infections, such as tetracyclines and macrolides. It is noteworthy that all chloramphenicol resistant strains in our study were additionally resistant to tetracycline or erythromycin or both. The genome sequence publication of S. suis strain BM407 described that the resistance determinants erm(B), tet(O), cat, tet(L) and tet(M) are all located within a 40 Kb DNA region of a conjugative mobile element in this strain [20]. This would support the hypothesis of co-selection of antimicrobial resistance determinants.
Conclusion
This study reported an increasing proportion of antimicrobial resistant S. suis isolates from adult patients with meningitis in southern Vietnam over 11 consecutive years. We demonstrated the presence and expression of tet(L) gene in S. suis. Our results imply that the simultaneous expression of the tet(L) gene and additional tetracycline resistance gene(s), encoding for the ribosomal protection protein(s), in S. suis is associated with higher MIC to tetracycline.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NTH designed the experiments, performed data analysis, interpretation and wrote the first draft of the manuscript. TTBC and PHA performed experiments and data analysis. HDTN and NTHM were involved in study design and clinical sample collection. MW performed statistical analysis and contributed in writing the manuscript. JC and SB contributed to data collection and writing of the manuscript. NVVC and TTH involved in study design and data collection. JF and CS contributed to study design, interpretation, coordination, and writing of the manuscript. All authors read and approved the final version of the manuscript.
Author's information
NTH is a microbiologist at OUCRU-HTD HCMC, Wellcome Trust Oversea Programme in Vietnam. Her main research interest is emerging zoonotic pathogens with the current focus on S. suis infection in humans and pigs.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Ngo T Hoa, Email: hoant@oucru.org.
Tran TB Chieu, Email: chieuttb@oucru.org.
Ho DT Nghia, Email: nghiahdt@oucru.org.
Nguyen TH Mai, Email: mainth@oucru.org.
Pham H Anh, Email: anhph@oucru.org.
Marcel Wolbers, Email: mwolbers@oucru.org.
Stephen Baker, Email: sbaker@oucru.org.
James I Campbell, Email: jcampbell@oucru.org.
Nguyen VV Chau, Email: chaunvv@oucru.org.
Tran T Hien, Email: hientt@oucru.org.
Jeremy Farrar, Email: jfarrar@oucru.org.
Constance Schultsz, Email: schultsz@gmail.com.
Acknowledgements
We acknowledge the technical staff of the diagnostic microbiology laboratory for their support in isolation and susceptibility testing. Dr. Thuy Le, Mr. Ngo Thanh Long and Dr. Thai Quoc Hieu are acknowledged for helpful discussion. This work is supported in part by the Wellcome Trust International Travelling Fellowship to N.T.H (064874) and by the Wellcome Trust Major Overseas Programme in Viet Nam.
References
- Wertheim HF, Nghia HD, Taylor W, Schultsz C. Streptococcus suis: an emerging human pathogen. Clin Infect Dis. 2009;48(5):617–625. doi: 10.1086/596763. [DOI] [PubMed] [Google Scholar]
- Yu H, Jing H, Chen Z, Zheng H, Zhu X, Wang H, Wang S, Liu L, Zu R, Luo L. et al. Human Streptococcus suis outbreak, Sichuan, China. Emerg Infect Dis. 2006;12(6):914–920. doi: 10.3201/eid1206.051194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk M, Segura M, Xu J. Streptococcus suis infections in humans: the Chinese experience and the situation in North America. Anim Health Res Rev. 2007;8(1):29–45. doi: 10.1017/S1466252307001247. [DOI] [PubMed] [Google Scholar]
- Mai NT, Hoa NT, Nga TV, Linh le D, Chau TT, Sinh DX, Phu NH, Chuong LV, Diep TS, Campbell J. et al. Streptococcus suis meningitis in adults in Vietnam. Clin Infect Dis. 2008;46(5):659–667. doi: 10.1086/527385. [DOI] [PubMed] [Google Scholar]
- Wertheim HF, Nguyen HN, Taylor W, Lien TT, Ngo HT, Nguyen TQ, Nguyen BN, Nguyen HH, Nguyen HM, Nguyen CT. et al. Streptococcus suis, an important cause of adult bacterial meningitis in northern Vietnam. PLoS One. 2009;4(6):e5973. doi: 10.1371/journal.pone.0005973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Wada A, Ikebe T, Ohnishi M, Mita K, Endo M, Matsuo H, Asatuma Y, Kuramoto S, Sekiguchi H. et al. Characteristics of Streptococcus suis isolated from patients in Japan. Jpn J Infect Dis. 2006;59(6):397–399. [PubMed] [Google Scholar]
- Chu YW, Cheung TK, Chu MY, Tsang VY, Fung JT, Kam KM, Lo JY. Resistance to tetracycline, erythromycin and clindamycin in Streptococcus suis serotype 2 in Hong Kong. Int J Antimicrob Agents. 2009. [DOI] [PubMed]
- Hendriksen RS, Mevius DJ, Schroeter A, Teale C, Jouy E, Butaye P, Franco A, Utinane A, Amado A, Moreno M. et al. Occurrence of antimicrobial resistance among bacterial pathogens and indicator bacteria in pigs in different European countries from year 2002-2004: the ARBAO-II study. Acta Vet Scand. 2008;50:19. doi: 10.1186/1751-0147-50-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel A, Baele M, Devriese LA, Goossens H, Wisselink HJ, Decostere A, Haesebrouck F. Prevalence and mechanism of resistance against macrolides and lincosamides in Streptococcus suis isolates. Vet Microbiol. 2001;83(3):287–297. doi: 10.1016/S0378-1135(01)00426-6. [DOI] [PubMed] [Google Scholar]
- Princivalli MS, Palmieri C, Magi G, Vignaroli C, Manzin A, Camporese A, Barocci S, Magistrali C, Facinelli B. Genetic diversity of Streptococcus suis clinical isolates from pigs and humans in Italy (2003-2007) Euro Surveill. 2009;14(33) doi: 10.2807/ese.14.33.19310-en. [DOI] [PubMed] [Google Scholar]
- Ye C, Bai X, Zhang J, Jing H, Zheng H, Du H, Cui Z, Zhang S, Jin D, Xu Y. et al. Spread of Streptococcus suis sequence type 7, China. Emerg Infect Dis. 2008;14(5):787–791. doi: 10.3201/eid1405.070437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Ning Y, Zhang Z, Song L, Qiu H, Gao H. In vitro antimicrobial susceptibility of Streptococcus suis strains isolated from clinically healthy sows in China. Vet Microbiol. 2008;131(3-4):386–392. doi: 10.1016/j.vetmic.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Cantin M, Harel J, Higgins R, Gottschalk M. Antimicrobial resistance patterns and plasmid profiles of Streptococcus suis isolates. J Vet Diagn Invest. 1992;4(2):170–174. doi: 10.1177/104063879200400209. [DOI] [PubMed] [Google Scholar]
- Takamatsu D, Osaki M, Sekizaki T. Chloramphenicol resistance transposable element TnSs1 of Streptococcus suis, a transposon flanked by IS6-family elements. Plasmid. 2003;49(2):143–151. doi: 10.1016/S0147-619X(02)00149-X. [DOI] [PubMed] [Google Scholar]
- Wasteson Y, Hoie S, Roberts MC. Characterization of antibiotic resistance in Streptococcus suis. Vet Microbiol. 1994;41(1-2):41–49. doi: 10.1016/0378-1135(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Manzin A, Palmieri C, Serra C, Saddi B, Princivalli MS, Loi G, Angioni G, Tiddia F, Varaldo PE, Facinelli B. Streptococcus suis meningitis without history of animal contact, Italy. Emerg Infect Dis. 2008;14(12):1946–1948. doi: 10.3201/eid1412.080679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzotti L, La Gioia F, Dellaglio F, Torriani S. Characterization of tetracycline-resistant Streptococcus thermophilus isolates from Italian soft cheeses. Appl Environ Microbiol. 2009;75(12):4224–4229. doi: 10.1128/AEM.01380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Avial I, Rodriguez-Avial C, Culebras E, Picazo JJ. Distribution of tetracycline resistance genes tet(M), tet(O), tet(L) and tet(K) in blood isolates of viridans group streptococci harbouring erm(B) and mef(A) genes. Susceptibility to quinupristin/dalfopristin and linezolid. Int J Antimicrob Agents. 2003;21(6):536–541. doi: 10.1016/S0924-8579(03)00062-1. [DOI] [PubMed] [Google Scholar]
- Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245(2):195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Holden MT, Hauser H, Sanders M, Ngo TH, Cherevach I, Cronin A, Goodhead I, Mungall K, Quail MA, Price C. et al. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One. 2009;4(7):e6072. doi: 10.1371/journal.pone.0006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghia HD, Hoa NT, Linh le D, Campbell J, Diep TS, Chau NV, Mai NT, Hien TT, Spratt B, Farrar J. et al. Human case of Streptococcus suis serotype 16 infection. Emerg Infect Dis. 2008;14(1):155–157. doi: 10.3201/eid1401.070534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montanari MP, Mingoia M, Giovanetti E, Varaldo PE. Differentiation of resistance phenotypes among erythromycin-resistant Pneumococci. J Clin Microbiol. 2001;39(4):1311–1315. doi: 10.1128/JCM.39.4.1311-1315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra-Kumar S, Lammens C, Piessens J, Goossens H. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother. 2005;49(11):4798–4800. doi: 10.1128/AAC.49.11.4798-4800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerso Y, Pedersen AG, Aarestrup FM. Identification of Tn5397-like and Tn916-like transposons and diversity of the tetracycline resistance gene tet(M) in enterococci from humans, pigs and poultry. J Antimicrob Chemother. 2006;57(5):832–839. doi: 10.1093/jac/dkl069. [DOI] [PubMed] [Google Scholar]
- Berthelot-Herault F, Marois C, Gottschalk M, Kobisch M. Genetic diversity of Streptococcus suis strains isolated from pigs and humans as revealed by pulsed-field gel electrophoresis. J Clin Microbiol. 2002;40(2):615–619. doi: 10.1128/JCM.40.2.615-6192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65(2):232–260. doi: 10.1128/MMBR.65.2.232-260.2001. second page, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Rev. 1996;19(1):1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- Melville CM, Scott KP, Mercer DK, Flint HJ. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob Agents Chemother. 2001;45(11):3246–3249. doi: 10.1128/AAC.45.11.3246-3249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HE, Veenbergen V, van der Velde J, Damman M, Wisselink HJ, Smits MA. The cps genes of Streptococcus suis serotypes 1, 2, and 9: development of rapid serotype-specific PCR assays. J Clin Microbiol. 1999;37(10):3146–3152. doi: 10.1128/jcm.37.10.3146-3152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisselink HJ, Joosten JJ, Smith HE. Multiplex PCR assays for simultaneous detection of six major serotypes and two virulence-associated phenotypes of Streptococcus suis in tonsillar specimens from pigs. J Clin Microbiol. 2002;40(8):2922–2929. doi: 10.1128/JCM.40.8.2922-2929.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15(4):209–215. doi: 10.1006/mcpr.2001.0363. [DOI] [PubMed] [Google Scholar]