Abstract
Background
Genome-wide linkage studies in multiple ethnic populations found chromosome 1q21-q25 was the strongest and most replicable linkage signal in the human chromosome. Studies in Pima Indian, Caucasians and African Americans identified several SNPs in DUSP12 and ATF6, located in chromosome 1q21-q23, were associated with type 2 diabetes.
Methods
We selected 19 single nucleotide polymorphisms (SNPs) that could tag 98% of the SNPs with minor allele frequencies over 0.1 within DUSP12-ATF6 region. These SNPs were genotyped in a total of 3,700 Chinese Han subjects comprising 1,892 type 2 diabetes patients and 1,808 controls with normal glucose regulation.
Results
None of the SNPs and haplotypes showed significant association to type 2 diabetes in our samples. No association between the SNPs and quantitative traits was observed either.
Conclusions
Our data suggests common SNPs within DUSP12-ATF6 locus may not play a major role in glucose metabolism in the Chinese.
Background
Type 2 diabetes is a complex disease caused by both genetic and environmental factors. Although recent genome-wide association studies have identified several novel, possibly causative genes, the contribution of them to disease risk is still very limited [1]. Thus the genetic architecture of type 2 diabetes remained largely unknown. Previous genome-wide linkage studies in multiple ethnic populations, including Caucasians, Chinese and Pima Indian, showed that chromosome 1q21-q25 was the strongest and most replicable linkage signal in the human chromosome [2-9]. Although genome-wide association studies show no strong association signal in this region, whether variants harbored in this region conferred modest effect to the disease are worthy to be analyzed.
Dual specificity phosphatase 12 (DUSP12) and activating transcription factor 6 (ATF6) were two neighbored genes locating on the chromosome 1q21-q23. DUSP12 is a glucokinase - associated protein identified from rat hepatic cDNA library through yeast two-hybrid, using glucokinase as bait. It may participate in glycolysis in the liver and pancreatic beta-cell through dephosphorylation of glucokinase in the cytoplasm [10]. ATF6 is a key sensor of endoplasmic reticulum stress. It activates unfolded protein response through regulating a group of genes encoding molecular chaperones and folding enzymes [11]. Previous studies identified several single nucleotide polymorphisms (SNPs) in this region associated with type 2 diabetes in different populations. Among them, rs2070150 (P145A) was firstly identified to be associated with type 2 diabetes in Pima Indian, while rs4579731, rs3820449 and rs10918215 were reported later in studies focusing on Caucasians and African Americans [12-15]. However, International Type 2 Diabetes 1q Consortium failed to detect any association signal on DUSP12 and ATF6 in a fine mapping study in multiethnic samples [16]. Even though, only 285 East Asian origin samples were included in the previous studies and no one analyzed the association between SNPs from this region and type 2 diabetes in large Asian samples. Therefore, we performed the present study, aiming to test if variants from DUSP12 and ATF6 played a role in the genetic susceptibility of type 2 diabetes in the Chinese.
Methods
Participants
In this case-control study, we recruited 3,700 unrelated Chinese Han subjects, including 1,892 case and 1,808 controls. All the subjects were eastern Chinese Han ancestry, residing in Shanghai and nearby region. In the present study, all case subjects were type 2 diabetes patients selected from Shanghai Diabetes Institute inpatient database. Control subjects were community-based populations enrolled from the Shanghai Diabetes Studies [17]. The inclusion and exclusion criteria for the cases and controls were described previously [17]. Briefly, all cases were type 2 diabetes patients defined according to 1999 WHO criteria (fasting plasma glucose ≥7.0 mmol/l and/or 2-h plasma glucose ≥11.1 mmol/l) and were treated with oral hypoglycemic agents and/or insulin. The control subjects were normal glucose tolerance defined based on fasting plasma glucose <6.1 mmol/l and 2-h plasma glucose <7.8 mmol/l. This study was approved by the institutional review board of Shanghai Jiao Tong University Affiliated Sixth People's Hospital. Written informed consent was obtained from each participant.
Clinical measurement
All subjects underwent detailed clinical investigations, as described previously [17]. Briefly, anthropometric parameters such as height, weight, waist and hip circumference (for the control subjects only) were measured. For the control subjects, blood samples were obtained at 0 and 120 min during the oral glucose tolerance tests (OGTTs) to measure plasma glucose and serum insulin levels. Lipid profiles such as total cholesterol and triglyceride were also obtained. Insulin resistance and pancreatic β-cell function were assessed by homeostasis model assessment (HOMA) [18]. HOMA-IR = fasting insulin × fasting plasma glucose ÷ 22.5, HOMA-B = 20 × fasting insulin ÷ (fasting plasma glucose ÷ 3.5).
SNPs selection, genotyping and quality control
We selected 19 SNPs that spanning 197 kb of DUPS12 and ATF6 region, from 10 kb 5' upstream the DUSP12 to 2 kb 3' downstream the ATF6. These SNPs could tag 98% of the SNPs with MAF over 0.1 derived from HapMap Phase III Chinese Han database under the threshold of r2 ≥0.7. Among them, 7 SNPs located in the coding region. The SNPs previously reported were either directly genotyped or in linkage disequilibrium (LD) with genotyped SNPs. All the SNPs were genotyped using Sequenom's MassARRAY iPLEX system (MassARRAY Compact Analyzer, Sequenom, San Diego, CA, USA). The key quality control requirements were: 1) sample call rates ≥75%; 2) SNP call rate ≥85%; 3) less than two discrepant genotypes of 100 duplicate samples; and 4) Hardy-Weinberg equilibrium test ≥0.05 in controls and cases respectively. After the quality control procedures of the genotypes, 71 individuals were excluded. And one SNP (rs3767635) failed Hardy-Weinberg equilibrium test. The average call rate for the remaining 18 SNPs was 97.5%, and the average concordance rate based on 100 duplicate comparisons for each SNP was 99.4%. Detailed information of the call rates and concordance rates for the SNPs was shown in the Additional file 1 Table S1.
Statistical analyses
Observed genotypes were tested for fit to the expectation of Hardy-Weinberg equilibrium using χ2 test. Pairwise LD was estimated from the combined data of cases and controls calulating |D'| and r2 using Haploview (version 4.1) http://www.broadinstitute.org/haploview/haploview[19]. Haplotype block structure was determined using confidence interval algorithm [20] and haplotype frequencies were estimated by Expectation-Maximization algorithm [21] using Haploview (v 4.1). Allele, genotype and haplotype frequencies for cases and controls were compared using χ2 test or Fisher's exact test. Odds ratios (ORs) with 95% confidence intervals (CIs) were presented. The genotype - disease association analyses were performed under the additive model adjusting age, gender and BMI as confounding factors by logistic regression. Quantitative traits with skewed distribution were natural logarithmically transformed to approximate univariate normality. Quantitative traits were analyzed under an additive genetic model by linear regression adjusted for age, sex, and BMI. All statistical analyses were performed by SAS (version 8.0; SAS Institute Inc., Cary, NC, USA) unless specified otherwise. A two-tailed P value <0.05 was considered significant. The allele frequencies in HapMap populations and statistic power of the SNPs were shown in the Additional file 2 Table S2.
Results
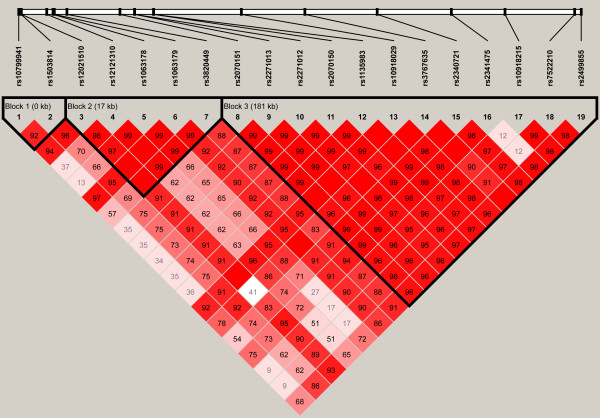
A total of 18 SNPs were successfully genotyped in 3,629 individuals in the present study. The LD pattern of these SNPs was shown in Figure 1. Three haplotype blocks were constructed in this region.
Figure 1.
Linkage disequilibrium plots for SNPs genotyped in DUSP12 - ATF6 locus in the Chinese samples. Shades of pink indicate the strength of pairwise LD based on |D'|. Number shown are |D'| of each SNP pair.
The single SNP association analysis showed that no SNP was significantly associated with type 2 diabetes in our samples. The minimum P value was 0.0954 for rs10799941. Logistic regression analysis adjusting age, gender and BMI as confounding factors also suggested no association between SNPs and type 2 diabetes (Table 1). For the haplotype analysis, we compared the haplotype distributions between cases and controls and observed that no haplotype was nominally associated with type 2 diabetes (Table 2).
Table 1.
Allele frequencies and association to type 2 diabetes for SNPs in the DUSP12-ATF6 locus
| Gene | SNP | Chromosome position | Major/minor allele | Risk allele | Risk allele frequency | OR (95%CI) | Pallele | Pgenotype | |
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | ||||||||
| DUSP12 | rs10799941 | 159974818 | T,G | T | 0.540 | 0.520 | 1.0821(0.9863-1.1873) | 0.0954 | 0.1122 |
| DUSP12 | rs1503814 | 159975743 | C,T | T | 0.329 | 0.316 | 1.0613(0.9616-1.1714) | 0.2373 | 0.3788 |
| DUSP12 | rs12021510 | 159975835 | A,G | A | 0.930 | 0.929 | 1.0141(0.8466-1.2147) | 0.8794 | 0.9472 |
| DUSP12 | rs12121310 | 159985839 | A,C | C | 0.379 | 0.367 | 1.0546(0.9572-1.1620) | 0.2823 | 0.5793 |
| DUSP12 | rs1063178 | 159988331 | C,T | T | 0.460 | 0.458 | 1.0074(0.9177-1.1060) | 0.8766 | 0.7165 |
| DUSP12 | rs1063179 | 159988828 | C,T | C | 0.772 | 0.762 | 1.0594(0.9498-1.1817) | 0.3002 | 0.3300 |
| DUSP12 | rs3820449 | 159993796 | C,T | C | 0.694 | 0.694 | 1.0002(0.9049-1.1056) | 0.9964 | 0.6854 |
| ATF6 | rs2070151 | 160014680 | C,T | T | 0.311 | 0.300 | 1.0512(0.9511-1.1619) | 0.3282 | 0.2568 |
| ATF6 | rs2271013 | 160020426 | A,G | G | 0.310 | 0.300 | 1.0506(0.9505-1.1612) | 0.3338 | 0.2515 |
| ATF6 | rs2271012 | 160020465 | C,T | T | 0.317 | 0.302 | 1.0706(0.9666-1.1858) | 0.1908 | 0.1373 |
| ATF6 | rs2070150 | 160027900 | G,C | C | 0.312 | 0.301 | 1.0568(0.9559-1.1685) | 0.2805 | 0.2015 |
| ATF6 | rs1135983 | 160027936 | C,T | T | 0.309 | 0.297 | 1.0563(0.9553-1.1680) | 0.2856 | 0.2305 |
| ATF6 | rs10918029 | 160062520 | G,A | G | 0.780 | 0.774 | 1.0360(0.9272-1.1575) | 0.5326 | 0.6230 |
| ATF6 | rs2340721 | 160116009 | A,C | C | 0.349 | 0.344 | 1.0197(0.9247-1.1244) | 0.6959 | 0.9399 |
| ATF6 | rs2341475 | 160145232 | G,A | G | 0.658 | 0.646 | 1.0531(0.9497-1.1677) | 0.3268 | 0.4704 |
| ATF6 | rs10918215 | 160166355 | A,G | G | 0.432 | 0.427 | 1.0223(0.9304-1.1234) | 0.6460 | 0.4317 |
| ATF6 | rs7522210 | 160193803 | C,G | G | 0.433 | 0.427 | 1.0258(0.9341-1.1264) | 0.5940 | 0.3425 |
| ATF6 | rs2499855 | 160196385 | A,G | A | 0.897 | 0.890 | 1.0753(0.9203-1.2563) | 0.3606 | 0.5117 |
Table 2.
Association analyses of haplotypes in DUSP12-ATF6 locus with type 2 diabetes
| Haplotype | Haplotype frequencies | P value | |
|---|---|---|---|
| Cases | Controls | ||
| Block 1 (rs10799941-rs1503814) | |||
| GC | 0.449 | 0.467 | 0.1199 |
| TT | 0.318 | 0.304 | 0.1919 |
| TC | 0.222 | 0.217 | 0.5799 |
| GT | 0.011 | 0.013 | 0.5919 |
| Block 2 (rs12021510-rs12121310-rs1063178-rs1063179-rs3820449) | |||
| ACTCT | 0.307 | 0.306 | 0.8928 |
| AACCC | 0.242 | 0.236 | 0.5579 |
| AACTC | 0.228 | 0.237 | 0.3592 |
| AATCC | 0.080 | 0.089 | 0.1618 |
| GACCC | 0.070 | 0.069 | 0.8911 |
| ACTCC | 0.074 | 0.064 | 0.0897 |
| Block 3 (rs2070151-rs10918029-rs7522210-rs2499855) | |||
| CGCA | 0.342 | 0.341 | 0.9407 |
| TGGA | 0.304 | 0.296 | 0.4348 |
| CACA | 0.218 | 0.226 | 0.3917 |
| CGGG | 0.100 | 0.107 | 0.3166 |
| CGGA | 0.021 | 0.017 | 0.3030 |
We then analyzed the association between SNPs and quantitative traits related to glucose metabolism in the individuals with normal glucose regulation. No SNP was associated with plasma glucose and serum insulin levels at fasting status as well as 2-h after glucose stimulation. No significant association was detected between insulin sensitivity and beta cell function either (Table 3).
Table 3.
Association between SNPs from DUSP12-ATF6 and clinical features related to glucose metabolism in the normal glucose regulation subjects
| SNP | Fasting glucose | 2 h glucose | Fasting insulin | HOMA-IR | HOMA-B | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P | Beta | SE | P | Beta | SE | P | Beta | SE | P | Beta | SE | P | |
| rs10799941 | 0.0065 | 0.0164 | 0.6933 | 0.0325 | 0.0387 | 0.4024 | -0.0087 | 0.0226 | 0.7019 | -0.0076 | 0.0237 | 0.7492 | -0.0164 | 0.0249 | 0.5116 |
| rs1503814 | 0.0098 | 0.0179 | 0.5833 | 0.0247 | 0.0425 | 0.5613 | 0.0023 | 0.0251 | 0.9273 | 0.0065 | 0.0262 | 0.8036 | -0.0135 | 0.0277 | 0.6264 |
| rs12021510 | 0.0073 | 0.0322 | 0.8203 | -0.0795 | 0.0760 | 0.2953 | 0.0039 | 0.0452 | 0.9319 | 0.0054 | 0.0473 | 0.9086 | -0.0021 | 0.0499 | 0.9670 |
| rs12121310 | 0.0288 | 0.0173 | 0.0963 | 0.0740 | 0.0411 | 0.0719 | -0.0066 | 0.0241 | 0.7858 | 0.0010 | 0.0253 | 0.9678 | -0.0400 | 0.0267 | 0.1346 |
| rs1063178 | 0.0298 | 0.0165 | 0.0722 | 0.0356 | 0.0392 | 0.3639 | 0.0083 | 0.0230 | 0.7192 | 0.0162 | 0.0240 | 0.5008 | -0.0259 | 0.0254 | 0.3068 |
| rs1063179 | -0.0178 | 0.0192 | 0.3541 | 0.0082 | 0.0454 | 0.8561 | -0.0241 | 0.0262 | 0.3590 | -0.0258 | 0.0274 | 0.3483 | -0.0090 | 0.0288 | 0.7552 |
| rs3820449 | 0.0216 | 0.0178 | 0.2256 | 0.0739 | 0.0421 | 0.0791 | -0.0179 | 0.0247 | 0.4687 | -0.0113 | 0.0259 | 0.6627 | -0.0489 | 0.0273 | 0.0737 |
| rs2070151 | -0.0187 | 0.0184 | 0.3094 | -0.0112 | 0.0435 | 0.7970 | 0.0023 | 0.0254 | 0.9295 | -0.0061 | 0.0266 | 0.8182 | 0.0328 | 0.0280 | 0.2426 |
| rs2271013 | -0.0203 | 0.0184 | 0.2704 | -0.0126 | 0.0435 | 0.7728 | 0.0014 | 0.0252 | 0.9559 | -0.0073 | 0.0264 | 0.7822 | 0.0330 | 0.0278 | 0.2367 |
| rs2271012 | -0.0196 | 0.0186 | 0.2934 | -0.0193 | 0.0440 | 0.6615 | 0.0086 | 0.0257 | 0.7376 | 0.0004 | 0.0269 | 0.9883 | 0.0376 | 0.0285 | 0.1866 |
| rs2070150 | -0.0177 | 0.0185 | 0.3397 | -0.0154 | 0.0438 | 0.7249 | 0.0062 | 0.0257 | 0.8106 | -0.0020 | 0.0269 | 0.9405 | 0.0349 | 0.0283 | 0.2178 |
| rs1135983 | -0.0168 | 0.0185 | 0.3633 | -0.0192 | 0.0438 | 0.6609 | 0.0128 | 0.0256 | 0.6180 | 0.0046 | 0.0268 | 0.8642 | 0.0427 | 0.0283 | 0.1314 |
| rs10918029 | 0.0075 | 0.0197 | 0.7027 | 0.0282 | 0.0467 | 0.5451 | -0.0102 | 0.0271 | 0.7066 | -0.0071 | 0.0284 | 0.8029 | -0.0177 | 0.0300 | 0.5561 |
| rs2340721 | -0.0023 | 0.0174 | 0.8958 | 0.0488 | 0.0410 | 0.2342 | -0.0074 | 0.0239 | 0.7581 | -0.0064 | 0.0250 | 0.7988 | -0.0125 | 0.0264 | 0.6346 |
| rs2341475 | 0.0202 | 0.0187 | 0.2810 | -0.0207 | 0.0445 | 0.6419 | 0.0021 | 0.0257 | 0.9350 | 0.0093 | 0.0269 | 0.7299 | -0.0226 | 0.0289 | 0.4332 |
| rs10918215 | 0.0022 | 0.0167 | 0.8941 | -0.0441 | 0.0396 | 0.2647 | 0.0120 | 0.0232 | 0.6054 | 0.0093 | 0.0243 | 0.7006 | 0.0193 | 0.0256 | 0.4501 |
| rs7522210 | 0.0001 | 0.0166 | 0.9945 | -0.0425 | 0.0394 | 0.2809 | 0.0095 | 0.0230 | 0.6810 | 0.0061 | 0.0241 | 0.8001 | 0.0213 | 0.0253 | 0.3991 |
| rs2499855 | 0.0153 | 0.0266 | 0.5654 | -0.0842 | 0.0630 | 0.1814 | 0.0172 | 0.0367 | 0.6391 | 0.0199 | 0.0384 | 0.6045 | 0.0023 | 0.0407 | 0.9546 |
Discussion
Genome-wide linkage studies in various populations suggested the existence of multiple susceptibility gene(s) for type 2 diabetes at chromosome 1q21-q24 [2-9]. Several specific genes in this region, such as LMNA, NOS1AP and ATF6, were identified that they might confer risk for diabetes in some populations [12-16,22-24]. Among these genes, ATF6 is a strong candidate by its biological function in endoplasmic reticulum stress and unfolded protein response, which linked insulin demand with beta cell failure and diabetes. ATF6 is also the binding target of WFS1, a known type 2 diabetes susceptible gene, and mediates its effect on endoplasmic reticulum stress [25]. However, although we performed the association study by analyzing 18 SNPs in 3700 Chinese Han, we failed to find any evidence of association between SNPs from this locus and traits related to glucose metabolism in our samples. One possible explanation might be that the statistical power of our samples was not enough to detect the effects of this locus in the Chinese population. Although we had over 80% power to detect the association at the 0.05 level based on the previously reported ORs in non-Asian populations (1.2 ~ 1.3) and allele frequencies of reported SNPs in our Chinese samples, we could not exclude the possibility the reported effect size was overestimated due to the "winner's curse" effect or novel associated SNPs with lower minor allele frequencies in the Chinese existed, in this case our samples may not have sufficient power. Secondly, the relatively loose criteria for tagging SNP selection, which is the limitation of the current study, missed information for a group of SNPs in this region. As we used r2 over 0.7 and minor allele frequency over 0.1 as SNPs selection criterion, we failed to capture 15(6.8%) SNPs if the stringent criterion r2 over 0.8 and minor allele frequency over 0.05 was adopted. Thirdly, the LD pattern and allele frequencies differed between Chinese Hans and previously studied populations, which suggested population differences in the genetic architecture between Chinese and other ethnic populations, may also partly explain the lack of association between this locus and previously reported phenotypes. Finally we cannot exclude the possibility that rare variants within this region participated in the pathogenesis of diabetes as we only focused on the common ones.
Conclusion
Our data suggests common variants within DUSP12 and ATF6 genes may not play a major role in glucose metabolism in the Chinese. However, due to the limitation of the current study, the effects of SNPs from this locus on type 2 diabetes need to be tested in further studies with larger East Asian origin samples and higher marker density.
List of abbreviations
ATF6: activating transcription factor 6; CI: confidence interval; DUSP12: dual specificity phosphatase 12; HOMA: homeostasis model assessment; LD: linkage disequilibrium; OGTT: oral glucose tolerance test; OR: odds ratio; SNP: single nucleotide polymorphism;
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CH designed the study, participated in genotyping, performed statistical analysis and drafted the manuscript. RZ prepared the DNA samples and participated in genotyping. CW participated in genotyping. XM participated in sample collection and clinical studies. JW participated in the clinical study and revised the manuscript. YB participated in clinical study and contributed to discussion. KX contributed to discussion. WJ supervised the study and revised the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Call rates and concordance rates of SNPs genotyped. This file contains detailed information of quality control analysis of the SNPs, including call rates and concordance rates.
Allele frequencies and statistic power of the SNPs. This file contains the allele frequencies of all SNPs in the HapMap populations and our samples. The statistic power of the SNPs in our samples was also shown in this file.
Contributor Information
Cheng Hu, Email: alfredhc@sjtu.edu.cn.
Rong Zhang, Email: rongzhang11@yahoo.com.cn.
Congrong Wang, Email: wcr601@hotmail.com.
Xiaojing Ma, Email: masister1996@sina.com.
Jie Wang, Email: jiediana113@126.com.
Yuqian Bao, Email: byq522@163.com.
Kunsan Xiang, Email: sphxiang@163.com.
Weiping Jia, Email: wpjia@sjtu.edu.cn.
Acknowledgements
This work was supported by research grants from Project of National Natural Science Foundation of China (30630061), Shanghai Rising-Star Program (09QA1404400), the National 863 project of China (2006AA02A409), Key Project of Science and Technology of Shanghai (09DZ1950202), Shanghai Municipal Hospitals' Project of Chronic Disease Prevention and Treatment (SHDC12007316) and National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK073490). We thank all nursing and medical staff at Shanghai Clinical Center for Diabetes for their dedication and professionalism.
References
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RL, Ehm MG, Pettitt DJ, Prochazka M, Thompson DB, Timberlake D, Foroud T, Kobes S, Baier L, Burns DK, Almasy L, Blangero J, Garvey WT, Bennett PH, Knowler WC. An autosomal genomic scan for loci linked to type II diabetes mellitus and body-mass index in Pima Indians. Am J Hum Genet. 1998;63(4):1130–1138. doi: 10.1086/302061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Hoffman MD, Teng K, Leppert MF, Hasstedt SJ. A genome-wide search for type 2 diabetes susceptibility genes in Utah Caucasians. Diabetes. 1999;48(5):1175–1182. doi: 10.2337/diabetes.48.5.1175. [DOI] [PubMed] [Google Scholar]
- Vionnet N, Hani EH, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Lepretre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P. Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am J Hum Genet. 2000;67(6):1470–1480. doi: 10.1086/316887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O'Rahilly S, Frayling TM, Bell JI, Lathrop GM, Bennett A, Dhillon R, Fletcher C, Groves CJ, Jones E, Prestwich P, Simecek N, Rao PV, Wishart M, Bottazzo GF, Foxon R, Howell S, Smedley D, Cardon LR, Menzel S, McCarthy MI. A genomewide scan for loci predisposing to type 2 diabetes in a U.K. population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet. 2001;69(3):553–569. doi: 10.1086/323249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh WC, St Jean PL, Mitchell BD, Pollin TI, Knowler WC, Ehm MG, Bell CJ, Sakul H, Wagner MJ, Burns DK, Shuldiner AR. Genome-wide and fine-mapping linkage studies of type 2 diabetes and glucose traits in the Old Order Amish: evidence for a new diabetes locus on chromosome 14q11 and confirmation of a locus on chromosome 1q21-q24. Diabetes. 2003;52(2):550–557. doi: 10.2337/diabetes.52.2.550. [DOI] [PubMed] [Google Scholar]
- Langefeld CD, Wagenknecht LE, Rotter JI, Williams AH, Hokanson JE, Saad MF, Bowden DW, Haffner S, Norris JM, Rich SS, Mitchell BD. Linkage of the metabolic syndrome to 1q23-q31 in Hispanic families: the Insulin Resistance Atherosclerosis Study Family Study. Diabetes. 2004;53(4):1170–1174. doi: 10.2337/diabetes.53.4.1170. [DOI] [PubMed] [Google Scholar]
- Ng MC, So WY, Cox NJ, Lam VK, Cockram CS, Critchley JA, Bell GI, Chan JC. Genome-wide scan for type 2 diabetes loci in Hong Kong Chinese and confirmation of a susceptibility locus on chromosome 1q21-q25. Diabetes. 2004;53(6):1609–1613. doi: 10.2337/diabetes.53.6.1609. [DOI] [PubMed] [Google Scholar]
- Xiang K, Wang Y, Zheng T, Jia W, Li J, Chen L, Shen K, Wu S, Lin X, Zhang G, Wang C, Wang S, Lu H, Fang Q, Shi Y, Zhang R, Xu J, Weng Q. Genome-wide search for type 2 diabetes/impaired glucose homeostasis susceptibility genes in the Chinese: significant linkage to chromosome 6q21-q23 and chromosome 1q21-q24. Diabetes. 2004;53(1):228–234. doi: 10.2337/diabetes.53.1.228. [DOI] [PubMed] [Google Scholar]
- Munoz-Alonso MJ, Guillemain G, Kassis N, Girard J, Burnol AF, Leturque A. A novel cytosolic dual specificity phosphatase, interacting with glucokinase, increases glucose phosphorylation rate. J Biol Chem. 2000;275(42):32406–32412. doi: 10.1074/jbc.M000841200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. Signaling the unfolded protein response from the endoplasmic reticulum. J Biol Chem. 2004;279(25):25935–25938. doi: 10.1074/jbc.R400008200. [DOI] [PubMed] [Google Scholar]
- Das SK, Chu WS, Hale TC, Wang X, Craig RL, Wang H, Shuldiner AR, Froguel P, Deloukas P, McCarthy MI, Zeggini E, Hasstedt SJ, Elbein SC. Polymorphisms in the glucokinase-associated, dual-specificity phosphatase 12 (DUSP12) gene under chromosome 1q21 linkage peak are associated with type 2 diabetes. Diabetes. 2006;55(9):2631–2639. doi: 10.2337/db05-1369. [DOI] [PubMed] [Google Scholar]
- Thameem F, Farook VS, Bogardus C, Prochazka M. Association of amino acid variants in the activating transcription factor 6 gene (ATF6) on 1q21-q23 with type 2 diabetes in Pima Indians. Diabetes. 2006;55(3):839–842. doi: 10.2337/diabetes.55.03.06.db05-1002. [DOI] [PubMed] [Google Scholar]
- Chu WS, Das SK, Wang H, Chan JC, Deloukas P, Froguel P, Baier LJ, Jia W, McCarthy MI, Ng MC, Damcott C, Shuldiner AR, Zeggini E, Elbein SC. Activating transcription factor 6 (ATF6) sequence polymorphisms in type 2 diabetes and pre-diabetic traits. Diabetes. 2007;56(3):856–862. doi: 10.2337/db06-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meex SJ, van Greevenbroek MM, Ayoubi TA, Vlietinck R, van Vliet-Ostaptchouk JV, Hofker MH, Vermeulen VM, Schalkwijk CG, Feskens EJ, Boer JM, Stehouwer CD, van der Kallen CJ, de Bruin TW. Activating transcription factor 6 polymorphisms and haplotypes are associated with impaired glucose homeostasis and type 2 diabetes in Dutch Caucasians. J Clin Endocrinol Metab. 2007;92(7):2720–2725. doi: 10.1210/jc.2006-2280. [DOI] [PubMed] [Google Scholar]
- Prokopenko I, Zeggini E, Hanson RL, Mitchell BD, Rayner NW, Akan P, Baier L, Das SK, Elliott KS, Fu M, Frayling TM, Groves CJ, Gwilliam R, Scott LJ, Voight BF, Hattersley AT, Hu C, Morris AD, Ng M, Palmer CN, Tello-Ruiz M, Vaxillaire M, Wang CR, Stein L, Chan J, Jia W, Froguel P, Elbein SC, Deloukas P, Bogardus C. et al. Linkage disequilibrium mapping of the replicated type 2 diabetes linkage signal on chromosome 1q. Diabetes. 2009;58(7):1704–1709. doi: 10.2337/db09-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Zhang R, Wang C, Ma X, Wang C, Fang Q, Bao Y, Xiang K, Jia W. A genetic variant of G6PC2 is associated with type 2 diabetes and fasting plasma glucose level in the Chinese population. Diabetologia. 2009;52(3):451–456. doi: 10.1007/s00125-008-1241-3. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71(5):1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen KR, Groves CJ, Hanson RL, Knowler WC, Shuldiner AR, Elbein SC, Mitchell BD, Froguel P, Ng MC, Chan JC, Jia W, Deloukas P, Hitman GA, Walker M, Frayling TM, Hattersley AT, Zeggini E, McCarthy MI. Common variation in the LMNA gene (encoding lamin A/C) and type 2 diabetes: association analyses in 9,518 subjects. Diabetes. 2007;56(3):879–883. doi: 10.2337/db06-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner L, Andersen G, Sparso T, Grarup N, Glumer C, Borch-Johnsen K, Jorgensen T, Hansen T, Pedersen O. Common variation in LMNA increases susceptibility to type 2 diabetes and associates with elevated fasting glycemia and estimates of body fat and height in the general population: studies of 7,495 Danish whites. Diabetes. 2007;56(3):694–698. doi: 10.2337/db06-0927. [DOI] [PubMed] [Google Scholar]
- Hu C, Wang C, Zhang R, Ng MC, Bao Y, So WY, Ma RC, Ma X, Chan JC, Xiang K, Jia W. Association of genetic variants of NOS1AP with type 2 diabetes in a Chinese population. Diabetologia. 2010;53(2):290–298. doi: 10.1007/s00125-009-1594-2. [DOI] [PubMed] [Google Scholar]
- Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, Ghosh R, Hayashi E, Ishihara H, Oka Y, Permutt MA, Urano F. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Invest. 2010;120(3):744–755. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Call rates and concordance rates of SNPs genotyped. This file contains detailed information of quality control analysis of the SNPs, including call rates and concordance rates.
Allele frequencies and statistic power of the SNPs. This file contains the allele frequencies of all SNPs in the HapMap populations and our samples. The statistic power of the SNPs in our samples was also shown in this file.