Abstract
The purpose of this study was to examine the expression of phospholipid scramblase 1 (PLSCR1) in tumor tissues and plasma specimens of patients with colorectal cancer (CRC), as well as analyze its association with clinical parameters. The expression levels of PLSCR1 protein in 104 matched CRC and adjacent normal tissue sections and 50 pairs of CRC tissue blocks were determined by use of immunohistochemical and Western blot analyses, respectively. To evaluate the diagnostic potential of PLSCR1, the plasma levels of PLSCR1 were investigated in 111 additional subjects (59 CRC patients and 52 healthy controls) by Western blot. PLSCR1 was overexpressed in malignant adenocarcinoma tissues compared with normal colorectal mucosa (P < 0.001). In addition, the plasma level of PLSCR1 was not only significantly elevated in CRC patients compared with healthy individuals (P < 0.001), but it was also substantially increased in early stage CRC (P < 0.001). Importantly, the overall sensitivity and specificity of PLSCR1 for CRC detection were 80% and 59.6%, respectively. The area under the ROC curve of PLSCR1 for CRC diagnosis is 0.75, which increases to 0.8 if combined with the measurement of carcinoembryonic antigen. Univariate analysis with the Cox regression model revealed that elevated PLSCR1 expression indicated a poor prognosis for CRC. This study showed that PLSCR1 protein levels were significantly elevated in both the cancer tissue and plasma of CRC patients. Moreover, the plasma levels of PLSCR1 were significantly elevated in patients with early stage CRC compared with healthy individuals, suggesting that PLSCR1 might be used as a noninvasive serological diagnostic and prognostic biomarker for CRC.
INTRODUCTION
Colorectal cancer (CRC), with an estimated 1 million new cases and 500,000 deaths annually, is the third most common cancer worldwide (1). CRC is thought to take several years to develop from a precancerous adenoma to a malignant carcinoma (2). Clinically, the stage of disease at initial diagnosis is the most important prognostic factor for CRC patients. Studies have shown that early detection of CRC and subsequent intervention during an early stage has the potential to reduce both the incidence and mortality of the disease (3–5). Currently, available screening methods include digital rectal examination, fecal occult blood test and colonoscopy (6,7). However, the diagnostic value of the currently most reliable noninvasive screening test, the fecal occult blood test, is limited in terms of its low sensitivity and lack of patient compliance (8,9). To overcome this problem, the identification of novel biomarkers that can aid the early detection of CRC is crucial.
Tumor markers are widely used for the detection and monitoring of cancer in clinical laboratory tests. Currently, few clinically verified markers are suggested to predict the biological behavior of CRC. Some potential colorectal tumor markers, such as carcinoembryonic antigen (CEA), CA 242, CA 19-9, CA 50, tissue plasminogen activator, tissue-polypeptide–specific antigen and tissue inhibitor of metalloproteinase 1 have been extensively studied. In clinical tests, none of these serological markers have demonstrated both the high sensitivity and high specificity needed to detect early stage CRC (10–13).
Phospholipid scramblase 1 (PLSCR1), a newly identified calcium-dependent plasma-membrane protein (14,15), has been revealed to be important in the transbilayer movement of phosphatidylserine and other aminophospholipids to the plasma membrane outer leaflet. This transbilayer movement usually occurs after a physiological event, such as cellular injury and apoptosis (16–18). Furthermore, studies have reported that PLSCR1 may be involved in the regulation of tumor cell proliferation (19). Silverman et al. have shown that overexpression of PLSCR1 suppresses the growth of ovarian carcinoma cells in vivo (20). Interestingly, in our previous membrane proteomics study, we used liquid chromatography–tandem mass spectrometry technology to quantify membrane proteins of CRC tissue (21) and found that the expression of PLSCR1 was upregulated in CRC tissue compared with normal tissue. Collectively, these reports suggest that PLSCR1 may play an important role in tumorigenesis. However, the biological functions and the expression levels of PLSCR1 in patients with CRC have not been well investigated. The aim of this study was to investigate the expression of PLSCR1 protein in CRC tissues and to test the possible clinical relevance of plasma PLSCR1 levels for the detection of CRC.
MATERIALS AND METHODS
SUBJECTS
All the study subjects and clinical specimens were consecutively collected from the Department of Colorectal Surgery, Chang Gung Memorial Hospital, Taiwan. All of these CRC patients had histologically verified adenocarcinoma of the colon or rectum that was confirmed by pathologists. Patient characteristics were obtained from pathology records: age, sex, tissue stage, clinical stage (TNM [tumor-node-metastasis] classification), lymph node status and presence of possible distant metastases. Patients with history of other malignant or infectious diseases and those who had surgery within 6 months of the beginning of this study were excluded from this retrospective study. For comparison of tissue PLSCR1 levels, a total of 104 pairs of colorectal carcinoma and noncancerous colorectal tissues were enrolled for this analysis. Among them, 50 paired tissues were randomly selected and examined first by Western blotting. Then, to further confirm the results of this preliminary observation, all of the 104 tissue specimens were sectioned and evaluated by use of immunohistochemical (IHC) staining. For plasma PLSCR1 detection, 111 fresh blood samples (59 from CRC patients and 52 from age-matched healthy controls) were obtained and stored at −80°C until use. Among them, the number of CRC patients with a TNM classification of early stage (I and II) and advanced stage (III and IV) CRC were 25 and 34, respectively. Written informed consent was obtained from all participants. This study was approved by the Medical Ethics and Human Clinical Trial Committee of Chang Gung Memorial Hospital.
Western Blot Analysis
Clinical tissue specimens were taken from freshly isolated surgical resections, snap frozen in liquid nitrogen, and then stored at −80°C until use. For the analysis of PLSCR1 expression in CRC tissues, frozen tissues were thawed and resuspended in lysis solution (0.25 mol/L sucrose, 10 mmol/L Tris-HCl pH 7.6, 1 mmol/L MgCl2, 1% sodium dodecyl sulfate [SDS]) with protease inhibitors (20 μg/μL aprotinin, 20 μg/μL leupeptin, and 1 mmol/L phenylmethanesulfonyl fluoride; protein:protein inhibitor was 100:1, v/v). After homogenization (Polytron System PT 1200 E, Luzernerstrasse, Switzerland) on ice, 40 μg of tissue lysate was subjected to Western blot analysis. For Western blot analysis of PLSCR1 in plasma, 1 μL of each plasma sample was fractionated through a 15% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel, and the proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA, USA). The membrane was blocked with 5% skim milk in Tris-buffered saline–Tween buffer (25 mmol/L Tris, 190 mmol/L NaCl, and 0.5% [v/v] Tween 20, pH 7.5) and then incubated with primary rabbit antihuman PLSCR1 polyclonal antibody (1:1000 dilution; ProteinTech, Chicago, IL, USA) at 4°C, overnight. After being washed, the membranes were incubated at 25°C for 1 h with peroxidase-conjugated mouse antirabbit IgG antibody (1:5,000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA). After being washed, the membranes were developed with a chemiluminescence reagent kit (Amersham Pharmacia Biotechnology, Piscataway, NJ, USA) and then photographed with Kodak Biomax light films (Eastman Kodak, Rochester, NY, USA). Immunoblot images were analyzed by an Imagemaster (Amersham Pharmacia Biotech), and the band intensities are presented in arbitrary units (AU).
Immunohistochemical Analysis
Tumor tissue blocks that were used for IHC were first fixed in 4% paraformaldehyde and then embedded in paraffin. To eliminate differences in the genetic background, malignant tissues and adjacent normal tissues were prepared from the same resection. Normal tissue was obtained from the distal edge of the resection at least 10 cm from the tumor. To reduce selection bias, two independent histopathologists reviewed areas of normal colonic epithelium, benign polyps and adenocarcinoma. Tissue sections (5-μm thick) were obtained from paraffin-embedded tissue blocks, mounted on silanized slides (Superfrost, Menzel, Braunschweig, Germany), deparaffinized with xylene (2 × 10 min) and rehydrated with an ethanol gradient as previously described (22). To eliminate endogenous peroxidase activity, the tissue sections were incubated with 3% H2O2 at room temperature for 30 min. The specimens were subsequently heated in a microwave oven for antigen retrieval (10 mmol/L citrate buffer, pH 6.0, 20 min, 700 W). To block nonspecific binding, the slides were preincubated with 10% nonimmune goat serum at 37°C for 30 min. The samples were then incubated with antihuman PLSCR1 monoclonal antibody (mouse, 1:1000; Lifespan, Seattle, WA, USA) for 30 min at room temperature. After being washed with phosphate-buffered saline (pH 7.4), the slides were incubated with horseradish peroxidase–labeled antimouse IgG secondary antibody (rabbit, 1:2000 dilution; Jackson ImmunoResearch Laboratories, West Grove, PA, USA) for 30 min at room temperature and then developed by using 3,3′-diaminobenzidine (Sigma, St. Louis, MO, USA). The sections were subsequently counterstained with hematoxylin, washed in running tap water, dehydrated and mounted in Neo-Mount (Merck, Darmstadt, Germany). Negative control reactions were conducted by omitting the primary antibody.
Scoring of Immunostaining
Immunostaining was evaluated and scored by two experienced pathologists who were blinded to the clinical and pathological parameters as well as the clinical outcome, as described previously (23,24). The definition of immunostaining intensity was as follows: score of 0 (negative), no brown particles in the tumor epithelial cell cytoplasm or plasma membrane; score of 1 (weak), light-brown particles in the tumor epithelial cell cytoplasm or plasma membrane; score of 2 (moderate), general brown particles in the tumor epithelial cell cytoplasm or plasma membrane; and score of 3 (strong), deep brown particles in the tumor epithelial cell cytoplasm or plasma membrane. The percentage of PLSCR1-positive cells was semiquantitatively determined by assessing the entire tumor section. The extent of immunostaining was semiquantitatively scored and the specimens were classified into five groups: 0, no positive epithelial cells were observed; 1, fewer than 25% PLSCR1-positive cells; 2, more than 25% and fewer than 50% PLSCR1-positive cells; 3, more than 50% and fewer than 75% PLSCR1-positive cells; and 4, more than 75% PLSCR1-positive cells. The immunoreactive score, also known as the staining index (SI), was calculated by multiplying the score of positive cells with the staining-intensity score. For this study, we defined an SI of 0 to indicate negative PLSCR1 protein expression and an SI of 1 or more to indicate positive expression.
Carcinoembryogenic Antigen Assay
Plasma carcinoembryogenic antigen (CEA) levels were assayed with a commercially available CEA ELISA kit (Roche Diagnostics, Mannheim, Germany) according to manufacturer’s instructions.
Statistical Analysis
For the analysis of tissue IHC results, the association between protein expression and clinicopathological characteristics was analyzed by using the Pearson χ2 test. For the analysis of plasma PLSCR1 levels, the differences between individual variables from two groups were analyzed by the independent t test. For determination of the factors related to overall survival, the Cox proportional hazards model was used, and the probability was calculated using the log-rank test by the Kaplan–Meier method. The diagnostic potential was evaluated by performing receiver operating characteristic curves (ROC) analysis, and the discriminative efficacy of the individual biomarker was calculated by the area under the ROC curve (AUC). All P values were derived from two-tailed statistical tests, and a value of 0.05 or less was regarded as statistically significant.
All supplementary materials are available online at www.molmed.org.
RESULTS
Expression of PLSCR1 in CRC Tissue
To test whether PLSCR1 proteins were overexpressed in malignant colorectal carcinoma tissues, we preliminarily used Western blot to examine the expression of PLSCR1 proteins in 50 paired CRC tissue lysates. Figure 1 shows that PLSCR1 proteins were expressed at higher levels in tumor tissues compared with their normal counterparts (63.2 ± 41 versus 29 ± 28.1, P < 0.001 by Student t test). To further verify this observation, we carried out IHC analysis on a larger panel of tissue sections that contained 104 colorectal tumor tissues. IHC staining results revealed that the expression levels of PLSCR1 protein in benign adenomatous polyps and malignant carcinoma tissues were substantially increased compared with normal colorectal mucosal epithelium (Figure 2).
Figure 1.
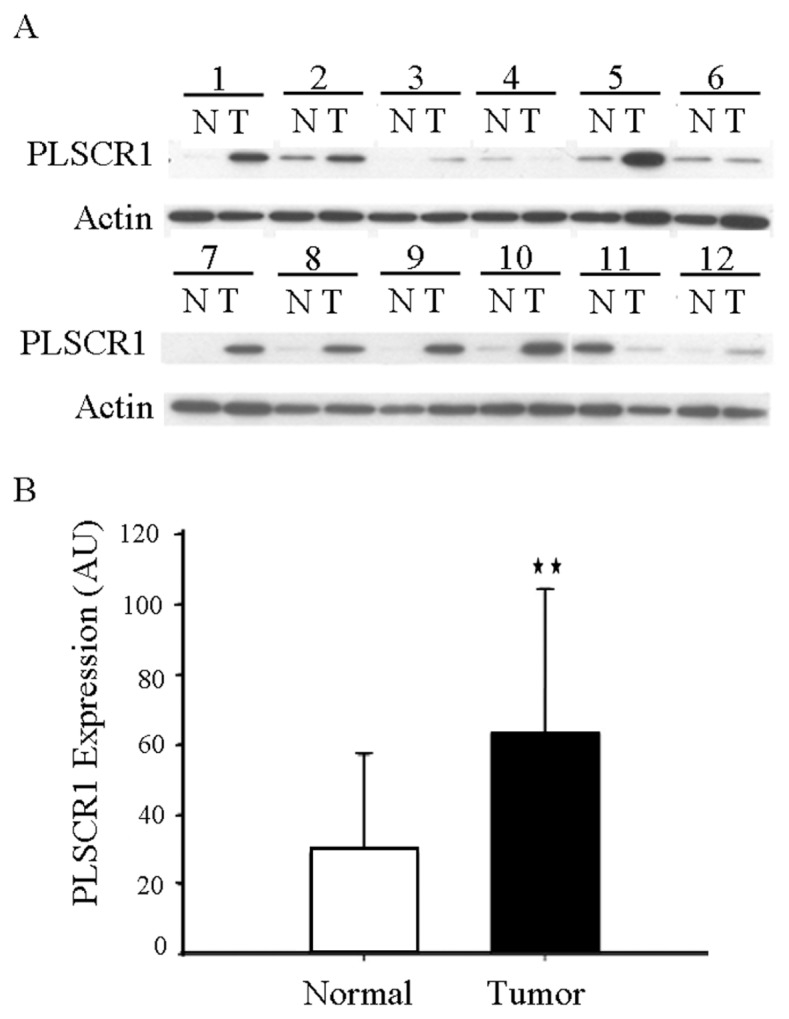
PLSCR1 overexpression in CRC tissues. (A) Representative Western blot for PLSCR1 comparing 12 pairs of CRC tissues ( T ) and matched adjacent normal tissues (N). β-Actin was used as a loading control. (B) PLSCR1 proteins were expressed at higher levels in tumor tissues compared to normal tissue.**P < 0.001 by Student t test.
Figure 2.
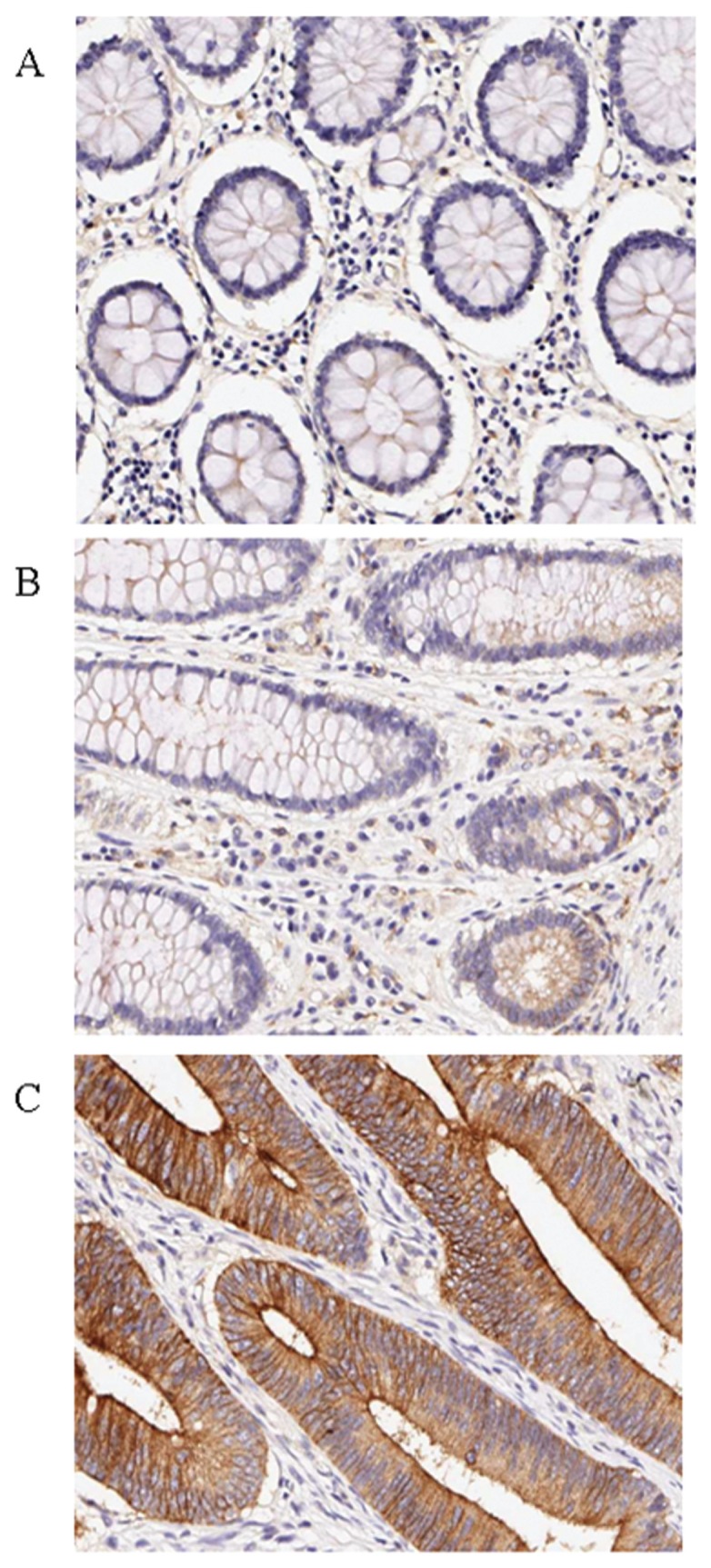
Immunohistochemical analysis of CRC tissues for PLSCR1 expression (brown color). The paired paraffin-embedded CRC tissue sections were stained with the anti-PLSCR1 antibody. A representative case is presented that contains normal colorectal mucosa, a benign adenomatous polyp and colorectal adenocarcinoma tissue. (A) Low or negative staining of PLSCR1 in normal colorectal mucosa. (B) Upregulated expression of PLSCR1 in benign colorectal polyp tissues. (C) High-level expression of PLSCR1 in colorectal adenocarcinoma tissues. All slides were counterstained with hematoxylin. All photographs are presented at 200× magnification.
Plasma Levels of PLSCR1 in CRC Patients
To test the level of PLSCR1 protein in plasma, we performed Western blot analysis on 111 plasma samples from 59 CRC patients and 52 healthy controls. Figure 3 shows the individual plasma PLSCR1 levels across the different groups. The results revealed that the levels of PLSCR1 protein in the plasma of patients with early stage CRC, as well as advanced stage CRC, were substantially increased compared with non-CRC controls. The mean plasma level of PLSCR1 was 2.4-fold higher in the cohort of CRC patients (44.0 AU) than the healthy controls (18.5 AU) (Student t test, P < 0.001). When 13.9 AU was set as the cutoff value, overexpression of PLSCR1 was found in 40% (21 of 52) of normal controls and 80% (47 of 59) of CRC patients, respectively (Table 1).
Figure 3.
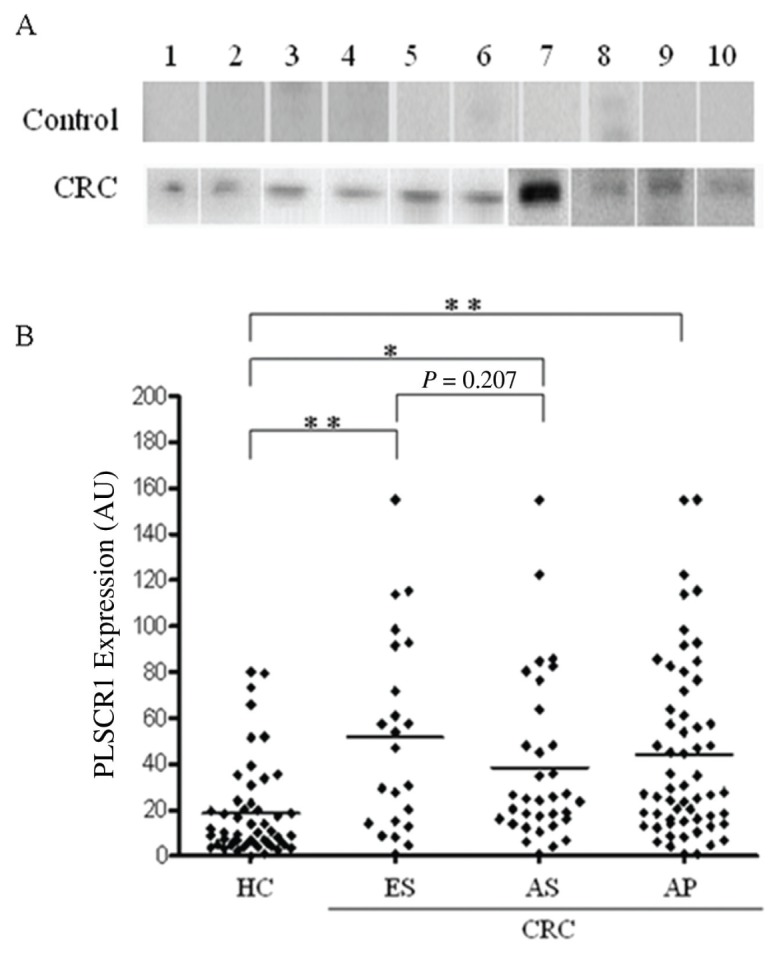
Western blot analysis of PLSCR1 expression in plasma. (A) Plasma samples (1 μL) from each tested specimen were denatured and loaded onto an SDS-PAGE gel. Each line represents an individual participant. Representative samples from healthy controls with negative or low PLSCR1 plasma levels and CRC patients with elevated PLSCR1 plasma levels are shown. (B) PLSCR1 was elevated in the plasma of CRC patients at different disease stages compared with plasma from healthy controls. HC, healthy controls; ES, early stage (TNM stage I and II); AS, advanced stage (TNM stage III and IV); AP, all patients; the horizontal bar represents the mean value of each group; *P < 0.01, **P < 0.001, Student t test.
Table 1.
Sensitivity values of CEA and PLSCR1 measurements in CRC.
| Sensitivity |
||||
|---|---|---|---|---|
| CRC | No. tested | CEAa (A) | PLSCR1b (B) | Combined (A) + (B) |
| Early stage | 25 | 4 (16%) | 20 (80%) | 20 (80%) |
| Advanced stage | 34 | 18 (53%) | 27 (79%) | 30 (88%) |
| All CRC patients | 59 | 22 (37%) | 47 (80%) | 50 (85%) |
Cutoff value ≥5 ng/mL.
Cutoff value ≥13.9 AU.
Association of PLSCR1 Levels with Clinicopathological Variables
We further examined the relationships between the level of PLSCR1 expression and clinicopathological variables including age, sex, histological grade, tumor stage, lymph node metastasis, distal metastasis and CEA concentration in CRC patients. Statistical analysis of both tissue and plasma samples revealed that no significant differences were observed between PLSCR1 expression and the clinicopathological parameters examined in the present study (Supplementary Table 1).
Diagnostic Value of Plasma PLSCR1 Protein
To evaluate the diagnostic potential of PLSCR1, both PLSCR1 and CEA plasma levels were evaluated in patients with CRC and in healthy controls. Figure 4 illustrates the relationship between PLSCR1 and CEA plasma levels in individual blood samples from CRC patients. As can be seen, when cutoff values of 5 ng/mL and 13.9 AU were applied for CEA and PLSCR1, respectively, the overall CRC diagnostic sensitivity and specificity of PLSCR1 were 80% and 60%, respectively, and those of CEA were 37% and 87%, respectively. When we combined PLSCR1 and CEA measurements, the diagnostic sensitivity increased to 85% and the specificity increased to 48% (Table 1). When the diagnostic parameters were calculated for early stage carcinoma only (TNM stage I and II), the sensitivities of CEA and PLSCR1 were 16% and 80%, respectively. The seropositivity ratios of PLSCR1 in the CEA-positive (n = 22) and CEA-negative (n = 37) subgroups were 86.4% (19:22) and 75.7% (28:37), respectively (Supplementary Figure 1). The clinical relevance of PLSCR1 and CEA for CRC diagnosis is represented by an ROC curve (Figure 4A). The AUC was 0.75 for PLSCR1 and 0.65 for CEA, respectively. The combination of both markers leads to a further increase in the AUC to 0.8.
Figure 4.
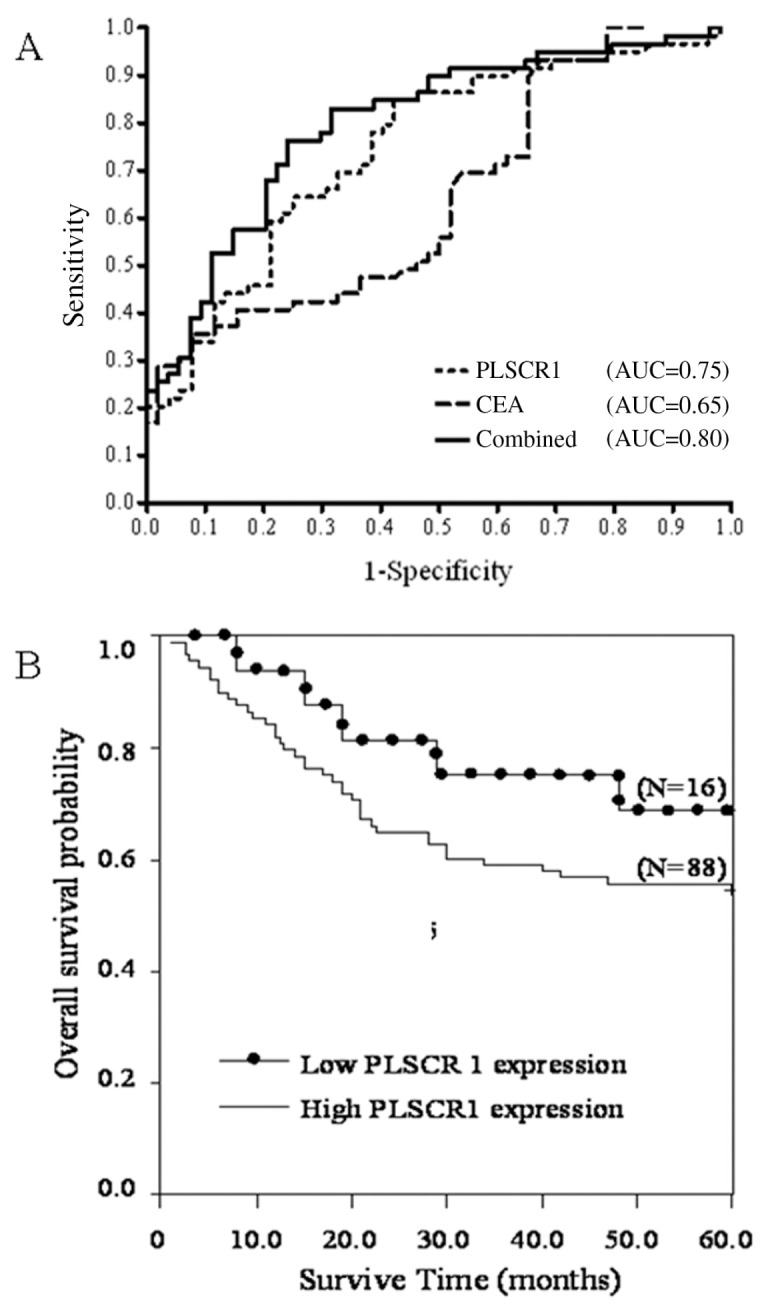
(A) ROC curves for CEA and PLSCR1. Plasma concentrations of CEA and PLSCR1 among 59 CRC samples and 52 healthy control samples were determined by Western blot. The relationships between the specificity and the sensitivity of PLSCR1 and CEA for the detection of CRC are represented by ROC curves. The AUC value was 0.65 for CEA, 0.75 for PLSCR1 and 0.8 for both analyzed in combination. (B) Prognostic values of PLSCR1 expression for CRC patients. The Kaplan–Meier curve shows the relationship between 5-year overall survival and PLSCR1 expression. There was a significant correlation between PLSCR1 expression and shorter survival (P < 0.05).
Prognostic Implications of Plasma PLSCR1 Levels
Using the Cox regression model, we performed univariate analysis of data from the 104 patients who had valid survival data. We found no difference in survival when the data were analyzed according to the patients’ age, sex, tumor histology or lymph node status. However, factors including tumor metastasis, TNM stage, CEA level and PLSCR1 expression were found to be independent prognostic indicators for poor overall survival (Table 2). For analysis of the association between the expression of PLSCR1 and survival, we divided the CRC patients into two groups according to the IHC SI value, that is, SI = 1 was the cutoff point. The mean survival period was 39.2 months in patients with high PLSCR1 expression, compared with 49.8 months in patients with low PLSCR1 expression. There was a significant correlation between high expression of PLSCR1 and poor survival. The group of patients with low PLSCR1 expression had longer overall survival (P = 0.047; relative risk, 0.438; 95% confidence interval: 0.306–0.552) compared with the high-expression group (Figure 4B).
Table 2.
Univariate Cox regression analysis of 5-year overall survival in CRC.
| Overall survival |
|||
|---|---|---|---|
| Clinicopathological parameter | Relative risk | 95% Confidence interval | P |
| Age, years | |||
| <60 versus ≥60 | 1.331 | 0.742–2.389 | 0.338 |
| Sex | |||
| Female versus male | 1.478 | 0.814–2.684 | 0.200 |
| Tumor stage | |||
| T1–T2 versus T3–T4 | 0.042 | 0.001–3.071 | 0.148 |
| Nodal stage | |||
| N0 versus N1–3 | 0.572 | 0.242–1.351 | 0.203 |
| Tumor metastasis | |||
| M0 versus M1 | 0.168 | 0.091–0.308 | <0.001 |
| TNM stage | |||
| I, II versus III, IV | 0.107 | 0.015–0.774 | 0.027 |
| Tumor grade (differentiation) | |||
| Well versus moderate, poor | 0.515 | 0.125–2.125 | 0.359 |
| CEA (5 ng/mL cutoff) | |||
| Low versus high | 0.531 | 0.294–0.961 | 0.036 |
| PLSCR1 expression | |||
| Low versus high | 0.438 | 0.306–0.552 | 0.047 |
DISCUSSION
To our knowledge, this study is the first to investigate the elevated expression of PLSCR1 protein in neoplastic tissues of the colorectum. Intriguingly, we also found that PLSCR1 overexpression is associated with different stages of tumor progression, from normal tissue to malignant lesions, and that PLSCR1 levels in the plasma have diagnostic potential for CRC.
In addition the known role of PLSCR1 as a scramblase, increasing evidence suggests that PLSCR1 plays a role in cell signaling, maturation and apoptosis and the growth of cancer cells (25,26). Recently, PLSCR1 has been reported to be a substrate of cellular protein kinases (17,27,28) and to potentiate the antiviral activity of interferon (29). Furthermore, results of several reported studies have revealed that PLSCR1 overexpression is associated with the differentiation of human myeloid leukemia cells into granulocytes (30) and the suppression of ovarian carcinoma cell growth (20). However, the role of PLSCR1 in the development of CRC remains to be clarified.
To further elucidate the correlation between PLSCR1 expression and the neoplastic transformation of epithelial cells in the colorectum, a more detailed IHC analysis of tissues from all tumor stages was performed. Overexpression of PLSCR1 was consistently observed in almost all CRC patients (84.6%; 88 of 104), including patients with colonic adenoma and CRC of every stage (Figure 2). However, our results did not demonstrate a strong correlation between PLSCR1 expression and the clinicopathological characteristics of patients with CRC (Supplementary Table 1). Given these and previous findings, we suggest that PLSCR1 overexpression is a common feature of tumor tissues and that it may play an important role during tumorigenesis.
Early detection of CRC is extremely meaningful because the survival rate for CRC is much better when the disease is diagnosed at an early, rather than an advanced, stage. Previous studies have shown that an adenoma polyp is a precancerous lesion of CRC. To further determine whether PLSCR1 was overexpressed in premalignant lesions of CRC, we compared the IHC staining score (SI) between normal and benign polyp tissues. Results demonstrated that adenoma polyp tissues express higher levels of PLSCR1 protein than their normal counterparts (Figure 2). Based on this observation, it is conceivable that overexpression of PLSCR1 is an early event in CRC development and that this overexpression could serve as a marker for the early detection of CRC. A larger scale clinical study is required to verify this speculation.
The identification of tumor-associated proteins that are differentially expressed between normal and malignant tissues may provide insights into tumorigenesis, and these proteins may also be useful biomarkers for CRC diagnosis or prognosis. Compared to traditional screening methods, serological biomarker for CRC detection and monitoring can be analyzed in a relatively noninvasive manner. Various serological biomarkers for CRC are currently available, such as nicotinamide N-methyltransferase, proteasome activator complex subunit 3, selenium-binding protein 1, and urokinase-type plasminogen activator antigen (31–37). However, only a few proteins have thus far been established to fulfill the requirements for detection of CRC (38). So far, CEA is the most well-established tumor marker for CRC (39). Authors of a recent review, in which they evaluated 19 studies, reported that the overall sensitivity of CEA for CRC varied between 43% and 69% (13). Thus, the potential clinical usefulness of PLSCR1 might be best discussed in the context of CEA.
Practically speaking, a protein that is overexpressed in cancer tissues and is secreted into the circulation has the potential to serve as a serological cancer marker. Our results demonstrated that PLSCR1 is overexpressed in CRC tissue. Thus, we speculated that PLSCR1 might be released from the tumor cells into the circulation, giving rise to elevated PLSCR1 levels in the plasma. To evaluate the diagnostic value of PLSCR1, we determined the levels of PLSCR1 in the plasma of CRC patients and their normal counterparts. As a benchmark, we also assessed CEA levels in the same plasma samples. As described above, sensitivity of PLSCR1 as a single marker was superior to that of CEA (80% versus 37%). In contrast to CEA, which exhibited higher sensitivity values only in advanced tumor stages, PLSCR1 seropositivity was elevated in early stage CRC, suggesting that it might be more relevant for the screening of asymptomatic patients at risk for the development of CRC. However, PLSCR1 had been reported to act as a positive acute-phase protein and to play roles in cell proliferation, differentiation, apoptosis and tumorigenesis (40–42). These findings indicate that PLSCR1 may have differential expression in various inflammatory conditions and neoplastic diseases and thus may not be specific to colon cancer. This preliminary observation should be validated in a larger patient population with CRC or diseases that have a high risk of developing into CRC, for example, chronic inflammatory bowel disease.
The combination of multiple biomarkers generally improves tumor diagnostic performance (10,43,44). In this study, the possible clinical relevance of PLSCR1 was further confirmed by establishing ROC curves. We then found that the simultaneous determination of both PLSCR1 and CEA led to greater diagnostic sensitivity in our group of CRC patients. This result is supported by a previous report of the combination of CEA with u-PA (urokinase-type plasminogen activator) or CA 19-9 for screening of CRC patients (37).
Results of previous studies indicated that PLSCR1 may have differential expression in various inflammatory conditions and neoplastic diseases (40,41). Although we found that the mean plasma levels of PLSCR1 in patients with early stage CRC were a little higher than those in patients with advanced stage CRC, the difference was not statistically significant. Furthermore, our data also revealed no significant differences in PLSCR1 levels between CRC tumors of various stages. These results may indicate that overexpression of PLSCR1 in malignant tissue is independent of tumor stage. In addition, a previous investigation has shown that high expression of PLSCR1 can be regarded as a significant prognostic factor for acute myelogenous leukemia (45). Similarly, we demonstrated that tumor metastasis, TNM stage, CEA level and PLSCR1 expression were independent prognostic factors for CRC by univariate analysis with overall survival as an endpoint. Further prospective, large-cohort studies with detailed follow-up are needed to understand the mechanisms leading to such an outcome.
CONCLUSION
In conclusion, our data reveal that PLSCR1 is overexpressed in malignant CRC tissues compared with normal colorectum epithelium. Moreover, we demonstrated that patients with low PLSCR1 expression have longer overall survival, compared with patients with high expression. In addition, we showed for the first time that plasma levels of PLSCR1 were significantly elevated in CRC patients, even at early stages of CRC, suggesting that PLSCR1 could be both a novel diagnostic biomarker and an important prognostic factor for CRC.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants (CMRPD180271 and CMRPG371431) from Chang Gung University and Chang Gung Memorial Hospital, Taiwan. CA Chang thanks the National Science Council of the Republic of China (Taiwan) for financial support (grant number NSC-98-2113-M-010-001-MY3).
Footnotes
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
Online address: http://www.molmed.org
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 3.Hardcastle JD, et al. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996;348:1472–7. doi: 10.1016/S0140-6736(96)03386-7. [DOI] [PubMed] [Google Scholar]
- 4.Kronborg O, Fenger C, Olsen J, Jorgensen OD, Sondergaard O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348:1467–71. doi: 10.1016/S0140-6736(96)03430-7. [DOI] [PubMed] [Google Scholar]
- 5.Walsh JM, Terdiman JP. Colorectal cancer screening: scientific review. JAMA. 2003;289:1288–96. doi: 10.1001/jama.289.10.1288. [DOI] [PubMed] [Google Scholar]
- 6.Winawer S, et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale—update based on new evidence. Gastroenterology. 2003;124:544–60. doi: 10.1053/gast.2003.50044. [DOI] [PubMed] [Google Scholar]
- 7.Inger DB. Colorectal cancer screening. Prim Care. 1999;26:179–87. doi: 10.1016/s0095-4543(05)70108-1. [DOI] [PubMed] [Google Scholar]
- 8.Ahlquist DA. Fecal occult blood testing for colorectal cancer: can we afford to do this. Gastroenterol Clin North Am. 1997;26:41–55. doi: 10.1016/s0889-8553(05)70282-x. [DOI] [PubMed] [Google Scholar]
- 9.Collins JF, Lieberman DA, Durbin TE, Weiss DG. Accuracy of screening for fecal occult blood on a single stool sample obtained by digital rectal examination: a comparison with recommended sampling practice. Ann Intern Med. 2005;142:81–5. doi: 10.7326/0003-4819-142-2-200501180-00006. [DOI] [PubMed] [Google Scholar]
- 10.Duffy MJ, et al. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43:1348–60. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 11.Levy M, Visokai V, Lipska L, Topolcan O. Tumor markers in staging and prognosis of colorectal carcinoma. Neoplasma. 2008;55:138–42. [PubMed] [Google Scholar]
- 12.Thomson DM, Krupey J, Freedman SO, Gold P. The radioimmunoassay of circulating carcinoembryonic antigen of the human digestive system. Proc Natl Acad Sci U S A. 1969;64:161–7. doi: 10.1073/pnas.64.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hundt S, Haug U, Brenner H. Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2007;16:1935–53. doi: 10.1158/1055-9965.EPI-06-0994. [DOI] [PubMed] [Google Scholar]
- 14.Basse F, Stout JG, Sims PJ, Wiedmer T. Isolation of an erythrocyte membrane protein that mediates Ca2+-dependent transbilayer movement of phospholipid. J Biol Chem. 1996;271:17205–10. doi: 10.1074/jbc.271.29.17205. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Zhou Q, Wiedmer T, Sims PJ. Level of expression of phospholipid scramblase regulates induced movement of phosphatidylserine to the cell surface. J Biol Chem. 1998;273:6603–6. doi: 10.1074/jbc.273.12.6603. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Q, et al. Molecular cloning of human plasma membrane phospholipid scramblase: a protein mediating transbilayer movement of plasma membrane phospholipids. J Biol Chem. 1997;272:18240–4. doi: 10.1074/jbc.272.29.18240. [DOI] [PubMed] [Google Scholar]
- 17.Frasch SC, et al. Regulation of phospholipid scramblase activity during apoptosis and cell activation by protein kinase Cdelta. J Biol Chem. 2000;275:23065–73. doi: 10.1074/jbc.M003116200. [DOI] [PubMed] [Google Scholar]
- 18.Sims PJ, Wiedmer T. Unraveling the mysteries of phospholipid scrambling. Thromb Haemost. 2001;86:266–75. [PubMed] [Google Scholar]
- 19.Kasukabe T, Okabe-Kado J, Honma Y. TRA1, a novel mRNA highly expressed in leukemogenic mouse monocytic sublines but not in nonleukemogenic sublines. Blood. 1997;89:2975–85. [PubMed] [Google Scholar]
- 20.Silverman RH, et al. Suppression of ovarian carcinoma cell growth in vivo by the interferon-inducible plasma membrane protein, phospholipid scramblase 1. Cancer Res. 2002;62:397–402. [PubMed] [Google Scholar]
- 21.Chen JS, et al. Comparison of membrane fraction proteomic profiles of normal and cancerous human colorectal tissues with gel-assisted digestion and iTRAQ labeling mass spectrometry. FEBS J. 2010;277:3028–38. doi: 10.1111/j.1742-4658.2010.07712.x. [DOI] [PubMed] [Google Scholar]
- 22.Cattoretti G, et al. Antigen unmasking on formalin-fixed, paraffin-embedded tissue sections. J Pathol. 1993;171:83–98. doi: 10.1002/path.1711710205. [DOI] [PubMed] [Google Scholar]
- 23.Cao WF, et al. Prognostic significance of stomatin-like protein 2 overexpression in laryngeal squamous cell carcinoma: clinical, histologic, and immunohistochemistry analyses with tissue microarray. Hum Pathol. 2007;38:747–52. doi: 10.1016/j.humpath.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Feng Han Q, et al. Expression of sFRP-4 and beta-catenin in human colorectal carcinoma. Cancer Lett. 2006;231:129–37. doi: 10.1016/j.canlet.2005.01.026. [DOI] [PubMed] [Google Scholar]
- 25.Zwaal RF, Comfurius P, Bevers EM. Surface exposure of phosphatidylserine in pathological cells. Cell Mol Life Sci. 2005;62:971–88. doi: 10.1007/s00018-005-4527-3. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, et al. Antileukemic roles of human phospholipid scramblase 1 gene, evidence from inducible PLSCR1-expressing leukemic cells. Oncogene. 2006;25:6618–27. doi: 10.1038/sj.onc.1209677. [DOI] [PubMed] [Google Scholar]
- 27.Pastorelli C, et al. IgE receptor type I-dependent tyrosine phosphorylation of phospholipid scramblase. J Biol Chem. 2001;276:20407–12. doi: 10.1074/jbc.M100790200. [DOI] [PubMed] [Google Scholar]
- 28.Sun J, et al. c-Abl tyrosine kinase binds and phosphorylates phospholipid scramblase 1. J Biol Chem. 2001;276:28984–90. doi: 10.1074/jbc.M102505200. [DOI] [PubMed] [Google Scholar]
- 29.Dong B, et al. Phospholipid scramblase 1 potentiates the antiviral activity of interferon. J Virol. 2004;78:8983–93. doi: 10.1128/JVI.78.17.8983-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamaki T, et al. Role of MmTRA1b/phospholipid scramblase1 gene expression in the induction of differentiation of human myeloid leukemia cells into granulocytes. Exp Hematol. 2002;30:421–9. doi: 10.1016/s0301-472x(02)00779-8. [DOI] [PubMed] [Google Scholar]
- 31.Kim H, et al. Suppression of human selenium-binding protein 1 is a late event in colorectal carcinogenesis and is associated with poor survival. Proteomics. 2006;6:3466–76. doi: 10.1002/pmic.200500629. [DOI] [PubMed] [Google Scholar]
- 32.Alfonso P, et al. Proteomic expression analysis of colorectal cancer by two-dimensional differential gel electrophoresis. Proteomics. 2005;5:2602–11. doi: 10.1002/pmic.200401196. [DOI] [PubMed] [Google Scholar]
- 33.Friedman DB, et al. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2004;4:793–811. doi: 10.1002/pmic.200300635. [DOI] [PubMed] [Google Scholar]
- 34.Roessler M, et al. Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res. 2005;11:6550–7. doi: 10.1158/1078-0432.CCR-05-0983. [DOI] [PubMed] [Google Scholar]
- 35.Stulik J, et al. Proteome study of colorectal carcinogenesis. Electrophoresis. 2001;22:3019–25. doi: 10.1002/1522-2683(200108)22:14<3019::AID-ELPS3019>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 36.Roessler M, et al. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol Cell Proteomics. 2006;5:2092–101. doi: 10.1074/mcp.M600118-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Huber K, et al. Clinical value of determination of urokinase-type plasminogen activator antigen in plasma for detection of colorectal cancer: comparison with circulating tumor-associated antigens CA 19–9 and carcinoembryonic antigen. Cancer Res. 1993;53:1788–93. [PubMed] [Google Scholar]
- 38.Bast RC, Jr, et al. update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol. 2001;2000;19:1865–78. doi: 10.1200/JCO.2001.19.6.1865. [DOI] [PubMed] [Google Scholar]
- 39.Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep. 2008;41:685–92. doi: 10.5483/bmbrep.2008.41.10.685. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Zhao Q, Chen GQ. Phospholipid scramblase 1. Acta Physiol Sin. 2006;58:501–10. [PubMed] [Google Scholar]
- 41.Sahu SK, Gummadi SN, Manoj N, Aradhyam GK. Phospholipid scramblases: an overview. Arch Biochem Biophys. 2007;462:103–14. doi: 10.1016/j.abb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Lu B, et al. Expression of the phospholipid scramblase (PLSCR) gene family during the acute phase response. Biochim Biophys Acta. 2007;1771:1177–85. doi: 10.1016/j.bbalip.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Mor G, et al. Serum protein markers for early detection of ovarian cancer. Proc Natl Acad Sci USA. 2005;102:7677–82. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider J, Bitterlich N, Schulze G. Improved sensitivity in the diagnosis of gastro-intestinal tumors by fuzzy logic-based tumor marker profiles including the tumor M2-PK. Anticancer Res. 2005;25:1507–15. [PubMed] [Google Scholar]
- 45.Yokoyama A, et al. MmTRA1b/phospholipid scramblase 1 gene expression is a new prognostic factor for acute myelogenous leukemia. Leuk Res. 2004;28:149–57. doi: 10.1016/s0145-2126(03)00189-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


