Acute hantavirus infection in humans triggers a rapid expansion and long-term persistence of NK cells.
Abstract
Natural killer (NK) cells are known to mount a rapid response to several virus infections. In experimental models of acute viral infection, this response has been characterized by prompt NK cell activation and expansion followed by rapid contraction. In contrast to experimental model systems, much less is known about NK cell responses to acute viral infections in humans. We demonstrate that NK cells can rapidly expand and persist at highly elevated levels for >60 d after human hantavirus infection. A large part of the expanding NK cells expressed the activating receptor NKG2C and were functional in terms of expressing a licensing inhibitory killer cell immunoglobulin-like receptor (KIR) and ability to respond to target cell stimulation. These results demonstrate that NK cells can expand and remain elevated in numbers for a prolonged period of time in humans after a virus infection. In time, this response extends far beyond what is considered normal for an innate immune response.
Several experimental models have demonstrated a role for NK cells in host responses against virus infections (Lodoen and Lanier, 2006; Lee et al., 2007). The perhaps most well characterized experimental model system in this respect is that of infection of mice with mouse CMV (Dokun et al., 2001; Lodoen and Lanier, 2006; Sun et al., 2009). In experimental mouse CMV infection, the NK cell response is characterized by proliferation of a specific subset of NK cells that peaks within a few days after infection. Subsequently, this NK cell population undergoes rapid contraction by apoptosis (Dokun et al., 2001; Robbins et al., 2004). To investigate more directly how results from studies of viral infections in experimental model systems compare with infections in humans, we have studied the NK cell response throughout the course of an acute virus infection in humans.
In humans, involvement of NK cells in host responses to viruses were first indicated by the finding that virus-induced IFN-α enhanced NK cell–mediated cytotoxicity (Santoli et al., 1978; Trinchieri et al., 1978). Subsequently, low NK cell cytotoxic activity was linked to increased sensitivity to severe disseminating herpesvirus infections (Ching and Lopez, 1979; Quinnan et al., 1982; Merino et al., 1986; Joncas et al., 1989). NK cell defects were also shown to occur at chronic stages of HIV infection (Bonavida et al., 1986; Katz et al., 1987). Perhaps the most convincing data, however, for a role of NK cells in host responses to viral infections in humans has come from studies of patients with primary immunodeficiencies affecting NK cell numbers and/or NK cell function (Biron et al., 1989; Orange, 2006; Bryceson et al., 2007). In addition, several studies have described different characteristics of NK cells in patients with chronic viral infections (Fauci et al., 2005; Rehermann and Nascimbeni, 2005). However, few studies have more directly followed the human NK cell response throughout an acute virus infection.
The opportunity to do so accompanied a Puumala hantavirus outbreak that occurred in Northern Sweden during 2007 (Pettersson et al., 2008). In humans, Puumala hantaviruses cause hemorrhagic fever with renal syndrome, a disease characterized by severe symptoms with occasional mortalities which stem from capillary leakage (Vapalahti et al., 2003; Schönrich et al., 2008). In infected individuals, virus replication has been documented in vascular endothelium, but the virus does not seem to cause direct cytopathic effects (Schönrich et al., 2008). The ensuing viremia that develops is normally cleared within 1–2 wk after the onset of symptoms (Schönrich et al., 2008). During the course of the present Puumala hantavirus outbreak, we prospectively collected clinical samples and followed NK cell responses in 16 patients from their first presentation at the emergency unit with acute symptoms until up to 15 mo after symptom debut. This enabled us to investigate in detail the NK cell response in virally infected humans, from the very first days of clinical symptoms until resolution of disease and beyond.
The results show that NK cells, in a majority of the studied patients, rapidly expand and remain at significantly elevated numbers for >2 mo thereafter. Possible mechanisms behind this finding were investigated and the functionality of responding cells was determined. The results are discussed in relation to NK cell memory and the possible role of previous virus infections the present responses.
RESULTS AND DISCUSSION
NK cells rapidly expand and persist at elevated levels during acute hantavirus infection in humans
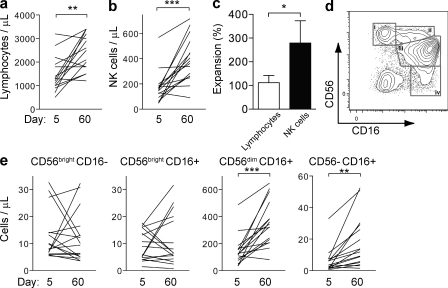
In blood samples from 16 patients infected with hantavirus (Table S1 and Fig. S1), absolute numbers of total lymphocytes, total NK cells, and individual NK cell subsets were determined at days 5 and 60 after the onset of symptoms. Surprisingly, at day 60 after onset of symptoms, total lymphocyte numbers were increased approximately twofold compared with those on day 5 (Fig. 1 a). Among lymphocytes, NK cell numbers increased three- to fourfold (Fig. 1, b and c). When specific NK cell subsets were studied, the numbers of CD56dim and atypical CD56neg (Gonzalez et al., 2009; Björkström et al., 2010b) NK cells had increased markedly, whereas the numbers of CD56bright NK cells did not change significantly (Fig. 1, d and e). The increases in absolute numbers of NK cells correlated well with heightened frequencies of total NK cells (Fig. S2 a) and specific NK cell subpopulations (Fig. S2 b).
Figure 1.
Increase in CD56dim NK cells in human hantavirus infection. PBMCs from patients with acute hantavirus infection were analyzed by flow cytometry. (a and b) Absolute numbers of lymphocytes (a) and NK cells (b) at days 5 and 60 after symptom debut. (c) Relative changes of total lymphocytes and NK cells from days 5–60 after symptom debut (mean ± SEM). (d) Definition of CD56brightCD16− (i), CD56brightCD16+ (ii), CD56dimCD16+ (iii), and CD56−CD16+ (iv) NK cells by flow cytometry from one representative patient with acute hantavirus infection. Cells were gated on the CD3−CD4−CD14−CD19− population within the single cell lymphocyte gate. (e) Absolute numbers of the different NK cell subsets at days 5 and 60. For a–e, n = 16. *, P < 0.05; **, P < 0.01; ***, P < 0.0001, paired Student’s t test.
These initial findings prompted us to perform a more detailed kinetic analysis of the increase in CD56dim NK cell numbers at the acute phase of hantavirus infection and throughout the convalescent period until 15 mo after onset of disease. After initial low NK cell numbers, possibly as a result of extravasation into tissues during the acute phase of infection (Terajima et al., 2007; Schönrich et al., 2008), NK cell numbers increased markedly and peaked at around 10 d after the onset of symptoms. CD56dim NK cell numbers then remained elevated for at least 60 d (Fig. 2 a). At 15 mo after infection, CD56dim NK cell numbers had declined to those in uninfected controls (Fig. 2 a).
Figure 2.
Rapid proliferation and sustained elevation of CD56dim NK cell numbers. (a) Numbers of CD56dimCD16+ NK cells (solid lines) evaluated longitudinally from day 5 until day 450 after onset of clinical symptoms in infected patients (n = 16) and uninfected controls (n = 26; **, P < 0.01, Mann-Whitney test; mean ± SEM; the dashed lines represent upper and lower SEM intervals for mean of CD56dimCD16+ NK cells in the control individuals). (b–e) Expression of intracellular Ki67 used as an indicator of cells that are dividing or have recently divided. (b) Representative examples of Ki67 expression in CD56dim NK cells analyzed by flow cytometry at days 5, 11, and 60 after onset of symptoms in one hantavirus-infected patient. (c) Frequency of Ki67-positive cells in controls (n = 5) and patients 5 d after symptoms debut (n = 6; **, P = 0.0043, Mann-Whitney test; mean ± SEM). (d and e) Frequency (d) and absolute numbers (e) of Ki67+ CD56dim NK cells evaluated longitudinally in the infected patients (n = 8; mean ± SEM). (f) Plasma levels of IL-15 in the hantavirus-infected patients (n = 16) and noninfected controls (n = 24). IL-15 levels were significantly increased during acute infection compared with the convalescent stage (day 450; **, P < 0.01, Mann-Whitney test; mean ± SEM). (g and h) Expression of Bcl-2 in CD8 T cells and NK cells from patients with acute hantavirus infection analyzed by flow cytometry. Representative staining of Ki67 and Bcl-2 on CD8 T cells (g) and CD56dim NK cells (h) from one patient at day 5 after onset of symptoms, as well as the mean expression level (MFI) of Bcl-2 in Ki67+ and Ki67− CD8 T cells (g) and CD56dim NK cells (h) at days 5, 11, and 450 after onset of symptoms (n = 8; *, P < 0.05, Mann-Whitney test; horizontal bars represent mean).
Increased NK cell numbers is a direct consequence of induced proliferation
To determine whether the increased numbers of CD56dim NK cells was a consequence of NK cell proliferation or, alternatively (but perhaps less likely), NK cell recruitment to the blood, expression of the proliferation marker Ki67 (Gerdes et al., 1984) was analyzed in the CD56dim NK cell population. Compared with uninfected individuals, where Ki67 expression is low in a majority of CD56dim NK cells, up to 50% of the CD56dim NK cells expressed Ki67 during the first 10 d of clinical symptoms after hantavirus infection (Fig. 2, b and c). Subsequently, the proportion of Ki67-expressing CD56dim NK cells declined, both with respect to relative frequencies and in absolute numbers (Fig. 2, d and e). Interestingly, the decline in frequency of Ki67+ NK cells was temporarily associated with the decline in viral load (Fig. S1).
IL-15 and Bcl-2 are required for NK cell proliferation and persistence
To identify possible factors contributing to NK cell proliferation and persistence, we first determined levels of plasma cytokines known to support NK cell proliferation and homeostasis. One key cytokine in this respect is IL-15 (Prlic et al., 2003; Ranson et al., 2003; Becknell and Caligiuri, 2005). Interestingly, we found that plasma levels of IL-15 were elevated up to 60 d after disease onset in most of the hantavirus-infected patients (Fig. 2 f). In contrast, levels of IL-2, IL-12, or IFN-α were not significantly altered in the infected patients over time (Fig. S3). Thus, the increased plasma levels of IL-15 could likely contribute to the observed CD56dim NK cell proliferation.
IL-15 may, however, not only contribute to NK cell proliferation but may also play a dominant role in promoting the survival of proliferating NK cells (Rodella et al., 2001). This may, at least in part, occur via up-regulation of the antiapoptotic molecule Bcl-2 (Prlic et al., 2003; Ranson et al., 2003). Therefore, we studied Bcl-2 expression in proliferating NK cells in hantavirus-infected patients and compared the results with observations in CD8 T cells. Indeed, consistent with published observations (Miller et al., 2008), we found that proliferating (Ki67+) CD8 T cells expressed significantly lower levels of Bcl-2 compared with nonproliferating (Ki67−) CD8 T cells (Fig. 2 g). In contrast, proliferating (Ki67+) NK cells expressed elevated levels of Bcl-2 compared with nonproliferating (Ki67−) NK cells (Fig. 2 h). These data suggest that Bcl-2 up-regulation may prevent early apoptosis of proliferating NK cells and contribute to an accumulation of these cells in the course of the infection and, possibly, even beyond its resolution.
Hantavirus-infected endothelial cells up-regulate the NKG2C-ligand HLA-E
Specific NK cell activation receptors have been shown to exhibit important roles in stimulating proliferation and contributing to the control of viral infections in experimental model systems (Dokun et al., 2001; Mandelboim et al., 2001; Arase et al., 2002; Gazit et al., 2006; Sun et al., 2009). These receptors have in common that they may recognize virus-associated and/or induced-self molecules on infected cells (Bryceson et al., 2006). To identify candidate NK cell activation receptors that might be of importance in recognition of target cells during human hantavirus infection, we infected primary human endothelial cells with hantavirus to assess changes in NK cell receptor-ligand expression (Fig. 3, a and b). In the course of these studies, we first observed an up-regulation of intercellular adhesion molecule (ICAM) 1, a ligand for lymphocyte function-associated antigen 1 (LFA-1), on infected endothelial cells (Fig. 3 c). LFA-1 has a well established role in promoting lymphocyte adhesion to endothelial cells (Robertson et al., 1990; Timonen, 1997). Furthermore, in NK cells, ICAM-1–LFA-1 interactions contribute to NK cell activation and are critical for polarization of NK cell lytic granules toward target cells (Bryceson et al., 2005). In contrast to ICAM-1, infected human endothelial cells did not show any marked up-regulation of ligands for NKG2D (MICA, MICB, or ULBP1-4) or DNAM-1 (CD112 or CD155). However, a significant up-regulation of HLA-E was observed (Fig. 3 c). This was interesting because HLA-E is a ligand for the activating NK cell receptor NKG2C (Braud et al., 1998), which has been previously implicated in host responses to CMV-infected cells (Gumá et al., 2004; Kuijpers et al., 2008).
Figure 3.
Hantavirus-infected endothelial cells up-regulate HLA-E. (a and b) Expression of ligands for NK cell receptors analyzed on hantavirus-infected primary human umbilical vein endothelial cells. (a) Immunofluorescence staining with patient serum on uninfected and infected cells 24 and 48 h after infection (hpi). Original magnification, 40×. Bars, 20 µm. (b) Kinetics of infection of endothelial cells. (c and d) Expression of ligands for NKG2D, NKG2A/C, DNAM-1, LFA-1, and KIRs assessed on infected (dashed lines) and uninfected (solid lines) endothelial cells 72 h after infection. Shaded histograms represent isotype control stainings. Co-culture of endothelial cells with UV-inactivated hantavirus for 72 h did not alter the expression of these ligands compared with uninfected cells (not depicted). Data in a–d are representative of three separate experiments.
Human CMV encodes UL40, a protein which in its signal sequence has a peptide motif identical to that of several HLA class I signal sequences which binds to and stabilizes HLA-E (Tomasec et al., 2000). To address whether a similar mechanism for increased expression of HLA-E exits in hantavirus-infected cells, we searched predicted HLA-E binding peptides motifs in the hantavirus proteins. Although we did identify one such peptide, this peptide did not stabilize HLA-E expression in vitro (unpublished data). Thus, virus-encoded HLA-E–binding peptides are a less likely explanation for the induced HLA-E expression. As HLA-E is dependent on signal sequences from other HLA class I molecules for its expression, another possible explanation for the up-regulation of HLA-E in hantavirus-infected cells is via an indirect mechanism linked to induced expression of classical HLA class I molecules. To test this hypothesis, we analyzed infected endothelial cells for expression of classical HLA class I molecules. Indeed, we did observe increased expression of classical HLA class I molecules in the infected cells, including HLA-A2 and HLA-C (Fig. 3 d). These may contribute peptide cargo for HLA-E, leading to increased cell surface expression (Braud et al., 1998). Additionally, it cannot be excluded that cytokines produced during the infection may also directly affect HLA-E expression.
Expanding and persisting NK cells are confined to an NKG2C-expressing subset
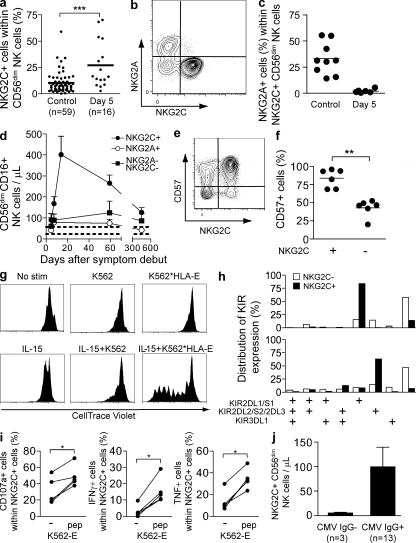
Because HLA-E is a ligand for the activating NK cell receptor NKG2C (Braud et al., 1998), which has been previously implicated in host responses to virus-infected cells (Gumá et al., 2004; Kuijpers et al., 2008), we examined more directly if this receptor might be involved in regulation of the NK cell response to human hantavirus-infected cells. Strikingly, we observed that the hantavirus-infected patients exhibited a three- to fourfold increase in the frequency of NKG2C+ NK cells already at day 5 compared with uninfected controls (Fig. 4 a). This observation could not be explained by cytokine-induced up-regulation of NKG2C expression on NKG2C− cells (unpublished data). In contrast, NKG2A (an inhibitory receptor binding to HLA-E) was present at a lower frequency at day 5 in the infected patients than in uninfected healthy subjects (unpublished data). Furthermore the expanded NKG2C+ NK cells did not co-express NKG2A (Fig. 4, b and c). When longitudinally assessing the numbers of NKG2C+ NK cells in infected patients, we observed an expansion and persistence of NKG2C+ NK cells for a prolonged period of time (Fig. 4 d). This specific expansion of NKG2C+ cells did account for a significant part of the overall NK cell expansion observed within the course of this infection. Phenotypically, a predominant proportion of these NKG2C+ cells expressed CD57 after expansion (Fig. 4, e and f), indicating that they may represent highly mature and, possibly, terminally differentiated CD56dim NK cells (Björkström et al., 2010c; Lopez-Vergès et al., 2010).
Figure 4.
Expansion of NKG2C+ NK cells. (a) Frequency of NKG2C+ cells within the CD56dim NK cell population on day 5 after onset of symptoms in hantavirus-infected patients compared with noninfected controls (n = 16 infected, 59 uninfected; ***, P < 0.0001, unpaired Student’s t test; mean). (b) Representative example of costaining for NKG2A and NKG2C on CD56dim NK cells in hantavirus-infected patient at day 5 after onset of symptoms. (c) Co-expression of NKG2C and NKG2A evaluated at day 5 in infected (n = 6) and uninfected control (n = 9) individuals (horizontal bars represent mean). (d) Numbers of NKG2C+, NKG2A+, and NKG2C−NKG2A− CD56dimCD16+ NK cells evaluated longitudinally from day 5 until day 450 after onset of symptoms in infected patients (n = 6; mean ± SEM). Dashed lines represent the upper and lower SEM intervals for mean of NKG2C+ NK cell numbers in control individuals (n = 26). (e) Representative example of costaining for NKG2C and CD57 on CD56dim NK cells in one hantavirus-infected patient at day 60 after onset of symptoms. (f) Expression of CD57 evaluated at day 60 after symptom debut (n = 6) in NKG2C+ and NKG2C− CD56dim NK cells (**, P = 0.0022, Mann-Whitney test; mean). (g) Proliferation of NKG2C+ NK cells after stimulation with IL-15 and/or target cells measured by dilution of CellTrace violet. Purified NK cells were incubated for 7 d with or without irradiated K562 cells or K562*HLA-E cells, and in the absence (top) or presence (bottom) of IL-15. One representative experiment out of two is shown. (h) Expression pattern of the three major KIRs on NKG2C+ and NKG2C− NK cells. In one representative donor, a majority of the NKG2C+ NK cells were single positive for KIR2DL1/S1 (top), whereas the second representative donor shows a selective expression of KIR2DL2/S2/2DL3 (bottom) in the NKG2C+ population (see Fig. S4 for separation between activation and inhibitory KIRs). Two representative donors out of five analyzed are shown. (i) Degranulation and cytokine production responses quantified in NKG2C+ CD56dim NK cells from patients in the convalescent phase of infection after triggering with K562-E cells with or without addition of an HLA-E stabilizing peptide (n = 5; *, P < 0.05, Mann-Whitney test). (j) Absolute numbers of NKG2C+ CD56dim NK cells in CMV IgG− and IgG+ infected individuals at day 5 (mean ± SEM).
Cytokines, such as IL-15, may likely contribute to expansion of NK cells. In support of this notion, studies in humanized mice have shown that trans-presentation of IL-15 is crucial for the development and proliferation of human NK cells (Ranson et al., 2003; Huntington et al., 2009; Strowig et al., 2010). A recent study also demonstrated a dramatic expansion of NK cells (and memory CD8 T cells) in the circulation of the rhesus macaques given IL-15 (Lugli et al., 2010). However, it is more difficult to envisage how cytokines such as IL-15 could drive a specific subpopulation of NK cells, unless these harbor specific memory characteristics (Cooper et al., 2009; Sun et al., 2009). To address if cytokines, directly or in combination with target cell recognition, could lead to expansion of NKG2C+ NK cells ex vivo, we exposed NKG2C+ NK cells to IL-15 in the absence or presence of target expressing HLA-E ligands. Exposure of NKG2C+ NK cells to IL-15 did not suffice to induce significant proliferation of the cells (Fig. 4 g). However, simultaneous exposure to IL-15 and target cells expressing HLA-E led to a marked proliferative response of the NKG2C+ NK cells (Fig. 4 g). Thus, based on these studies, it is possible that recognition of infected target cells in the presence of IL-15 might have contributed to the expansion of NKG2C+ NK cells observed in hantavirus-infected patients.
Expanding NKG2C+ NK cells are licensed and functionally competent cells
The data presented thus far demonstrate a significant expansion of predominantly NKG2C+ CD56dim NK cells during acute hantavirus infection but do not disclose the functional capacity of the expanded cells. To address this, we determined the licensing status by inhibitory killer cell Ig-like receptor (KIR)/HLA expression (Kim et al., 2005; Anfossi et al., 2006) and functionality of the expanding cells upon stimulation with target cells expressing HLA-E.
NK cells expressing inhibitory receptors for self-MHC class I molecules are rendered licensed, that is, functionally more responsive to stimulation (Kim et al., 2005; Anfossi et al., 2006). In five patients examined in detail, the NKG2C+ NK cells uniformly expressed a single inhibitory HLA-C binding KIR (Fig. 4 h; and Fig. S4, a and b). In contrast, the NKG2C− NK cells in the same individuals showed a characteristic KIR expression pattern with variegated distribution of KIRs (Fig. 4 h). KIR and HLA genotyping revealed that the KIR expressed on the NKG2C+ NK cells was a KIR known to mediate NK cell licensing in four of these five patients (Table S2). This pattern observed may contribute to responsiveness to infected target cells. However, in this context, it is noteworthy that in models of experimental CMV infection, unlicensed NK cells contribute significantly to the control of infection (Orr et al., 2010). The observation of expression of a single licensing KIR on a dominant proportion of NKG2C+ cells in some of the patients studied is interesting, as it resembles some degree of clonality in the response. The driving forces behind this pattern are unknown.
To address functionality of the NKG2C+ NK cells directly, we stimulated patient NK cells with K562 cells that had been transfected with HLA-E. HLA-E was stabilized on the surface by adding an HLA-E–stabilizing peptide. Stimulation of NKG2C+ NK cells with HLA-E–expressing target cells increased degranulation, as well as IFN-γ and TNF production, both quantitatively and qualitatively (Fig. 4 i and not depicted). These results demonstrate a skewing of the NK cell compartment with the accumulation of numerous licensed multifunctional NKG2C+ NK cells after hantavirus infection.
CMV infection and NKG2C+ NK cell expansion in hantavirus infection
Most humans are infected with CMV, an infection normally occurring early in life which is associated with establishment of latency and sporadic reactivations. CMV seropositivity has been associated with higher frequencies of NKG2C+ NK cells in peripheral blood as compared with frequencies in uninfected (CMV seronegative) individuals (Gumá et al., 2004; Kuijpers et al., 2008). We therefore addressed if the elevated levels of NKG2C+ NK cells in hantavirus-infected patients could be explained by CMV reactivation. However, we did not detect any CMV DNA in plasma from infected patients, and the absolute numbers of CD8 T cells specific for the CMV pp65 epitope were not altered throughout the course of hantavirus infection (unpublished data). Thus, we have no evidence supporting the possibility that the increase in NKG2C+ NK cells could be a result of CMV reactivation during human hantavirus infection. However, it is noteworthy that 3 out of 16 patients included in the present cohort were CMV IgG negative. Interestingly, these patients had lower absolute numbers of NKG2C+ NK cells than the majority of patients being CMV IgG positive at day 5, and no subsequent expansion occurred in the IgG-negative patients (Fig. 4 j). Based on this small number of patients, the possibility arises that previous CMV infection may have primed a population of NKG2C+ NK cells for efficient expansion after hantavirus infection (Cooper et al., 2009; Sun et al., 2009). This intriguing possibility needs to be addressed further in future studies, pending the emergence of new epidemics and availability of similar clinical material.
Conclusions
The proliferation of a specific subset of NKG2C+ NK cells in a majority of patients infected with hantavirus in humans resembles results described in experimental models of CMV infection where Ly49H+ NK cells rapidly expand as a consequence of infection (Dokun et al., 2001; Robbins et al., 2004; Sun et al., 2009). However, distinct from what has been observed in mouse models, the expansion of NKG2C+ NK cells was maintained for >60 d. Consistent with previous reports on NK cell memory-like features in mice (O’Leary et al., 2006; Sun et al., 2009), one may speculate that some NKG2C+ NK cells in CMV-positive individuals harbor such features and are among the cells that rapidly proliferate in response to hantavirus infection. The present observations suggest that the human NK cell population inherently may possess features not classically attributed to the innate immune response (Sun and Lanier, 2009), including long-term persistence of specific subsets of cells and, possibly, memory-like features. Whether this represents an adaptation of the NK cell repertoire to future infections with the same or similar pathogens is unclear. Given the fact that NK cells are parts of the innate immune system, these findings merit redefinition of the possible features of an innate immune response.
MATERIALS AND METHODS
Study design and human material.
Peripheral blood was prospectively obtained from 16 patients infected with hantavirus. The following inclusion criteria were used: (a) Verified diagnosis of acute hantavirus infection. Infection was verified by an immunofluorescence test for hantavirus-reactive IgM and IgG antibodies in sera from the patients or viral load quantification by real-time PCR from patient plasma as previously described (Evander et al., 2007). (b) Access to a first sample drawn at an early time point after symptom debut (typically 3–5 d). (c) Sequential acquisition of peripheral blood during acute and convalescent phases of infection according to a defined sampling schedule with weekly samples taken during the first three weeks and later follow up samples. 35 uninfected blood donors, age- and sex-matched with the infected patients, were included as a control cohort. For isolation of PBMC, whole blood from infected patients was collected in CPT tubes (BD), centrifuged, and washed. PBMCs were frozen in 90% human albumin (Octapharma), 10% DMSO (WAK-Chemie Medical), and 50 IE heparin (LEO Pharma) and stored at −150°C for later analysis. The study was approved by the Regional Ethics Committee of Umeå University (approval number 04-113M). Written and oral informed consent was obtained from all study subjects. All clinical data, including lymphocyte count, were obtained through standard clinical procedures.
Antibodies for flow cytometry.
The following mAbs were used: anti-CD3 Pacific blue and anti-CD3 Cascade yellow (Dako); anti-CD56 PE-Cy7, anti-CD14 APC-Cy7, anti-CD16 Pacific blue, anti-CD4 biotin, anti-Ki67 FITC, anti-Bcl-2 PE, anti–ICAM-1 PE, anti-KIR3DL1 (Dx9 clone) FITC, anti-CD107a FITC, and anti-CD19 APC-Cy7 (BD); anti-CD4 biotin, visualized with Streptavidin Qdot 605 (Invitrogen); anti-KIR3DL1 (Dx9 clone) Alexa Fluor 700 (BioLegend); anti-NKG2A, anti-KIR2DL1/S1 (EB6 clone) APC, and anti-CD155 (Beckman Coulter); anti-NKG2A, conjugated with Pacific blue using a mAb labeling kit (Invitrogen); anti-KIR2DL2/S2/2DL3 (Gl183 clone; Beckman Coulter), biotinylated with FluoReporter Mini-Biotin-XX Protein Labeling kit (Invitrogen) and detected with Streptavidin PerCP (BD); anti-NKG2C PE, anti-MICA, anti-MICB, anti-ULBP1, anti-ULBP2, anti-ULBP3, anti-ULBP4, anti-KIR2DL3 (180701 clone) FITC, and anti-KIR2DL1 (143211 clone) FITC (R&D Systems); anti–HLA-E (eBioscience); and anti-CD112 (RDI). Intracellular cytokines were visualized with anti-TNF Alexa Fluor 647 (eBioscience) and anti–IFN-γ FITC (BD). HLA-A2 expression was evaluated with anti–HLA-A2 PE (clone BB7.2; BD) and HLA-C expression was evaluated with the L31 hybridoma (provided by L. Berg, Karolinska Insitutet, Stockholm, Sweden) after acid-wash treatment of the cells. Unconjugated mABs were visualized using a secondary APC mAB (BD). CD8 T cells specific for CMV were identified and enumerated using APC-conjugated HLA-A2 tetrameric complexes refolded with the CMV pp65 epitope NLVPMVATV (Beckman Coulter).
Flow cytometry.
Cell surface staining of purified PBMC or HUVECs (human umbilical cord endothelial cells; Lonza) was performed as previously described (Björkström et al., 2010a). For intracellular staining of PBMC with anti-Ki67, anti–BCL-2, or cytokines, cells were permeabilized with Cytofix/Cytoperm (BD). Samples were acquired on a CyAn ADP nine-color flow cytometer (Beckman Coulter) equipped with a 25-mW 405-nm laser, a 20-mW 488-nm laser, and a 25-mW 635-nm laser as previously described (Björkström et al., 2010a). Single-stained polystyrene beads (BD) were used for compensation purposes. Software-based compensation was performed using the compensation platform in FlowJo software version 8 (Tree Star, Inc.).
Infection of endothelial cells.
Pooled HUVECs were grown according to the manufacturer’s instructions using EGM-2 BulletKit (Lonza). Before infections, cells were seeded in cell culture plates and grown without supplementing the EGM-2 medium with hydrocortisone until 90% confluency. The Hantaan hantavirus (HTNV) strain 76–118 was used in the present study. Propagation and titration of HTNV were performed on Vero E6 cells as previously described (Stoltz et al., 2007). Cells were infected, or treated with the same amount of UV-inactivated virus as a control for nonreplicating virus, or with medium alone as a negative control.
Detection of virus-infected cells.
At 24, 48, 72, and 96 h after HTNV infection, HUVECs were fixed in methanol for 10 min at room temperature, followed by an incubation for 1 h at 37°C with convalescent human anti-hantavirus serum diluted 1:40 in PBS. After rinsing three times with PBS, cells were incubated for 1 h at 37°C with FITC-conjugated goat anti–human IgG (Sigma-Aldrich) diluted 1:50 and 5 µg/ml DAPI (Sigma-Aldrich) in PBS.
KIR and HLA genotyping.
Genomic DNA was isolated from 100 µl of peripheral blood using DNase Blood and Tissue KIT (QIAGEN). KIR genotyping was done as previously described using PCR-SPP technology and a KIR typing kit (Olerup SPP; Fauriat et al., 2008). The KIR ligand -Bw4, -Cw3 (C1), and -Cw4 (C2) motifs were determined using the KIR HLA ligand kit (Olerup-SPP).
Cell lines and surface stabilization of HLA-E.
K562 cells transfected with HLA-E*01033 (K562-E; provided by K. Söderström, Novo Nordisk A/S, Copenhagen, Denmark) were maintained in RPMI 1640 medium supplemented with 100 µg/ml l-glutamine, 10% heat-inactivated FCS, 100 U/ml penicillin G, 100 µg/ml streptomycin, and 1 mg/ml geneticin. Before functional experiments with K562-E cells, HLA-E expression was stabilized by pulsing with 100 µM of the synthetic HLA-G*0101 signal peptide VMAPRTLFL at 26°C and 5% CO2 for 15 h.
NK cell functional assays.
PBMCs were thawed and rested overnight in complete medium at a concentration of 106 cells/ml in 37°C and 5% CO2. The next day, 0.2 × 106 PBMCs were mixed with target cells at a ratio of 10:1 in V-bottom 96-well plates in a final volume of 200 µl and incubated for 6 h at 37°C in 5% CO2. When intracellular cytokine staining was performed, Brefeldin A (GolgiPlug; BD) was included at a dilution of 1:250 after 1 h of co-culture. After incubation, cells were surface stained and evaluated for CD107a, IFN-γ, and TNF expression as previously described (Bryceson et al., 2010).
NK cell proliferation assay.
To assess cytokine and/or HLA-E–induced proliferation of NKG2C+ cells, NK cells were isolated from PBMC of healthy individuals using an NK cell isolation kit (Miltenyi Biotec), labeled with CellTrace violet (Invitrogen), and incubated for 7 d with irradiated (90 Gy) K562 cells or K562*HLA-E cells (NK cell to target cell ratio of 1:1) in the presence or absence of 20 ng/ml human recombinant IL-15 (PeproTech). Proliferation was assessed by analyzing dilution of CellTrace violet in NKG2C+ CD56dim NK cells by flow cytometry. The K562*HLA-E transfectant (clone 2B4), constitutively expressing stabilized HLA-E (Falk et al., 2002), was provided by C.S. Falk (University of Heidelberg, Heidelberg, Germany).
Statistics.
Data were statistically analyzed using Prism software (GraphPad Software, Inc.). P-values of <0.05 were considered significant. For comparisons between groups having <15 observations, nonparametric statistical tests were used, for example, the Mann Whitney test. For comparisons between groups having >15 observations, parametric statistical tests were used, for example, paired and nonpaired Student’s t tests. If nothing else is noted, bars in the figures represent SEM.
Online supplemental material.
Fig. S1 shows clinical and virological data from the hantavirus-infected patients. Fig. S2 shows the relative frequency of NK cells of total lymphocytes in the infected patients and healthy controls, as well as the relative frequencies of CD56brightCD16−, CD56brightCD16+, CD56dimCD16+, and CD56−CD16+ NK cells out of total NK cells in the infected patients and healthy controls. Fig. S3 presents data on the levels of NK cell stimulatory cytokines from the infected patients. Fig. S4 shows the FACS gating algorithm used to identify single-, double-, and triple-KIR+ NK cells and representative FACS plots for the algorithm used to dissect expression of KIR2DL1 and KIR2DS1, as well as KIR2DL3 and KIR2DL2/S2 on NK cells. Table S1 shows clinical characteristics of patients included in the study. Table S2 shows KIR and KIR-ligand genotyping of infected patients. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100762/DC1.
Acknowledgments
We thank patients and blood donors who have contributed clinical material to this study. We are also thankful to the staff at the Department of Infectious Diseases, Laboratory of Clinical Hematology and Blood Bank, Umeå University Hospital, for assistance with the collection of clinical material.
This work was supported by grants from the Swedish Foundation for Strategic Research, the Swedish Research Council, the Swedish Cancer Society, the Royal Swedish Academy of Sciences, the Cancer Society of Stockholm, the Karolinska Institutet, the Karolinska University Hospital, the County of Västerbotten, the Medical Faculty of Umeå University, and the County Councils of Northern Sweden.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ICAM
- intercellular adhesion molecule
- KIR
- killer cell Ig-like receptor
- LFA-1
- lymphocyte function-associated antigen 1
References
- Anfossi N., André P., Guia S., Falk C.S., Roetynck S., Stewart C.A., Breso V., Frassati C., Reviron D., Middleton D., et al. 2006. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 25:331–342 10.1016/j.immuni.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Arase H., Mocarski E.S., Campbell A.E., Hill A.B., Lanier L.L. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 296:1323–1326 10.1126/science.1070884 [DOI] [PubMed] [Google Scholar]
- Becknell B., Caligiuri M.A. 2005. Interleukin-2, interleukin-15, and their roles in human natural killer cells. Adv. Immunol. 86:209–239 10.1016/S0065-2776(04)86006-1 [DOI] [PubMed] [Google Scholar]
- Biron C.A., Byron K.S., Sullivan J.L. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320:1731–1735 10.1056/NEJM198906293202605 [DOI] [PubMed] [Google Scholar]
- Björkström N.K., Fauriat C., Bryceson Y.T., Sandberg J.K., Ljunggren H.G., Malmberg K.J. 2010a. Analysis of the KIR repertoire in human NK cells by flow cytometry. Methods Mol. Biol. 612:353–364 10.1007/978-1-60761-362-6_24 [DOI] [PubMed] [Google Scholar]
- Björkström N.K., Ljunggren H.G., Sandberg J.K. 2010b. CD56 negative NK cells: origin, function, and role in chronic viral disease. Trends Immunol. 31:401–406 10.1016/j.it.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Björkström N.K., Riese P., Heuts F., Andersson S., Fauriat C., Ivarsson M.A., Björklund A.T., Flodström-Tullberg M., Michaëlsson J., Rottenberg M.E., et al. 2010c. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 116:3853–3864 10.1182/blood-2010-04-281675 [DOI] [PubMed] [Google Scholar]
- Bonavida B., Katz J., Gottlieb M. 1986. Mechanism of defective NK cell activity in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. I. Defective trigger on NK cells for NKCF production by target cells, and partial restoration by IL 2. J. Immunol. 137:1157–1163 [PubMed] [Google Scholar]
- Braud V.M., Allan D.S., O’Callaghan C.A., Söderström K., D’Andrea A., Ogg G.S., Lazetic S., Young N.T., Bell J.I., Phillips J.H., et al. 1998. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 391:795–799 10.1038/35869 [DOI] [PubMed] [Google Scholar]
- Bryceson Y.T., March M.E., Barber D.F., Ljunggren H.G., Long E.O. 2005. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J. Exp. Med. 202:1001–1012 10.1084/jem.20051143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson Y.T., March M.E., Ljunggren H.G., Long E.O. 2006. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol. Rev. 214:73–91 10.1111/j.1600-065X.2006.00457.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson Y.T., Rudd E., Zheng C., Edner J., Ma D., Wood S.M., Bechensteen A.G., Boelens J.J., Celkan T., Farah R.A., et al. 2007. Defective cytotoxic lymphocyte degranulation in syntaxin-11 deficient familial hemophagocytic lymphohistiocytosis 4 (FHL4) patients. Blood. 110:1906–1915 10.1182/blood-2007-02-074468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryceson Y.T., Fauriat C., Nunes J.M., Wood S.M., Björkström N.K., Long E.O., Ljunggren H.G. 2010. Functional analysis of human NK cells by flow cytometry. Methods Mol. Biol. 612:335–352 10.1007/978-1-60761-362-6_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching C., Lopez C. 1979. Natural killing of herpes simplex virus type 1-infected target cells: normal human responses and influence of antiviral antibody. Infect. Immun. 26:49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M.A., Elliott J.M., Keyel P.A., Yang L., Carrero J.A., Yokoyama W.M. 2009. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA. 106:1915–1919 10.1073/pnas.0813192106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokun A.O., Kim S., Smith H.R., Kang H.S., Chu D.T., Yokoyama W.M. 2001. Specific and nonspecific NK cell activation during virus infection. Nat. Immunol. 2:951–956 10.1038/ni714 [DOI] [PubMed] [Google Scholar]
- Evander M., Eriksson I., Pettersson L., Juto P., Ahlm C., Olsson G.E., Bucht G., Allard A. 2007. Puumala hantavirus viremia diagnosed by real-time reverse transcriptase PCR using samples from patients with hemorrhagic fever and renal syndrome. J. Clin. Microbiol. 45:2491–2497 10.1128/JCM.01902-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk C.S., Mach M., Schendel D.J., Weiss E.H., Hilgert I., Hahn G. 2002. NK cell activity during human cytomegalovirus infection is dominated by US2-11-mediated HLA class I down-regulation. J. Immunol. 169:3257–3266 [DOI] [PubMed] [Google Scholar]
- Fauci A.S., Mavilio D., Kottilil S. 2005. NK cells in HIV infection: paradigm for protection or targets for ambush. Nat. Rev. Immunol. 5:835–843 10.1038/nri1711 [DOI] [PubMed] [Google Scholar]
- Fauriat C., Andersson S., Björklund A.T., Carlsten M., Schaffer M., Björkström N.K., Baumann B.C., Michaëlsson J., Ljunggren H.G., Malmberg K.J. 2008. Estimation of the size of the alloreactive NK cell repertoire: studies in individuals homozygous for the group A KIR haplotype. J. Immunol. 181:6010–6019 [DOI] [PubMed] [Google Scholar]
- Gazit R., Gruda R., Elboim M., Arnon T.I., Katz G., Achdout H., Hanna J., Qimron U., Landau G., Greenbaum E., et al. 2006. Lethal influenza infection in the absence of the natural killer cell receptor gene Ncr1. Nat. Immunol. 7:517–523 10.1038/ni1322 [DOI] [PubMed] [Google Scholar]
- Gerdes J., Lemke H., Baisch H., Wacker H.H., Schwab U., Stein H. 1984. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 133:1710–1715 [PubMed] [Google Scholar]
- Gonzalez V.D., Falconer K., Björkström N.K., Blom K.G., Weiland O., Ljunggren H.G., Alaeus A., Sandberg J.K. 2009. Expansion of functionally skewed CD56-negative NK cells in chronic hepatitis C virus infection: correlation with outcome of pegylated IFN-alpha and ribavirin treatment. J. Immunol. 183:6612–6618 10.4049/jimmunol.0901437 [DOI] [PubMed] [Google Scholar]
- Gumá M., Angulo A., Vilches C., Gómez-Lozano N., Malats N., López-Botet M. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 104:3664–3671 10.1182/blood-2004-05-2058 [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Legrand N., Alves N.L., Jaron B., Weijer K., Plet A., Corcuff E., Mortier E., Jacques Y., Spits H., Di Santo J.P. 2009. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 206:25–34 10.1084/jem.20082013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncas J., Monczak Y., Ghibu F., Alfieri C., Bonin A., Ahronheim G., Rivard G. 1989. Brief report: killer cell defect and persistent immunological abnormalities in two patients with chronic active Epstein-Barr virus infection. J. Med. Virol. 28:110–117 10.1002/jmv.1890280211 [DOI] [PubMed] [Google Scholar]
- Katz J.D., Mitsuyasu R., Gottlieb M.S., Lebow L.T., Bonavida B. 1987. Mechanism of defective NK cell activity in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. II. Normal antibody-dependent cellular cytotoxicity (ADCC) mediated by effector cells defective in natural killer (NK) cytotoxicity. J. Immunol. 139:55–60 [PubMed] [Google Scholar]
- Kim S., Poursine-Laurent J., Truscott S.M., Lybarger L., Song Y.J., Yang L., French A.R., Sunwoo J.B., Lemieux S., Hansen T.H., Yokoyama W.M. 2005. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 436:709–713 10.1038/nature03847 [DOI] [PubMed] [Google Scholar]
- Kuijpers T.W., Baars P.A., Dantin C., van den Burg M., van Lier R.A., Roosnek E. 2008. Human NK cells can control CMV infection in the absence of T cells. Blood. 112:914–915 10.1182/blood-2008-05-157354 [DOI] [PubMed] [Google Scholar]
- Lee S.H., Miyagi T., Biron C.A. 2007. Keeping NK cells in highly regulated antiviral warfare. Trends Immunol. 28:252–259 10.1016/j.it.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Lodoen M.B., Lanier L.L. 2006. Natural killer cells as an initial defense against pathogens. Curr. Opin. Immunol. 18:391–398 10.1016/j.coi.2006.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Vergès S., Milush J.M., Pandey S., York V.A., Arakawa-Hoyt J., Pircher H., Norris P.J., Nixon D.F., Lanier L.L. 2010. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 116:3865–3874 10.1182/blood-2010-04-282301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli E., Goldman C.K., Perera L.P., Smedley J., Pung R., Yovandich J.L., Creekmore S.P., Waldmann T.A., Roederer M. 2010. Transient and persistent effects of IL-15 on lymphocyte homeostasis in nonhuman primates. Blood. 116:3238–3248 10.1182/blood-2010-03-275438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelboim O., Lieberman N., Lev M., Paul L., Arnon T.I., Bushkin Y., Davis D.M., Strominger J.L., Yewdell J.W., Porgador A. 2001. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature. 409:1055–1060 10.1038/35059110 [DOI] [PubMed] [Google Scholar]
- Merino F., Henle W., Ramírez-Duque P. 1986. Chronic active Epstein-Barr virus infection in patients with Chediak-Higashi syndrome. J. Clin. Immunol. 6:299–305 10.1007/BF00917330 [DOI] [PubMed] [Google Scholar]
- Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D., Murali-Krishna K., Mahar P.L., Edupuganti S., Lalor S., et al. 2008. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 28:710–722 10.1016/j.immuni.2008.02.020 [DOI] [PubMed] [Google Scholar]
- O’Leary J.G., Goodarzi M., Drayton D.L., von Andrian U.H. 2006. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat. Immunol. 7:507–516 10.1038/ni1332 [DOI] [PubMed] [Google Scholar]
- Orange J.S. 2006. Human natural killer cell deficiencies. Curr. Opin. Allergy Clin. Immunol. 6:399–409 10.1097/ACI.0b013e3280106b65 [DOI] [PubMed] [Google Scholar]
- Orr M.T., Murphy W.J., Lanier L.L. 2010. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat. Immunol. 11:321–327 10.1038/ni.1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson L., Boman J., Juto P., Evander M., Ahlm C. 2008. Outbreak of Puumala virus infection, Sweden. Emerg. Infect. Dis. 14:808–810 10.3201/eid1405.071124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prlic M., Blazar B.R., Farrar M.A., Jameson S.C. 2003. In vivo survival and homeostatic proliferation of natural killer cells. J. Exp. Med. 197:967–976 10.1084/jem.20021847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinnan G.V., Jr, Kirmani N., Rook A.H., Manischewitz J.F., Jackson L., Moreschi G., Santos G.W., Saral R., Burns W.H. 1982. Cytotoxic t cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N. Engl. J. Med. 307:7–13 10.1056/NEJM198207013070102 [DOI] [PubMed] [Google Scholar]
- Ranson T., Vosshenrich C.A., Corcuff E., Richard O., Müller W., Di Santo J.P. 2003. IL-15 is an essential mediator of peripheral NK-cell homeostasis. Blood. 101:4887–4893 10.1182/blood-2002-11-3392 [DOI] [PubMed] [Google Scholar]
- Rehermann B., Nascimbeni M. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 5:215–229 10.1038/nri1573 [DOI] [PubMed] [Google Scholar]
- Robbins S.H., Tessmer M.S., Mikayama T., Brossay L. 2004. Expansion and contraction of the NK cell compartment in response to murine cytomegalovirus infection. J. Immunol. 173:259–266 [DOI] [PubMed] [Google Scholar]
- Robertson M.J., Caligiuri M.A., Manley T.J., Levine H., Ritz J. 1990. Human natural killer cell adhesion molecules. Differential expression after activation and participation in cytolysis. J. Immunol. 145:3194–3201 [PubMed] [Google Scholar]
- Rodella L., Zamai L., Rezzani R., Artico M., Peri G., Falconi M., Facchini A., Pelusi G., Vitale M. 2001. Interleukin 2 and interleukin 15 differentially predispose natural killer cells to apoptosis mediated by endothelial and tumour cells. Br. J. Haematol. 115:442–450 10.1046/j.1365-2141.2001.03055.x [DOI] [PubMed] [Google Scholar]
- Santoli D., Trinchieri G., Koprowski H. 1978. Cell-mediated cytotoxicity against virus-infected target cells in humans. II. Interferon induction and activation of natural killer cells. J. Immunol. 121:532–538 [PubMed] [Google Scholar]
- Schönrich G., Rang A., Lütteke N., Raftery M.J., Charbonnel N., Ulrich R.G. 2008. Hantavirus-induced immunity in rodent reservoirs and humans. Immunol. Rev. 225:163–189 10.1111/j.1600-065X.2008.00694.x [DOI] [PubMed] [Google Scholar]
- Stoltz M., Ahlm C., Lundkvist A., Klingström J. 2007. Lambda interferon (IFN-lambda) in serum is decreased in hantavirus-infected patients, and in vitro-established infection is insensitive to treatment with all IFNs and inhibits IFN-gamma-induced nitric oxide production. J. Virol. 81:8685–8691 10.1128/JVI.00415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T., Chijioke O., Carrega P., Arrey F., Meixlsperger S., Rämer P.C., Ferlazzo G., Münz C. 2010. Human NK cells of mice with reconstituted human immune system components require preactivation to acquire functional competence. Blood. 116:4158–4167 10.1182/blood-2010-02-270678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Lanier L.L. 2009. Natural killer cells remember: an evolutionary bridge between innate and adaptive immunity? Eur. J. Immunol. 39:2059–2064 10.1002/eji.200939435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.C., Beilke J.N., Lanier L.L. 2009. Adaptive immune features of natural killer cells. Nature. 457:557–561 10.1038/nature07665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terajima M., Hayasaka D., Maeda K., Ennis F.A. 2007. Immunopathogenesis of hantavirus pulmonary syndrome and hemorrhagic fever with renal syndrome: Do CD8+ T cells trigger capillary leakage in viral hemorrhagic fevers? Immunol. Lett. 113:117–120 10.1016/j.imlet.2007.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timonen T. 1997. Natural killer cells: endothelial interactions, migration, and target cell recognition. J. Leukoc. Biol. 62:693–701 [DOI] [PubMed] [Google Scholar]
- Tomasec P., Braud V.M., Rickards C., Powell M.B., McSharry B.P., Gadola S., Cerundolo V., Borysiewicz L.K., McMichael A.J., Wilkinson G.W. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 287:1031 10.1126/science.287.5455.1031 [DOI] [PubMed] [Google Scholar]
- Trinchieri G., Santoli D., Koprowski H. 1978. Spontaneous cell-mediated cytotoxicity in humans: role of interferon and immunoglobulins. J. Immunol. 120:1849–1855 [PubMed] [Google Scholar]
- Vapalahti O., Mustonen J., Lundkvist A., Henttonen H., Plyusnin A., Vaheri A. 2003. Hantavirus infections in Europe. Lancet Infect. Dis. 3:653–661 10.1016/S1473-3099(03)00774-6 [DOI] [PubMed] [Google Scholar]