Thymus-specific serine protease expression in stromal as well as hematopoietic cells in the thymus is needed for diversification of the endogenous repertoire of TCRs specific for a particular protein antigen.
Abstract
Thymus-specific serine protease (TSSP) is a novel protease that may contribute to the generation of the peptide repertoire presented by MHC class II molecules in the thymus. Although TSSP deficiency has no quantitative impact on the development of CD4 T cells expressing a polyclonal T cell receptor (TCR) repertoire, the development of CD4 T cells expressing the OTII and Marilyn transgenic TCRs is impaired in TSSP-deficient mice. In this study, we assess the role of TSSP in shaping the functional endogenous polyclonal CD4 T cell repertoire by analyzing the response of TSSP-deficient mice to several protein antigens (Ags). Although TSSP-deficient mice responded normally to most of the Ags tested, they responded poorly to hen egg lysozyme (HEL). The impaired CD4 T cell response of TSSP-deficient mice to HEL correlated with significant alteration of the dominant TCR-β chain repertoire expressed by HEL-specific CD4 T cells, suggesting that TSSP is necessary for the intrathymic development of cells expressing these TCRs. Thus, TSSP contributes to the diversification of the functional endogenous CD4 T cell TCR repertoire in the thymus.
The functional T cell repertoire is shaped in the thymus by positive and negative selection through interactions with MHC–peptide complexes expressed by thymic epithelial cells (TECs) and BM-derived APCs. Consequently, the size of the CD4 T cell compartment and its diversity are determined by the complexity of the self-peptide repertoire presented by MHC class II molecules in the thymus (Marrack and Kappler, 1997; Ebert et al., 2009; Lo et al., 2009).
The peptides for loading on MHC class II molecules are generated by sequential proteolysis of endosomal proteins. At present, only cysteine proteases, including cathepsin (Cat)-S, -L, -F, and -H and asparaginyl endopeptidase (AEP), and the aspartyl proteases Cat-D and -E have been linked to antigen (Ag) processing (Hsing and Rudensky, 2005). However, some indirect evidence suggests that serine proteases and metalloproteases may also contribute to this latter process (Musson et al., 2003, 2006).
Although the different Ag-processing enzymes present broad specificities, the generation of some peptides may be strictly dependent on a given protease. Thus, Cat-S appeared necessary for the generation of some class II epitopes of myoglobin (Myo), hen egg lysozyme (HEL), OVA, and KLH (Nakagawa et al., 1999; Shi et al., 1999; Plüger et al., 2002). Large-scale analysis of the I-Ab–bound peptides in embryonic fibroblast lines expressing either Cat-S or Cat-L also suggested that these two enzymes have some substrate specificities (Hsieh et al., 2002). Similarly, AEP enhances processing of tetanus toxin for presentation by human EBV-B cell lines but destroys a dominant epitope of myelin basic protein (Manoury et al., 1998, 2002). Whether and how these substrate specificities impact on the selection of the CD4 T cell repertoire in the thymus remain unknown.
Thymus-specific serine protease (TSSP) is a putative serine protease that was initially described as a gene linked to a diabetes susceptibility locus in humans (Lie et al., 1999). Subsequent studies showed that TSSP was predominantly expressed in the endosomal compartment of cortical TECs (cTECs; Bowlus et al., 1999; Carrier et al., 1999). This latter observation led to the hypothesis that TSSP may contribute to the generation of the peptide repertoire presented by MHC class II molecules in the thymus and consequently shape the CD4 T cell repertoire. TSSP-deficient (Tssp−/−) B6 and nonobese diabetic (NOD) mice show normal thymic development of CD4 T cells expressing polyclonal TCR (Cheunsuk et al., 2005; Gommeaux et al., 2009; unpublished data). However, TSSP is necessary for the development of CD4 T cells expressing the class II–restricted OTII and Marilyn transgenic TCRs (Gommeaux et al., 2009). These different observations suggested that TSSP has likely no generic role in MHC class II presentation but may contribute to the diversification of the peptide repertoire presented by MHC class II molecules in the thymus. In this study, we evaluated the impact of this putative peptide-editing function of TSSP on the development of the functional polyclonal CD4 T cell repertoire of Tssp−/− NOD mice.
RESULTS AND DISCUSSION
Tssp−/− mice are immunocompetent
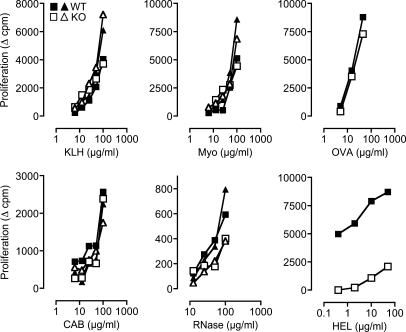
Because TSSP is thought to have a role in type 1 diabetes, we focused our study on Tssp−/− NOD mice. To assess the role of TSSP in the development of a functional CD4 T cell repertoire, we immunized Tssp−/− and WT control NOD mice with different protein Ags in CFA and analyzed the in vitro proliferative response of purified CD4 T cells upon restimulation with the immunizing Ag and WT APC. We found that the CD4 T cell responses of Tssp−/− and WT control mice to KLH, equine Myo, chicken OVA, chicken conalbumin, and bovine RNase (RNase) were similar (Fig. 1). In sharp contrast, the CD4 T cell response of Tssp−/− mice to HEL was dramatically reduced as compared with that of WT control mice (Fig. 1). Collectively, these results show that Tssp−/− mice are immunocompetent and respond to most of the protein Ags tested to levels comparable with those of WT mice. However, Tssp−/− mice are hyporesponsive to HEL.
Figure 1.
Reduced CD4 T cell proliferative response to HEL in Tssp−/− mice. Tssp−/− (KO) and WT control mice were immunized with the indicated protein. 10–11 d later, CD4 T cells were isolated from the draining LN and restimulated in vitro with WT spleen APCs and graded concentrations of the immunizing protein. For each Ag, one representative experiment out of at least two performed with one or two mice of each genotype is shown. Each symbol corresponds to one individual mouse. CAB, conalbumin.
The reduced response of Tssp−/− mice to HEL does not result from defective processing of the HEL protein
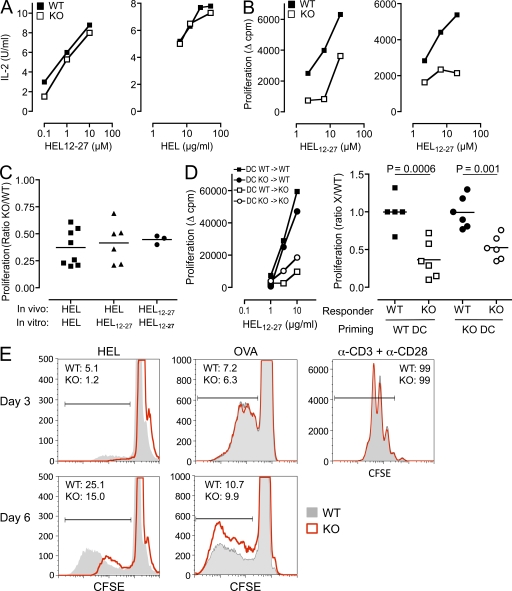
Because TSSP is a putative protease of the MHC class II pathway, it was possible that the defective response of Tssp−/− mice to HEL may result from inefficient processing of the protein by APCs in vivo and consequently inefficient priming of the HEL-specific CD4 T cells. To address this issue, we first analyzed the presentation of HEL protein, and as control the HEL12–27 peptide, by WT and Tssp−/− spleen APCs using the BW-HEL clone that expresses mouse CD4 and an IAg7-restricted TCR specific for the NOD immunodominant epitope HEL12–27. Both types of APCs presented equally well the HEL protein and HEL12–27 peptide to the BW-HEL clone, indicating that expression of TSSP by spleen APCs is not required for generation of the HEL immunodominant epitope in vitro (Fig. 2 A). To further determine whether TSSP was not required for HEL processing in vivo, we immunized Tssp−/− and WT control mice with either the HEL protein or the HEL12–27 peptide and restimulated in vitro the primed CD4 T cells with the HEL12–27 peptide. We found that regardless of the nature of Ag used for the in vivo priming, the proliferative response of CD4 T cells from Tssp−/− mice was reduced as compared with that of WT CD4 T cells (Fig. 2, B and C). Finally, we compared the in vivo priming efficacy of peptide-loaded BM-derived WT and Tssp−/− DCs and found that the two DC populations were equivalent at inducing high response in WT control and low response in Tssp−/− mice (Fig. 2 D).
Figure 2.
The reduced response of Tssp−/− mice to HEL is a CD4 T cell property. (A) IL-2 production by the HEL-specific T cell transfectant BW-HEL upon stimulation with splenic APCs from Tssp−/− (KO) or WT control mice loaded with graded concentrations of HEL12–27 peptide or HEL protein, as indicated. One representative experiment out of four performed is shown. (B) WT or Tssp−/− (KO) mice were primed with HEL protein (left) or HEL12–27 peptide (right). 11 d later, CD4 T cells were isolated from the draining LN and restimulated in vitro with WT spleen APCs and graded concentrations of HEL12–27 peptide. (C) WT and Tssp−/− mice were primed in vivo, and their CD4 T cells were restimulated in vitro with WT spleen APCs and either HEL protein or HEL12–27 peptide, as indicated. Results are expressed as the ratio of the proliferation of TSSP-deficient T cells divided by that of WT control. The figure compiles the results of three to five independent experiments. Each symbol corresponds to one mouse. Horizontal bars represent the mean. One-sample t test showed that the responses of WT and Tssp−/− mice are significantly different for all stimulation conditions (P < 0.002). (D) WT and Tssp−/− (KO) mice were primed in vivo with either WT or TSSP-deficient DCs pulsed with 40 ng HEL12–27 peptide, and their CD4 T cells were restimulated in vitro with WT spleen APCs and a titrated amount of HEL12–27 peptide. One representative experiment is presented in the left panel. The right panel combines results of three independent experiments expressed as the ratio of the proliferation of WT or TSSP-deficient T cells (X) divided by the mean value of the WT controls in the same experiment. Each symbol corresponds to one mouse. Horizontal bars represent the mean. Significant p-values are shown. (E) Purified LN CD4 T cells from HEL- or OVA-immunized WT and Tssp−/− mice were labeled with CFSE and stimulated with WT splenocytes together with HEL12–27 peptide, OVA protein, or anti-CD3/CD28 antibodies, as indicated. Cell division was evaluated by analyzing CFSE dilution on gated CD4 T cells at day 3 and 6 of activation. The percentage of divided WT or TSSP-deficient (KO) CD4 T cells is shown. One representative experiment out of four performed is shown.
To gain more insight into the reduced response of Tssp−/− mice, we examined the proliferative profile of CFSE-labeled HEL-primed CD4 T cells upon in vitro restimulation. We found that at both day 3 and 6 after in vitro restimulation, CD4 T cells from Tssp−/− deficient mice accomplished fewer divisions than those from WT mice (Fig. 2 E). No such difference was observed in response to OVA or anti-CD3 stimulation (Fig. 2 E). These results strongly suggest that in mice lacking TSSP, the HEL-specific CD4 T cells might display a lower affinity than their WT counterparts.
Impaired thymic selection of HEL-specific CD4 T cells in Tssp−/− mice
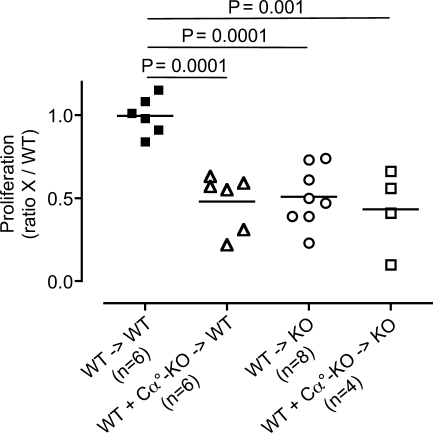
The aforementioned results suggested that TSSP might be necessary for the intrathymic development of high affinity HEL-specific CD4 T cells. In agreement, we found that the thymic development of CD4 T cells expressing a given HEL-specific IAg7-restricted TCR was impaired in Tssp−/− mice as compared with WT NOD mice (Fig. S1). We next examined whether TSSP deficiency may likewise impact on the generation of a polyclonal HEL-specific CD4 T cell repertoire using BM chimeras. TSSP is highly expressed by cTEC (Bowlus et al., 1999; Carrier et al., 1999; Cheunsuk et al., 2002) but also, at lower levels, by thymic DCs (unpublished data). Therefore, we examined the effect of TSSP deficiency in radio-resistant TECs on the differentiation of some HEL-specific CD4 T cells by injecting WT BM cells into lethally irradiated Tssp−/− NOD-Cα° mice (WT → KO) or, as control, into NOD-Cα° host (WT → WT). Conversely, to assess the role of TSSP-deficient BM-derived thymic APCs in deleting some HEL-specific CD4 T cells, we reconstituted irradiated NOD-Cα° mice with a mix of WT and NOD-Cα° TSSP-deficient BM cells (WT + Cα°-KO → WT). We found that both WT → KO and WT + Cα°-KO → WT chimeras showed a reduced CD4 T cell response to HEL that was comparable with that observed in Tssp−/− mice or control chimera, in which both the TECs and thymic APCs lack TSSP expression (Fig. 3).
Figure 3.
Impaired thymic selection of HEL-specific CD4 T cells in Tssp−/− mice. WT or Tssp−/− (KO) mice were lethally irradiated before reconstitution with WT BM cells (WT) or a 1:1 mix of WT and NOD-Cα° TSSP-deficient (Cα°-KO) BM cells, as indicated (BM → host). 8–10 wk after reconstitution, chimeras were immunized with HEL protein, and 7 d later, their CD4 T cell proliferative response to 10 µg/ml HEL12–27 pulsed WT APCs was analyzed. The results of three independent experiments are expressed as the ratio of the proliferation of WT or TSSP-deficient CD4 T cells (X) divided by the mean value of the WT control chimeras in the same experiment. Each symbol corresponds to one individual chimera. Horizontal bars represent the mean. Significant p-values are shown.
Collectively, these results indicate that hyporesponsiveness of Tssp−/− mice to HEL reflects a CD4 T cell property that is acquired during thymic differentiation. The observation that in addition to TEC thymic DCs lacking TSSP can also induce hyporesponsiveness to HEL reveals a new function of TSSP in the thymus. Indeed, in this latter case, TSSP-deficient DCs likely induce the deletion of some high affinity HEL-reactive CD4 T cells through the presentation of high affinity TCR ligands. Thus, the protease TSSP would prevent the generation of this HEL mimotope. Although we do not detect TSSP messenger RNA in Tssp−/− thymi, we cannot formally exclude the trivial possibility that the recombination events during gene targeting of the Tssp locus led to a truncated polypeptide containing an HEL12–27 mimotope. Nonetheless, our observations further questioned the mechanism by which TSSP-deficient TECs, and more likely cTECs, may alter the HEL-specific CD4 T cell repertoire. Indeed, cTECs can induce positive selection but also negative selection of some self-reactive T cells (Goldman et al., 2005; McCaughtry et al., 2008; Klein et al., 2009). Therefore, it is possible that cTECs lacking TSSP may induce negative selection of some HEL-specific CD4 T cells either directly or indirectly through cross-presentation of self-determinants by thymic DCs.
Distinct TCR repertoire features of HEL-responsive CD4 T cells from Tssp−/− mice
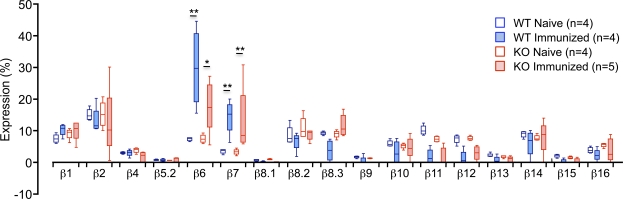
The aforementioned results suggested that TSSP deficiency might lead to modification of the TCR repertoire of HEL-responsive CD4 T cells. To address this question, we first examined whether TSSP deficiency may alter the Vβ chain usage of HEL12–27-specific CD4 T cells. For this experiment, we isolated CD4 T cells from HEL-primed mice, labeled them with CFSE, and stimulated them in vitro with the HEL12–27 peptide and syngeneic APC. 5–6 d later, the HEL-responsive CD4 T cells were FACS sorted based on CFSE dilution, and expression of the different Vβ gene segments was analyzed by real-time PCR. Parallel experiments showed that the Vβ usage of naive CD4 T cells from unprimed WT and Tssp−/− mice were similar (Fig. 4). We found in the HEL-specific CD4 T cells from WT mice a marked amplification of the Vβ6 segment, representing 29.9 ± 13.2% (mean ± SD; n = 4) of the total Vβ gene usage and a fourfold increase compared with naive CD4 T cells (Fig. 4). Among other Vβ segments, only Vβ7 values were significantly increased in HEL12–27-responsive CD4 T cells from all WT mice relative to CD4 T cells from naive mice (fourfold increase). However, Vβ7 usage by HEL-specific CD4 T cells was not significantly different from that of other Vβ segments. Therefore, in NOD mice, HEL12–27-specific CD4 T cells preferentially use Vβ6 segments. Importantly, such Vβ6 bias was not observed when the same CD4 T cells were stimulated with anti-CD3/CD28 mAbs (Table S1).
Figure 4.
Vβ gene segment usage by naive and HEL12–27-responsive CD4 T cells from Tssp−/− and WT control mice. CD4 T cells isolated from HEL-primed WT or Tssp−/− mice were CFSE labeled and stimulated with HEL12–27 peptide in the presence of irradiated WT splenocytes. After 5–6 d of culture, dividing CD4 T cells were FACS sorted before RT-coupled real-time PCR analysis. Alternatively, LN CD4 T cells were isolated from 9–13-wk-old naive mice. Box and whiskers graph shows the Vβ gene usage for naive or HEL-specific CD4 T cells isolated from WT or Tssp−/− (KO) mice (n = number of mice). Horizontal bars show the median values, boxes show the 25th and 75th percentile, and bars show the minimal and maximal values. In WT mice, Vβ6 usage was significantly higher than that of all other Vβ genes, except Vβ7 (P < 0.05). The p-value of those Vβ with an increased representation in CD4 T cells from primed mice as compared with that of naive mice is shown (*, P < 0.05; **, P < 0.01).
This prominent Vβ6 segment usage was not observed for HEL12–27-responsive CD4 T cells from HEL-primed Tssp−/− mice. Indeed, for this CD4 T cell population, the representation of Vβ6 segments (17.8 ± 7.9%; n = 5) was not significantly higher than that of Vβ1, Vβ2, Vβ7, Vβ8.2, and Vβ8.3 segments (Fig. 4). However, in this case too, Vβ6 and Vβ7 gene usage by HEL-specific CD4 T cells was significantly increased as compared with that of naive CD4 T cells (2.4- and 3.7-fold, respectively). Thus, the reduced CD4 T cell response of Tssp−/− mice to HEL12–27 correlated with a lowered usage of the Vβ6 gene segment.
To further examine the impact of TSSP deficiency on the HEL-specific TCR repertoire, we cloned and sequenced their Vβ6-Jβ-Cβ region. The analysis of 132 clones derived from the aforementioned four WT and five Tssp−/− mice identified 54 distinct CDR3 sequences that could be ordered into three groups (Table I). A first group included sequences found only in WT CD4 T cells (42% of sequences; sequences 1–23), and a second group included those found only in CD4 T cells from Tssp−/− mice (37% of the sequences; sequences 35–54). Finally, 20% of sequences could be amplified from both types of cells (sequences 24–34).
Table I.
Vβ6-Cβ joint of HEL12–27-specific CD4 T cells
| Sequence no. | Sequences (aa) | Jβ | L | nCh | WT | Tssp−/− | |||||||
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 5 | |||||
| 1 | SIWQGRV | 1.1 | 7 | 1 | 1 | ||||||||
| 2 | SIGGNTEV | 1.1 | 8 | −1 | 1 | ||||||||
| 3 | WDRGGATEV | 1.1 | 9 | −1 | 1 | ||||||||
| 4 | SDRVHSDY | 1.2 | 8 | 0 | 1 | ||||||||
| 5 | SIDTSGNTL | 1.3 | 9 | −1 | 1 | 1 | |||||||
| 6 | SIRTGGFEQ | 2.1 | 9 | 0 | 1 | ||||||||
| 7 | SRTDYAEQ | 2.1 | 8 | −1 | 1 | ||||||||
| 8 | SIWGGNYAEQ | 2.1 | 10 | −1 | 1 | ||||||||
| 9 | SSRDRNYAEQ | 2.1 | 10 | 0 | 1 | ||||||||
| 10 | SIRDRNYAEQ | 2.1 | 10 | 0 | 1 | ||||||||
| 11 | SVRDKNTGQL | 2.2 | 10 | 1 | 1 | ||||||||
| 12 | SIWGSAETL | 2.3 | 9 | −1 | 1 | 1 | |||||||
| 13 | SRLAHRAETL | 2.3 | 10 | 2 | 1 | ||||||||
| 14 | SLGTAETL | 2.3 | 8 | −1 | 1 | ||||||||
| 15 | TPAGSSAETL | 2.3 | 10 | −1 | 1 | ||||||||
| 16 | SIVGGRNTL | 2.4 | 9 | 1 | 1 | ||||||||
| 17 | SGTQDTQ | 2.5 | 7 | −1 | 1 | ||||||||
| 18 | SIDWGNQDTQ | 2.5 | 10 | −2 | 1 | ||||||||
| 19 | SIGGTGGYEQ | 2.7 | 10 | −1 | 1 | ||||||||
| 20 | SIAWTGGYEQ | 2.7 | 10 | −1 | 1 | ||||||||
| 21 | SIEDRYEQ | 2.7 | 8 | −2 | 1 | ||||||||
| 22 | SIADRYEQ | 2.7 | 8 | −1 | 2 | ||||||||
| 23 | SINRGEQ | 2.7 | 7 | 0 | 1 | ||||||||
| 24 | SINERL | 1.4 | 6 | 0 | 1 | 2 | |||||||
| 25 | SIGRGERL | 1.4 | 8 | 1 | 1 | 1 | |||||||
| 26 | SIGTISNERL | 1.4 | 10 | 0 | 1 | 1 | |||||||
| 27 | SISNYAEQ | 2.1 | 8 | −1 | 1 | 2 | 2 | 1 | |||||
| 28 | RGQGNNYAEQ | 2.1 | 10 | 0 | 1 | 1 | |||||||
| 29 | TPDRNTGQL | 2.2 | 9 | 0 | 1 | 2 | 1 | 2 | |||||
| 30 | SIISAETL | 2.3 | 8 | −1 | 1 | 1 | 2 | ||||||
| 31 | SLESAETL | 2.3 | 8 | −2 | 1 | 1 | 2 | 1 | 2 | ||||
| 32 | IFDRGSAETL | 2.3 | 10 | −1 | 3 | 8 | 7 | 7 | |||||
| 33 | SILWGPYEQ | 2.7 | 9 | −1 | 1 | 3 | |||||||
| 34 | SSGLSYEQ | 2.7 | 8 | −1 | 2 | 1 | |||||||
| 35 | SIYRGHGNTL | 1.3 | 10 | 2 | 1 | ||||||||
| 36 | SIGFSNERL | 1.4 | 9 | 0 | 1 | ||||||||
| 37 | TLTPGAEQ | 2.1 | 8 | −1 | 1 | ||||||||
| 38 | SILGGNYAEQ | 2.1 | 10 | −1 | 4 | 4 | 2 | ||||||
| 39 | SIVWGGIDAEQ | 2.1 | 11 | −2 | 1 | ||||||||
| 40 | SPDWGRNYAEQ | 2.1 | 11 | −1 | 1 | ||||||||
| 41 | RQDTQ | 2.2 | 4 | 0 | 1 | ||||||||
| 42 | SLLQNTGQL | 2.2 | 9 | 0 | 1 | ||||||||
| 43 | SLDRNTGQL | 2.2 | 9 | 0 | 1 | ||||||||
| 44 | SMGDITGQL | 2.2 | 9 | −1 | 2 | ||||||||
| 45 | SWDFLNTGQL | 2.2 | 10 | −1 | 2 | 4 | 2 | ||||||
| 46 | SIRTGNTGQL | 2.2 | 10 | 1 | 1 | ||||||||
| 47 | TGAGSSAETL | 2.3 | 10 | −1 | 1 | ||||||||
| 48 | WQTDQ | 2.5 | 5 | −1 | 1 | ||||||||
| 49 | SRLGGDTQ | 2.5 | 8 | 0 | 1 | ||||||||
| 50 | SIGIEQ | 2.7 | 6 | −1 | 2 | ||||||||
| 51 | SIVGAGEQ | 2.7 | 8 | −1 | 1 | ||||||||
| 52 | SRGLGDYEQ | 2.7 | 9 | −1 | 1 | ||||||||
| 53 | SIRRGHYEQ | 2.7 | 9 | 2 | 4 | 1 | |||||||
| 54 | SMGWGGLSYEQ | 2.7 | 11 | −1 | 1 | ||||||||
Compilation of 54 distinct CDR3 sequences of Vβ6 TCR chain from purified HEL-responsive CD4 T cells from four WT (1–4) and five Tssp−/− (1–5) HEL-immunized mice. For each sequence, the relevant Jβ segment, CDR3 length (L), and CDR3 net charge (nCh) are indicated. Negatively and positively charged amino acids are shown underlined and in bold, respectively. The number of times each sequence occurred in individual mice is shown and appears when corresponding to dominant CDR3 sequences. The 54 individual sequences correspond to a total of 132 clones analyzed. .
There were no significant differences in the Jβ segment usage between Vβ6 chains found in HEL12–27-responsive CD4 T cells from WT or Tssp−/− mice. Analysis of all the Vβ6 CDR3 sequences points to several interesting findings. First, although HEL-responsive CD4 T cells from Tssp−/− mice had accomplished fewer divisions, they showed less CDR3 diversity. Indeed, repeated occurrences of few Vβ6 rearrangements were found for TSSP-deficient but not for WT CD4 T cells. Thus, the percentage of unique sequences among the total number of CDR3 sequences for WT mice was 73, 100, 85, and 73% (mice 1–4, respectively), and for Tssp2/2 mice it was 33, 37%, 73, 83, and 42% (mice 1–5, respectively; Table I). Because these clones showed the same nucleotide sequence (unpublished data), they likely derive from the same naive CD4 T cell precursor that had a selective advantage. Second, among CDR3 sequences shared by both types of cells, one of them (sequence 32, IFDRGSAETL) was clearly more frequently used by CD4 T cells from Tssp−/− mice: found 22 times among three out of five Tssp−/− mice versus 3 times for one out of four WT mice (Table I). This CDR3 was thus overtly more available within the CD4 T cell repertoire of Tssp−/− mice and presumably did not allow for a robust CD4 T cell response to HEL challenge. Finally, although no dominant CDR3 rearrangements were revealed by this analysis in WT mice, two additional dominant CDR3 rearrangements were found in Tssp−/− mice (sequence 38, SILGGNYAEQ; sequence 45, SWDFLNTGQL).
To further compare the HEL-specific TCR repertoire of WT and Tssp−/− mice, we amplified and sequenced Vβ6-Jβ2.1 joints, a rearrangement which was frequently used by HEL12–27-responsive CD4 T cells in both strains of mice. The analysis of 92 clones revealed that Vβ6-Jβ2.1 joints found only in WT CD4 T cells were characterized by a high frequency of positively charged residues and prevalence of an arginine residue, relative to their counterparts from Tssp−/− mice (Table II). This CDR3 feature correlated with a marked increase in Dβ1.1 gene segment usage (10/12 CDR3 sequences), suggesting that these rearrangements may confer a better fit. This analysis also revealed the dominant repertoire of HEL-specific CD4 T cells in WT mice (SRTDYAEQ, SISNYAEQ, and SILGGNYAEQ). Only one of these rearrangements (SILGGNYAEQ) was also dominant and public in Tssp−/− mice (Table II). To confirm these observations, we estimated, by real-time RT-PCR, the distribution of these three clonotypes among HEL-specific CD4 T cells in WT and Tssp−/− mice. We first amplified the Vβ6-Cβ segments and used clonotype-specific primers for the real-time PCR amplification. Although the SILGGNYAEQ and SRTDYAEQ sequences were public and dominant in WT control mice, only the former was also public and dominant in Tssp−/− mice (Fig. S2 B). Furthermore, the representation of the SRTDYAEQ clonotype was very low in Tssp−/− mice, representing <10% of that detected in WT mice (Fig. S2 A). Regarding the SISNYAEQ rearrangement, the combined sequence analysis (Table II) and real-time PCR analysis (Fig. S2) indicated that this rearrangement is public but not dominant in both strains of mice. Overall, these results show that one of the public rearrangements found in WT mice (SRTDYAEQ) was barely used by Tssp−/− mice.
Table II.
Vβ6-Jβ2.1 joint of HEL12–27-specific CD4 T cells
| Sequences (aa) | L | nCh | WT | Tssp−/− | |||||||
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 5 | |||
| SRTDYAEQ | 8 | −1 | 2 | 4 | 1 | ||||||
| SIRTGGFEQ | 9 | 0 | 1 | ||||||||
| SRQGGDTQY | 9 | 0 | 1 | ||||||||
| SIWGGNYAEQ | 10 | −1 | 1 | ||||||||
| SSRDRNYAEQ | 10 | 0 | 1 | ||||||||
| SIRDRNYAEQ | 10 | 0 | 1 | ||||||||
| FPRIPGKNYAEQ | 12 | 1 | 1 | ||||||||
| SISNYAEQ | 8 | −1 | 1 | 1 | 8 | 8 | 1 | 1 | 1 | 1 | |
| SIWDRYAEQ | 9 | −1 | 2 | 2 | 1 | ||||||
| STGGNYAEQ | 9 | −1 | 2 | 3 | |||||||
| SILGGNYAEQ | 10 | −1 | 2 | 4 | 3 | 6 | 4 | 5 | 2 | 2 | |
| RGQGNNYAEQ | 10 | 0 | 2 | 1 | 1 | 1 | |||||
| TLTPGAEQ | 8 | −1 | 1 | ||||||||
| RQQNYAEQ | 8 | 0 | 1 | ||||||||
| SIWDGYAEQ | 9 | −2 | 1 | ||||||||
| PIQENYAEQ | 9 | −2 | 2 | ||||||||
| SILWGPYEQY | 10 | −1 | 1 | ||||||||
| RGQGNRCAEQ | 10 | 1 | 1 | ||||||||
| SIVWGGIDAEQ | 11 | −2 | 1 | ||||||||
| SPDWGRNYAEQ | 11 | −1 | 2 | 4 | |||||||
Compilation of 18 distinct CDR3 sequences derived from 92 clones of Vβ6-Jβ2.1 joints from purified HEL-responsive CD4 T cells from four WT (1–4) and five Tssp−/− (1–5) HEL-immunized mice. For each sequence, the CDR3 length (L) and CDR3 net charge (nCh) are indicated. Negatively and positively charged amino acids are shown underlined and in bold, respectively. The number of times each sequence occurred in individual mice is shown.
Altogether, the aforementioned data indicate that hyporesponsiveness of Tssp−/− mice to HEL challenge is associated with alteration of the TCR repertoire of the HEL-specific CD4 T cells. Thus, TSSP is necessary for the intrathymic development of CD4 T cells that confer robust anti-HEL response but dispensable for the development of CD4 T cells specific for KLH, Myo, OVA, conalbumin, and RNase. Although this analysis is still limited, these results suggest that the development of the functional CD4 T cell repertoire corresponding to roughly 15% of the Ags tested requires TSSP function in the thymus. Thus, TSSP contributes substantially to the diversification of the functional CD4 T cell repertoire.
Although the function of TSSP has not yet been proven experimentally, the demonstration that TSSP is necessary for the selection of some CD4 T cell specificities strongly suggests that TSSP is a novel protease of the class II presentation pathway (Gommeaux et al., 2009; this study). Compared with other class II Ag-processing enzymes, such as the Cat family or AEP, TSSP would then be the first example of a protease of this pathway with a restricted but significant impact on CD4 T cell repertoire diversification and immune responsiveness.
MATERIALS AND METHODS
Mice.
TSSP-deficient NOD mice and WT control littermates were generated by backcrossing Tssp+/− B6 mice (Gommeaux et al., 2009) onto the NOD background for 10 generations. All experiments involving animals were performed in accordance with national and European regulations and Institut National de la Santé et de la Recherche Médicale institutional guidelines.
Immunization and DC generation.
Mice were immunized by subcutaneous injections into the footpad and tail base of 20 µg HPLC-purified (>95% purity) HEL12–27 peptide (GeneCust) or 100 µg of protein emulsified in CFA (Sigma-Aldrich). DCs were derived from BM culture in the presence of GM-CSF (10% conditioned media). At day 6, BM DCs were activated for 18 h with 1 µg/ml polyinosinic-polycytidylic acid and 50 ng/ml TNF and then loaded with 40 nM HEL12–27 for 2 h at 37°C and washed three times before subcutaneous injections into the footpad and tail base (2 × 106/mouse). Proliferation of draining LN CD4 T cells was analyzed at day 11 after immunization.
BM chimeras.
6–8-wk-old control or TSSP-deficient NOD mice on a TCR Cα-deficient (Cα°) background were irradiated (9.5 Gy) the day before i.v. injection of T cell–depleted BM cells that consisted of either WT cells (5–6 × 106) or a 1:1 mix of WT and TSSP-deficient NOD-Cα° BM cells. After 7–10 wk, the chimeras were i.p. injected with 200 µg HEL protein emulsified in adjuvant system (Sigma-Aldrich). The proliferative response of spleen CD4 T cells was analyzed 7 d later.
T cell proliferation assays.
CD4 T cells were isolated from draining LNs or spleen by negative selection using anti–CD8-α (H58.55.8), anti-FcγRb (2.4G2), anti-CD11b (M1/70), and anti-B220 (RA3-6B2) mAbs and anti–rat IgG–coated magnetic beads (Dynabeads; Invitrogen). 4 × 105 purified CD4 T cells were stimulated in the presence of the appropriate Ag along with 106 irradiated (20 Gy) NOD splenocytes. After 3–4 d, the cultures were pulsed with 1 µCi/well of [3H]thymidine (GE Healthcare). Background values corresponding to cultures without Ag were subtracted to the experimental values (Δcpm). Alternatively, 2 × 106 purified CD4 T cells were CFSE labeled and stimulated with 10 µM HEL12–27 peptide or 50 µg/ml OVA protein or 1 µg/ml anti-CD3 and anti-CD28 antibodies and 2 × 106 T cell–depleted spleen APCs. 3 and 6 d later, the cells were stained with APC-conjugated anti-CD4 antibody and propidium iodide to exclude dead cells.
Ag presentation assays.
The BW-HEL clone was generated by transfecting BWα−β− thymoma with mouse CD4 and the TCR-α and TCR-β chain of a HEL12–27-specific T cell clone (Burton et al., 2008). 5 × 105 T cell–depleted spleen APCs were pulsed in vitro with graded doses of Ag and incubated with 5 × 104 BW-HEL cells for 24 h. IL-2 production was tested by CTL.L assay.
Analysis of Vβ usage by real-time PCR and sequencing of CDR3 segments.
CFSE-labeled CD4 T cells were stimulated with 10 µM HEL12–27 peptide as described in T cell proliferation assays. WT and TSSP-deficient cells were cultured for 5 d and 6 d, respectively. Cells that had accomplished more than one division were FACS sorted based on CFSE dilution and CD4 expression using an Epics ALTRA flow cytometer (Beckman Coulter). RNA was extracted from either naive CD4 T cells or in vitro stimulated CD4 T cells using the RNeasy Micro kit (QIAGEN), and cDNA was produced using SuperScript II enzyme (Invitrogen) according to manufacturers’ instructions. Vβ expression pattern was assessed by real-time PCR using Vβ-specific primers and probes as described previously that allowed for the detection of 24 different Vβ (Lim et al., 2002; Pannetier et al., 1993). The probe sequence is 5′-AAATGTGACTCCACCCAAGGTCTCCTTGTT-3′.
The relative usage of each Vβ expressed as a percentage was calculated using the following equation:
in which Ct(x) is the fluorescence threshold cycle number measured for Vβx.
For sequencing, the PCR reaction was made using Phusion High-Fidelity DNA polymerase (Finnzymes) and Vβ6 forward primers together with Cβ or Jβ2.1 reversed primers. The PCR product was directly subcloned using TOPO TA cloning kit for sequencing (Invitrogen).
Statistics.
Unpaired t tests were calculated using Prism software (GraphPad Software, Inc.). Unpaired values are shown. For Fig. 2 C, one-sample t tests were run using Prism software.
Online supplemental material.
Fig. S1 shows the intrathymic development of CD4 T cells expressing a given HEL-specific IAg7-restricted TCR in WT and Tssp−/− thymic environment. Fig. S2 shows the distribution, among HEL-specific CD4 T cells from WT and Tssp−/− mice, of the three dominant clonotypes shown in Table II (SRTDYAEQ, SISNYAEQ, and SILGGNYAEQ). Table S1 shows the Vβ gene usage of anti-CD3/CD28–stimulated CD4 T cells from WT and Tssp−/− mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100027/DC1.
Acknowledgments
We thank J.-C. Guery, J. van Meerwijk, and L. Pelletier for helpful discussion and critical reading of the manuscript and the personnel of the Institut Fédératif de Recherche 150 animal facility and flow cytometry facility for technical assistance.
This work was supported in part by the Institut National de la Santé et de la Recherche Médicale and the Centre National de la Recherche Scientifique and by grants from the European Foundation for the Study of Diabetes/Novo Nordisk Program in Diabetes Research and Juvenile Diabetes Research Foundation.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- AEP
- asparaginyl endopeptidase
- Ag
- antigen
- Cat
- cathepsin
- cTEC
- cortical TEC
- HEL
- hen egg lysozyme
- Myo
- myoglobin
- NOD
- nonobese diabetic
- TEC
- thymic epithelial cell
- TSSP
- thymus-specific serine protease
References
- Bowlus C.L., Ahn J., Chu T., Gruen J.R. 1999. Cloning of a novel MHC-encoded serine peptidase highly expressed by cortical epithelial cells of the thymus. Cell. Immunol. 196:80–86 10.1006/cimm.1999.1543 [DOI] [PubMed] [Google Scholar]
- Burton A.R., Vincent E., Arnold P.Y., Lennon G.P., Smeltzer M., Li C.S., Haskins K., Hutton J., Tisch R.M., Sercarz E.E., et al. 2008. On the pathogenicity of autoantigen-specific T-cell receptors. Diabetes. 57:1321–1330 10.2337/db07-1129 [DOI] [PubMed] [Google Scholar]
- Carrier A., Nguyen C., Victorero G., Granjeaud S., Rocha D., Bernard K., Miazek A., Ferrier P., Malissen M., Naquet P., et al. 1999. Differential gene expression in CD3epsilon- and RAG1-deficient thymuses: definition of a set of genes potentially involved in thymocyte maturation. Immunogenetics. 50:255–270 10.1007/s002510050601 [DOI] [PubMed] [Google Scholar]
- Cheunsuk S., Hsu T., Gershwin M.E., Bowlus C.L. 2002. Analysis of the IDDM candidate gene Prss16 in NOD and NON mice. Dev. Immunol. 9:183–186 10.1080/10446670310001593497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheunsuk S., Lian Z.X., Yang G.X., Gershwin M.E., Gruen J.R., Bowlus C.L. 2005. Prss16 is not required for T-cell development. Mol. Cell. Biol. 25:789–796 10.1128/MCB.25.2.789-796.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert P.J., Jiang S., Xie J., Li Q.J., Davis M.M. 2009. An endogenous positively selecting peptide enhances mature T cell responses and becomes an autoantigen in the absence of microRNA miR-181a. Nat. Immunol. 10:1162–1169 10.1038/ni.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman K.P., Park C.S., Kim M., Matzinger P., Anderson C.C. 2005. Thymic cortical epithelium induces self tolerance. Eur. J. Immunol. 35:709–717 10.1002/eji.200425675 [DOI] [PubMed] [Google Scholar]
- Gommeaux J., Grégoire C., Nguessan P., Richelme M., Malissen M., Guerder S., Malissen B., Carrier A. 2009. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur. J. Immunol. 39:956–964 10.1002/eji.200839175 [DOI] [PubMed] [Google Scholar]
- Hsieh C.S., deRoos P., Honey K., Beers C., Rudensky A.Y. 2002. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J. Immunol. 168:2618–2625 [DOI] [PubMed] [Google Scholar]
- Hsing L.C., Rudensky A.Y. 2005. The lysosomal cysteine proteases in MHC class II antigen presentation. Immunol. Rev. 207:229–241 10.1111/j.0105-2896.2005.00310.x [DOI] [PubMed] [Google Scholar]
- Klein L., Hinterberger M., Wirnsberger G., Kyewski B. 2009. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 9:833–844 10.1038/nri2669 [DOI] [PubMed] [Google Scholar]
- Lie B.A., Todd J.A., Pociot F., Nerup J., Akselsen H.E., Joner G., Dahl-Jørgensen K., Rønningen K.S., Thorsby E., Undlien D.E. 1999. The predisposition to type 1 diabetes linked to the human leukocyte antigen complex includes at least one non-class II gene. Am. J. Hum. Genet. 64:793–800 10.1086/302283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A., Baron V., Ferradini L., Bonneville M., Kourilsky P., Pannetier C. 2002. Combination of MHC-peptide multimer-based T cell sorting with the Immunoscope permits sensitive ex vivo quantitation and follow-up of human CD8+ T cell immune responses. J. Immunol. Methods. 261:177–194 10.1016/S0022-1759(02)00004-2 [DOI] [PubMed] [Google Scholar]
- Lo W.L., Felix N.J., Walters J.J., Rohrs H., Gross M.L., Allen P.M. 2009. An endogenous peptide positively selects and augments the activation and survival of peripheral CD4+ T cells. Nat. Immunol. 10:1155–1161 10.1038/ni.1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoury B., Hewitt E.W., Morrice N., Dando P.M., Barrett A.J., Watts C. 1998. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 396:695–699 10.1038/25379 [DOI] [PubMed] [Google Scholar]
- Manoury B., Mazzeo D., Fugger L., Viner N., Ponsford M., Streeter H., Mazza G., Wraith D.C., Watts C. 2002. Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat. Immunol. 3:169–174 10.1038/ni754 [DOI] [PubMed] [Google Scholar]
- Marrack P., Kappler J. 1997. Positive selection of thymocytes bearing alpha beta T cell receptors. Curr. Opin. Immunol. 9:250–255 10.1016/S0952-7915(97)80144-6 [DOI] [PubMed] [Google Scholar]
- McCaughtry T.M., Baldwin T.A., Wilken M.S., Hogquist K.A. 2008. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J. Exp. Med. 205:2575–2584 10.1084/jem.20080866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musson J.A., Walker N., Flick-Smith H., Williamson E.D., Robinson J.H. 2003. Differential processing of CD4 T-cell epitopes from the protective antigen of Bacillus anthracis. J. Biol. Chem. 278:52425–52431 10.1074/jbc.M309034200 [DOI] [PubMed] [Google Scholar]
- Musson J.A., Morton M., Walker N., Harper H.M., McNeill H.V., Williamson E.D., Robinson J.H. 2006. Sequential proteolytic processing of the capsular Caf1 antigen of Yersinia pestis for major histocompatibility complex class II-restricted presentation to T lymphocytes. J. Biol. Chem. 281:26129–26135 10.1074/jbc.M605482200 [DOI] [PubMed] [Google Scholar]
- Nakagawa T.Y., Brissette W.H., Lira P.D., Griffiths R.J., Petrushova N., Stock J., McNeish J.D., Eastman S.E., Howard E.D., Clarke S.R., et al. 1999. Impaired invariant chain degradation and antigen presentation and diminished collagen-induced arthritis in cathepsin S null mice. Immunity. 10:207–217 10.1016/S1074-7613(00)80021-7 [DOI] [PubMed] [Google Scholar]
- Pannetier C., Cochet M., Darche S., Casrouge A., Zöller M., Kourilsky P. 1993. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc. Natl. Acad. Sci. USA. 90:4319–4323 10.1073/pnas.90.9.4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plüger E.B., Boes M., Alfonso C., Schröter C.J., Kalbacher H., Ploegh H.L., Driessen C. 2002. Specific role for cathepsin S in the generation of antigenic peptides in vivo. Eur. J. Immunol. 32:467–476 [DOI] [PubMed] [Google Scholar]
- Shi G.P., Villadangos J.A., Dranoff G., Small C., Gu L., Haley K.J., Riese R., Ploegh H.L., Chapman H.A. 1999. Cathepsin S required for normal MHC class II peptide loading and germinal center development. Immunity. 10:197–206 10.1016/S1074-7613(00)80020-5 [DOI] [PubMed] [Google Scholar]