Abstract
Like other DNA viruses that replicate in the nucleus, herpes simplex virus 1 (HSV-1) regulates the association of histones with its genome to promote viral replication and gene expression. We previously demonstrated that SNF2H, a member of the ISWI family of chromatin-remodeling factors, is concentrated in HSV-1 replication compartments in the nuclei of infected cells, suggesting that this cellular enzyme plays a role in viral replication. We show here that small interfering RNA (siRNA)-mediated knockdown of SNF2H in HEp-2 cells resulted in an approximately 20-fold decrease in HSV-1 replication, arguing that SNF2H promotes efficient HSV-1 replication. Decreases in HSV-1 replication were observed with multiple SNF2H-specific siRNAs, and the extent of the replication decrease correlated with the amount of SNF2H knockdown, indicating that the phenotype resulted from decreased SNF2H levels rather than off-target effects of the siRNAs. We also observed a decrease in the accumulation of immediate-early (IE) gene products in HSV-1-infected cells in which SNF2H was knocked down. Histone H3 occupancy on viral promoters was increased in HSV-1-infected cells that were transfected with SNF2H-specific siRNAs, suggesting that SNF2H promotes removal of histones from viral promoters during infection. Furthermore, chromatin immunoprecipitation (ChIP) studies showed that SNF2H associated with the HSV-1 genome during infection, which suggests that SNF2H may directly remodel viral chromatin. We hypothesize that SNF2H is recruited to viral promoters during HSV-1 infection, where it can remodel the chromatin state of the viral genome, facilitate the transcription of immediate-early genes, and enhance viral replication.
IMPORTANCE
It is becoming increasingly appreciated that regulation of the state of chromatin is a major determinant in control of gene expression. It has also become clear that the state of chromatin of the herpes simplex virus type 1 (HSV-1) genome is dynamically regulated during both productive and latent stages of infection. In addition, multiple viral gene products have been reported to play roles in regulating the viral chromatin state. However, the cellular chromatin-remodeling factors involved in altering nucleosome occupancy at viral genes remain largely unknown. The results in this report represent the first evidence that cellular chromatin-remodeling proteins, and SNF2H in particular, can play important roles in regulating the chromatin state of the HSV-1 genome during infection. This work also further establishes HSV-1 infection as a useful model to study chromatin control of gene expression and suggests that disrupting the regulation of viral chromatin states can possibly be exploited as a novel antiviral therapeutic target.
INTRODUCTION
DNA viruses that replicate in the nuclei of their host cells are faced with the challenge of regulating the association of histones with viral DNA as the cell defense mechanisms attempt to silence the incoming “foreign” DNA. Nuclear DNA viruses generally encode gene products that function to counteract this inhibitory cellular response by coordinating the modification of these histones and/or by remodeling the viral chromatin state through the redirection of cellular enzymes to the viral genome to allow for efficient viral replication and gene expression.
The double-stranded DNA genome of herpes simplex virus 1 (HSV-1) associates with histones to form chromatin rapidly following infection of a host cell, and the resulting chromatin structure is dynamically regulated throughout productive HSV-1 infection (1–3). During productive infection, viral chromatin is generally in a euchromatic state that is conducive to active transcription (4, 5), and the nucleosomes associated with HSV-1 DNA are unstable (6). Additionally, the association of histones with the HSV-1 genome is decreased at late times postinfection (1, 3). Several viral gene products have been implicated in playing a role in generation of the euchromatin-like state of the viral genome (1, 2, 7, 8), suggesting that HSV-1 encodes multiple activities to ensure that its genome remains competent for transcription despite the cellular response that attempts to silence foreign gene expression.
One viral gene product that has been shown to play a role in regulating the chromatin state of the viral genome is VP16. VP16 is a potent transcriptional activator that enters the host cell within the virion and stimulates the expression of viral immediate-early (IE) genes. VP16 forms a complex with the cellular HCF-1 and Oct-1 proteins, which is termed the activator complex. This complex associates with the histone acetyltransferase enzymes CBP and p300 and the chromatin-remodeling factors Brg1 and BRM and recruits these cellular proteins to viral promoters (2). Cells infected with VP16 mutant viruses have increased levels of histone H3 at viral immediate-early promoters, and these histones have lower levels of acetylation (2). However, experiments with HeLa cells or human foreskin fibroblasts (HFF) utilizing RNA interference (RNAi) knockdown of cellular proteins, such as CBP, p300, Brg1, or BRM, or mutant or knockout cell lines suggest that these enzymes are dispensable for expression of immediate-early genes (9). Other cellular proteins that modify histones associated with HSV-1 DNA are Set1 (4), which adds methyl marks associated with euchromatin, and LSD1 (10), which removes methyl marks associated with heterochromatin.
A second viral gene product that plays a role in regulating the state of viral chromatin during productive infection is the IE gene product ICP0 (1, 7). ICP0 is a virus gene-encoded transcriptional transactivator that stimulates the expression of many viral genes (11, 12). Cells infected with an ICP0 mutant virus exhibit increased occupancy of histone H3 at viral promoters and also a decrease in the proportion of histone H3 that is acetylated (1). The viral genome of ICP0 mutants does not show reduced association with histones at late times postinfection (1), indicating that ICP0 plays a role in histone removal from the viral genome. The mechanism by which ICP0 results in a decrease in the occupancy of histone H3 at viral promoters is not known.
Multiple cellular chromatin-remodeling factors and histone-modifying enzymes are recruited to HSV-1 replication compartments (RCs) (13), which are nuclear domains where viral DNA replication and late gene transcription occur (14, 15). This previous study from our laboratory (13) provided some of the first evidence that these cellular proteins may play an important role in HSV-1 replication. The most abundantly identified chromatin-remodeling protein in viral RCs was SNF2H (13), which is a member of the ISWI family of chromatin-remodeling enzymes, and is the ATPase catalytic subunit of several chromatin-remodeling complexes. This observation led us to hypothesize that SNF2H could remodel viral chromatin and that this chromatin-remodeling activity was important for efficient HSV-1 replication.
Similar to other catalytic subunits of chromatin-remodeling complexes, SNF2H by itself exhibits little substrate specificity. Instead, the target specificity of SNF2H-containing complexes is generally conferred by the nonenzymatic subunits that associate with SNF2H in these complexes. The different chromatin-remodeling complexes containing SNF2H or SNF2H homologs appear to have different and distinct biological activities, including DNA replication through regions of heterochromatin (16, 17), and both stimulation (18–23) and repression (24, 25) of gene expression.
It is clear that regulation of the state of chromatin of the viral genome is important for HSV-1 replication. Most current models involve viral gene products redirecting cellular enzymes to modify the chromatin associated with HSV-1 genes during productive infection. However, the roles of ATP-dependent chromatin-remodeling factors in regulating viral chromatin state have been less well defined, and there are currently no known chromatin-remodeling proteins that are required for efficient HSV-1 replication and gene expression. In this report, we investigated the hypothesis that the chromatin-remodeling factor SNF2H is required for efficient HSV-1 replication by knocking down the levels of SNF2H using RNAi and assaying the effects on viral replication, gene expression, and chromatin state.
RESULTS
SNF2H is required for efficient HSV-1 replication.
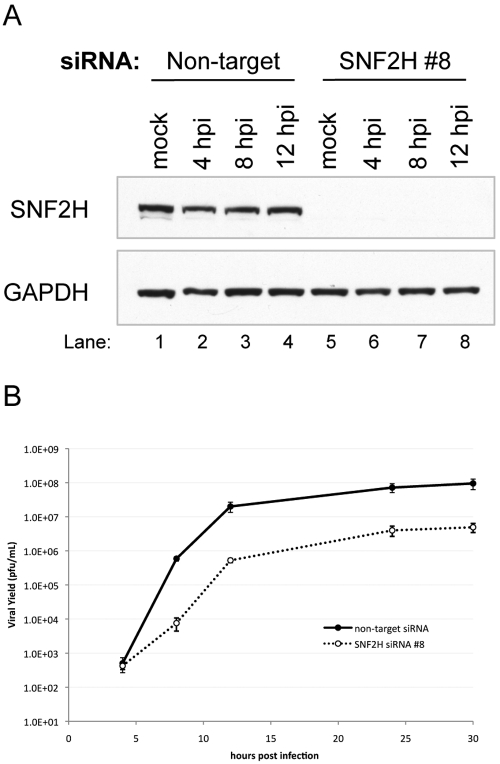
To investigate the role of the cellular chromatin-remodeling factor SNF2H in HSV-1 replication, we reduced its levels using RNAi. We transfected HEp-2 cells with small interfering RNAs (siRNAs) that are specific for SNF2H mRNA or nonspecific siRNAs that are known not to target any human or mouse mRNAs. We observed a substantial decrease in the accumulation of SNF2H in cells transfected with the SNF2H-specific siRNAs relative to cells transfected with the nonspecific control siRNAs (Fig. 1A). The nonspecific control siRNAs did not reduce SNF2H levels relative to mock-transfected or nontransfected controls (data not shown). SNF2H levels were also not changed over the course of HSV-1 infection (Fig. 1A, lanes 1 to 4). The levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were not altered following transfection with either siRNA (Fig. 1A), demonstrating the specificity of the SNF2H siRNAs. There were usually 10 to 25% fewer cells in the cultures transfected with the SNF2H siRNAs at the time of infection compared to nontarget siRNA-transfected cells, but from this time on, both populations of cells divided at similar rates (data not shown). Additionally, there were no obvious signs of cell death observed in cells transfected with either siRNA (not shown).
FIG 1 .
Decreased SNF2H levels result in reduced HSV-1 replication. (A) Immunoblot measuring levels of SNF2H following transfection of HEp-2 cells with either SNF2H-specific siRNA or nontarget control siRNA. siRNA-transfected cells were either mock infected or infected with HSV-1 strain KOS at an MOI of 10 for the time indicated (hours postinfection [hpi]). The cellular GAPDH gene product serves as a recovery and loading control. (B) SNF2H knockdown following transfection of SNF2H-specific siRNA resulted in a decrease in HSV-1 replication relative to viral replication in cells transfected with nontarget control siRNA. siRNA-transfected cells were infected with HSV-1 at an MOI of 10, and at the times indicated, samples were harvested, and viral yield (PFU/ml) was determined by plaque assay on Vero cells. Results are averages of at least four independent experiments, and the error bars represent the standard errors of the means.
To determine whether knocking down SNF2H levels had an effect on HSV-1 replication, we performed a single-cycle viral replication assay with HEp-2 cells that were transfected with either SNF2H-specific siRNAs or nontarget control siRNAs. siRNA-transfected cells were infected with HSV-1 for various times, and the viral yield at each time point was determined by plaque assay on Vero cells. At 30 h postinfection, by which time the viral yield had plateaued, we observed an approximately 20-fold reduction in replication in the cells transfected with SNF2H-specific siRNAs relative to the cells transfected with the control siRNAs (Fig. 1B). These results suggested that SNF2H promotes efficient HSV-1 replication.
HSV-1 replication reduction is due to decreased SNF2H levels.
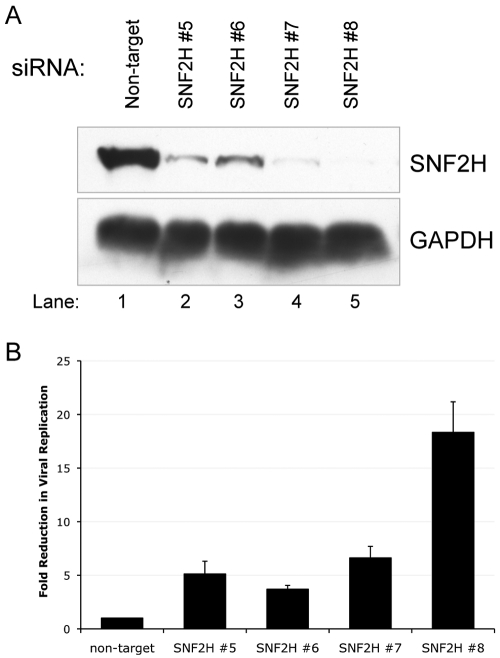
To determine whether the reduction in HSV-1 replication was a result of reduced levels of SNF2H, rather than nonspecific, off-target effects of the siRNAs, we knocked down SNF2H with four different siRNAs individually and assayed the effect of each on SNF2H accumulation and HSV-1 replication. We observed that all four individual SNF2H-specific siRNAs resulted in a decrease in the levels of SNF2H when transfected into HEp-2 cells (Fig. 2A), with SNF2H siRNA 8 resulting in the greatest decrease in SNF2H accumulation, followed by siRNA 7, siRNA 5, and then SNF2H siRNA 6 (Fig. 2A). SNF2H levels remained high in cells transfected with the nontarget control siRNA (Fig. 2A), and none of the siRNAs altered the levels of GAPDH (Fig. 2A), demonstrating the specificity of the SNF2H knockdowns.
FIG 2 .
The extent of SNF2H knockdown correlates with reduction of HSV-1 replication. (A) Immunoblot evaluating the efficiency of SNF2H knockdown following transfection of HEp-2 cells with different siRNAs targeting SNF2H or a control nontargeting siRNA. The cellular GAPDH protein serves as a recovery and loading control. (B) The efficiency of SNF2H knockdown correlates with the extent of the observed decrease in HSV-1 replication. siRNA-transfected HEp-2 cells were infected with HSV-1 at an MOI of 10. At 24 hours postinfection, cultures were harvested, and viral yield was determined by plaque assay on Vero cells. Data are presented as the fold decreases in viral replication relative to the level of viral replication observed in HEp-2 cells transfected with nontarget control siRNA. Results are the averages of three independent experiments, and the error bars represent the standard errors of the means.
We next examined HSV-1 replication following knockdown of SNF2H with each individual siRNA. We observed that transfection of each of the individual SNF2H-specific siRNAs resulted in a reduction in HSV-1 replication relative to transfection of the control siRNA (Fig. 2B). Because all four individual siRNAs knocked down SNF2H accumulation and also decreased HSV-1 replication, the HSV-1 replication defect is most likely a result of lower SNF2H levels rather than nonspecific, off-target effects of the siRNAs, as all four siRNAs would not be expected to have the same off-target effects. In addition, the amount of SNF2H knockdown correlated very well with the extent of the decrease in HSV-1 replication, i.e., SNF2H siRNA 8 resulted in the most efficient knockdown of SNF2H and also resulted in the greatest decrease in viral replication, and siRNA 6 knocked down SNF2H the least efficiently and also resulted in the smallest decrease in viral replication. These results further indicated that the decrease in HSV-1 replication was a result of reduced SNF2H levels, rather than nonspecific, off-target effects of the siRNAs.
SNF2H is required for efficient viral immediate-early gene expression.
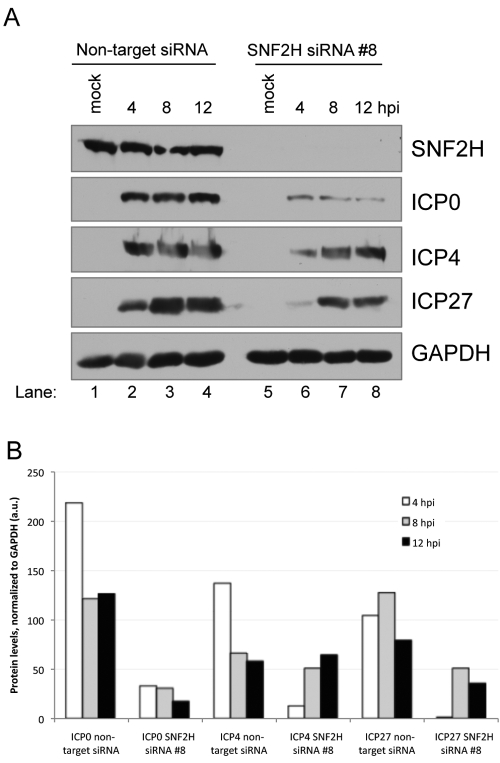
To investigate the mechanism(s) responsible for the observed reduction in HSV-1 replication when SNF2H levels were knocked down, we measured the accumulation of the viral immediate-early gene products ICP0, ICP4, and ICP27 in siRNA-transfected cells. HEp-2 cells that were transfected with either siRNA specific for SNF2H or the nontarget control siRNA were infected with HSV-1, and lysates were prepared for immunoblots at various times postinfection. In these assays, SNF2H levels were efficiently decreased throughout HSV-1 infection following transfection with the SNF2H-specific siRNAs but remained high in cells transfected with the nontarget siRNAs (Fig. 3A). Accumulation of the viral immediate-early gene product ICP0 was reduced in SNF2H siRNA-transfected cells relative to nontarget control siRNA-transfected cells (Fig. 3A). ICP0 expression in HSV-1-infected cells in which SNF2H levels were knocked down was reduced approximately 7-fold at 4 h postinfection, approximately 4-fold at 8 h postinfection, and greater than 7-fold at 12 h postinfection (Fig. 3B). These results indicated that efficient expression of ICP0 was dependent on SNF2H. We also observed a reduction in the accumulation of ICP4 in the cells transfected with SNF2H-specific siRNAs at early times postinfection relative to cells transfected with the nonspecific control siRNAs (Fig. 3A). The decrease in ICP4 accumulation was approximately 10-fold at 4 h postinfection, but the levels were comparable with both siRNA treatments at 8 and 12 hours postinfection (Fig. 3B). Similar to the case for ICP4, accumulation of ICP27 was dependent on SNF2H at early times postinfection but less so at later times (Fig. 3A and B). Similar results were observed in three additional independent experiments (not shown). We also observed a decrease in IE mRNA levels in RNA hybridization assays when SNF2H levels were knocked down (results not shown).
FIG 3 .
Reducing SNF2H levels results in decreased ICP0 accumulation. (A) Immunoblot measuring the levels of HSV-1 immediate-early proteins in SNF2H-specific siRNA-transfected HEp2 cells relative to nontarget control siRNA-transfected cells. The cells were either mock infected or infected with HSV-1 at an MOI of 10 for the time indicated. (B) Quantification of the immunoblot in panel A, as described in Materials and Methods. a.u. arbitrary units.
We also observed reductions in the accumulation of the early ICP8 and thymidine kinase (TK) proteins, and the late glycoprotein C (gC) when SNF2H levels were reduced relative to the control siRNA-transfected cells (results not shown). In addition, we observed a defect in viral DNA replication in HSV-1-infected cells in which SNF2H levels were knocked down relative to nontarget siRNA-transfected cells (results not shown). While expression of the early and late gene products and replication of viral DNA may directly depend on SNF2H, it is more likely that these defects were a downstream consequence of the reduced levels of IE gene products when SNF2H levels were knocked down.
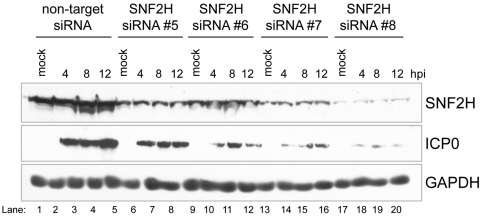
To address whether the decreases observed in the accumulation of immediate-early gene products were a consequence of decreased levels of SNF2H, we assayed the levels of ICP0 following infection of cells that were previously transfected with each of the four individual siRNAs that target SNF2H. We observed a correlation between the extent of SNF2H knockdown and the severity of the decrease in ICP0 accumulation. In agreement with the results in Fig. 3, we observed that accumulation of the immediate-early gene product ICP0 was dependent on SNF2H, and the accumulation of ICP0 correlated with the levels of SNF2H (Fig. 4). For example, SNF2H siRNA 8 knocked down the level of SNF2H to the greatest extent and had the greatest effect on the accumulation of ICP0, followed by siRNA 7, then siRNA 5, and then siRNA 6 (Fig. 4). These results suggested that the decrease in the accumulation of ICP0 when SNF2H was knocked down was due specifically to the lower levels of SNF2H, rather than nonspecific off-target effects following transfection of siRNAs.
FIG 4 .
The extent of SNF2H knockdown correlates with the reduction of ICP0 levels. Immunoblot measuring HSV-1 ICP0 protein levels in cells transfected with SNF2H-specific or -nonspecific siRNAs. HEp-2 cells were transfected with either one of the four individual siRNAs targeting SNF2H, or the nontargeting negative-control siRNA and were then either mock infected or infected with HSV-1 at an MOI of 10 for the time indicated. The cellular GAPDH protein serves as a recovery and loading control.
Association of SNF2H with the HSV-1 genome.
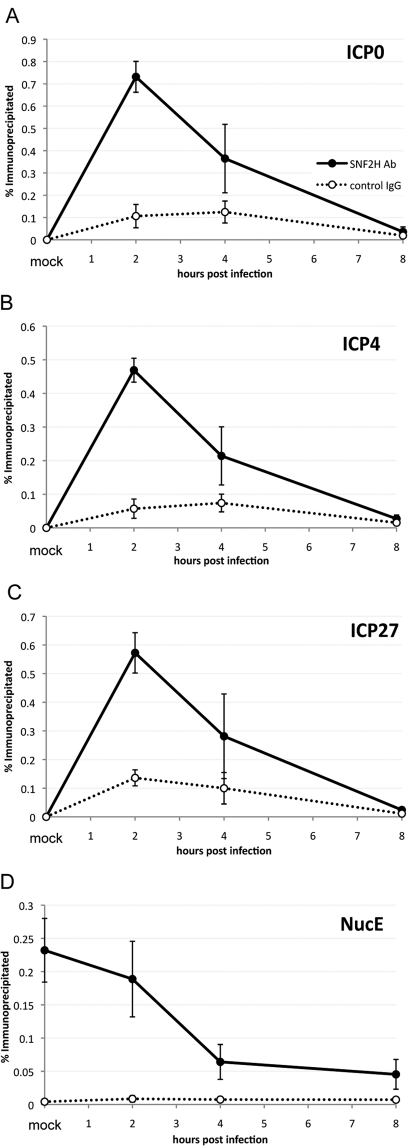
Because reduction of SNF2H levels resulted in decreased levels of ICP0 expression, we hypothesized that SNF2H might directly remodel viral chromatin. To determine whether SNF2H was associated with viral chromatin, we performed a chromatin immunoprecipitation (ChIP) assay using an antibody specific for SNF2H and assayed the immunoprecipitated chromatin by real-time PCR using primers specific for viral promoter regions. We detected SNF2H associating with viral promoters at 2 h postinfection (Fig. 5A to C), and the levels were highest on the ICP0 promoter (Fig. 5A), consistent with ICP0 being most dependent on SNF2H for its expression. The level of SNF2H associated with the viral genome decreased by 4 h postinfection and was nearly reduced to background levels by 8 h postinfection (Fig. 5A to C). These results provided evidence that SNF2H associates directly with the viral genome and this association is detected at viral promoters. SNF2H association with the nucleosome E (NucE) region of the cellular pS2 promoter was also decreased during the course of infection (Fig. 5D), suggesting that in addition to recruiting SNF2H to viral promoters, HSV-1 infection may also reduce the association of SNF2H with cellular targets.
FIG 5 .
SNF2H associates with the HSV-1 genome. Chromatin immunoprecipitation assay measuring SNF2H levels on viral promoters in HeLa cells that were transfected with a plasmid expressing SNF2H. Cells were infected with HSV-1 at an MOI of 20 for the time indicated and fixed with formaldehyde. Chromatin was prepared and subjected to immunoprecipitation using either an antibody specific for SNF2H (SNF2H Ab) or control rabbit IgG. Immunoprecipitated DNA was measured by real-time PCR. (A) ICP0 gene promoter; (B) ICP4 gene promoter; (C) ICP27 gene promoter; (D) NucE region of the cellular pS2 promoter. Data are presented as the percentages of DNA immunoprecipitated, and the values are the averages of three independent experiments. Error bars represent the standard errors of the means.
Histone H3 levels are increased on viral promoters when SNF2H levels are knocked down.
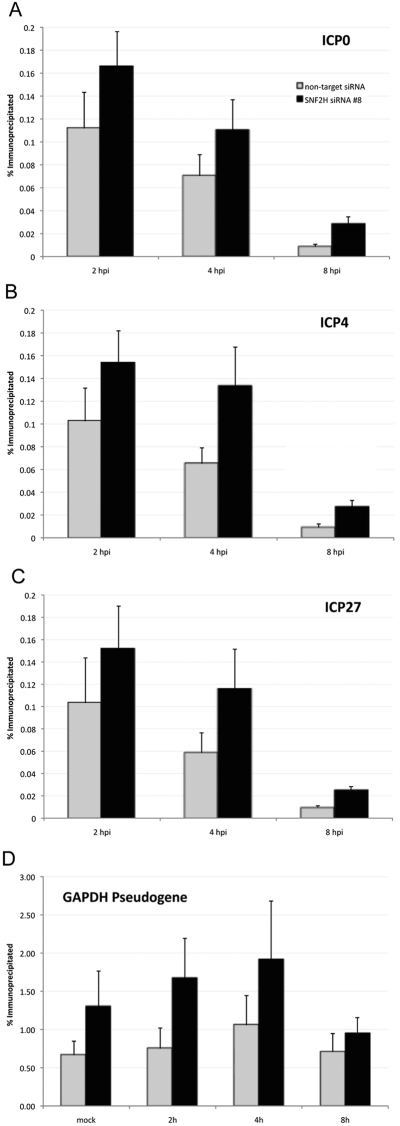
To address whether SNF2H plays a role in regulating the state of chromatin of the viral genome, we performed chromatin immunoprecipitation assays on cells that were transfected with either an SNF2H-specific siRNA or a nontarget control siRNA. In these ChIP assays, we used an antibody specific for histone H3, which we utilized as a marker for total histone levels. We assayed the H3-immunoprecipitated chromatin for levels of the viral immediate-early ICP0, ICP4, and ICP27 gene promoters using quantitative real-time PCR to quantify the association of histone H3 with these viral promoters during HSV-1 infection. We observed higher levels of DNA associated with histone H3 for all of the viral promoters tested in infected cells when SNF2H levels were knocked down relative to control siRNA-transfected cells (Fig. 6A to C).
FIG 6 .
Histone H3 levels on viral promoters increase when SNF2H levels are reduced. Chromatin immunoprecipitation assay measuring histone H3 levels on viral promoters in HEp-2 cells that were transfected with SNF2H-specific or nontarget control siRNA. The cells were infected with HSV-1 at an MOI of 10 for the time indicated, and then the cells were fixed. Chromatin was prepared and subjected to immunoprecipitation using either an antibody specific for histone H3 or normal rabbit IgG. Immunoprecipitated DNA was measured by real-time PCR. (A) ICP0 gene promoter; (B) ICP4 gene promoter; (C) ICP27 gene promoter; (D) GAPDH pseudogene. Data are presented as the percentages of the DNA immunoprecipitated with background signal from the normal rabbit IgG subtracted. The values are averages of at least five independent experiments. Error bars represent the standard errors of the means.
For any individual combination of conditions (any one gene at any one time point), we observed the trend of increased histone H3 occupancy when the levels of SNF2H were reduced by RNAi, but the small sample size and scatter in the data rendered them statistically underpowered to detect small effects once Bonferroni’s correction for multiple comparisons was applied. However, when data were pooled across genes (combining values for one gene at multiple time points), across time points (combining values for multiple genes at one time point), or across the whole sample (combining values for all genes at all time points) in a pairwise manner, these effects were highly statistically significant (P << 0.001 for each of these comparisons by a two-tailed Wilcoxon rank sum test, using paired sample testing with Bonferroni’s correction for multiple testing). Therefore, the increase in the association of histone H3 with viral gene promoters when SNF2H levels were reduced was statistically significant. These results argue that SNF2H normally acts to remodel viral chromatin during HSV-1 infection by removing histones from these promoters, either directly or indirectly.
We also observed a comparable increase in the amount of histone H3 associated with the promoter region of a cellular GAPDH pseudogene, which we used as an internal cellular control, following the histone H3 immunoprecipitation from SNF2H siRNA-transfected cells relative to nontarget control siRNA-transfected cells (Fig. 6D). These results suggested that the chromatin state at this cellular locus is also remodeled by SNF2H.
We have previously observed that histone H3 loses its association with the viral genome between 4 and 8 h postinfection, in a viral DNA replication-independent manner (1). In this study, we also observed a loss of association between histone H3 and the viral genome by 8 h postinfection (Fig. 6A to C). This decrease in histone levels was independent of SNF2H because it still occurred when SNF2H levels were knocked down (Fig. 6A to C), suggesting that SNF2H is likely not the chromatin-remodeling factor important for removing histones from the viral genome at later times postinfection.
Our results indicate that SNF2H promotes efficient HSV-1 replication and immediate-early gene expression. Furthermore, our results argue that SNF2H associates directly with the viral genome, perhaps to remodel the chromatin composition of the ICP0 promoter to facilitate expression of this critical viral regulatory gene product.
DISCUSSION
HSV-1 DNA becomes associated with histones at early times postinfection (1, 3), and the histones acquire euchromatic modifications (4, 5); however, the levels of histones associated with the viral genome decrease at later times postinfection (1, 3). The LSD1 demethylase is required for efficient viral IE gene expression (10), demonstrating that euchromatic histone modifications are required for optimal viral IE gene transcription. The roles of chromatin remodeling and nucleosome positioning in viral gene expression and replication are less clear. Although VP16 plays a role in recruiting BRM and BRG1 to viral IE promoters (2), those chromatin-remodeling proteins are not required for viral gene expression (9). In this study we have found that the SNF2H chromatin-remodeling factor is required for optimal viral gene expression. This argues that chromatin remodeling or removal or repositioning of nucleosomes at viral IE genes, in addition to histone modification, is needed for efficient IE gene expression.
Requirement for SNF2H for viral gene expression and replication.
The decrease in HSV-1 replication and the lower levels of ICP0 when SNF2H levels are knocked down argue that SNF2H is required for maximal levels of viral gene expression and replication. SNF2H may be required to directly remodel viral chromatin, or SNF2H may stimulate the expression of another cellular gene that is directly required for efficient HSV-1 replication. Because SNF2H associates with the HSV-1 genome in ChIP assays and because there is an increase in the occupancy of histone H3 at viral promoters when SNF2H is knocked down, we favor a model in which SNF2H directly remodels viral chromatin and we think that this activity is important for HSV-1 replication and gene expression. We attribute the decrease in viral replication when SNF2H levels were reduced by RNAi to the observed lower levels of IE gene products, and particularly to the decrease in ICP0 accumulation.
Because the block in viral replication was not complete when SNF2H levels were knocked down, it seems that either HSV-1 replication is not completely dependent on SNF2H or that SNF2H is still present in sufficient amounts to allow reduced levels of viral replication to occur. Alternatively, there may be other redundant or homologous chromatin-remodeling pathways that are able to partially compensate for the lower levels of SNF2H that result after transfection of SNF2H-specific siRNA.
Mechanism of SNF2H regulation of HSV-1 gene expression.
The accumulation of ICP0 was highly dependent on SNF2H in HSV-1-infected HEp-2 cells. However, knocking down the levels of SNF2H did not have as great an effect on the accumulation of the other immediate-early gene products, ICP4 and ICP27, especially at later times postinfection. These results raise the intriguing possibility that the immediate-early genes can differ in their requirement for cellular chromatin-modifying factors to facilitate their expression. Consistent with this, other studies have shown differences in the regulation of the different immediate-early gene promoters (26–28).
The decrease in ICP0 accumulation during HSV-1 infection when SNF2H is knocked down may be a result of the lack of SNF2H-dependent chromatin remodeling at the ICP0 promoter. The observed association of SNF2H with the ICP0 promoter and the increased levels of histone H3 at the ICP0 promoter when SNF2H levels are knocked down are consistent with this model. However, the promoters of the other immediate-early genes investigated, the ICP4 and ICP27 promoters, also exhibit increased histone H3 occupancy when SNF2H levels were knocked down and associate with SNF2H in ChIP assays, albeit to slightly lower levels, suggesting that SNF2H may exhibit different chromatin-remodeling activities on different viral promoters. For example, SNF2H may be able to preferentially remodel nucleosomes containing modifications associated with repression of transcription specifically at the ICP0 promoter. Another possibility is that remodeling of chromatin at the ICP0 promoter allows specific transcriptional transactivators to recognize unique sequences at this gene. It is also possible that the expression of ICP0 is more sensitive to higher levels of nucleosomes present at its promoter than other immediate-early genes. SNF2H is known to have different chromatin-remodeling activities on different cellular genes (29), so it is perhaps not surprising that SNF2H can also regulate viral genes differentially.
It has recently been reported that nucleosomes that are remodeled by ISWI family chromatin-remodeling enzymes can associate with and be modified by SET domain-containing histone methyltransferase enzymes, while unmodified nucleosomes or those remodeled by enzymes in the SWI/SNF family cannot (30). During productive infection, histone H3 at HSV-1 promoters is trimethylated on lysine 4, which is a mark of active transcription, by the SET domain of the cellular enzyme Set1 (4). Therefore, the remodeling of nucleosomes on the viral genome by SNF2H may be required for these histones to be methylated by Set1. We are currently investigating this hypothesis and are further characterizing the chromatin state of the viral genome when SNF2H levels are reduced by RNAi.
Recruitment of SNF2H activity to the viral genome.
To compete with the vastly larger number of cellular genes in the nucleus for SNF2H association, HSV-1 genes likely encode mechanisms to preferentially recruit SNF2H to the viral genome. The virion protein VP16 and associated host proteins are leading candidates to direct SNF2H to the HSV-1 IE promoters. VP16 enters the infected cells along with the virion (31) and stimulates expression of IE genes. In addition, the VP16 activator complex is known to associate with some cellular chromatin-remodeling factors (2), and perhaps it also interacts with SNF2H. The VP16 activator complex also interacts with histone acetyltransferase and histone methyltransferase enzymes and recruits these proteins to viral promoters to stimulate viral gene expression (2). This raises the possibility that VP16 recruits multiple types of chromatin-remodeling and histone-modifying factors to viral immediate-early genes to ensure their expression, and perhaps VP16 recruits different complexes to specific promoters, which may each differ in their requirement for cellular factors.
We have shown that the viral early ICP8 gene product associates and colocalizes with SNF2H during infection (13), and therefore, ICP8 may recruit SNF2H specifically to viral promoters or other regions of the HSV-1 genome. ICP8 binds to single-stranded viral DNA as it is being replicated, raising the possibility that ICP8 may direct SNF2H to replication forks to remodel viral chromatin and to facilitate HSV-1 DNA replication or to remodel viral chromatin following HSV-1 DNA replication. We have shown that ICP8 plays a role in stimulating the expression of late genes (32), but the mechanism of this activity remains unknown. ICP8 may promote the expression of late genes by directing the chromatin-remodeling activity of SNF2H to the promoters of these late genes. However, because ICP8 is expressed after ICP0 and is not a virion component, ICP8 is probably not the viral factor responsible for mediating the SNF2H-dependent expression of ICP0.
In this study we found that the association between SNF2H and HSV-1 immediate-early promoters was highest at early times postinfection and that this association decreased over the course of infection. A similar pattern was observed for the association between histone H3 and the viral genome (1, 3). We are currently investigating the mechanism(s) involved in regulating the association between SNF2H and HSV-1 chromatin, including determining whether viral proteins or DNA replication play a role in this process.
In addition to associating with SNF2H and recruiting this enzyme to the HSV-1 genome, viral factors might facilitate the association of SNF2H with the viral genome by disrupting the interactions between SNF2H and its cellular targets by displacing or modifying cellular cofactors that normally interact with SNF2H. We observed a decrease in the levels of SNF2H associated with the cellular pS2 promoter during the course of HSV-1 infection, which is consistent with this possibility. We are currently investigating which SNF2H-containing complex is responsible for stimulating viral immediate-early gene expression and also whether HSV-1 infection modifies or alters the composition of SNF2H complexes.
Additional activities of chromatin-remodeling complexes in HSV-1 productive infection.
We and others have shown that the HSV-1 genome associates with histones rapidly following infection and that the association of viral DNA with histones decreases at later times postinfection (1, 3). The consequences of this change in histone association with the viral DNA are not understood. At this time, the mechanisms and factors involved in the decrease of histone association with the viral genome are also not well defined. It is unlikely that SNF2H plays a major role in this activity because we observed a dramatic decrease in the levels of histone H3 association with the viral genome between 4 and 8 h postinfection in cells with SNF2H levels reduced by RNAi. We are currently investigating whether other cellular chromatin-remodeling factors are responsible for the decreased association of the HSV-1 genome with nucleosomes at later times postinfection. Viral DNA that is packaged into virions does not contain nucleosomes, so it is possible that the mechanism responsible for the decreased association of viral DNA with nucleosomes at late times postinfection also ensures that packaged genomes are histone free.
During the course of this study, we observed decreased expression of viral early and late genes, as well as decreased levels of HSV-1 DNA replication, when SNF2H levels were lowered by RNAi. We attributed these observations to downstream consequences of reduced viral immediate-early gene expression. We cannot exclude the possibility that SNF2H and/or other chromatin-remodeling factors play a direct role in regulating all of the classes of viral gene expression as well as viral DNA replication.
The work described in this report establishes SNF2H as a cellular factor that plays a role in remodeling the chromatin state of the HSV-1 genome and that promotes efficient viral gene expression and replication. This work also sheds new light on the mechanisms involved in regulating chromatin dynamics during HSV-1 infection and in regulating the expression of viral genes. In particular, this is the first report of an ATP-dependent chromatin-remodeling enzyme being required for efficient HSV-1 gene expression and demonstrates that nucleosome positioning, in addition to histone modification, may play a key role in regulating the expression of viral genes. Furthermore, these results further establish HSV-1 infection as a useful model system to investigate mechanisms involved in chromatin-mediated regulation of gene expression, specifically to investigate the mechanisms by which SNF2H remodels chromatin.
MATERIALS AND METHODS
Cells and viruses.
HEp-2, HeLa, and Vero cells were obtained from American Type Cell Culture (Manassas, VA). The cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% heat-inactivated fetal bovine serum and 5% heat-inactivated newborn calf serum (NCS).
All experiments were performed with HSV-1 wild-type strain KOS (33). This virus was propagated and titrated on Vero cells following standard procedures.
siRNA transfections.
Double-stranded SNF2H-specific and nontarget control siRNAs were purchased from Dharmacon. The siRNAs (nontarget siRNA 1, 5′-UGGUUUACAUGUCGACUAA-3′; SNF2H siRNA 5, 5′-GGAAUGGUAUACUCGGAUA-3′; SNF2H siRNA 6, 5′-GGGCAAAUAGAUUCGAGUA-3′; SNF2H siRNA 7, 5′-GGAUUUACCAAUUGGAAUA-3′; SNF2H siRNA 8, 5′-GUUCUUUCCUCCACGUUUA-3′) were transfected into HEp-2 cells using the HiPerFect transfection reagent (Qiagen) at a final siRNA concentration of 5 nM according to the manufacturer’s instructions. At 3 days following the siRNA transfection, the cells were passaged and transfected a second time following the same protocol. Three days following the second transfection, cells were assayed for SNF2H levels by immunoblotting and/or infected with HSV-1.
Immunoblotting.
Cell monolayers were washed with phosphate-buffered saline supplemented with calcium and magnesium (PBS-ABC), and lysates were prepared by scraping cells into 2× SDS-PAGE loading buffer and boiling for 5 min. Polypeptides were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked for 1 h at room temperature with 5% milk in Tris-buffered saline with 0.1% Tween 20 (TBST). Blocked membranes were reacted with primary antibodies diluted in 5% milk in TBST. The antibodies were SNF2H rabbit polyclonal antibody (Bethyl Laboratories) (at a dilution of 1:2,500), GAPDH mouse monoclonal antibody (Applied Biological Materials Inc.) (1:20,000), ICP0 mouse monoclonal antibody (East Coast Bio) (1:5,000), ICP27 mouse monoclonal antibody (Abcam) (1:20,000), and ICP4 58S mouse monoclonal antibody (gift from Neal DeLuca [34]) (1:10,000). Horseradish peroxidase (HRP)-conjugated secondary antibodies (Santa Cruz Biotechnology) were diluted 1:5,000 to 1:20,000 in TBST with 5% milk. Immunoblots were developed using ECL reagents (Pierce).
Viral replication assays.
Cells were counted prior to infection to allow for accurate determination of the multiplicity of infection (MOI). Cell monolayers were rinsed with PBS-ABC supplemented with 1% inactivated calf serum and 0.1% glucose (PBS-ICS) and were then infected with HSV-1 at an MOI of 10 PFU/cell in PBS-ICS in a 37°C shaking incubator. Following a 1-h adsorption, the monolayers were washed with acid wash solution (40 mM citric acid, 10 mM potassium chloride, 135 mM sodium chloride) and with DMEM supplemented with 1% inactivated calf serum. After the washes, DMEM with 1% serum was added to the cells, and infection was allowed to proceed for the amount of time indicated. The cultures were harvested by scraping the infected-cell monolayer followed by freezing the scraped cells and media from the culture. Following the freeze-thaw cycle, the lysate was clarified by centrifugation, and viral yields were determined by plaque assay on Vero cells.
Immunoblot quantification.
Lysates from nontarget siRNA-transfected HEp-2 cells infected with HSV-1 at an MOI of 10 for 12 h were prepared as described above and were diluted to generate a standard curve. These diluted samples were resolved and immunoblotted in parallel with the other infected-cell samples, as described above. Values for the intensity of each band on scanned films were obtained using Quantity One software (Bio Rad). The standard curve dilutions were assigned arbitrary concentrations and were used to determine concentrations for each unknown sample.
Plasmid construction and transfection.
pFB.SNF2h.c-FLAG (35), which is a plasmid used to generate recombinant baculovirus expressing FLAG-tagged SNF2H, was kindly provided by R. Kingston. The SNF2H-coding region was isolated by digesting pFB.SNF2h.c-FLAG with BamHI, blunting the sticky ends with the Klenow fragment of DNA polymerase I, and digesting with XbaI. The fragment containing SNF2H sequences was purified using the QIAquick gel extraction kit (Qiagen) and ligated to the pCIΔ vector (which is pCI [Promega] digested with AflII and religated to remove the intron [36]) that was digested with EcoRI, blunted with Klenow fragment, and digested with XbaI. The resulting plasmid, pCIΔ.SNF2H.FLAG, was transfected into HeLa cells in 10-cm dishes using Effectene transfection reagents (Qiagen) according to the manufacturer’s instructions.
ChIP assays.
The SNF2H chromatin immunoprecipitation (ChIP) assay was performed as described previously (1), but with some modifications. pCIΔ.SNF2H.FLAG-transfected HeLa cells in 10-cm dishes were infected with HSV-1 at an MOI of 20. At the times postinfection indicated, the cells were fixed and cross-linked by adding formaldehyde to a final concentration of 1% and incubated at room temperature for 20 min with gentle shaking. Formaldehyde was quenched by adding glycine to a final concentration of 125 mM and incubating at room temperature for 5 min with gentle shaking. The fixed-cell monolayer was then washed twice with cold PBS-ABC and scraped into fresh cold PBS-ABC containing 1× Complete protease inhibitors (Roche), and the cells were collected by centrifugation. The cell pellets were frozen at −80°C and later thawed on ice, lysed in 250 µl of radioimmunoprecipitation assay (RIPA) lysis buffer (10 mM sodium phosphate, 150 mM sodium chloride, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 2 mM EDTA, 1× Complete protease inhibitors) on ice for 20 min, and chromatin was fractionated by sonication using a Bioruptor 200 (Diagenode) for three cycles of 15 s on and 45 s off for 10 min, followed by three cycles of 30 s on and 30 s off for 5 min on the high-power setting. The majority of the DNA fragments were less than 1 kbp. The chromatin was subjected to centrifugation, and the supernatant containing the soluble chromatin in each sample was divided into two 100-µl aliquots, one for the SNF2H ChIP and the other for a control rabbit IgG ChIP. The chromatin was then diluted 1:10 in ChIP dilution buffer (150 mM sodium chloride, 10 mM sodium phosphate, 2 mM EDTA, 1.1% Triton X-100, 0.1% SDS, 1× Complete protease inhibitors) to a total volume of 1 ml. Five percent of each sample was removed to use to generate standard curves for real-time PCR analysis (see below) and to determine the input value for each sample. SNF2H rabbit polyclonal antibody (Bethyl) or control rabbit IgG (Millipore) was added (2 µg), along with 20 µl of Magna ChIP protein A magnetic beads (Millipore), and samples were incubated overnight on a rotating incubator at 4°C. Immune complexes were washed twice with ChIP dilution buffer, twice with lithium chloride wash buffer (50 mM HEPES [pH 7.5], 250 mM lithium chloride, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 0.7% sodium deoxycholate, 1× Complete protease inhibitors), and twice with Tris-EDTA (TE) buffer. The chromatin was eluted from the beads by adding elution buffer (1% SDS, 100 mM sodium bicarbonate) and heating to 65°C. Sodium chloride was added to a final concentration of 200 mM, and the cross-linking was reversed by heating to 95°C for 15 min. Samples were treated with 1 µg RNase A for 30 min at 37°C and then with proteinase K for 1 h at 45°C, and the eluted chromatin was purified using the QIAquick PCR purification kit (Qiagen).
The histone H3 chromatin immunoprecipitation assay was performed similarly to the SNF2H ChIP, with minor changes described below. siRNA-transfected HEp-2 cells were infected with HSV-1 at an MOI of 10. At the times postinfection indicated, the cells were fixed, cross-linked, and harvested as described above for the SNF2H ChIP assay. The cell pellets were frozen at −80°C, thawed on ice, and lysed with SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1], 1× Complete protease inhibitors) on ice for 20 min. Chromatin was prepared, and samples were incubated with antibodies as described above. The chromatin was diluted 1:10 in ChIP dilution buffer (150 mM sodium chloride, 10 mM sodium phosphate, 2 mM EDTA, 1.1% Triton X-100, 0.01% SDS, 1× Complete protease inhibitors), and diluted chromatin was precleared with single-stranded DNA (ssDNA)-protein A agarose beads. An aliquot containing 5% of each sample was removed to use to generate standard curves for real-time PCR analysis and to determine the input value for each sample. ChIP-grade histone H3 rabbit polyclonal antibody (Abcam) or control rabbit IgG (Millipore) (1.5 µg) was added to each sample, and the samples were incubated overnight on a rotating incubator at 4°C. Immune complexes were isolated by collection on ssDNA-protein A agarose beads. The beads were washed twice with ChIP dilution buffer, once in ChIP dilution buffer with 500 mM sodium chloride, once with lithium chloride wash buffer (250 mM lithium chloride, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, 20 mM Tris [pH 8.1], 1× Complete protease inhibitors), and twice with TE buffer. The chromatin was eluted from the beads, cross-links were reversed, and DNA was purified as described above.
Real-time PCR.
Real-time PCR was performed as described previously (1) using Power SYBR green master mix and a Prisma 7300 sequence detection system (Applied Biosystems) according to the manufacturer’s recommendations. The final volume of each reaction mixture was 25 µl, which contained a final concentration of 100 nM of the primers specified (the primer sequences were as follows: for ICP0, 5′-CGCCTTCCCGAAGAAACTCA-3′ [forward] and 5′-CGCTCAATGAACCCGCATT-3′ [reverse]; for ICP4, 5′-GCGCTCCGTGGGACGAT-3′ [forward] and 5′-CGGCCCCTGGGACTATATGA-3′ [reverse]; for ICP27, 5′-CCGCCGGCCTGGATGTGACG-3′ [forward] and 5′-CGTGGTGGCCGGGGTGGTGCTC-3′ [reverse]; for the GAPDH pseudogene, 5′-TTCGACAGTCAGCCGCATCTTCTT-3′ [forward] and 5′-CAGGCGCCCAATACGACCAAATC-3′ [reverse]; for the NucE region of the pS2 gene, 5′-CCGGCCATCTCTCACTATGAA-3′ [forward] and 5′-CCTTCCCGCCAGGGTAAATAC-3′ [reverse]). The specificity of each PCR product was evaluated by performing a dissociation curve for each reaction. In each PCR run, the samples were measured in duplicate, and their relative abundance was determined by comparison with values obtained for a standard curve that was generated by serial 10-fold dilutions of the HSV-1-infected input samples. The real-time PCR results for the ChIP experiments are presented as the percent immunoprecipitated, which was calculated as the signal with the specific antibody (histone H3 or SNF2H) divided by the value obtained from 1% input of the corresponding sample. For the histone H3 ChIPs, the control IgG background was subtracted from the specific histone H3 signal.
Statistical analysis.
Statistical analysis was performed using the R package of statistical software (http://cran.r-project.org). Significance testing was done by applying a two-tailed Wilcoxon rank sum test, using paired-sample testing with Bonferroni’s correction for multiple testing.
ACKNOWLEDGMENTS
This work was funded by NIH grant AI 063106 (D.M.K.). K.F.B. was supported by NIH institutional NRSA training grant AI 007245 and NIH individual NRSA fellowship AI 081477.
We thank members of the Knipe laboratory for advice and helpful discussions, especially Anna Cliffe for help with ChIP assays and for designing many of the primer pairs used in the real-time PCR assays. We thank Catherine Sodroski for initial work setting up SNF2H transfections and ChIP assays. We thank Robert Kingston for providing the pFB.SNF2h.c-FLAG plasmid.
Footnotes
Citation Bryant, K. F., R. C. Colgrove, and D. M. Knipe. 2011. Cellular SNF2H chromatin-remodeling factor promotes herpes simplex virus 1 immediate-early gene expression and replication. mBio 2(1):e00330-10. doi:10.1128/mBio.00330-10.
REFERENCES
- 1. Cliffe A. R., Knipe D. M. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Herrera F. J., Triezenberg S. J. 2004. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 78:9689–9696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oh J., Fraser N. W. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82:3530–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang J., Kent J. R., Placek B., Whelan K. A., Hollow C. M., Zeng P. Y., Fraser N. W., Berger S. L. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 80:5740–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kent J. R., Zeng P. Y., Atanasiu D., Gardner J., Fraser N. W., Berger S. L. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 78:10178–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lacasse J. J., Schang L. M. 2010. During lytic infections, herpes simplex virus type 1 DNA is in complexes with the properties of unstable nucleosomes. J. Virol. 84:1920–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gu H., Roizman B. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. U. S. A. 104:17134–17139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hancock M. H., Cliffe A. R., Knipe D. M., Smiley J. R. 2010. Herpes simplex virus VP16, but not ICP0, is required to reduce histone occupancy and enhance histone acetylation on viral genomes in U2OS osteosarcoma cells. J. Virol. 84:1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kutluay S. B., DeVos S. L., Klomp J. E., Triezenberg S. J. 2009. Transcriptional coactivators are not required for herpes simplex virus type 1 immediate-early gene expression in vitro. J. Virol. 83:3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang Y., Vogel J. L., Narayanan A., Peng H., Kristie T. M. 2009. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 15:1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai W., Schaffer P. A. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J., Silverstein S. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J. Virol. 66:2916–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taylor T. J., Knipe D. M. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Bruyn Kops A., Knipe D. M. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857–868 [DOI] [PubMed] [Google Scholar]
- 15. Knipe D. M., Senechek D., Rice S. A., Smith J. L. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bozhenok L., Wade P. A., Varga-Weisz P. 2002. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 21:2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Collins N., Poot R. A., Kukimoto I., Garcia-Jimenez C., Dellaire G., Varga-Weisz P. D. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627–632 [DOI] [PubMed] [Google Scholar]
- 18. Badenhorst P., Voas M., Rebay I., Wu C. 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16:3186–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Deuring R., Fanti L., Armstrong J. A., Sarte M., Papoulas O., Prestel M., Daubresse G., Verardo M., Moseley S. L., Berloco M., Tsukiyama T., Wu C., Pimpinelli S., Tamkun J. W. 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5:355–365 [DOI] [PubMed] [Google Scholar]
- 20. Ito T., Bulger M., Pazin M. J., Kobayashi R., Kadonaga J. T. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145–155 [DOI] [PubMed] [Google Scholar]
- 21. LeRoy G., Orphanides G., Lane W. S., Reinberg D. 1998. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282:1900–1904 [DOI] [PubMed] [Google Scholar]
- 22. Levenstein M. E., Kadonaga J. T. 2002. Biochemical analysis of chromatin containing recombinant Drosophila core histones. J. Biol. Chem. 277:8749–8754 [DOI] [PubMed] [Google Scholar]
- 23. Mizuguchi G., Tsukiyama T., Wisniewski J., Wu C. 1997. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol. Cell 1:141–150 [DOI] [PubMed] [Google Scholar]
- 24. Fazzio T. G., Kooperberg C., Goldmark J. P., Neal C., Basom R., Delrow J., Tsukiyama T. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goldmark J. P., Fazzio T. G., Estep P. W., Church G. M., Tsukiyama T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423–433 [DOI] [PubMed] [Google Scholar]
- 26. Elshiekh N. A., Harris-Hamilton E., Bachenheimer S. L. 1991. Differential dependence of herpes simplex virus immediate-early gene expression on de novo-infected cell protein synthesis. J. Virol. 65:6430–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gelman I. H., Silverstein S. 1987. Herpes simplex virus immediate-early promoters are responsive to virus and cell trans-acting factors. J. Virol. 61:2286–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kushnir A. S., Davido D. J., Schaffer P. A. 2010. Role of nuclear factor Y in stress-induced activation of the herpes simplex virus type 1 ICP0 promoter. J. Virol. 84:188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Corona D. F., Tamkun J. W. 2004. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim. Biophys. Acta 1677:113–119 [DOI] [PubMed] [Google Scholar]
- 30. Krajewski W. A., Reese J. C. 2010. SET domains of histone methyltransferases recognize ISWI-remodeled nucleosomal species. Mol. Cell. Biol. 30:552–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heine J. W., Honess R. W., Cassai E., Roizman B. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14:640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao M., Knipe D. M. 1991. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J. Virol. 65:2666–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schaffer P., Vonka V., Lewis R., Benyesh-Melnick M. 1970. Temperature-sensitive mutants of herpes simplex virus. Virology 42:1144–1146 [DOI] [PubMed] [Google Scholar]
- 34. Showalter S. D., Zweig M., Hampar B. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aalfs J. D., Narlikar G. J., Kingston R. E. 2001. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem. 276:34270–34278 [DOI] [PubMed] [Google Scholar]
- 36. Murphy C. G., Lucas W. T., Means R. E., Czajak S., Hale C. L., Lifson J. D., Kaur A., Johnson R. P., Knipe D. M., Desrosiers R. C. 2000. Vaccine protection against simian immunodeficiency virus by recombinant strains of herpes simplex virus. J. Virol. 74:7745–7754 [DOI] [PMC free article] [PubMed] [Google Scholar]