Summary
Neuregulin (NRG) signaling through the receptor tyrosine kinase, ERBB3, is required for embryonic development, and dysregulated signaling has been associated with cancer progression. Here we show that NRG1/ERBB3 signaling inhibits melanocyte (MC) maturation and promotes undifferentiated, migratory and proliferative cellular characteristics. Embryonic analyses demonstrated that initial MC specification and distribution were not dependent on ERBB3 signaling. However NRG1/ERBB3 signaling was both necessary and sufficient to inhibit differentiation of later stages of MC development in culture. Analysis of tissue arrays of human melanoma samples suggests that ERBB3 signaling may also contribute to metastatic progression of melanoma as ERBB3 was phosphorylated in primary tumors compared to nevi or metastatic lesions. NRG1-treated MCs demonstrated increased proliferation and invasion and altered morphology concomitant with decreased levels of differentiation genes, increased levels of proliferation genes and altered levels of melanoma progression and metastases genes. ERBB3 activation in primary melanomas suggests that NRG1/ERBB3 signaling may contribute to the progression of melanoma from benign nevi to malignancies. We propose that targeting ERBB3 activation and downstream genes identified in this study may provide novel therapeutic interventions for malignant melanoma.
Keywords: Erbb3, neuregulin, melanocytes, melanoma, neural crest
Introduction
Melanocytes (MCs) are specialized cells that produce and store melanin, a pigment that provides protection against ultraviolet-induced DNA damage and is responsible for coloration of the hair, skin, and eye. The precursor cells to MCs, melanoblasts (Mbs), can first be distinguished from the multipotent neural crest (NC) stem cells by the expression of the early MC marker microphthalmia-associated transcription factor (MITF) around embryonic day (E) 10.0 in mice (Hou et al., 2000, Silver et al., 2006). Once specified, Mbs proliferate and migrate along a dorso-lateral pathway through the mesenchymal layer of the skin. Starting at E13.5, a subset of Mbs migrate from the mesenchymal layer into overlying epidermis where some colonize the developing hair. Starting at E15.5 Mbs initiate terminal differentiation, synthesize melanin and become MCs.
Various signaling pathways, including those mediated by receptor tyrosine kinases such as KIT, have been implicated in Mb expansion, survival, and migration (Bennett, 1991, Bennett et al., 1998, Matsui et al., 1990). Other receptor tyrosine kinases whose function in MC development has not been fully explored include members of the EGFR superfamily of receptor tyrosine kinases which are expressed in various tissues of epithelial, mesenchymal, and neuronal origin. Their expression has also been observed in human MC cultures. Despite this suggestive expression data, a functional role for EGFR family-mediated signaling in MC development and disease has not been clearly established.
The EGFR superfamily of receptor tyrosine kinases consists of four distinct members: ERBB1 (EGFR/HER1), ERBB2 (HER2), ERBB3 (HER3), and ERBB4 (HER4). Activation of these ERBB receptors is controlled by the spatial and temporal expression of the receptor combinations and of their ligands, the neuregulins (NRGs). NRGs belong to a group of EGF-like growth and differentiation factors that specifically bind to the ERBB3 and ERBB4 receptors (Wen et al., 1994, Carraway et al., 1994). Binding of NRG1 to the extracellular domain of ERBB3 or ERBB4 alters receptor conformation and promotes dimerization with another member of the family, usually ERBB2. Receptor heterodimerization leads to trans- and autophosphorylation of the cytoplasmic tyrosine residues, resulting in the activation of downstream signaling. In the case of ERBB3, heterodimerization is especially critical, as ERBB3 has no kinase activity due to a substitution of critical residues in the cytoplasmic domain (Guy et al., 1994, Carraway et al., 1994, Sierke et al., 1997), and can only become phosphorylated in trans- by heterodimerization with ERBB2 or ERBB4 (Kim et al., 1998). Particularly, ERBB2/3 receptor pairs have been reported to elicit strong mitogenic responses upon ligand binding (Sliwkowski et al., 1994).
Expression of the ERBB2 and ERBB3 receptors in human primary MC cultures and melanoma cell lines has been reported, however the role these receptors play in MC development and disease is unclear. In the current study we show that NRG1/ERBB3 signaling is not essential for early Mb development. However it inhibits MC differentiation and is activated in primary human melanomas. Consistent with these observations, we show that activation of ERBB3 decreases expression of differentiation genes while increasing expression of genes associated with cellular proliferation and genes that promote migration and metastases, indicating that NRG1/ERBB3 signaling may be a potential therapeutic target for melanoma treatment.
Materials and Methods
Mouse husbandry
ErbBmsp1 mice originated on a BALB/cJ and C57BL/6J mixed genetic background and are maintained by crossing to C57BL/6J. Erbb3KO1/+ and Erbb3KO2/+ mice from a mixed background were re-derived and maintained on C57BL/6J. Mice were housed in the National Human Genome Research Institute animal facility according to the NIH guidelines. Noon on the day of vaginal plug observation was designated E0.5 for timed pregnancies. Whole-mount in situ hybridization was performed as previously described (Loftus et al., 2002).
Cell culture and RNA isolation
Melan-Ink4a cells were grown at 37°C with 5% CO2 in RPMI medium 1640 supplemented with 10% Fetal Bovine serum (FBS) (Gibco,Scotland, UK), 200 nM 12-O-Tetradecanoylphorbol 13-acetate (TPA) (Sigma-Aldrich, St Louis, MO, USA) and 200 pM cholera toxin (CT) (Sigma-Aldrich). Medium was changed three times a week. When indicated 10 nM NRG1-β1 (NRG1) (R&D Systems, #377-HB) was added to the cells at time of passage. Total RNA was isolated form 90% confluent cells as reported by Loftus et al., (Loftus et al., 2002) with one modification: following the Trizol reagent (Invitrogen) lysis of the cell, a 5 min, 65°C incubation was added.
Cell Cycle Analysis
Cell cycle distribution was determined by flow cytometric analysis of propidium iodide-labeled cells. Melan-Ink4a cells were seeded at 2.5×105 cells/60-mm plate and treated with 10 nM NRG1-β1(R&D Systems, Minneapolis, MN) in the presence or absence of TPA and CT for six to eight days, respectively. Cells were harvested in 0.05% Trysin-EDTA 1X (Gibco), washed in 1X PBS and equal number of cells was incubated in propidium iodide (NuCycl PI, # 7033, Exalpha Biologicals, Inc.) for 20 minutes at 37°C. Cell cycle analysis was performed by FACSCalibur (BD Biosciences), and data analyzed using Flowjo software (Tree Star, Inc. Ashland, OR).
Invasion Assay
In vitro invasion assays were performed using 8μm transwell polycarbonate membrane coated with a layer of polymerized basement membrane matrix (InnoCyte Cell Assay Kit, Calbiochem, San Diego, CA). Melan-Ink4a cells were serum, CT, and TPA starved overnight. Inserts were rehydrated as described by manufacturer, and 2×104 cells were seeded into invasion inserts. NRG1-β1 treated cells were stimulated with 10nM NRG1-β1 at time of plating into invasion chambers. Cells were allowed 48 hours to invade before being stained with Hema 3 Stat Pack (Fisher Scientific, Kalamazoo, MI). Experiments were performed in two independent assays and in duplicate for each assay.
Microarray processing and analysis
Samples were prepared according to Affymetrix protocols (Affymetrix, Inc). RNA quality and quantity was ensured using the Bioanalyzer (Agilent, Inc) and NanoDrop (Thermo Scientific, Inc) respectively. Per RNA labeling, 200 nanograms of total RNA was used in conjunction with the Affymetrix recommended protocol for the GeneChip 1.0 ST chips. The hybridization cocktail containing the fragmented and labeled cDNAs were hybridized to The Affymetrix Mouse GeneChip® 1.0 ST chips. The chips were washed and stained by the Affymetrix Fluidics Station using the standard format and protocols as described by Affymetrix. The probe arrays were stained with streptavidin phycoerythrin solution (Molecular Probes, Carlsbad, CA) and enhanced by using an antibody solution containing 0.5 mg/mL of biotinylated anti-streptavidin (Vector Laboratories, Burlingame, CA). An Affymetrix Gene Chip Scanner 3000 was used to scan the probe arrays. Gene expression intensities were calculated using the Gene Chip Operating software 1.2 (GCOS 1.2, Affymetrix). Each cell line was done in triplicate labeling and hybridization. Cell files generated by the Affymetrix GCOS program were imported in the Affymetrix Expression Consol program and RMA (Robust Microarray Analysis) normalization was performed to generate the .Chp files. GeneSifter software (VizX Labs, Seattle, WA) and Pairwise comparisons were performed on the .Chp files and t-test (p < 0.05) and Benjamin and Hochberg correction (= 0.05) were applied.
Ingenuity Pathway analysis
Gene expression perturbed by NRG1 treatment was evaluated using Ingenuity Pathway analysis (IPA) software (Ingenuity Systems version 7.0-1802). Genes with a minimal 1.5-fold change and p < 0.05 were uploaded into the IPA software to generate functional networks and perform pathway analysis.
Neural tube explant cultures of neural crest
Timed pregnancies between Erbb3msp1/+ animals were established and trunk region neural tubes were surgically isolated from E9.5 old embryos using trypsin digestion. The remaining portion of the embryo was stored for DNA isolation and genotyped for the Erbb3msp1 mutation. Each neural tube was placed in a well of a 8-well chamber slide containing DMEM supplemented with 10% fetal bovine serum, glutamine, penicillin/streptomycin (GIBCO, Bethesda, MD), 100 nM Endothelin-3 (EDN3) (Sigma), 50 ng/ml stem cell factor (SCF) (R&D Systems, Minneapolis, MN), 1 ng/ml Wnt3a (R&D Systems) and when indicated 10 nM NRG1. The culture media was changed three times a week. Cultures were grown for 14 days and then assayed for pigment production using light microscopy. For immunohistochemistry 12 day cultures were trypsinized, and re-plated into 8-well chamber slides and the following day were fixed in 4% PFA for 20 minutes and immunostaining was performed as described by (1). Mouse monoclonal MITF antibody was used at a concentration of 1:800. Rabbit polyclonal TYRP1 antibody was used at 1:400 dilution.
Immunohistochemistry
Tissue arrays were obtained from the Tissue Array Research Program of the National Cancer Institute, Bethesda, MD. Deparaffinization, antigen-retrieval and staining of tissue arrays were performed as previously described (Weeraratna et al., 2004). Phosho-ERBB3 antibody was obtained from Cell Signaling and was used at a concentration of 1:100. Scoring of tissue arrays was performed by a pathologist (SMH).
Immunoblot analysis
Cell lysates containing equal amounts of protein (BCA protein assay kit, Pierce, Rockford, IL) were separated on 4-12% Tris-Glycine SDS gel (Invitrogen, Carlsbad, CA), transferred to polyvinylidene fluoride membrane (Invitrogen) and blocked with 5% milk in TBST [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Tween 20] for 1 hour at room temperature. The membranes were incubated with primary antibody against ERBB3 (1:1000 diluted in 5% milk in TBST) (Santa Cruz Biotechnology; C-17), phospho-ERBB3 (1:500 diluted in 5% BSA in TBST) (Cell Signaling, Santa Cruz, CA; 21D3), AKT (1:1000 diluted in 5% BSA in TBST) (Cell Signaling), phospho-AKT (Ser473) (1:500 diluted in 5% BSA in TBST) (Cell signaling), p44/42 MAPK (1:1000 diluted in 5% milk in TBST) (Cell Signaling), phopsho-p44/42 MAPK (1:1000 diluted in 5% BSA in TBST) (Cell Signaling) and MITF (1:1000 diluted in 2% milk in TBST) at 4°C overnight. The membranes were also incubated with TYR (1:10,000), SL (1: 2000), DCT (1: 5000) and TYRP1 (1: 8000) antibodies for 1 hour at room temperature. All four antibodies were kindly provided by Dr. Vincent Hearing (National Cancer Institute, Bethesda, MD). After washing in TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000) (Amersham Biosciences, Little Chalfont, UK) for 1 hour at room temperature. The protein bands were detected using an enhanced chemiluminescence kit (Millipore, Billerica, MA).
Results
Mb specification and migration in the absence of ERBB3 signaling
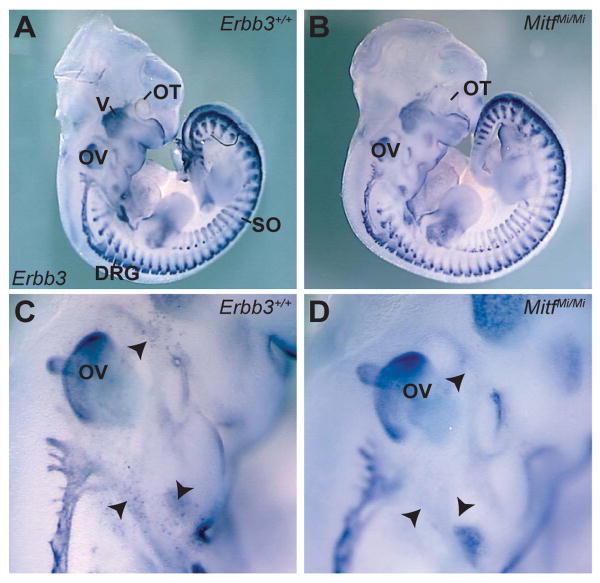
To evaluate a role for Erbb3 in MC development, in situ hybridization was utilized to examine Erbb3 mRNA in mouse embryos. At E11.5, Erbb3 expression was detected in previously described locations: dorsal root ganglia, sympathetic ganglia, somites, and in cranial ganglia and nerves (Meyer et al., 1997) (Figure 1A). A previously uncharacterized population of Erbb3-expressing cells was also observed in locations consistent with them being Mbs (Figure 1C). Examination of Erbb3 expression in MitfMi/Mi mutant embryos that are devoid of Mbs (Nakayama et al., 1998, Opdecamp et al., 1997) showed this cell population was absent, demonstrating that these Erbb3-expressing cells were Mbs (Figure 1B and D).
Figure 1. Erbb3is expressed in MC precursors during embryonic development.
Lateral view of Erbb3 whole-mount in situ hybridization in wild-type (A, C) and MitfMi/Mi (B, D) embryos at E11.5. In both wild-type embryos and MitfMi/Mi mutants, Erbb3 expression is observed in the cranial ganglia and nerves, DRG and somites. In wild-type embryos (C), a population of Erbb3-expressing cells (black arrowheads) is detected in the vicinity of the otic vesicle, the location where Mbs are normally found. These Erbb3-expressing cells were absent in Mb-deficient mutants (D), indicating that these cells are Mbs. Labeled structures are otocyst (OT) trigeminal ganglia (V), dorsal root ganglia (DRG), otic vesicle (OV) and somites (SO).
To investigate whether Erbb3 is required for Mb development, the distribution of Mbs was examined using in situ analysis of two Mb markers, Silver (Si) and Dopachrome Tautomerase (Dct) (Baxter and Pavan, 2003) in embryos containing functionally-inactive allele of Erbb3, Erbb3msp1/msp1 (Buac et al., 2008). At E10.5 when Mbs are specified and begin to express Mb markers, similar numbers of Mbs were observed in control and mutant embryos in the expected locations in the head, adjacent to otic vesicle and near the forelimb (Figure S1). While variability was observed between embryos as expected, the average number of Si-expressing cells was not noticeably different between Erbb3 mutants and control embryos indicating that Mb specification is not affected by impaired ERBB3 signaling. Similar results were seen using two additional alleles of ERBB3 (Erbb3KO1/KO1 and Erbb3KO2/KO2) (data not shown) (Riethmacher et al., 1997).
To determine if lack of ERBB3 signaling affects Mb expansion and migration, we evaluated Si and Dct expression at E11.5 and E12.5, as during these later stages Mbs proliferate and migrate along the dorso-lateral pathway through the mesenchyme along the lateral edge of the body. At E11.5 and E12.5, no difference in Mb distribution and density was observed between Erbb3+/+(Figure S2A, C and E), Erbb3msp1/msp1 (Figure S2B, D and F) or two other Erbb3- null embryos (Figure S3).
To address if the KIT receptor tyrosine kinase pathway compensated for ERBB3 signaling deficiency, we analyzed the effect of the Erbb3msp1 allele on Mb development in a Kit haploinsufficient background (KitW-LacZ/+) (Bernex et al., 1996). No apparent difference in the distribution of Mbs was noted in KitW-LacZ/+; Erbb3msp1/msp1embryos compared to KitW-LacZ/+ (Figure S4) demonstrating that redundancy from the KIT receptor tyrosine kinase is not likely compensating for absence of ERBB3 signaling. Taken together, these results indicate that while ERBB3 is expressed in Mbs, ERBB3 signaling does not play an essential role in initial specification of Si+ and Dct+ Mbs or in their migration along the dorso-lateral pathway during these early stages.
NRG1/ERBB3 signaling inhibits MC differentiation and melanogenesis
While impaired ERBB3 signaling does not disrupt initial Mb specification, expansion and migration, we could not rule out the possibility that ERBB3 is required at later stages of MC development such as differentiation and melanin production. Since Erbb3 null embryos die prior to E13.5, an in vivo analysis of MC development beyond E12.5 was precluded (Buac et al., 2008, Riethmacher et al., 1997). Therefore, we tested whether a NC cell culture system in which NC cells were derived from isolated neural tubes, could be used to assess NRG1/ERBB3 signaling. We demonstrated that ligand-induced ERBB3 phosphorylation and activation of its downstream pathways (AKT and MAPK) was robust in control (wildtype and Erbb3msp1/+) but not in mutant (Erbb3msp1/msp1) cultures (Figure S5), indicating that this system could be used to evaluate a requirement of ERBB3 signaling in later stages of MC differentiation.
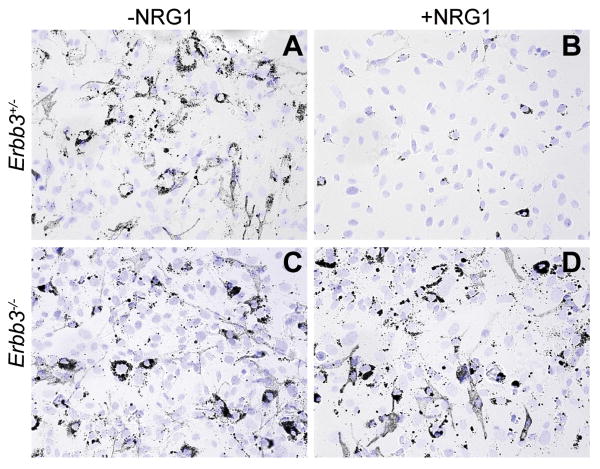
To evaluate the ability of Erbb3-null NC cells to generate pigmented MCs, neural tubes were monitored for onset and intensity of pigmentation. The Erbb3msp1/msp1 mutant cultures (n=5) consistently exhibited pigmented MCs earlier than control cultures (n=9). Moreover, the MCs from Erbb3msp1/msp1 mutant cultures (Figure 2C) appeared more pigmented than those from control cultures (Figure 2A), suggesting that ERBB3 signaling may be required to inhibit MC development and differentiation.
Figure 2. MC differentiation in NC cultures.
Bright field images of Erbb3+/- (A,B) and Erbb3-/- (C,D) NC cultures grown in the absence (A,C) or presence (B,D) of NRG1. Cells were cultured for two weeks after neural tube isolation, replated and DAPI stained one day later. Although differentiated MCs (dark cells) were detected in both genotypes, pigmented MCs from Erbb3 mutant cultures (C) were darker. When exogenous NRG1 was added to Erbb3+/- NC cultures (B), a dramatic reduction in pigmented MCs was observed. No such difference was noted when Erbb3 mutant cultures (D) were grown with NRG1, suggesting that activation of NRG1/ERBB3 signaling inhibits MC differentiation.
To explore whether exogenous NRG1 has an effect on MC differentiation and melanin production, Erbb3+/- cultures were grown in the presence of 10 nM NRG1. After two weeks, the formation of melanin-containing cells was assayed. Strikingly, control cultures grown in the presence of NRG1 exhibited a severe reduction in the number of pigmented MCs (Figure 2B) compared to cultures grown without exogenous NRG1 (Figure 2A) where differentiated, melanin-containing MCs were observed. Analysis of immunostaining for Mb marker (MITF) and later stage differentiation marker, tyrosinase-related protein 1 (TYRP1) indicated that Erbb3+/- cultures treated with NRG1 contained fewer TYRP1+/MITF+ cells (Figure S6). Quantification of MITF+ and TYRP1+ cells showed that the percentage of TYRP1/MITF-double positive cells was significantly lower in NRG1-treated control cultures (P = 0.002) (Figure S6E), providing a strongly evidence that NRG1 negatively regulates MC differentiation and melanin synthesis.
Contrary to the inhibitory effects observed in control cultures, in Erbb3 null cultures pigmented MCs were formed both in the presence and absence of exogenous NRG1 (Figure 2C and D). No striking differences were observed in the amount of pigmentation or the proportion of TYRP1+/MITF+ cells in NRG1 treated and untreated mutant MCs (Figure S6). The similar appearance of NRG1-treated and untreated mutant MCs suggests that the effect of NRG1 on these cultures is through ERBB3. Taken together, these results indicate that NRG1/ERBB3 signaling suppresses MC differentiation and pigment production even in the presence of growth factors such as WNT3a, EDN3 and SCF, which strongly promote MC differentiation.
ERBB3 is frequently activated in primary melanoma
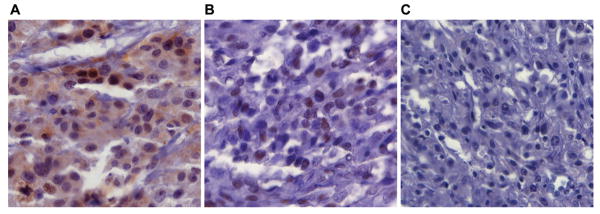
Given the inhibitory effects of NRG1/ERBB3 signaling on MC differentiation that we observed in NC cultures we hypothesized that ERBB3 activation might be associated with progression of human melanoma from the more benign, differentiated state of MCs in nevi to the more metastatic, less differentiated state seen in malignant melanoma cells. To assess this we evaluated the activation status of ERBB3 by immunostaining for phospho-ERBB3 on tissue arrays of human melanoma specimens (Figure 3 and Table 1). The 124 surgically obtained samples on the array consisted of melanocytic lesions ranging from melanocytic nevi to primary and metastatic melanoma. Striking patterns of phospho-ERBB3 expression were observed frequently in these samples. Phospho-ERBB3 staining was present in 64% of nevi (21/33), 76% of primary melanoma (25/33), 27% of lymph node metastases (7/19) and 8% of visceral metastases (3/39) (Table 1). Interestingly, while the majority of the staining observed was nuclear, only samples from primary melanomas (18%) also demonstrated cytoplasmic staining indicative of highly active ERBB3 (Figure 3). Taken together, these data demonstrate that ERBB3 is frequently activated in human melanomas.
Figure 3. Phospho-ERBB3 staining in melanoma tissue arrays.
Representative photomicrographs of immunohistochemistry for phospho-ERBB3. All images taken at 320× magnification. (A) Nuclear and cytoplasmic staining of phospho-ERBB3 in a primary melanoma. (B) Nuclear staining of phospho-ERBB3 in a melanoma metastatic to a lymph node. (C) A metastatic melanoma that is negative for staining of phospho-ERBB3.
Table 1. Phospho-ERBB3 expression in melanoma.
| Nevi | Primary Melanomas | Lymph Node Metastasis | Other Metastasis | |
|---|---|---|---|---|
| Nuclear | 64% | 58% | 27% | 8% |
| Cyto/Nuclear | 0 | 18%* | 0 | 0 |
| Negative | 36% | 24% | 63% | 92% |
p-values (Fisher's exact test) of cyto/nuclear staining in primary melanomas compared to nevi: p = 1 × 10-5; to lymph node metastasis: p = 4 × 10-9; and to other metastasis: p = 4 × 10-9; show statistical significance.
NRG1 increases proliferation and invasion of MCs
To understand how activation of ERBB3 signaling may modulate melanoma progression, we examined the effects of NRG1 on MC morphology. The melan-Ink4a cell line (MC-derived, deficient for Ink4a) was used as it allows stable propagation in culture (Sviderskaya et al., 1995). Western blot analysis showed that melan-Ink4a cells expressed the ERBB3 and ERBB2 receptors and were able to respond to NRG1 stimuli by phosphorylating ERBB3 and activating the AKT and MAPK pathways (Figure S7).
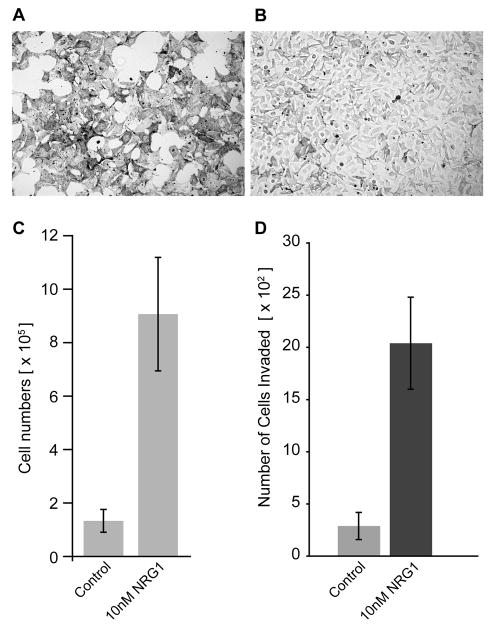
Growth of melan-Ink4a cells requires addition of the PKC agonist tetradecanoyl phorbol acetate (TPA) and the cAMP elevator cholera toxin (CT) (Sviderskaya et al., 1995). Within two days of TPA and CT withdrawal, melan-Ink4a cells stop dividing and dramatically change their morphology, exhibiting a large, flattened and heavily-pigmented appearance (Figure 4A and C). When grown without TPA and CT but in the presence of NRG1, both the proliferation and morphology were rescued, resembling cultures treated with TPA/CT (Figure 4B and C). Flow cytometric analysis of the cell cycle revealed that NRG1 treatment reduced the number of cells in G0-G1 and increased the proportion of cells in S and G2/M phases (Figure S8). To determine if ERBB3 signaling alters invasive ability of melan-Ink4a cells, control and NRG1 treated cells were grown in Boyden chambers and assessed for their ability to invade through basement membrane matrix (Figure 4D). NRG1 treatment resulted in a greater 5-fold increase in the number of cells that were able to invade the matrix (P < 0.0003). These results indicate that NRG1 can substitute for TPA and CT ny promoting the growth, expansion and invasion of melan-Ink4a cells.
Figure 4. NRG1 increases proliferation and invasion of melan-Ink4a cells.
Bright filed images of melan-Ink4a cells (A, B) in media lacking TPA and CT (A) and in media lacking TPA and CT but with 10nM NRG1 added (B). (C) Cell doublings in the absence of TPA and CT (control) and in media containing NRG1 without TPA and CT (10 nM NRG1). Cells were plated at equal numbers (2.5 × 105) and total numbers of cells were counted four days after the indicated treatments using hemocytometer. While melan-Ink4a grown in the absence of TPA/CT stopped proliferating, addition of NRG1 increased number of Melan-Ink4a cells. D) Increased invasion in the presence of NRG1. Control and NRG1-treated Melan-Ink4a cells were grown in Boyden chambers and assessed for their ability to invade through basement membrane matrix. For quantification, stained cells were photographed on Zeiss Axiovert 135 microscope and cells were counted in at least 16 random fields. Columns represent the absolute number of cells that invaded the matrix 48 hours after seeding (n=4).
When grown with NRG1 for eight days, melan-Ink4a cells appeared smaller and more bipolar compared to non-NRG1 treated cells (Figure S9). Interestingly, NRG1 addition also resulted in a dramatic pigment reduction, determined by the color of the MC pellet (Figure S9), also suggestive of a less differentiated state. Taken together these results demonstrate that NRG1 signaling in MCs results in morphological changes and decreased pigmentation consistent with a less differentiated state and increased proliferation and invasive capacity.
NRG1 expression correlates with dedifferentiation, increased proliferation and alteration in cancer pathways
To examine the molecular mechanisms by which the morphological changes, decreased pigmentation and increased proliferation of MCs occur and to gain insight into the implication of increased ERBB3 activity in melanomas, we used gene expression profile analysis to examine the effects of NRG1 treatment of melan-Ink4a cells. At the 1.5-fold change level, an analysis revealed 1316 genes were differentially expressed in the presence of NRG1 relative to untreated cells (p<0.05) with 374 significantly downregulated and 942 upregulated (Table S1). To evaluate the distribution of differentially expressed genes into functional networks and to gain insight into cellular and molecular pathways altered by NRG1 treatment we used Ingenuity Pathway Analysis (IPA) software. Genes with minimum 1.5-fold change and p<0.05 were mapped to functional networks and ranked by score (Table S2). The functional categories included in the top 4 networks were cell cycle, cellular movement, cellular assembly and organization, DNA replication, recombination and repair, cancer and amino acid metabolism. At the 1.2-fold change level, expression profile analysis revealed 4241 differentially expressed genes, with 1396 genes downregulated and 2845 genes upregulated by NRG1. Of the 55 genes in this set that are involved in MC development and pigmentation (Baxter et al., 2007), 40% were downregulated in the presence of NRG1 (Table S3).
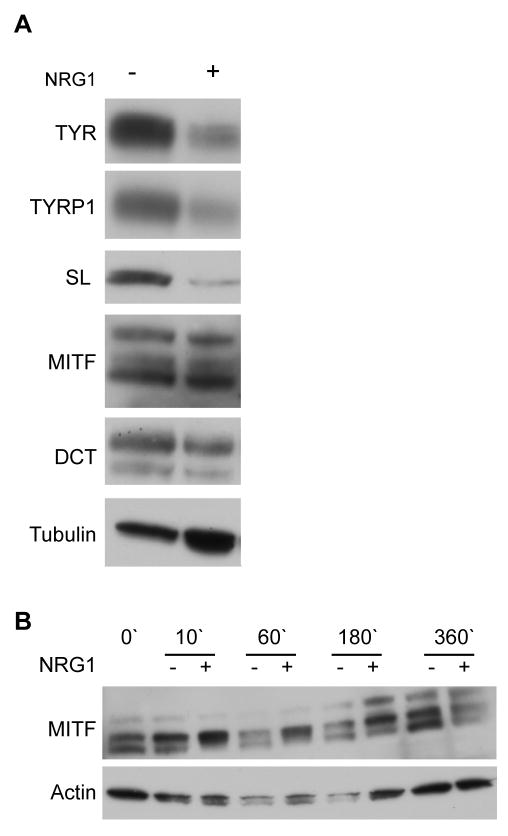
Consistent with this data, decreases in the protein levels of Tyrosinase (TYR), TYRP1, and SILVER (SL) were detected in NRG1-treated cultures (Figure 5A). While no significant change in MITF was noted in 6 day NRG1 treatment, a time course study revealed a NRG1 induced increase in MITF phosphorylation, and a subsequent reduction in total MITF protein levels at six hours after NRG1 addition (Figure 5B). These results were consistent with a NRG1 induced inhibition of differentiation and melanin synthesis that could be regulated at the level of MITF activity. Striking alterations were also observed in neural crest stem cell genes (Table S4) and in genes previously associated with melanoma initiation, invasion and metastasis (Tables S5 and S6). Taken together, these changes in gene expression are consistent with the observed hypopigmentation, proliferation and invasion phenotypes, suggesting that melan-Ink4a cells can be used to model the NRG1/ERBB3 signaling in melanoma and to test therapeutic interventions.
Figure 5. NRG1 effects expression of melanogenic enzymes.
(A) Protein levels of Tyrosinase (TYR), Tyrosinase-related protein 1 (TYRP1) and Silver (SL) were downregulated in NRG1-treated cultures (22 days after treatment). MITF and DCT expression was not visibly altered by NRG1. (B) A time-course analysis of MITF expression at 10, 60, 180, and 360 minutes after addition of NRG1. MITF phosphorylation (upper MITF band) was increased by NRG1 at 180′ and protein levels were decreased by NRG1 after 360 minutes, indicating that activation of ERBB3 pathway can regulate MITF activity levels.
Discussion
In this report, we investigated the role of NRG1/ERBB3 signaling in MC development and melanoma. We found that ERBB3 signaling negatively regulates MC differentiation and pigmentation while promoting proliferation. Furthermore, we showed that ERBB3 is frequently phosphorylated in primary melanomas implicating activation of this pathway in the transition from nevi to metastatic melanomas. These findings highlight the importance of NRG1/ERBB3 activation and its downstream targets as potential therapeutic approaches in the treatment of metastatic melanomas.
Expression analyses demonstrated that the numbers and distribution patterns of Mbs were similar in Erbb3 mutant and control embryos at E10.5 – E11.5, indicating that Mb specification, and initial proliferation, survival and migration are independent of functional ERBB3 signaling. An in vivo analysis of ERBB3 signaling at later stages of MC development was precluded by embryonic lethality of Erbb3 null mice. However, utilizing in vitro primary cultures, Erbb3 mutants were shown to produce pigmented MCs at an earlier time of onset and with a higher degree of pigmentation. These findings suggested that ERBB3 signaling inhibited MC differentiation. This hypothesis was further supported by exogenous NRG1 inducing a reduction in pigmented, TYRP1+ MCs. Interestingly, a recent analysis of a zebrafish mutant homozygous for an erbb3b null allele showed normal early larva pigment development, similar to our findings in mouse Erbb3 mutants, but impaired adult pigment pattern formation (Budi et al., 2008). Consistent with our hypothesis that NRG1/ERBB3 signaling inhibits MC differentiation, mice deficient for ADAM17, a protease that is required to cleave and thus activate membrane bound NRG1, demonstrate densely pigmented hair follicles and disorganized pigment granules in the hair (Horiuchi et al., 2005). Additional studies utilizing conditional knockouts are required to determine the functional relevance of NRG1/ERBB signaling in vivo at later stages of MC development and skin pigmentation.
In addition to MC development our studies indicate that ERBB3 plays a role in melanoma progression. A high percentage of nevi and primary melanomas exhibit activated ERBB3 signaling. ERBB3 overexpression has been previously associated with breast, lung, prostate and ovarian cancer, and several studies have also reported alterations of ERBB3 expression in melanoma (Sithanandam and Anderson, 2008). However these studies assessed total ERBB3 levels, whereas our analysis of tumor tissue arrays examined phosphorylated ERBB3, indicative of activated signaling, in various stages of melanoma. We found significant activation in nevi and primary melanomas with predominantly nuclear phospho-ERBB3, while only primary melanomas showed cytoplasmic staining.
As one of the major differences between nevi and primary melanoma involves a switch from a senescent to a proliferative state, these data support our hypothesis that ERBB3 activation may promote a release from senescence during tumorigenesis in nevi and primary melanomas. In support of this, recent data show that activation of ERBB3 in a murine metastatic melanoma cell line contributed to metastasis of melanoma cells (Ueno et al., 2008). Our results of reduced activation in metastatic lesions also support this theory, since often metastatic lesions are not highly proliferative, and increases in the metastatic capacity of melanoma have been linked with decreases in proliferation (Hoek et al., 2008, Hoek et al., 2006). The high frequency of phospho-ERBB3 in nevi and primary melanomas suggests that this pathway may be useful as a marker for disease progression and outcome.
The variation in subcellular localization of phospho-ERBB3 we observed in melanoma is consistent with reports of the EGFR family in other cancers (Wells and Marti, 2002, Carpenter, 2003, Lo and Hung, 2006). Recently, several studies have reported the nuclear localization of ERBB3 in prostate and breast cancer (Frogne et al., 2009, Koumakpayi et al., 2007, Koumakpayi et al., 2006, Kawano et al., 2008). While the nuclear role of EGFR family is not clearly understood, evidence exists linking it to transcriptional regulation, chromatin remodeling and nucleolar function. Future studies will be needed to understand the roles of activated ERBB3 in each cellular compartment and to understand how these influence melanoma progression.
Expression profiles of NRG1 treated MCs demonstrated changes in many genes previously associated in the progression of nevi to metastatic melanoma. Activating mutations in Nras and cyclin D1, both upregulated by NRG1 treatment, have been identified in melanomas and are thought to provide an initiating step in clonal expansion (Curtin et al., 2005, Bennett, 2008). Furthermore, activation of AKT pathway has been shown to promote a neoplastic phenotype by stimulating cell cycle progression, increasing antiapoptotic functions, and enhancing tumor cell invasion (Bennett, 2008, Stahl et al., 2004b, Stahl et al., 2004a). Interestingly, genes associated with epithelial-mesenchymal transition and melanoma invasion were also significantly altered. For instance, we noted NRG1 reduced expression of E-cadherin, an adhesion molecule whose loss of expression frequently coincides with malignant transformation of MCs, and also increased expression of N-cadherin, which has been linked with increased motility and invasiveness of tumor cells (Haass et al., 2005). NRG1 treatment also increased expression of matrix metalloproteinases 2 (MMP2) and Melanoma cell adhesion molecule (Mcam), molecules associated with melanoma invasion (Kondratiev et al., 2008, Vaisanen et al., 2008, Vaisanen et al., 1998, McGary et al., 2002, Xie et al., 1997), as well as Nedd9, which is overexpressed in human metastasis and shown to enhance invasion and migration (Kim et al., 2006). Also of note was the increased expression of the small GTPase RhoC which has been directly correlated with the metastatic potential of many cancers making it an attractive therapeutic target (Wenandy et al., 2008). Furthermore, NRG1 signaling dramatically reduced the expression of PlexinC1, which was recently identified as a potential melanoma tumor suppressor (Scott et al., 2008) and whose expression has an inverse correlation with melanoma depth of invasion. The ability of NRG1 to differentially regulate expression of genes linked to melanoma invasion and migration identifies melan-Ink4a cells as a useful tool to study molecular mechanisms of NRG1/ERBB3 signaling in melanoma.
Expression patterns were also consistent with the striking decreases in MC differentiation and pigmentation, showing both increases in neural crest and stem cell marker genes and decreases in genes associated with MC differentiation and pigmentation. While increased pigmentation is often observed in primary melanoma, there has been no study to determine if those pigmented lesions are more aggressive than those that are not, or importantly if the metastases arising from those pigmented lesions are less pigmented than the primary tumor of origin. Our findings would argue for less pigmented lesions being more aggressive. In support of this, high levels of melanin have been shown to deplete levels of immunosuppressive cytokines (Siegfried and Hendricks, 1994), and induce inflammation (Chan et al., 1994), implying that highly pigmented tumors are more susceptible to destruction by the immune system. Furthermore, two recent studies indicate that increased expression of differentiation antigens such as MART-1, which is involved in pigment production, may indicate decreased aggression in melanoma (Dissanayake et al., 2008). In the most recent study, increasing Wnt/β-catenin signaling increased melanogenesis in melanoma cells, and decreased their proliferation. Further, inhibitors of β-catenin increased the proliferation and in vivo metastasis of melanoma cells, while simultaneously decreasing pigmentation of these cells (Chien et al., 2009).
One possible mechanism by which NRG1 could inhibit differentiation and pigmentation while promoting proliferation is through MITF, a transcription factor known to be both responsive to MAP kinase signaling and to regulate MC survival, proliferation, migration and differentiation. In support of this, MITF mRNA was reduced in expression profile analysis and MITF protein was reduced upon short-term treatment with NRG1. Because MITF regulates many aspects of MC biology, a fine balance between its levels of mRNA expression and protein activity exists such that variation in levels of MITF function lead to differences in MC cell cycle, invasion, differentiation and melanogenesis (Hou and Pavan, 2008, Vance and Goding, 2004, Levy et al., 2006). Our findings suggest that NRG1/ERBB3 signaling is sufficient to induce changes in MITF expression, which in turn reduces the expression of melanogenic enzymes while altering the expression of genes involved in maintaining cells in a less differentiated state and promoting cell division.
In summary, we demonstrated that while dispensable for Mb development, NRG1/ERBB3 signaling impairs MC differentiation and melanin synthesis. Increased phosphorylation of ERBB3 in primary melanocytic lesions suggests that activation of NRG1/ERBB3 signaling may contribute to the progression of melanoma from benign nevi to metastatic malignancies. In support of this hypothesis, microarray analysis indicated that NRG1 alters expression of multiple genes associated with cancer initiation, invasion and metastasis, identifying NRG1/ERBB3 signaling as a potential therapeutic target for melanoma treatment.
Significance.
Identifying the cellular pathways that promote and inhibit the development of differentiated tissues from precursor cells can provide insights into developing diagnostics and treatments of human diseases including malignant cancers. In this study we show that signaling through the ERBB3 receptor is involved in inhibiting the differentiation of skin color-producing melanocytes from precursor cells. We also show that activation of the ERBB3 pathway is associated with the onset of melanoma, a malignant cancer arising from transformed melanocytes. Our findings suggest that the inhibition of ERBB3 signaling and manipulation of altered downstream genes could be used as therapeutic intervention in melanoma.
Supplementary Material
Supplemental Figure 1. Expression analysis of Mb-specific markers in control and Erbb3 mutant embryos at E10.5.Si expression analysis in control and Erbb3 mutant embryos at E10.5. Lateral view of Si whole-mount in situ hybridization in Erbb3+/+(A, C) and Erbb3msp1/msp1 (B) embryos. Si-expressing cells (black arrowheads) were detected in the RPE of the eye and the region below the otic vesicle (OV) in all four genotypes analyzed. Quantification of Si-expressing cells in Erbb3+/+, Erbb3msp1/msp1 (E) embryos at E10.5. Si-expressing cells were counted in whole-mount embryos using dissection microscope. The numbers of Si-expressing cells from three different embryos of each genotype were averaged and standard deviations calculated. There was no significant difference in the number of Si-expressing cells between Erbb3msp1/msp1 and Erbb3+/+ (unpaired t-test; p = 0.92)
Supplemental Figure 2. Expression analysis of Mb-specific markers in control and Erbb3 mutant embryos. At E11.5, Si-expressing cells are detected in the head region, the RPE of the eye, and along the rostral caudal axis in Erbb3+/+(A) and Erbb3msp1/msp1 (B) embryos. At E12.5, Dct expression is observed in the head (C, D) and caudal region (E, F) of Erbb3+/+(C, E) and Erbb3msp1/msp1 (D, F) embryos. No difference in the expression pattern of Si- and Dct-expressing cells was observed between Erbb3+/+ and Erbb3msp1/msp1 embryos at these two stages.
Dct and Si are normally expressed in Erbb3 null embryos. At E11.5 Dct-expressing cells (black arrowheads) were observed in the head and along the rostra-caudal axis in Erbb3+/+ (A), Erbb3KO1/KO1 (B), and Erbb3KO2/KO2 (C) embryos. Si-expressing cells (black arrowheads) showed similar expression pattern in Erbb3+/+ (D), Erbb3KO1/KO1 (E), and Erbb3KO2/KO2 (F) embryos at E11.5. No apparent difference in the expression pattern of these two markers was observed between control and Erbb3 null alleles.
Embryos generated from KitW-LacZ/+; Erbb3msp1/+ double heterozygote crosses were collected at E11.5 and stained for β-galactosidase activity. In KitW-LacZ/+; Erbb3msp1/+, β-gal-expressing cells (black arrowheads) were detected in the head (A), trunk (B), and caudal region (C) of the developing embryo. A similar β-gal expression pattern was noted in KitW-LacZ/+; Erbb3msp1/msp1 embryos (D, E, F). No major difference in the expression or distribution of β-gal+ cells was observed between KitW-LacZ/+; Erbb3msp1/+ and KitW-LacZ/+; Erbb3msp1/msp1 embryos.
(A) Derivation of primary MCs from neural tube explant cultures. Neural tubes generated from Erbb3msp1 heterozygous matings were isolated from E9.5 embryos and cultured in the presence of growth factors that promote Mb differentiation for 10 days (D). On day 10, individual cultures were trypsinized, and the same genotypes were pooled together and re-plated. Cells were grown under the same culture conditions as described above for an additional two days. After overnight starvation in 2% serum without growth factors, cells were stimulated with or without NRG1 for 15 minutes and lysed in lysis buffer. (B) Neural tube cell extracts (3 μg) were subjected to immunoblot analysis, and membranes were probed with anti-phospho-ERBB3 and total ERBB3 antibodies. NRG1-induced phosphorylation of ERBB3 was observed in Erbb3+/+ and Erbb3msp1/+ MC cultures, but not in Erbb3msp1/msp1 mutant cultures.
Immunostaining of MITF+ (red) and TYRP1+ (green) cells in Erbb3+/- (A and B) and Erbb3msp1/msp1 mutant (C and D) cultures grown in the absence of NRG1 (A and C) and in the presence of NRG1 (B and D). (E) Bar diagram depicting the percentage of TYRP1+/MITF+ cells in control and Erbb3 null NC cultures treated with or without NRG1. Error bars denote standard deviation. The proportion of TYRP1+/MITF+ cells was significantly reduced (p = 0.002; Student t-test) in NRG1-treated control cells compared to untreated cultures. The proportion of double positive cells in Erbb3 mutant cultures treated with NRG1 was at the comparable levels with untreated mutant cultures.
(A) Whole-cell lysates (30 μg) were subjected to immunoblotting using anti-ERBB2, anti-ERBB3, anti-ERBB4, anti-EGFR and anti-NRG1 antibodies. 293T lysates were used to confirm the specificity of the antibodies. Immunoblots were probed with antibody to α-Tubulin to monitor equal protein loading. Expression of ERBB2 and ERBB3 was observed in melan-Ink4a cell extracts. Protein lysates were devoid of EGFR, ERBB4 and NRG1 expression. (B) Serum-starved melan-Ink4a cells were stimulated with or without NRG1 for 15 minutes and cell lysates analyzed by immunoblotting using anti-phospho-ERBB3 (Tyr 1284), anti-phospho-AKT (Ser475), and anti-phospho-p44/42 MAPK antibody. After stripping, membranes were re-probed with antibodies against unphosphorylated forms to evaluate the amount of total proteins. Addition of NRG1 induced ERBB3 phosphorylation, which resulted in activation of the AKT and MAPK pathways as measured by AKT and MAPK phosphorylation.
Flow cytometric analysis of cells grown under three different conditions. −TPA/CT cells were arrested in G0-G1 stage. +NRG1-TPA/CT cells showed similar cell cycle progression as control cells (+TPA/CT).
Bright field images of melan-Ink4a cells grown in the presence of TPA and CT (A) and in the presence of NRG1, TPA and CT (B). Control cells contained large number of flat, pigment- containing MCs (dark spots), while NRG1-treated cells appeared less pigmented. Also, NRG1-treated cells were smaller in size and bipolar. Cell pellet was less pigmented in NRG1-treated cells. Equal number of cells was collected, spun down, and color of the cell pellet was evaluated between control and NRG1 treated cells.
Acknowledgments
We thank Dieter Riethmacher for providing Erbb3KO1/+ and Erbb3KO2/+ mice; Dorothy Bennett for Melan-Ink4a cells; Heinz Arnheiter for MITF antibody; Vincent Hearing for TYR, TYRP1, DCT and SL antibodies. Thanks to Stacy Anderson for cell cycle analysis and Julia Fekecs for graphic design. This work was supported by the National Human Genome Research Institute's Intramural Research Program and the Intramural Research Program of the National Institute on Aging.
References
- Baxter LL, Hsu BJ, Umayam L, Wolfsberg TG, Larson DM, Frith MC, Kawai J, Hayashizaki Y, Carninci P, Pavan WJ. Informatic and genomic analysis of melanocyte cDNA libraries as a resource for the study of melanocyte development and function. Pigment Cell Res. 2007;20:201–9. doi: 10.1111/j.1600-0749.2007.00372.x. [DOI] [PubMed] [Google Scholar]
- Baxter LL, Pavan WJ. Pmel17 expression is Mitf-dependent and reveals cranial melanoblast migration during murine development. Gene Expr Patterns. 2003;3:703–7. doi: 10.1016/j.modgep.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Bennett DC. Colour genes, oncogenes and melanocyte differentiation. J Cell Sci. 1991;98(Pt 2):135–9. doi: 10.1242/jcs.98.2.135. [DOI] [PubMed] [Google Scholar]
- Bennett DC. How to make a melanoma: what do we know of the primary clonal events? Pigment Cell Melanoma Res. 2008;21:27–38. doi: 10.1111/j.1755-148X.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- Bennett DC, Trayner ID, Piao X, Easty DJ, Kluppel M, Alexander WS, Wagner EF, Bernstein A. recessive spotting: a linked locus that interacts with W/Kit but is not allelic. Genes Cells. 1998;3:235–44. doi: 10.1046/j.1365-2443.1998.00184.x. [DOI] [PubMed] [Google Scholar]
- Bernex F, De Sepulveda P, Kress C, Elbaz C, Delouis C, Panthier JJ. Spatial and temporal patterns of c-kit-expressing cells in WlacZ/+ and WlacZ/WlacZ mouse embryos. Development. 1996;122:3023–33. doi: 10.1242/dev.122.10.3023. [DOI] [PubMed] [Google Scholar]
- Buac K, Watkins-Chow DE, Loftus SK, Larson DM, Incao A, Gibney G, Pavan WJ. A Sox10 expression screen identifies an amino acid essential for Erbb3 function. PLoS Genet. 2008;4:e1000177. doi: 10.1371/journal.pgen.1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budi EH, Patterson LB, Parichy DM. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development. 2008;135:2603–14. doi: 10.1242/dev.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr Opin Cell Biol. 2003;15:143–8. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- Carraway KL, 3rd, Sliwkowski MX, Akita R, Platko JV, Guy PM, Nuijens A, Diamonti AJ, Vandlen RL, Cantley LC, Cerione RA. The erbB3 gene product is a receptor for heregulin. J Biol Chem. 1994;269:14303–6. [PubMed] [Google Scholar]
- Chan CC, Hikita N, Dastgheib K, Whitcup SM, Gery I, Nussenblatt RB. Experimental melanin-protein-induced uveitis in the Lewis rat. Immunopathologic processes. Ophthalmology. 1994;101:1275–80. doi: 10.1016/s0161-6420(94)31199-7. [DOI] [PubMed] [Google Scholar]
- Chien AJ, Moore EC, Lonsdorf AS, Kulikauskas RM, Rothberg BG, Berger AJ, Major MB, Hwang ST, Rimm DL, Moon RT. Activated Wnt/beta-catenin signaling in melanoma is associated with decreased proliferation in patient tumors and a murine melanoma model. Proc Natl Acad Sci U S A. 2009;106:1193–8. doi: 10.1073/pnas.0811902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, Leboit PE, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–47. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Dissanayake SK, Olkhanud PB, O'connell MP, Carter A, French AD, Camilli TC, Emeche CD, Hewitt KJ, Rosenthal DT, Leotlela PD, et al. Wnt5A regulates expression of tumor-associated antigens in melanoma via changes in signal transducers and activators of transcription 3 phosphorylation. Cancer Res. 2008;68:10205–14. doi: 10.1158/0008-5472.CAN-08-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frogne T, Laenkholm AV, Lyng MB, Henriksen KL, Lykkesfeldt AE. Determination of HER2 phosphorylation at tyrosine 1221/1222 improves prediction of poor survival for breast cancer patients with hormone receptor-positive tumors. Breast Cancer Res. 2009;11:R11. doi: 10.1186/bcr2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy PM, Platko JV, Cantley LC, Cerione RA, Carraway KL., 3rd Insect cell-expressed p180erbB3 possesses an impaired tyrosine kinase activity. Proc Natl Acad Sci U S A. 1994;91:8132–6. doi: 10.1073/pnas.91.17.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass NK, Smalley KS, Li L, Herlyn M. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18:150–9. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–6. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, Weber BL, Nathanson KL, Phillips DJ, Herlyn M, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Zhou HM, Kelly K, Manova K, Blobel CP. Evaluation of the contributions of ADAMs 9, 12, 15, 17, and 19 to heart development and ectodomain shedding of neuregulins beta1 and beta2. Dev Biol. 2005;283:459–71. doi: 10.1016/j.ydbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Hou L, Panthier JJ, Arnheiter H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development. 2000;127:5379–89. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res. 2008;18:1163–76. doi: 10.1038/cr.2008.303. [DOI] [PubMed] [Google Scholar]
- Kawano O, Sasaki H, Endo K, Suzuki E, Haneda H, Yukiue H, Kobayashi Y, Yano M, Fujii Y. ErbB3 mRNA expression correlated with specific clinicopathologic features of Japanese lung cancers. J Surg Res. 2008;146:43–8. doi: 10.1016/j.jss.2007.05.030. [DOI] [PubMed] [Google Scholar]
- Kim HH, Vijapurkar U, Hellyer NJ, Bravo D, Koland JG. Signal transduction by epidermal growth factor and heregulin via the kinase-deficient ErbB3 protein. Biochem J. 1998;334(Pt 1):189–95. doi: 10.1042/bj3340189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, Brennan C, Hahn WC, Cordon-Cardo C, Wagner SN, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Kondratiev S, Gnepp DR, Yakirevich E, Sabo E, Annino DJ, Rebeiz E, Laver NV. Expression and prognostic role of MMP2, MMP9, MMP13, and MMP14 matrix metalloproteinases in sinonasal and oral malignant melanomas. Hum Pathol. 2008;39:337–43. doi: 10.1016/j.humpath.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Koumakpayi IH, Diallo JS, Le Page C, Lessard L, Filali-Mouhim A, Begin LR, Mes-Masson AM, Saad F. Low nuclear ErbB3 predicts biochemical recurrence in patients with prostate cancer. BJU Int. 2007;100:303–9. doi: 10.1111/j.1464-410X.2007.06992.x. [DOI] [PubMed] [Google Scholar]
- Koumakpayi IH, Diallo JS, Le Page C, Lessard L, Gleave M, Begin LR, Mes-Masson AM, Saad F. Expression and nuclear localization of ErbB3 in prostate cancer. Clin Cancer Res. 2006;12:2730–7. doi: 10.1158/1078-0432.CCR-05-2242. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–14. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006;94:184–8. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Larson DM, Baxter LL, Antonellis A, Chen Y, Wu X, Jiang Y, Bittner M, Hammer JA, 3rd, Pavan WJ. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci U S A. 2002;99:4471–6. doi: 10.1073/pnas.072087599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Zsebo KM, Hogan BL. Embryonic expression of a haematopoietic growth factor encoded by the Sl locus and the ligand for c-kit. Nature. 1990;347:667–9. doi: 10.1038/347667a0. [DOI] [PubMed] [Google Scholar]
- Mcgary EC, Lev DC, Bar-Eli M. Cellular adhesion pathways and metastatic potential of human melanoma. Cancer Biol Ther. 2002;1:459–65. doi: 10.4161/cbt.1.5.158. [DOI] [PubMed] [Google Scholar]
- Meyer D, Yamaai T, Garratt A, Riethmacher-Sonnenberg E, Kane D, Theill LE, Birchmeier C. Isoform-specific expression and function of neuregulin. Development. 1997;124:3575–86. doi: 10.1242/dev.124.18.3575. [DOI] [PubMed] [Google Scholar]
- Nakayama A, Nguyen MT, Chen CC, Opdecamp K, Hodgkinson CA, Arnheiter H. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech Dev. 1998;70:155–66. doi: 10.1016/s0925-4773(97)00188-3. [DOI] [PubMed] [Google Scholar]
- Opdecamp K, Nakayama A, Nguyen MT, Hodgkinson CA, Pavan WJ, Arnheiter H. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development. 1997;124:2377–86. doi: 10.1242/dev.124.12.2377. [DOI] [PubMed] [Google Scholar]
- Riethmacher D, Sonnenberg-Riethmacher E, Brinkmann V, Yamaai T, Lewin GR, Birchmeier C. Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature. 1997;389:725–30. doi: 10.1038/39593. [DOI] [PubMed] [Google Scholar]
- Scott GA, Mcclelland LA, Fricke AF, Fender A. Plexin C1, A Receptor for Semaphorin 7A, Inactivates Cofilin and Is a Potential Tumor Suppressor for Melanoma Progression. J Invest Dermatol. 2009;129:954–63. doi: 10.1038/jid.2008.329. [DOI] [PubMed] [Google Scholar]
- Siegfried CJ, Hendricks RL. Melanin can deplete immunosuppressive substances from the aqueous humor. Curr Eye Res. 1994;13:869–73. doi: 10.3109/02713689409015088. [DOI] [PubMed] [Google Scholar]
- Sierke SL, Cheng K, Kim HH, Koland JG. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J. 1997;322(Pt 3):757–63. doi: 10.1042/bj3220757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver DL, Hou L, Pavan WJ. The genetic regulation of pigment cell development. Adv Exp Med Biol. 2006;589:155–69. doi: 10.1007/978-0-387-46954-6_9. [DOI] [PubMed] [Google Scholar]
- Sithanandam G, Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008;15:413–48. doi: 10.1038/cgt.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwkowski MX, Schaefer G, Akita RW, Lofgren JA, Fitzpatrick VD, Nuijens A, Fendly BM, Cerione RA, Vandlen RL, Carraway KL., 3rd Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J Biol Chem. 1994;269:14661–5. [PubMed] [Google Scholar]
- Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004a;64:7002–10. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- Stahl S, Bar-Meir E, Friedman E, Regev E, Orenstein A, Winkler E. Genetics in melanoma. Isr Med Assoc J. 2004b;6:774–7. [PubMed] [Google Scholar]
- Sviderskaya EV, Wakeling WF, Bennett DC. A cloned, immortal line of murine melanoblasts inducible to differentiate to melanocytes. Development. 1995;121:1547–57. doi: 10.1242/dev.121.5.1547. [DOI] [PubMed] [Google Scholar]
- Ueno Y, Sakurai H, Tsunoda S, Choo MK, Matsuo M, Koizumi K, Saiki I. Heregulin-induced activation of ErbB3 by EGFR tyrosine kinase activity promotes tumor growth and metastasis in melanoma cells. Int J Cancer. 2008;123:340–7. doi: 10.1002/ijc.23465. [DOI] [PubMed] [Google Scholar]
- Vaisanen A, Kallioinen M, Taskinen PJ, Turpeenniemi-Hujanen T. Prognostic value of MMP-2 immunoreactive protein (72 kD type IV collagenase) in primary skin melanoma. J Pathol. 1998;186:51–8. doi: 10.1002/(SICI)1096-9896(199809)186:1<51::AID-PATH131>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Vaisanen AH, Kallioinen M, Turpeenniemi-Hujanen T. Comparison of the prognostic value of matrix metalloproteinases 2 and 9 in cutaneous melanoma. Hum Pathol. 2008;39:377–85. doi: 10.1016/j.humpath.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Vance KW, Goding CR. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res. 2004;17:318–25. doi: 10.1111/j.1600-0749.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- Weeraratna AT, Becker D, Carr KM, Duray PH, Rosenblatt KP, Yang S, Chen Y, Bittner M, Strausberg RL, Riggins GJ, et al. Generation and analysis of melanoma SAGE libraries: SAGE advice on the melanoma transcriptome. Oncogene. 2004;23:2264–74. doi: 10.1038/sj.onc.1207337. [DOI] [PubMed] [Google Scholar]
- Wells A, Marti U. Signalling shortcuts: cell-surface receptors in the nucleus? Nat Rev Mol Cell Biol. 2002;3:697–702. doi: 10.1038/nrm905. [DOI] [PubMed] [Google Scholar]
- Wen D, Suggs SV, Karunagaran D, Liu N, Cupples RL, Luo Y, Janssen AM, Ben-Baruch N, Trollinger DB, Jacobsen VL, et al. Structural and functional aspects of the multiplicity of Neu differentiation factors. Mol Cell Biol. 1994;14:1909–19. doi: 10.1128/mcb.14.3.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenandy L, Sorensen RB, Svane IM, Thor Straten P, Andersen MH. RhoC a new target for therapeutic vaccination against metastatic cancer. Cancer Immunol Immunother. 2008;57:1871–8. doi: 10.1007/s00262-008-0517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Luca M, Huang S, Gutman M, Reich R, Johnson JP, Bar-Eli M. Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res. 1997;57:2295–303. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Expression analysis of Mb-specific markers in control and Erbb3 mutant embryos at E10.5.Si expression analysis in control and Erbb3 mutant embryos at E10.5. Lateral view of Si whole-mount in situ hybridization in Erbb3+/+(A, C) and Erbb3msp1/msp1 (B) embryos. Si-expressing cells (black arrowheads) were detected in the RPE of the eye and the region below the otic vesicle (OV) in all four genotypes analyzed. Quantification of Si-expressing cells in Erbb3+/+, Erbb3msp1/msp1 (E) embryos at E10.5. Si-expressing cells were counted in whole-mount embryos using dissection microscope. The numbers of Si-expressing cells from three different embryos of each genotype were averaged and standard deviations calculated. There was no significant difference in the number of Si-expressing cells between Erbb3msp1/msp1 and Erbb3+/+ (unpaired t-test; p = 0.92)
Supplemental Figure 2. Expression analysis of Mb-specific markers in control and Erbb3 mutant embryos. At E11.5, Si-expressing cells are detected in the head region, the RPE of the eye, and along the rostral caudal axis in Erbb3+/+(A) and Erbb3msp1/msp1 (B) embryos. At E12.5, Dct expression is observed in the head (C, D) and caudal region (E, F) of Erbb3+/+(C, E) and Erbb3msp1/msp1 (D, F) embryos. No difference in the expression pattern of Si- and Dct-expressing cells was observed between Erbb3+/+ and Erbb3msp1/msp1 embryos at these two stages.
Dct and Si are normally expressed in Erbb3 null embryos. At E11.5 Dct-expressing cells (black arrowheads) were observed in the head and along the rostra-caudal axis in Erbb3+/+ (A), Erbb3KO1/KO1 (B), and Erbb3KO2/KO2 (C) embryos. Si-expressing cells (black arrowheads) showed similar expression pattern in Erbb3+/+ (D), Erbb3KO1/KO1 (E), and Erbb3KO2/KO2 (F) embryos at E11.5. No apparent difference in the expression pattern of these two markers was observed between control and Erbb3 null alleles.
Embryos generated from KitW-LacZ/+; Erbb3msp1/+ double heterozygote crosses were collected at E11.5 and stained for β-galactosidase activity. In KitW-LacZ/+; Erbb3msp1/+, β-gal-expressing cells (black arrowheads) were detected in the head (A), trunk (B), and caudal region (C) of the developing embryo. A similar β-gal expression pattern was noted in KitW-LacZ/+; Erbb3msp1/msp1 embryos (D, E, F). No major difference in the expression or distribution of β-gal+ cells was observed between KitW-LacZ/+; Erbb3msp1/+ and KitW-LacZ/+; Erbb3msp1/msp1 embryos.
(A) Derivation of primary MCs from neural tube explant cultures. Neural tubes generated from Erbb3msp1 heterozygous matings were isolated from E9.5 embryos and cultured in the presence of growth factors that promote Mb differentiation for 10 days (D). On day 10, individual cultures were trypsinized, and the same genotypes were pooled together and re-plated. Cells were grown under the same culture conditions as described above for an additional two days. After overnight starvation in 2% serum without growth factors, cells were stimulated with or without NRG1 for 15 minutes and lysed in lysis buffer. (B) Neural tube cell extracts (3 μg) were subjected to immunoblot analysis, and membranes were probed with anti-phospho-ERBB3 and total ERBB3 antibodies. NRG1-induced phosphorylation of ERBB3 was observed in Erbb3+/+ and Erbb3msp1/+ MC cultures, but not in Erbb3msp1/msp1 mutant cultures.
Immunostaining of MITF+ (red) and TYRP1+ (green) cells in Erbb3+/- (A and B) and Erbb3msp1/msp1 mutant (C and D) cultures grown in the absence of NRG1 (A and C) and in the presence of NRG1 (B and D). (E) Bar diagram depicting the percentage of TYRP1+/MITF+ cells in control and Erbb3 null NC cultures treated with or without NRG1. Error bars denote standard deviation. The proportion of TYRP1+/MITF+ cells was significantly reduced (p = 0.002; Student t-test) in NRG1-treated control cells compared to untreated cultures. The proportion of double positive cells in Erbb3 mutant cultures treated with NRG1 was at the comparable levels with untreated mutant cultures.
(A) Whole-cell lysates (30 μg) were subjected to immunoblotting using anti-ERBB2, anti-ERBB3, anti-ERBB4, anti-EGFR and anti-NRG1 antibodies. 293T lysates were used to confirm the specificity of the antibodies. Immunoblots were probed with antibody to α-Tubulin to monitor equal protein loading. Expression of ERBB2 and ERBB3 was observed in melan-Ink4a cell extracts. Protein lysates were devoid of EGFR, ERBB4 and NRG1 expression. (B) Serum-starved melan-Ink4a cells were stimulated with or without NRG1 for 15 minutes and cell lysates analyzed by immunoblotting using anti-phospho-ERBB3 (Tyr 1284), anti-phospho-AKT (Ser475), and anti-phospho-p44/42 MAPK antibody. After stripping, membranes were re-probed with antibodies against unphosphorylated forms to evaluate the amount of total proteins. Addition of NRG1 induced ERBB3 phosphorylation, which resulted in activation of the AKT and MAPK pathways as measured by AKT and MAPK phosphorylation.
Flow cytometric analysis of cells grown under three different conditions. −TPA/CT cells were arrested in G0-G1 stage. +NRG1-TPA/CT cells showed similar cell cycle progression as control cells (+TPA/CT).
Bright field images of melan-Ink4a cells grown in the presence of TPA and CT (A) and in the presence of NRG1, TPA and CT (B). Control cells contained large number of flat, pigment- containing MCs (dark spots), while NRG1-treated cells appeared less pigmented. Also, NRG1-treated cells were smaller in size and bipolar. Cell pellet was less pigmented in NRG1-treated cells. Equal number of cells was collected, spun down, and color of the cell pellet was evaluated between control and NRG1 treated cells.