Abstract
Copy-number variation in the human genome can be disease-causing or phenotypically neutral. This type of genetic rearrangement associated with human chromosome 21 (Hsa21) underlies partial Monosomy 21 and Trisomy 21. Mental retardation is a major clinical manifestation of partial Monosomy 21. To model this human chromosomal deletion disorder, we have generated novel mouse mutants carrying heterozygous deletions of the 2.3- and 1.1-Mb segments on mouse chromosome 10 (Mmu10) and Mmu17, respectively, which are orthologous to the regions on human 21q22.3, using Cre/loxP-mediated chromosome engineering. Alterations of the transcriptional levels of genes within the deleted intervals reflect gene-dosage effects in the mutant mice. The analysis of cognitive behaviors shows that the mutant mice carrying the deletion on either Mmu10 or Mmu17 are impaired in learning and memory. Therefore, these mutants represent mouse models for Monosomy 21-associated mental retardation, which can serve as a powerful tool to study the molecular mechanism underlying the clinical phenotype and should facilitate efforts to identify the haploinsufficient causative genes.
INTRODUCTION
Copy-number variation of chromosome regions exists in mammalian genomes (Adams et al. 2005; Freeman et al. 2006; Sebat et al. 2004; Zhang et al. 2009). Although most copy-number variations do not cause discernible phenotypes, a subset of these alterations are not neutral and are responsible for developmental and/or physiological abnormalities. The copy-number alterations associated with Hsa21 result in two human syndromes: Trisomy 21 (Down syndrome) and partial Monosomy 21. While Trisomy 21 is the most frequent live-born human aneuploidy, de novo deletions associated with partial Monosomy 21 occur at a relatively low rate in the human population. Partial Monosomy 21 causes various developmental anomalies, including mental retardation and cardiac defects in patients (Barnicoat et al. 1996; Bartsch et al. 1994; Chettouh et al. 1995; Ehling et al. 2004; Huret et al. 1995; Korenberg et al. 1991; Lyle et al. 2009; Nielsen and Tranebjaerg 1984; Theodoropoulos et al. 1995; Valero et al. 1999; Yamamoto et al. 1979). However, the underlying mechanism of partial Monosomy 21 is unknown.
In an attempt to map the critical genomic regions for the disease phenotypes of partial Monosomy 21, several groups examined relatively large numbers of human partial Monosomy 21 cases (Chettouh et al. 1995; Huret et al. 1995; Katzenstein et al. 2009; Lyle et al. 2009). In these studies, the data based on partial Monosomy 21 patients were used to map genomic regions associated with the disease phenotypes, including mental retardation. Partial Monosomy 21 cases due to interstitial deletions are most desirable for genetic mapping. Unfortunately, some patients with partial Monosomy 21 also carry anomalies involving other human chromosomes, such as segmental trisomies and deletions (Chettouh et al. 1995; Huret et al. 1995; Katzenstein et al. 2009; Lyle et al. 2009). The potential impact of other genomic alterations on phenotypes complicates the interpretation of the genotype-phenotype correlations inferred in some of these studies. While studies of human cases have been useful, causative genes have not been isolated. This is due principally to the small number of patients and to the resulting lack of a complete and informative set of partial Monosomy 21 cases (Lyle et al. 2009).
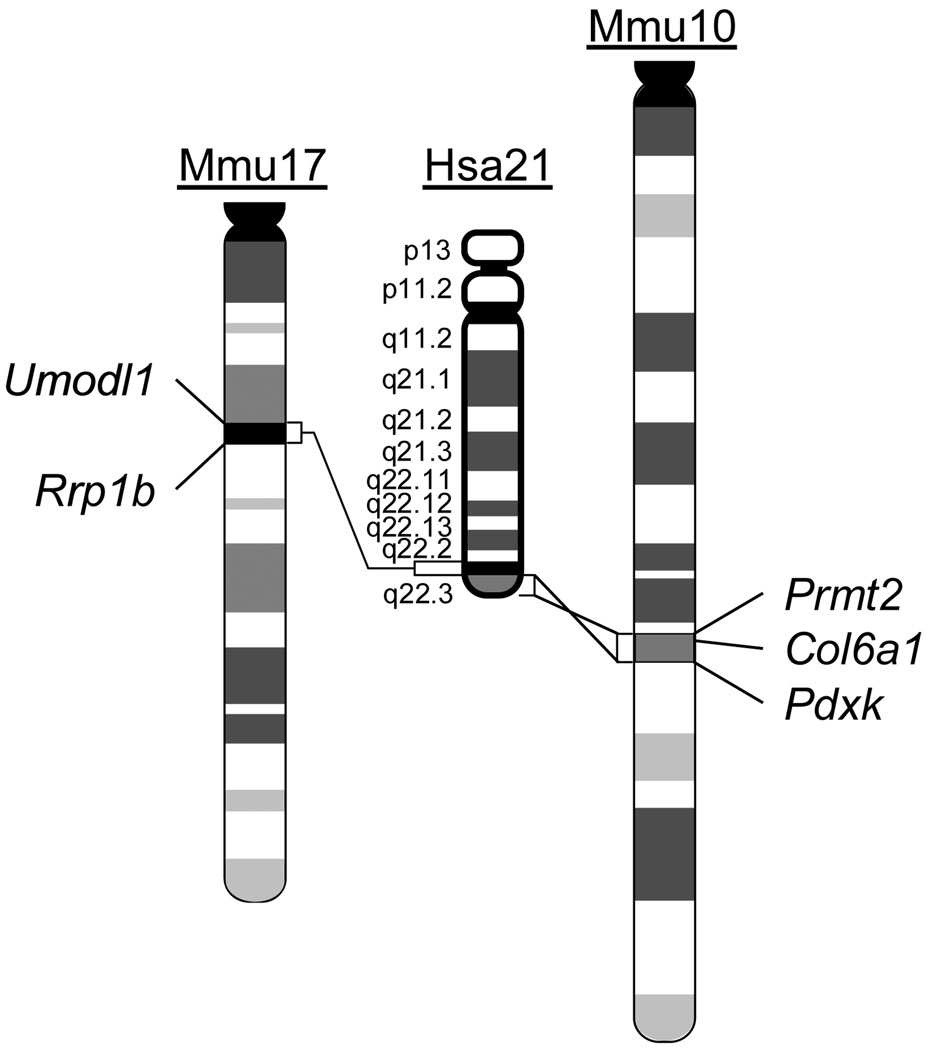
Given the difficulty in identifying causative genes by relying on patient studies, mouse-based approaches could be employed, which are based on syntenic conservation between the regions on Hsa21 and three regions located on Mmu10, Mmu16 and Mmu17. As gene-rich genomic segments, the syntenic regions for the proximal and distal parts of human 21q22.3 are located on Mmu17 and Mmu10, respectively (Fig. 1; Supplementary Tables 1, 2) (www.ensembl.org). Deletions of key genes in these regions may cause perturbations of the critical biological processes in various organs, which may in turn lead to abnormal phenotypes associated with partial Monosomy 21, such as cognitive deficits. To test these predictions and to establish a genotype-phenotype relationship, we generated and characterized two mutant mouse strains that carry deletions containing all the orthologs of the Hsa21 genes in the syntenic regions of Mmu10 and Mmu17.
Fig. 1.
Hsa21 and the syntenic regions on Mmu10 and Mmu17. The endpoints of engineered chromosomal deletions in the mouse mutants are indicated.
MATERIALS AND METHODS
Generation of Df(10)1Yey/+ and Df(17)1Yey/+ mice using Cre/loxP-mediated chromosome engineering
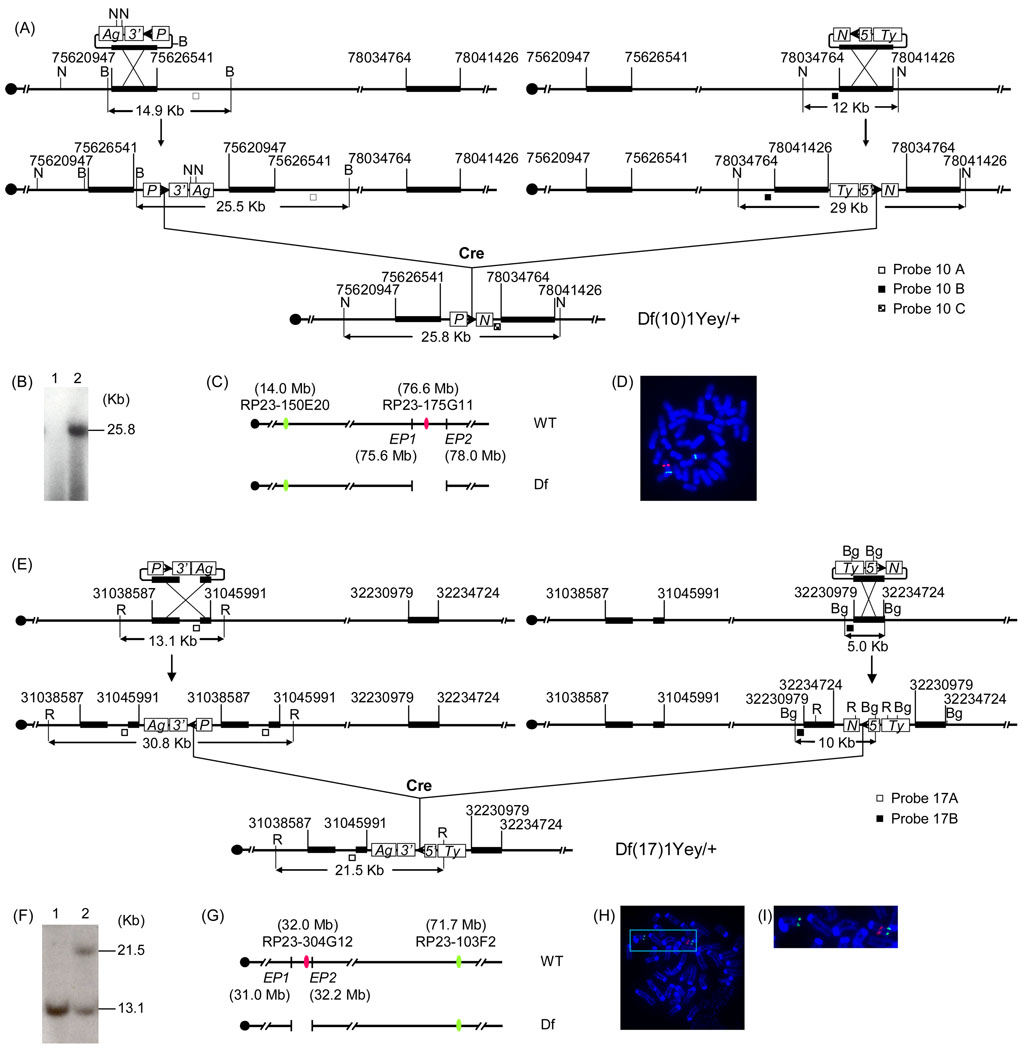
It is required that Hprt-deficient mouse embryonic stem (ES) cells be used for Cre/loxP-mediated chromosome engineering. Currently, only the ES cell lines isolated from 129 mice carry Hprt-null alleles, such as AB2.2 and HM1 (Bradley et al., 1999; Magin et al., 1992). In this study, we used AB2.2 cells isolated from 129SvEv mice. To generate Df(10)1Yey, the targeting vector pTV(10)1EP1 (Fig. 2a) was isolated from the 3’HPRT vector genomic library (Zheng et al. 1999) using PCR primers 5’-GCTGGCCTCTGA CTGTCTTGTTCC-3’ (forward) and 5’-GGGGCCATGTCACTGTCCTGCTA-3’ (reverse). The targeting vector pTV(10)1EP2 (Fig. 2a) was isolated from the 5 ’HPRT vector genomic library using PCR primers 5’-TCTGGAGCCATCTTCATAAAGCTG-3’ (forward) and 5’-AACTCACCGGTGATTGTGAAGA-3’ (reverse). LoxP was targeted by pTV(10)1EP1 and pTV(10)1EP2 to the regions proximal to Prmt2 and distal to Pdxk, respectively (Figs. 1 and 2a). To generate Df(17)1Yey in mouse ES cells, the targeting vector pTV(17)1EP1 was generated by inverting the orientation of the mouse genomic insert in MICER clone MHPP309a23 using AscI that flanks the insert (Fig. 2e) (Adams et al. 2004). To generate a gap probe (Probe 17B) for Southern blot analysis, a 2.7-kb AflII-AflII fragment was also removed from the mouse genomic insert in pTV(17)1EP1. We used MICER clone MHPN353c19 as the targeting vector pTV(17)1EP2 (Adams et al. 2004). pTV(17)1EP1 and pTV(17)1EP2 were used for inserting loxP into the regions proximal and distal to Abcg1 and Rrp1b, respectively (Figs. 1 and 2e). ES cell culture, gene-targeting and induction of Cre/loxP-mediated recombination, Southern blot analysis and injection of ES cells into blastocysts were performed as described previously (Yu et al. 2006).
Fig. 2.
Development of Df(10)1Yey/+ and Df(17)1Yey/+ mice using Cre/loxP-mediated chromosome engineering. (a) Strategy to generate Df(10)1Yey. B, BamHI; N, NheI; 5’, 5’HPRT fragment; 3’, 3’HPRT fragment; N, neomycin-resistance gene; P, puromycin-resistance gene; Ty, Tyrosinase transgene; Ag, Agouti transgene; arrowhead, loxP site. (b) Southern blot analysis of NheI-digested mouse tail DNA using Probe 10C. Lane 1, the wild-type mouse; lane 2, Df(10)1Yey/+ mouse. (c) Schematic representation of the genomic locations of BAC probes for FISH analysis; EP1 and EP2, endpoint 1 and endpoint 2, respectively. (d) FISH analysis of metaphase chromosomes prepared from embryonic fibroblasts carrying Df(10)1Yey/+. (e) Strategy to generate Df(17)1Yey. R, EcoRI; Bg, BglII. (f) Southern blot analysis of EcoRI-digested mouse tail DNA using Probe 17A. Lane 1, the wild-type mouse; lane 2, Df(17)1Yey/+ mouse. (g) Schematic representation of the genomic locations of BAC probes for FISH analysis; EP1 and EP2, endpoint 1 and endpoint 2, respectively. (h) FISH analysis of metaphase chromosomes prepared from embryonic fibroblasts carrying Df(17)1Yey/+. (i) Higher magnification of the area boxed in (h).
Fluorescent in situ hybridization (FISH) and real-time quantitative PCR
FISH analysis was performed as described previously (Yu et al. 2006). To detect the chromosomal deletion between Prmt2 and Pdxk, BAC clone RP23-175G11 was labeled with digoxigenin and detected with anti-digoxigenin–rhodamine antibody. BAC clone RP23-150E20 was labeled with biotin and detected with fluorescin isothiocyanate-avidin (Fig. 2c, d). To detect the chromosomal deletion between Abcg1 and Rrp1b, BAC clone RP23-304G12 was labeled with digoxigenin and detected with anti-digoxigenin–rhodamine antibody. BAC clone RP23-103F2 was labeled with biotin and detected with fluorescin isothiocyanate-avidin (Fig. 2g–i). Real-time quantitative PCR was carried out as previously reported (Li et al. 2007).
Behavioral tests
The mutant mice and their littermates were maintained at a temperature- and humidity-controlled animal facility. All mice used in the experiments were 2–4 months old. Before behavioral experiments, each mouse was pre-handled for 2 minutes every day for a week. The experimental procedures were approved by the Institutional Animal Care and Use Committee.
Open field observations were carried out in a 40 × 40 × 30 cm Plexiglas box. Each mouse was individually placed in the center of an open field and the following behavioral parameters were recorded during a 5-min test interval: 1) total distance traveled; 2) time spent moving; 3) number of rearing events (vertical activity). Average speed was calculated by dividing the total distance by the time spent moving. Behavioral parameters in the open field were analyzed using HVS Field 2020, an imaging-tracking and analysis system (HVS Image Ltd., Twickenham, Middlesex, UK). The number of rearings was counted manually.
A standard Morris water maze test was carried out in a circular pool (152 cm in diameter) of water at 25 ± 1° C (Clapcote et al. 2005; Clapcote and Roder 2004; D'Hooge et al. 1997; McIlwain et al. 2001). The experimental data were collected and analyzed using HVS Water 2020, an imaging-tracking and analysis system (HVS Image Ltd.). Each mouse had four trials each day. Visible-platform and hidden-platform training trials were carried out on day 1 and days 2–8, respectively. The amount of time spent finding the platform (latency), the distance traveled (path-length) and swimming speed were recorded. On day 9, a probe test was performed in which the platform was removed from the water and each mouse was allowed 60 sec to search the pool. The time spent in each quadrant was measured.
The contextual and cued fear conditioning test was performed as described previously (Clapcote et al. 2005; Lu et al. 1997) using the Fear Conditioning Video Tracking System (Med Associates Inc., St. Albans, VT). The contextual tests were performed 24 hrs as well as 72 hrs after the mice were first exposed to the fear conditioning test chamber.
The foot-shock sensitivity test was performed using the fear conditioning test chamber. A foot-shock was delivered every 10 sec starting at 0.05 mA with a 0.05 mA increment between each shock (Rosa et al. 2007). The minimal level of current needed to elicit flinching or vocalizing was recorded.
Statistical methods
The data from the open field, Morris water maze probe test, contextual and cued fear conditioning tests and foot-shock sensitivity test were subjected to a one-way ANOVA between genotypes. ANOVA did not detect any effects from gender in all the behavioral tests, so the data from males and females were pooled and analyzed together for these experiments. Data from the seven-day training trials of the Morris water maze hidden-platform version were analyzed using a two-way (genotype×day) ANOVA with the genotype as a between-subjects factor and the day as a repeated-measures factor. All values reported in the text and figures were expressed as means ± S.E.M.
RESULTS AND DISCUSSION
The entire Hsa21 syntenic region on Mmu10 spans approximately 2.3 Mb and contains approximately 41 orthologs, with the Prmt2 and Pdxk genes located at the proximal and distal ends, respectively. The entire Hsa21 syntenic region on Mmu17 spans approximately 1.1 Mb and contains approximately 19 orthologs, with the Abcg1 and Rrp1b genes located at the proximal and distal ends, respectively. (Fig. 1; Supplementary Tables 1, 2) (http://www.ensembl.org). To generate a deletion of the syntenic region on Mmu10, loxP was targeted to regions proximal to Prmt2 and distal to Pdxk in the genome of AB2.2 mouse ES cells (Fig. 2a). Df(Prmt2-Pdxk)1Yey, abbreviated as Df(10)1Yey, was generated by transfecting pOG231, a Cre-expression vector, into double-targeted ES cells (Fig. 2a; see Materials and Methods). A similar strategy was used to generate Df(Abcg1-Rrp1b)1Yey, abbreviated as Df(17)1Yey, by targeting loxP to regions proximal to Abcg1 and distal to Rrp1b, in AB2.2 ES cells (Fig. 2e; see Materials and Methods). Chimeras were generated by injecting the mutant ES cells into blastocysts isolated from C57BL/6J females, as described previously (Bradley 1987). The chimeric males were first mated with C57BL/6J females to assess germline transmission efficiency of the engineered deletions based on coat-color chimerism and Southern blot-based genotyping of the agouti progeny (Fig. 2b, f). The genotypes of the desired mutant mice were also confirmed by FISH analysis (Fig. 2c, d, g–i). The chimeric males with a high efficiency of germline transmission of Df(10)1Yey were identified, and they were mated with 129SvEv females to generate Df(10)1Yey/+ mice in the 129SvEv strain background. However, the chimeric males generated using the ES cells carrying Df(17)1Yey have a very low efficiency of germline transmission, so Df(17)1Yey/+ mice from the chimeras were established only in the 129SvEvxC57BL/6JF1 strain background. These Df(17)1Yey/+ mice were backcrossed to wild-type 129SvEv mice for seven generations, and the Df(17)1Yey/+ mice were then maintained by sibling mating. Both Df(10)1Yey/+ mice and Df(17)1Yey/+ mice appeared normal, with body weights similar to their wild-type littermates. Heterozygous mutant fetuses at E18.5 and postnatal mice from mating Df(10)1Yey/+ males or Df(17)1Yey/+ males to wild-type females were present at normal Mendelian ratios, suggesting that no haploinsufficient lethal gene is located in the regions.
We used real-time quantitative PCR to analyze the mRNA levels for five and two genes located within the deleted intervals in Df(10)1Yey/+ mice and Df(17)1Yey/+ mice, respectively. Our results showed that the deletions led to reduced mRNA levels in the brain for these genes (Table 1), indicating gene-dosage decreases associated with the deletions.
Table 1.
Normalized relative values (RQ) of expression in the brains*
| Gene name | RQ ± S.E.M. | |
|---|---|---|
| Df(10)1Yey/+ over +/+ | Adar2 | 0.48±0.02 |
| Dnmt3l | 0.52±0.06 | |
| Itgb2 | 0.52±0.01 | |
| S100b | 0.48±0.01 | |
| Trpc7 | 0.53±0.02 | |
| Df(17)1Yey/+ over +/+ | Cbs | 0.44 ± 0.01 |
| Pknox1 | 0.62 ± 0.03 | |
The values represent the means of triplicated samples. Gapdh was used as an internal control and is disomic in all strains.
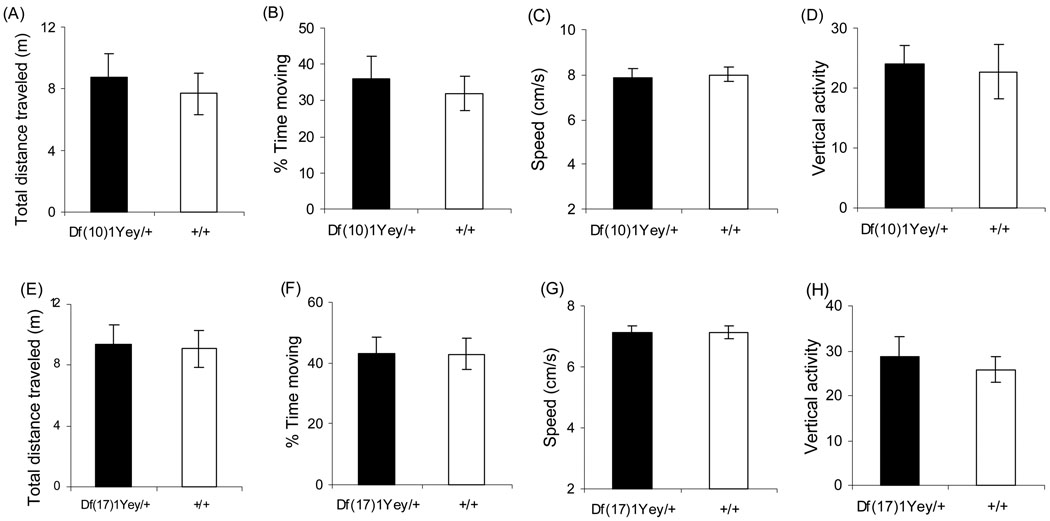
We used Df(10)1Yey/+ mice (n=16; 7 males and 9 females), the wild-type littermates of Df(10)1Ye/+ mice (n=16; 5 males and 11 females), Df(17)1Yey/+ mice (n=15; 10 males and 5 females) and the wild-type littermates of Df(17)1Yey/+ mice (n=15; 10 males and 5 females) for behavioral analysis. Although 129 mice have a reputation as inferior performers in the behavioral test, the 129SvEv substrain has been shown to be an excellent strain background for the Morris water maze test as well as the contextual and cued fear conditioning tests based on the data from Jacqueline Crawley’s laboratory (Crawley 2000; Holmes et al. 2002) and one of our own laboratories (Clapcote and Roder, 2004). We first performed a simple assessment of the mutant mice and their wild-type littermates in the open field and found they exhibited normal general activity level, gross locomotor activity and exploration habits (p>0.05) (Fig. 3). To examine the effect of Df(10)1Yey/+ and Dp(17)1Yey/+ on learning and memory, we compared the mutant mice to their wild-type littermates in the Morris water maze tasks. We first compared the mice using the visible platform version of the Morris water maze, which does not test hippocampal function. There were no differences between the mutant mice and their wild-type littermates in latency and path-length in finding the visible-cued platform (Fig. 4). This result indicates that the mutant mice appeared to be normal with respect to vision and motivation in finding the platform. We next compared the mice in the hidden platform version, which is a spatial navigation task that requires the mice to swim in a pool of opaque water until they locate a submerged platform. This type of learning has been considered hippocampus-dependent because of its sensitivity to hippocampal lesions (Sarnyai et al. 2000). Mean values for the mutant mice and their wild-type littermates in the acquisition phase of Morris water maze training in the hidden platform version are shown in Fig. 4. The mean latency for each genotype significantly decreased with training, but both Df(10)1Yey/+ and Df(17)1Yey/+ mice had a longer average latency than their wild-type littermates. The increased latency by Df(17)1Yey/+ mice could be explained by slower swimming speed (Fig. 4g). However, the abnormal latency of the Df(10)1Yey/+ mice could be caused by impaired spatial learning since Df(10)1Yey/+ mice showed increased swimming speed (Fig. 4a, c). Such possibilities are supported by the observations that Df(10)1Yey/+ mice, but not Df(17)1Yey/+ mice, took a longer path-length in locating the platform (p<0.001). In the probe test conducted on the day after the training period, both Df(10)1Yey/+ and Df(17)1Yey/+ mice spent a shorter time in the target quadrant (Northeast, NE) than their wild-type littermates (p=0.0006 and p=0.086, respectively). These results provide conclusive evidence that Df(10)1Yey/+ mice are impaired in spatial learning and memory.
Fig. 3.
Open field observation. Df(10)1Yey/+ mice (n=16) and Df(17)1Yey/+ mice (n=15) as well as their wild-type littermates with equal sample sizes were assessed by (a, e) total distance traveled (m, meter), (b, f) percentage of time spent moving, (c, g) average moving speed (cm/s, centimeter/second) and (d, h) vertical activity (number of rearings).
Fig. 4.
Behavioral analysis using Morris water maze. Df(10)1Yey/+ mice (n=16) (a–d) and Df(17)1Yey/+ mice (n=15) (e–h) as well as their wild-type littermates with equal sample sizes were examined, as described in Materials and Methods. (a, e) Latency to locate the platform (s, second), (b, f) path-length to locate the platform (m, meter) and (c, g) swimming speed (m/s, meter/second) during the learning trials of Morris water maze test. (d, h) The relative amount of time spent in different quadrants in the probe test on the second day after the end of the learning trials.
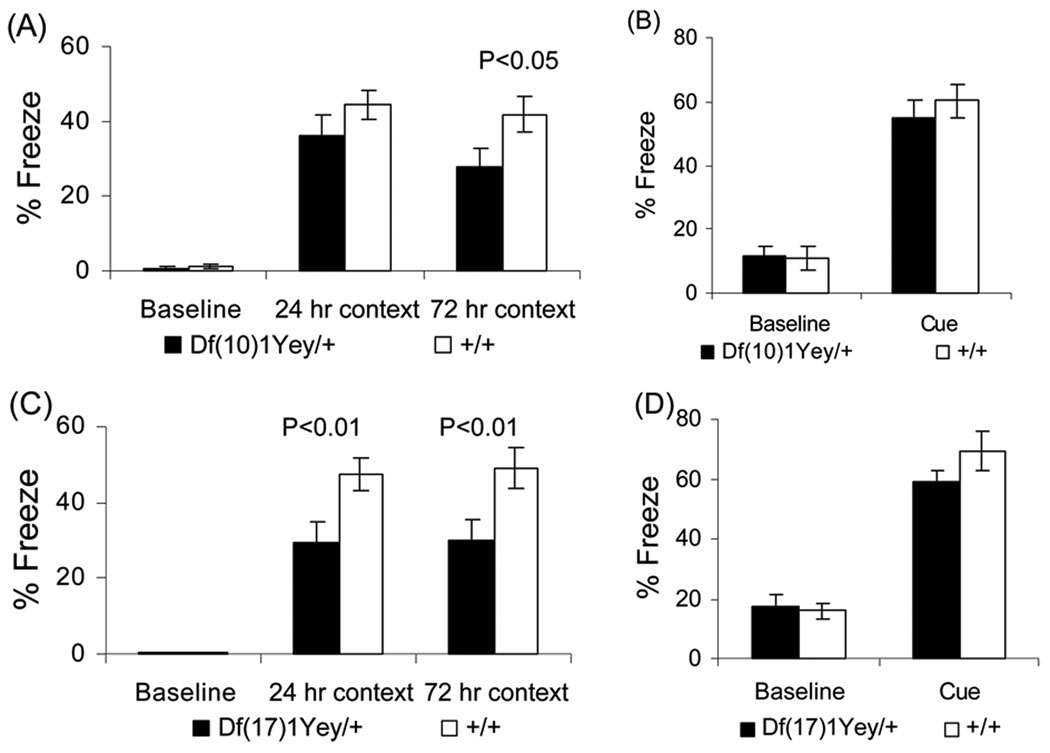
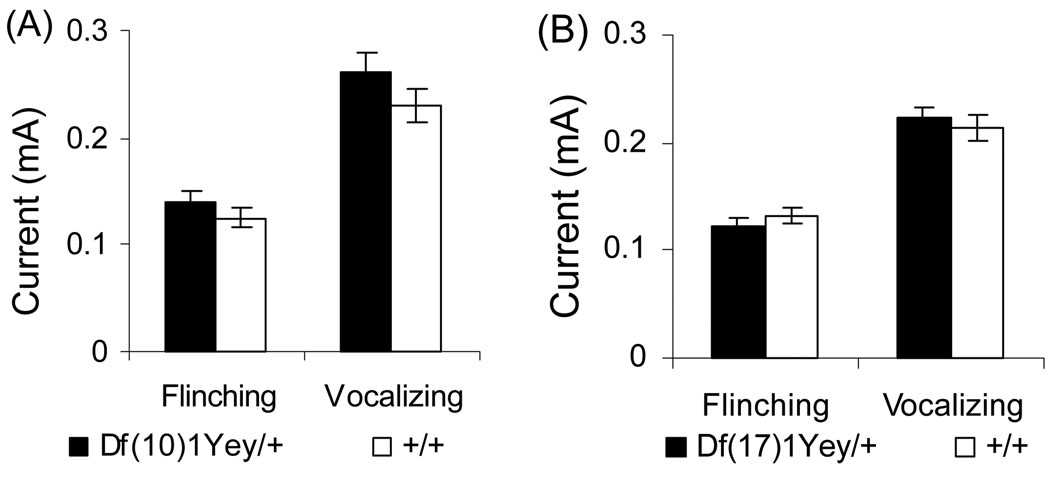
We performed additional assessments of the mutant mice’s learning and memory based on contextual and cued fear conditioning. Cued fear conditioning tests the amygdalar function, whereas contextual fear conditioning tests both the amygdalar and hippocampal functions (Holland and Bouton 1999; Logue et al. 1997; Phillips and LeDoux 1992). Mice of each genotype had a baseline (test-naïve) freezing level of under 2% prior to the presentation of the audible tone and foot-shock (Fig. 5a, c). There was no difference between the mutant mice and their wild-type littermates in baseline freezing. Both the mutant mice and the wild-type littermates increased their freezing in the 24-hr context tests relative to baseline freezing. However, Df(17)1Yey/+ mice froze less than their wild-type littermates (p<0.01) (Fig. 5c). In the 72-hr context tests, both Df(10)1Yey/+ mice and Df(17)1Yey/+ mice exhibited less freezing when compared to their wild-type littermates (p<0.05 and p<0.01, respectively) (Fig. 5a, c). These results suggest that the mutant mice are impaired in context-associated learning. After altering the context, the freezing levels of all strains prior to the presentation of the audible tone were similar (p>0.05) and were significantly lower than their freezing levels earlier when they were exposed to the original context (Fig. 5b, d), suggesting that none of the mouse strains exhibited generalized freezing in all conditions (Balogh et al. 2002). During presentation of the audible tone, all strains increased their freezing relative to the initial freezing in the altered context, and there was no significant difference between the mutant mice and the wild-type littermates in this measure of auditory cue-associated learning (Fig. 5b, d). To facilitate accurate interpretation of the fear conditioning test outcomes, we performed a foot-shock sensitivity test. Our results show that there was no difference in the mean threshold of the current to elicit flinching or vocalizing between the mutant mice and their wild-type littermates (Fig. 6).
Fig. 5.
Behavioral analysis using contextual and cued fear conditioning. Df(10)1Yey/+ mice (n=16) (a, b) and Df(17)1Yey/+ mice (n=15) (c, d) as well as their wild-type littermates with equal sample sizes were examined as described in Materials and Methods. (a, c) The percentage of time spent freezing before the foot-shock (baseline) as well as during the 24- and 72-hr contextual tests is shown. (b, d) The percentage of time spent freezing in the altered chamber before the auditory tone cue was delivered (baseline) as well as during the cue delivery is shown.
Fig. 6.
Analysis of sensitivity to electric foot-shock. The minimal levels of currents (mA) needed to elicit a response, either flinching or vocalizing, from Df(10)1Yey/+ mice (n=16) and Df(17)1Yey/+ mice (n=15) as well as their wild-type littermates with equal sample sizes are shown.
The aforementioned phenotypic studies provide conclusive evidence that the heterozygous deletion of the Hsa21 syntenic region on Mmu10 or Mmu17 results in impairment in learning and memory in mice. These results are consistent with data generated from human genetic studies in which patients carrying heterozygous deletions of the human syntenic regions 21q22.3 exhibit the mental retardation phenotype (Ehling et al. 2004; Estabrooks et al. 1990; Katzenstein et al. 2009; Lyle et al. 2009). The smallest deletions in these patients span approximately 7.9 Mb on 21q22.2-q22.3 (Ehling et al., 2004). Therefore, Df(10)1Yey/+ mice and Df(17)1Yey/+ mice can serve as genetic models for partial Monosomy 21-associated mental retardation.
The prevailing hypothesis on the phenotypic consequences of chromosomal abnormalities is that a specific phenotype is caused by dosage alteration of a specific gene or genes located within the rearranged interval. Many chromosomal abnormalities cause mental retardation-related phenotypes in humans and mice, suggesting the central nervous system is sensitive to dosage alterations of many genes. On the other hand, if the rearranged interval does not contain dosage sensitive genes, deletion or duplication should not lead to mental retardation-related phenotypes. Such cases have been observed in humans and mice (Besson et al. 2007; Gilmore et al. 2001; Zarate et al. 2007). Also, in our preliminary study, the duplication of human chromosome 21 syntenic region on Mmu10 did not cause impairment in the Morris water maze test or the contextual fear conditioning test (Yu et al., unpublished data). Therefore, the mental retardation-related phenotypes observed in Df(10)1Yey/+ and Df(17)1Yey/+ mice are caused by haploinsufficiencies of specific genes in the regions.
In contrast to patient-based studies, which are limited by the availability of partial Monosomy 21 cases carrying informative chromosomal deletions, the establishment of desired mouse models will make it feasible to perform in-depth genetic analysis of the syntenic genomic regions. Recently, Besson et al. reported that a 0.5-Mb heterozygous deletion between Prmt2 and Col6a1 in the Hsa21 syntenic region on Mmu10 did not cause impairment in learning and memory in Ms1Yah mice (Fig. 1; Supplementary Table 1) (Besson et al. 2007). Since the Prmt2-Col6a1 region is located within the deleted interval of Df(10)1Yey, 13 Hsa21 gene orthologs deleted in Ms1Yah mice can be excluded as the causative genes for the mutant phenotype observed in Df(10)1Yey/+ mice. Therefore, the causative genes for partial Monosomy 21-associated mental retardation are among 28 and 19 Hsa21 gene orthologs located on Mmu10 and Mmu17, respectively. Eighteen of these candidate genes have been mutagenized by using a gene-targeting approach (http://www.informatics.jax.org) but none of these targeted mutations has been reported to give a heterozygous cognitive phenotype related to mental retardation. To search for the causative genes, we could first generate and analyze subdeletions of the associated genomic regions using Cre/loxP-mediated chromosome engineering, which would be facilitated by ready-made targeting vectors from the Mouse Insertional and Chromosome Engineering Resource (MICER) (Adams et al. 2004). This effort should significantly further narrow down the critical genomic regions and reduce the number of candidate genes. The identities of the causative genes for Monosomy 21-associated mental retardation could be established by analyzing knockout mice carrying null alleles of these candidate genes. The null alleles generated from ongoing public knockout mouse projects, including those from the International Gene Trapping Consortium and from BAC-recombineering-based gene targeting pipeline, will provide timely-reagents for such a genetic analysis effect (Austin et al. 2004; Chan et al. 2007; Testa et al. 2004; Wurst 2005).
For mutational analysis of the mouse genome, heterozygous deletions with defined endpoints are important reagents because they facilitate rapid genetic mapping and maintenance of randomly generated mutations, such as those generated during ethylnitrosourea mutagenesis screens (Rinchik 2000; Rinchik and Russell 1990). Therefore, as viable heterozygous deletion mutants, Df(10)1Yey/+ and Df(17)1Yey/+ mice can also be used for functional analysis of the deleted genomic intervals.
Engineered mouse mutants have been instrumental in fruitful molecular genetic studies of several major human chromosomal deletion disorders, including DiGeorge syndrome (Lindsay et al. 1999; Lindsay et al. 2001; Merscher et al. 2001), Prader-Willi syndrome (Tsai et al. 1999) and Smith-Magenis syndrome (Walz et al. 2003). Similarly, Df(10)1Yey/+ and Df(17)1Yey/+ mutants developed from this study will play a critical role in genetic dissection of Monosomy 21-associated mental retardation, which in turn may lead to unraveling of the molecular mechanism underlying the clinical phenotype of this disorder.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Paul Szurek and Jeffrey LaDuca for their assistance. This project was supported in part by grants to Y.E. Yu from the Louis Sklarow Memorial Fund, the Jerome Lejeune Foundation and the National Heart, Lung and Blood Institute (R01HL091519).
Contributor Information
Tao Yu, Genetics Program and Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, NY 14263, USA.
Steven J. Clapcote, Institute of Membrane and Systems Biology, University of Leeds, Leeds LS2 8JT, UK
Zhongyou Li, Genetics Program and Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, NY 14263, USA.
Chunhong Liu, Genetics Program and Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, NY 14263, USA.
Annie Pao, Genetics Program and Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, NY 14263, USA.
Allison R. Bechard, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, University of Toronto, Toronto, ON M5G 1X5, Canada
Sandra Carattini-Rivera, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Sei-Ichi Matsui, Genetics Program and Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, NY 14263, USA.
John C. Roder, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, University of Toronto, Toronto, ON M5G 1X5, Canada
Antonio Baldini, Institute of Biosciences and Technology, Texas A&M University Health Science Center, Houston, TX 77030, USA.
William C. Mobley, Department of Neurosciences, University of California at San Diego, School of Medicine, La Jolla, CA 92093, USA
Allan Bradley, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, UK..
Y. Eugene Yu, Genetics Program and Department of Cancer Genetics, Roswell Park Cancer Institute, Buffalo, NY 14263, USA.
REFERENCES
- 1.Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, Jonkers J, Smith J, Plumb B, Taylor R, Nishijima I, Yu Y, Rogers J, Bradley A. Mutagenic insertion and chromosome engineering resource (MICER) Nat Genet. 2004;36:867–871. doi: 10.1038/ng1388. [DOI] [PubMed] [Google Scholar]
- 2.Adams DJ, Dermitzakis ET, Cox T, Smith J, Davies R, Banerjee R, Bonfield J, Mullikin JC, Chung YJ, Rogers J, Bradley A. Complex haplotypes, copy number polymorphisms and coding variation in two recently divergent mouse strains. Nat Genet. 2005;37:532–536. doi: 10.1038/ng1551. [DOI] [PubMed] [Google Scholar]
- 3.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. The knockout mouse project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balogh SA, Radcliffe RA, Logue SF, Wehner JM. Contextual and cued fear conditioning in C57BL/6J and DBA/2J mice: context discrimination and the effects of retention interval. Behav Neurosci. 2002;116:947–957. doi: 10.1037//0735-7044.116.6.947. [DOI] [PubMed] [Google Scholar]
- 5.Barnicoat AJ, Bonneau JL, Boyd E, Docherty Z, Fennell SJ, Huret JL, King M, Maltby EL, McManus S, Pilz DT, Shafei-Benaissa E, Super M, Tolmie J. Down syndrome with partial duplication and del (21) syndrome: study protocol and call for collaboration. Study I: Clinical assessment. Clin Genet. 1996;49:20–27. doi: 10.1111/j.1399-0004.1996.tb04319.x. [DOI] [PubMed] [Google Scholar]
- 6.Bartsch O, Petersen MB, Stuhlmann I, Mau G, Frantzen M, Schwinger E, Antonarakis SE, Mikkelsen M. "Compensatory" uniparental disomy of chromosome 21 in two cases. J Med Genet. 1994;31:534–540. doi: 10.1136/jmg.31.7.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besson V, Brault V, Duchon A, Togbe D, Bizot JC, Quesniaux VF, Ryffel B, Herault Y. Modeling the monosomy for the telomeric part of human chromosome 21 reveals haploinsufficient genes modulating the inflammatory and airway responses. Hum Mol Genet. 2007;16:2040–2052. doi: 10.1093/hmg/ddm152. [DOI] [PubMed] [Google Scholar]
- 8.Bradley A. Production and analysis of chimaeric mice. In: Robertson E, editor. Teratocarcinomas and Embryonic Stem Cells - A Practical Approach. IRL Press; 1987. pp. 113–151. [Google Scholar]
- 9.Chan W, Costantino N, Li R, Lee SC, Su Q, Melvin D, Court DL, Liu P. A recombineering based approach for high-throughput conditional knockout targeting vector construction. Nucleic Acids Res. 2007;35:e64. doi: 10.1093/nar/gkm163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chettouh Z, Croquette MF, Delobel B, Gilgenkrants S, Leonard C, Maunoury C, Prieur M, Rethore MO, Sinet PM, Chery M, et al. Molecular mapping of 21 features associated with partial monosomy 21: involvement of the APP-SOD1 region. Am J Hum Genet. 1995;57:62–71. [PMC free article] [PubMed] [Google Scholar]
- 11.Clapcote SJ, Lazar NL, Bechard AR, Roder JC. Effects of the rd1 mutation and host strain on hippocampal learning in mice. Behav Genet. 2005;35:591–601. doi: 10.1007/s10519-005-5634-5. [DOI] [PubMed] [Google Scholar]
- 12.Clapcote SJ, Roder JC. Survey of embryonic stem cell line source strains in the water maze reveals superior reversal learning of 129S6/SvEvTac mice. Behav Brain Res. 2004;152:35–48. doi: 10.1016/j.bbr.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Crawley JN. What's wrong with my mouse? New York: Wiley-liss; 2000. Of unicorns and chimeras; p. 18. Table 2.1. [Google Scholar]
- 14.D'Hooge R, Nagels G, Franck F, Bakker CE, Reyniers E, Storm K, Kooy RF, Oostra BA, Willems PJ, De Deyn PP. Mildly impaired water maze performance in male Fmr1 knockout mice. Neuroscience. 1997;76:367–376. doi: 10.1016/s0306-4522(96)00224-2. [DOI] [PubMed] [Google Scholar]
- 15.Ehling D, Kennerknecht I, Junge A, Prager B, Exeler R, Behre B, Horst J, Schmitt-John T, Bartsch O, Wirth J. Mild phenotype in two unrelated patients with a partial deletion of 21q22.2-q22.3 defined by FISH and molecular studies. Am J Med Genet A. 2004;131:265–272. doi: 10.1002/ajmg.a.30361. [DOI] [PubMed] [Google Scholar]
- 16.Estabrooks LL, Rao KW, Donahue RP, Aylsworth AS. Holoprosencephaly in an infant with a minute deletion of chromosome 21(q22.3) Am J Med Genet. 1990;36:306–309. doi: 10.1002/ajmg.1320360312. [DOI] [PubMed] [Google Scholar]
- 17.Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 18.Gilmore L, Cuskelly M, Jobling A, Smith S. Deletion of 8p: a report of a child with normal intelligence. Dev Med Child Neurol. 2001;43:843–846. doi: 10.1017/s0012162201001530. [DOI] [PubMed] [Google Scholar]
- 19.Holland PC, Bouton ME. Hippocampus and context in classical conditioning. Curr Opin Neurobiol. 1999;9:195–202. doi: 10.1016/s0959-4388(99)80027-0. [DOI] [PubMed] [Google Scholar]
- 20.Holmes A, Wrenn CC, Harris AP, Thayer KE, Crawley JN. Behavioral profiles of inbred strains on novel olfactory, spatial and emotional tests for reference memory in mice. Genes, Brain, & Behavior. 2002;1:55–69. doi: 10.1046/j.1601-1848.2001.00005.x. [DOI] [PubMed] [Google Scholar]
- 21.Huret JL, Leonard C, Chery M, Philippe C, Schafei-Benaissa E, Lefaure G, Labrune B, Gilgenkrantz S. Monosomy 21q: two cases of del(21q) and review of the literature. Clin Genet. 1995;48:140–147. doi: 10.1111/j.1399-0004.1995.tb04074.x. [DOI] [PubMed] [Google Scholar]
- 22.Katzenstein JM, Oghalai JS, Tonini R, Baker D, Haymond J, Caudle SE. Neurocognitive functioning of a child with partial trisomy 6 and monosomy 21. Neurocase. 2009;15:97–100. doi: 10.1080/13554790802631910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korenberg JR, Kalousek DK, Anneren G, Pulst SM, Hall JG, Epstein CJ, Cox DR. Deletion of chromosome 21 and normal intelligence: molecular definition of the lesion. Hum Genet. 1991;87:112–118. doi: 10.1007/BF00204163. [DOI] [PubMed] [Google Scholar]
- 24.Li Z, Yu T, Morishima M, Pao A, LaDuca J, Conroy J, Nowak N, Matsui S, Shiraishi I, Yu Y. Duplication of the entire 22.9-Mb human chromosome 21 syntenic region on mouse chromosome 16 causes cardiovascular and gastrointestinal abnormalities. Human Molecular Genetics. 2007;16:1359–1366. doi: 10.1093/hmg/ddm086. [DOI] [PubMed] [Google Scholar]
- 25.Lindsay EA, Botta A, Jurecic V, Carattini-Rivera S, Cheah YC, Rosenblatt HM, Bradley A, Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 26.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 27.Logue SF, Paylor R, Wehner JM. Hippocampal lesions cause learning deficits in inbred mice in the Morris water maze and conditioned-fear task. Behavior Neurosci. 1997;111:104–113. doi: 10.1037//0735-7044.111.1.104. [DOI] [PubMed] [Google Scholar]
- 28.Lu YM, Jia Z, Janus C, Henderson JT, Gerlai R, Wojtowicz JM, Roder JC. Mice lacking metabotropic glutamate receptor 5 show impaired learning and reduced CA1 long-term potentiation (LTP) but normal CA3 LTP. J Neurosci. 1997;17:5196–5205. doi: 10.1523/JNEUROSCI.17-13-05196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyle R, Bena F, Gagos S, Gehrig C, Lopez G, Schinzel A, Lespinasse J, Bottani A, Dahoun S, Taine L, Doco-Fenzy M, Cornillet-Lefebvre P, Pelet A, Lyonnet S, Toutain A, Colleaux L, Horst J, Kennerknecht I, Wakamatsu N, Descartes M, Franklin JC, Florentin-Arar L, Kitsiou S, Ait Yahya-Graison E, Costantine M, Sinet PM, Delabar JM, Antonarakis SE. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur J Hum Genet. 2009;17:454–466. doi: 10.1038/ejhg.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McIlwain KL, Merriweather MY, Yuva-Paylor LA, Paylor R. The use of behavioral test batteries: effects of training history. Physiol Behav. 2001;73:705–717. doi: 10.1016/s0031-9384(01)00528-5. [DOI] [PubMed] [Google Scholar]
- 31.Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen F, Tranebjaerg L. A case of partial monosomy 21q22.2 associated with Rieger's syndrome. J Med Genet. 1984;21:218–221. doi: 10.1136/jmg.21.3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips RG, LeDoux JE. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behavior Neurosci. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- 34.Rinchik EM. Developing genetic reagents to facilitate recovery, analysis, and maintenance of mouse mutations. Mamm Genome. 2000;11:489–499. doi: 10.1007/s003350010095. [DOI] [PubMed] [Google Scholar]
- 35.Rinchik EM, Russell LB. In: Genome Analysis. Davies K, Tilghman S, editors. Cold Spring Harbor: CSH Laboratory Press; 1990. pp. 121–158. [Google Scholar]
- 36.Rosa EF, Takahashi S, Aboulafia J, Nouailhetas VL, Oliveira MG. Oxidative stress induced by intense and exhaustive exercise impairs murine cognitive function. J Neurophysiol. 2007;98:1820–1826. doi: 10.1152/jn.01158.2006. [DOI] [PubMed] [Google Scholar]
- 37.Sarnyai Z, Sibille EL, Pavlides C, Fenster RJ, McEwen BS, Toth M. Impaired hippocampal-dependent learning and functional abnormalities in the hippocampus in mice lacking serotonin(1A) receptors. Proc Natl Acad Sci U S A. 2000;97:14731–14736. doi: 10.1073/pnas.97.26.14731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 39.Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 40.Theodoropoulos DS, Cowan JM, Elias ER, Cole C. Physical findings in 21q22 deletion suggest critical region for 21q- phenotype in q22. Am J Med Genet. 1995;59:161–163. doi: 10.1002/ajmg.1320590209. [DOI] [PubMed] [Google Scholar]
- 41.Tsai TF, Jiang YH, Bressler J, Armstrong D, Beaudet AL. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Hum Mol Genet. 1999;8:1357–1364. doi: 10.1093/hmg/8.8.1357. [DOI] [PubMed] [Google Scholar]
- 42.Valero R, Marfany G, Gil-Benso R, Ibanez MA, Lopez-Pajares I, Prieto F, Rullan G, Sarret E, Gonzalez-Duarte R. Molecular characterisation of partial chromosome 21 aneuploidies by fluorescent PCR. J Med Genet. 1999;36:694–699. [PMC free article] [PubMed] [Google Scholar]
- 43.Walz K, Caratini-Rivera S, Bi W, Fonseca P, Mansouri DL, Lynch J, Vogel H, Noebels JL, Bradley A, Lupski JR. Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol Cell Biol. 2003;23:3646–3655. doi: 10.1128/MCB.23.10.3646-3655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wurst W. Mouse geneticists need European strategy too. Nature. 2005;433:13. doi: 10.1038/433013c. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto Y, Ogasawara N, Gotoh A, Komiya H, Nakai H, Kuroki Y. A case of 21q--syndrome with normal SOD-1 activity. Hum Genet. 1979;48:321–327. doi: 10.1007/BF00272832. [DOI] [PubMed] [Google Scholar]
- 46.Yu YE, Morishima M, Pao A, Wang DY, Wen XY, Baldini A, Bradley A. A deficiency in the region homologous to human 17q21.33-q23.2 causes heart defects in mice. Genetics. 2006;173:297–307. doi: 10.1534/genetics.105.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zarate YA, Kogan JM, Schorry EK, Smolarek TA, Hopkin RJ. A new case of de novo 11q duplication in a patient with normal development and intelligence and review of the literature. Am J Med Genet A. 2007;143:265–270. doi: 10.1002/ajmg.a.31519. [DOI] [PubMed] [Google Scholar]
- 48.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng B, Mills AA, Bradley A. A system for rapid generation of coat color-tagged knockouts and defined chromosomal rearrangements in mice. Nucleic Acids Res. 1999;27:2354–2360. doi: 10.1093/nar/27.11.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.