Abstract
Expression of the antigrowth factor myostatin (MSTN) differs between fast and slow skeletal muscles and is increased in nearly every form of muscle atrophy, but the contribution of transcriptional vs. posttranscriptional mechanisms to its differing expression in these states has not been defined. We show here that levels of mature MSTN mRNA were sixfold greater in fast vs. slow muscle and were increased twofold in fast muscle in response to dexamethasone (Dex) injection in vivo and in C2C12 myotubes following Dex treatment in vitro, but that levels of MSTN pre-mRNA, a readout of transcription, only minimally and nonsignificantly differed in these states. Moreover, Dex treatment with or without cotransfection with a glucocorticoid receptor expression construct had only modest effects on mouse MSTN promoter activity in C2C12 myotubes. We therefore explored the potential contribution of posttranscriptional mechanisms, and the role of the microRNAs miR-27a and b in particular, on MSTN expression. The MSTN 3′-untranslated region (UTR) contains a putative recognition sequence for miR-27a and b that is conserved across a wide range of vertebrate species. Cotransfection of a MSTN 3′-UTR-luciferase construct with a miR-27b expression construct significantly attenuated by approximately half while mutation of the miR-27 recognition sequence significantly increased by approximately twofold the activity of a MSTN 3′-UTR construct and decreased mRNA degradation of a luciferase reporter construct in C2C12 myotubes. Expression of miR-27a and b was almost sixfold greater in slow-twitch than in fast-twitch muscle in vivo, and miR-27a expression was significantly decreased by nearly half by glucocorticoid treatment in vitro. Finally, the miR-27a and b promoters were activated by cotransfection with the slow-specific signaling molecules calcineurin and peroxisome proliferator-activated receptor-γ coactivator-1α. The present data represent the first demonstration that posttranscriptional mechanisms involving miR-27a and b may contribute to fast-specific and glucocorticoid-dependent myostatin expression in muscle.
Keywords: atrophy, luciferase, mRNA stability, myomir, transcription
one of the most dynamic properties of skeletal muscle fibers is their ability to dramatically change their cross-sectional area and thus the absolute amount of force they can produce. Muscle fiber size is influenced by a wide range of physiological inputs, including activity levels, nutrient levels, and levels of various growth hormones, cytokines, and other secreted factors. At any given moment it is the balance of these various pro- and antigrowth inputs that determines whether a muscle fiber is undergoing atrophy, hypertrophy, or is in a steady state.
One of the main antigrowth inputs determining skeletal muscle fiber size is the level of myostatin (MSTN) activity. MSTN is an autocrine/paracrine factor produced predominantly by skeletal muscle which acts back on muscle to dramatically inhibit its growth. Muscle-specific transgenic overexpression of MSTN is sufficient to cause muscle fiber atrophy (49), while both naturally occurring and genetically engineered inactivating mutations to the MSTN gene result in dramatic muscle hypertrophy in several different species (27, 42, 43, 45, 54). Together, these data demonstrate that factors influencing the level of MSTN expression have profound effects on muscle fiber size.
Moreover, expression of MSTN is increased in a wide array of atrophy models (10, 13, 18, 46), including several models of glucocorticoid-dependent muscle atrophy (5, 6, 35, 40), suggesting that changes in endogenous MSTN expression may contribute to the decreased muscle growth observed in these states. In addition, MSTN mRNA levels have been demonstrated by us (1, 2) and others (10) to be greater in fast-twitch than in slow-twitch muscles and thus may play a role in regulating differences in fiber size and/or fiber type between different muscles. However, at present the mechanisms governing increased MSTN expression during these states have not been sufficiently addressed. In particular, despite a wealth of in vitro studies on the transcriptional regulation of the MSTN gene (1–4, 6, 20, 39), it is not completely clear that MSTN gene expression is regulated exclusively or even predominantly at the level of transcription. We recently demonstrated that MSTN pre-mRNA is significantly increased in muscle from food-deprived mice and that this increase appears to be at least partly transcriptional (6) and dependent on glucocorticoid-induced activation of the CCAAT/enhancer-binding protein (C/EBP) family of transcriptional activators (6), but beyond this there is little evidence from either pre-mRNA quantification, nuclear run-on assays, or treatment with transcription inhibitors suggesting that the MSTN gene is transcriptionally regulated.
Transcriptional mechanisms are not the only determinant of gene expression, and, in particular, posttranscriptional regulation of mRNA stability or translation can be a major contributor to differences in gene expression in different tissue types and across different physiological states (59). Recently, much attention has focused on the potential role of microRNAs or miRs in the regulation of gene expression. MiRs are short single-stranded RNAs that are processed from longer hairpin precursors and that are complementary to sequences typically found in the untranslated regions (UTRs) of mRNAs. MiRs bind complementarily to these recognition sequences and can target the mRNAs for translational repression or for degradation by the RNA-induced silencing complex or RISC (12). Since their discovery in Caenorhabditis elegans in 1993 (36), miRs have subsequently been demonstrated to play a prominent role in the regulation of gene expression in a wide variety of organisms and in a nearly all tissues examined to date. Muscle gene expression in particular appears to be strongly governed by miR regulation, and several so-called “myomirs,” including miR-1-1, miR-133a and b, and miR-206, are highly enriched in skeletal muscle and regulate expression of key genes involved in muscle stem cell proliferation and differentiation (63).
The purpose of the present work was to determine the contribution of transcriptional vs. posttranscriptional mechanisms to the regulation of MSTN expression in two well-defined physiological states characterized by differential MSTN expression, during glucocorticoid-mediated muscle atrophy and in fast vs. slow muscles. Our evidence suggests that transcriptional changes are insufficient to account for the changes in MSTN expression in either of these states, but it supports the hypothesis that changes in the levels of two related miRs, miR-27a and/or miR-27b, may contribute to changes in MSTN mRNA levels in both cases. These data are thus the first to identify miR-27a and b as potential regulators of MSTN expression.
METHODS
Experimental animals.
All experimental procedures involving animals were approved by the Institutional Animal Care and Use Committee of the University of Colorado, Boulder, and complied with the guidelines of the American Physiological Society on the use of laboratory animals. Male wild-type C57/BL6J (C57) mice were obtained from our breeding colony in the Department of Integrative Physiology at the University of Colorado, Boulder. Tibialis anterior (TA) and soleus (Sol) muscles were isolated from wild-type mice (n = 5), frozen in liquid nitrogen, and stored at −80°C. For the dexamethasone (Dex) injection studies, wild-type C57 mice were injected subcutaneously once daily for 2 days with either vehicle alone (n = 4) or 500 μl of 50 μg/ml Dex (n = 5) for a final concentration of 1 mg/kg as described by Frost et al. (23). Twenty-four hours after the second injection, mice were killed and the TA and Sol muscles were isolated, frozen, and stored as described above.
Semiquantitative reverse transcription-polymerase chain reaction.
Levels of MSTN mRNA and pre-mRNA were evaluated using semiquantitative one-step RT-PCR with the Qiagen OneStep RT-PCR kit following the manufacturer's instructions. Total RNA was isolated by homogenizing muscles in 1 ml of TRIzol reagent (Invitrogen) followed by chloroform extraction and ethanol precipitation as described previously (1–6). RNA concentration was determined spectrophotometrically at A260 and brought to a concentration of 50 ng/μl. MSTN pre-mRNA was cleaned up to eliminate possible DNA contamination using the technique described by Elferink and Reiners (21) and Haddad, Baldwin, and coworkers (32). Briefly, 30 μg of total RNA was then incubated with 5 units RQ1 RNase-free DNase (Promega) for 30 min at 37°C to eliminate contaminating DNA. RNA was then reextracted with 150 μl of RNase-free H2O, 750 μl of TRIzol, and 250 μl of chloroform. RNA was pelleted by addition of 800 μl of 100% isopropanol and microfuged at maximum speed for 10 min. The pellet was washed twice with 75% ethanol, air dried for 15 min, frozen overnight at −40°C, then resuspended in 20 μl of RNAse-free H2O.
The reverse transcription reaction was carried out using 250 ng of RNA, 15 pmol each primer, 2.5 mM dNTPs, 5× buffer, and OneStep enzyme mix in a 25-μl reaction volume. The RT reaction was run for 30 min at 50°C, followed by the initial PCR activation step of 95°C for 25 min. Approximately 25–30 cycles of denaturation (1 min, 94°C), annealing (45 s, 55°C), and extension (45 s, 72°C) was followed by 10 min of final extension at 72°C. The primer sequences for MSTN mRNA consisted of two exonic primers, 5′-GGAACTGATCGATCAGTACG-3′ and 5′-AATCCAGTCCCATCCAAAGG-3′. The primer sequences for MSTN pre-mRNA consisted of a forward exonic primer (5′-TAGGGCCATGAAAGGAAAAATGAAGTCTA-3′) and a reverse intronic primer (5′-TCTCCGGGACCTCTTGGGTGTG-3′); as a control, primers for GAPDH were used (forward: 5′-ATCACCATCTTCCAGGAGCGA-3′; reverse: 5′-AGTCTTCTGGGTGGCAGTGAT-3′). Five microliters of 6× loading buffer were added to each sample, and the entire reaction was run on a 2% agarose gel containing 10 μg/ml ethidium bromide and visualized and photographed using a gel documentation system. Gel photos were scanned, and the bands for MSTN mRNA, MSTN pre-mRNA, and GAPDH were quantified by densitometry using NIH Image software.
RT-PCR was also used to quantify luciferase mRNA levels. C2C12 myotubes were plated on 12-well plates and transfected with either the wild-type or the miR-27 mutant (miR-27mut) version of the MSTN promoter plus both UTRs-luciferase reporter construct. After differentiating in low-serum medium for 2 days, myotubes were treated with 5 μg/ml actinomycin D for 0, 12, or 24 h to inhibit transcription. Total RNA was isolated using the TRIzol method and used as template for one-step RT-PCR as described above using the luciferase primers 5′-GCCTGAACGCTCTGATTAAGT-3′ and 5′-CACCTGCGTCGAAG-3′. RT-PCR products were run on a 2% agarose gel containing ethidium bromide, and gels were scanned and bands were quantified using NIH Image software.
Quantitative real-time RT-PCR.
RNA was isolated from skeletal muscle samples as described above. The reverse transcription reaction was carried out using 0.5 μg of RNA using the High Capacity cDNA Reverse Transcription kit from Applied Biosystems and RNA-specific probes according to the manufacturer's protocol. Primer and probe sets for miR-27a, miR-27b, miR-499, and U6 RNA were obtained from Applied Biosystems. All real-time PCR procedures were run in triplicate to correct for variances in loading. In addition, a standard curve ranging from 10- to 0.001-μg dilutions of mouse TA cDNA was run in duplicate for every assay to produce a standard curve for quantification. All values are expressed as the mean of the triplicate measure for the experimental (MSTN) divided by the mean of the triplicate measure of β-actin for each sample.
MicroRNA microarray.
A microRNA detection microarray was used to examine global microRNA expression in C2C12 myotubes and TA skeletal muscle. Briefly, RNA was isolated from three different platings of C2C12 myotubes or from four different TA muscles using the miRNeasy microRNA isolation kit from Qiagen and pooled to minimize idiosyncratic effects of any single experiment on the results. RNA was labeled with Cy5 dye and run on an LC Biosciences microRNA microarray using μParaflo technology and containing probes to miRBase 13.0 microRNAs. The signal values are derived by background subtraction and normalization. To be considered detectable, a microRNA transcript had to meet two conditions: 1) it had to have a signal intensity higher than 3× (background standard deviation) and 2) its spot coefficient of variation (CV) had to be <0.5. CV was calculated by (standard deviation)/(signal intensity). In addition, when repeating probes were present on the array, a transcript was listed as detectable only if the signals from at least 50% of the repeating probes were above detection level.
Cloning and mutagenesis.
The 1,200-bp mouse MSTN upstream promoter region was cloned from mouse genomic DNA using PCR as described previously (1). Briefly, ∼1,200 bp of the mouse MSTN upstream promoter region including the entire 5′-UTR to the translation initiation site were amplified using primers containing a MluI and an XhoI site at the 5′- and 3′-ends, respectively. The resulting PCR product was cut and then ligated into the pGL3basic luciferase expression plasmid (Promega) at these sites, and positive clones were screened by restriction digestion. Similarly, the 2.5-kb mouse MSTN promoter construct was cloned using a similar approach with the following primers: 5′-TAGTGGACGCGTAACCTTTTTAAGTCCTAAGTC-3′ and 5′-TAGTGGCTCGAGTGTATACTGTATTCCAAGTGGCTT-3′. The 300-bp mouse MSTN promoter was cloned from the 1,200-bp MSTN promoter construct using inverse PCR and the following primers: 5′-primer, 5′-TAGTGGCCTAGGTCTTTCATGATTTGGGGATTT-3′; 3′-primer, 5′-TAGTGGCCTAGGAAGAGCTCGGTACCTATCGAT-3′. The MSTN 3′-UTR expression construct was cloned as described previously (3). Briefly, the complete 1,448-bp MSTN 3′-UTR from the end of the coding region to the final, major poly(A) sequence reported previously (http://www.ncbi.nlm.nih.gov/nuccore/AY204900.1?ordinalpos=4&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum) was cloned using primers containing a 5′-XbaI and a 3′-SalI site and placed downstream of the luciferase expression cassette in pGL3 already containing the MSTN 1,200-bp promoter plus 5′-UTR described above; this construct is referred to as “MSTN promoter + both UTRs.” Inverse PCR was then used to remove everything except a minimal TATA box and the 5′-UTR from the 5′ flanking end of luciferase; this construct is referred to as “MSTN both UTRs.” Finally, the p100α 3′-UTR was also cloned into the pGL3basic vector using the XbaI and SalI sites as above.
Mutagenesis to alter the conserved glucocorticoid response element-2 (GRE2) in the MSTN 1,200-bp promoter was carried out using inverse PCR. The primers used were as follows: 5′-primer, 5′-TAGTGGCCTAGGTTTATATTAGTACACAGACTT-3′; 3′-primer, 5′-TAGTGGCCTAGGTTAAAGCTTATTAATCTCTGT-3′. Amplification results in a change of the GRE2 sequence from 5′-AGTACATTTATATTA-3′ to 5′-ccTAggTTTATATTA-3′ (altered bases in lowercase). All positive colonies were screened using digestion with AvrII. Finally, mutagenesis was used to mutate the miR-27 recognition sequence within the MSTN 3′-UTR. The primers used were 5′-CTTCCCCTCAATTTCGAAgCTGgGAGTTCAAGCACCACAGGCTGTAGG-3′ and its complement (mutated nucleotides in lowercase).
The region upstream of the mouse miR-23a/miR-27a/miR-24–2 cluster was cloned from mouse genomic DNA using the following primers: 5′-primer, 5′-TAGTGGACGCGTTGGTACTGGGTGCTAAATACC-3′; 3′-primer, 5′-TAATAACTCGAGCCCTGGCAATGTGATTTGTGA-3′. The −1,767 upstream promoter region for the mouse C9orf3 gene, within which miR-23b/miR-27b/miR-24–1 are found as a mirtron cluster, was kindly provided by Dr. Masanobu Satake (Tohoku University, Sendai, Japan).
The glucocorticoid receptor (GR) expression plasmid pSG5-GR was kindly provided by Dr. Stoney Simons (National Institutes of Health, Bethesda, MD). The miR-27b, miR-124, and null miR expression constructs were obtained from Cell Biolabs. The Cignal GRE Reporter Assay Kit from SA Biosciences, which contains a luciferase reporter construct driven by five copies of the glucocorticoid response element (GRE), was used to quantify GRE activity in response to Dex.
Cell culture and transfection.
C2C12 myoblasts were plated on 0.75% gelatin-coated six-well plates in proliferation medium consisting of Dulbecco's modified Eagle's medium (DMEM) supplemented with 20% fetal bovine serum (FBS) and 1% penicillin-streptomycin (pen/strep). Cells were split after reaching 90% confluence onto four to six gelatin-coated 24-well plates and were transfected with Lipofectamine 2000 as previously described (1, 2). Briefly, for each well, 1.5 μl of Lipofectamine 2000 and 1.0 μg of DNA was mixed in 100 μl of FBS-free and pen/strep-free DMEM and allowed to complex for 30 min. The transfection mix was added to wells containing proliferation medium and allowed to remain on cells for 1–2 days until they reached confluence, at which time the medium was removed and replaced with differentiation medium consisting of DMEM plus 1% horse serum for 2 days to induce differentiation into myotubes, at which time >90% of cells had differentiated into myotubes. After 2 days of differentiation, myotubes were treated with either dimethyl sulfoxide vehicle or with 1 μm Dex for 24–48 h. At the end of the treatment period, myotubes were washed once with sterile PBS and then lysed in passive lysis buffer (Promega). Luciferase activity was then determined using a luminometer and firefly luciferase reagent (Promega) as previously described (1, 2, 4).
Luciferase activity was also used as a readout of message decay as previously described (28, 55, 62, 65). C2C12 myoblasts were plated, split, and transfected with the MSTN UTR constructs and were then allowed to differentiate for 2 days in low-serum medium. Cells were then treated with low-serum medium containing 5 μg/ml actinomycin D. Wells were harvested at 0, 6, 12, and 24 h posttreatment and used to quantify luciferase activity as above.
Statistical analysis.
All in vivo studies represent an n = 4–7 animals per condition, and data from these studies are reported as means ± SE. All in vitro studies represent an n = 3 independent experiments consisting of 4–8 wells for each condition per experiment, and data from these studies are reported as means ± SE. Statistically significant differences between fed, fasted, and refeeding or fed, fasted, and RU486-treated body mass, tissue mass, or mRNA expression were determined by analysis of variance (ANOVA) and Fisher's post hoc test, with an α-level of 0.05 taken as significant. Differences between vehicle and Dex-injected values were determined by an independent t-test, with an α-level of 0.05 taken as significant. Statistically significant differences between green fluorescent protein (GFP) vs. pSG5-GR transfected with and without Dex treatment were determined by ANOVA and Fisher's post hoc test, with an α-level of 0.05 taken as significant. Differences between-wild type and GRE-mutated MSTN promoter activity were determined by an independent t-test, with an α-level of 0.05 taken as significant.
RESULTS
MSTN mRNA and pre-MRNA levels.
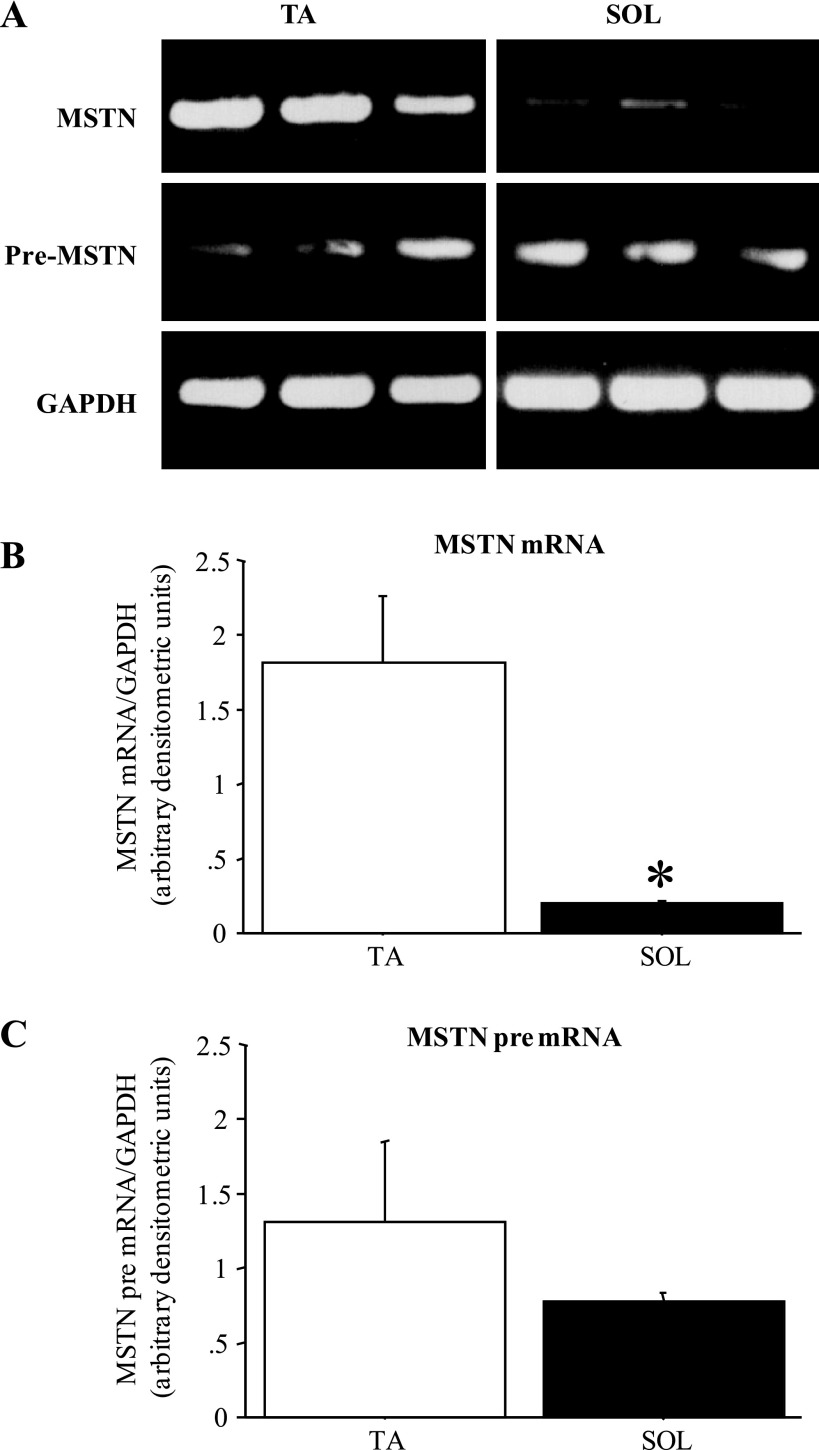
Consistent with what we (1, 2) and others (10) have shown previously, MSTN mRNA levels differ substantially between fast and slow skeletal muscles (Fig. 1). MSTN mRNA levels in the fast-twitch TA were 5- to 10-fold greater than those observed in the slow-twitch Sol (Fig. 1, A and B), but MSTN pre-mRNA levels were modestly but not significantly different between TA and Sol (Fig. 1, A and C).
Fig. 1.
Myostatin (MSTN) mRNA but not pre-mRNA levels differ between fast and slow muscles. A: gel demonstrating the expression pattern of MSTN mRNA, MSTN pre-mRNA, and GAPDH from 3 wild-type C57/BL6J (C57) tibialis anterior muscles (TA) and 3 soleus muscles (Sol). All images were taken at the same intensity. B: quantification of MSTN mRNA levels relative to GAPDH levels for TA and Sol muscles. Each bar represents the mean ± SE for n = 4 muscles. *Significantly different from TA muscle, P < 0.05. C: quantification of MSTN pre-mRNA levels relative to GAPDH levels for TA and Sol muscles. Each bar represents the mean ± SE for n = 4 muscles. MSTN pre-mRNA levels were not significantly different between TA and Sol.
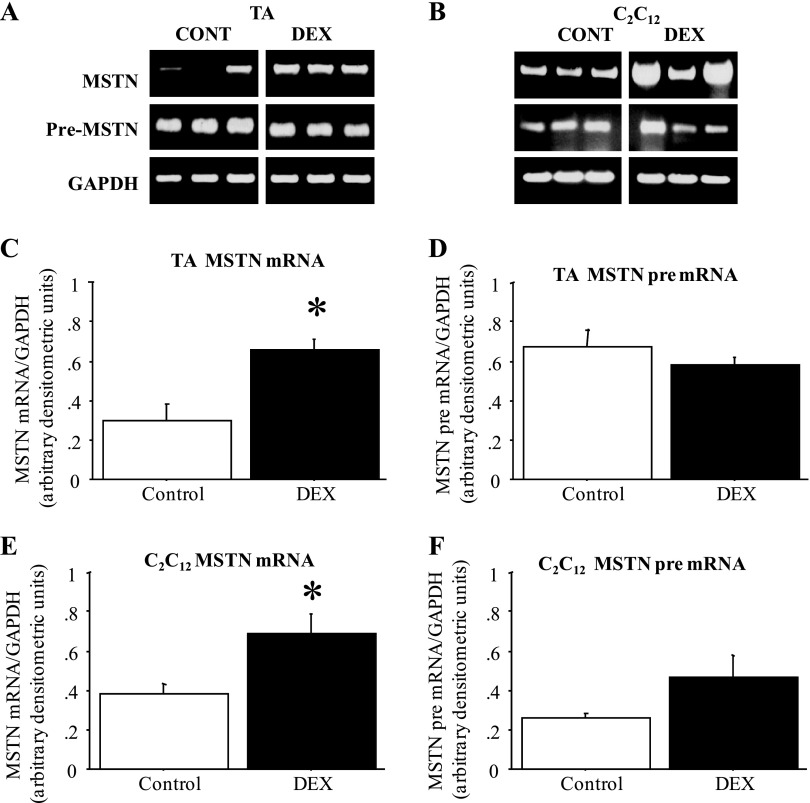
Moreover, in the TA muscle of mice injected daily for 2 days with Dex, MSTN mRNA levels increased approximately twofold compared with vehicle-injected controls, but MSTN pre-mRNA levels were again not significantly different in Dex-treated vs. control TA (Fig. 2, A, C, and D). Similarly, treatment of C2C12 myotubes with 1 μM Dex for 2 days also significantly increased MSTN mRNA levels while MSTN pre-mRNA levels were only modestly and nonsignificantly altered by Dex treatment (Fig. 2, B, E, and F). Thus, though MSTN mRNA levels were greater in fast than slow muscle and were increased both in vitro and in vivo by Dex treatment, changes in MSTN pre-mRNA levels, a readout of transcription, do not appear to account for the differences in MSTN expression in these states.
Fig. 2.
MSTN mRNA but not pre-mRNA levels are increased by dexamethasone (Dex) treatment. A: gel demonstrating the expression pattern of MSTN mRNA, MSTN pre-mRNA, and GAPDH from TA muscles from 3 vehicle-injected and 3 Dex-injected mice. All images were taken at the same intensity. Cont, control. B: gel demonstrating the expression pattern of MSTN mRNA, MSTN pre-mRNA, and GAPDH from TA muscles from 3 vehicle-treated and 3 Dex-treated plates of C2C12 myotubes. All images were taken at the same intensity. C: quantification of MSTN mRNA levels relative to GAPDH levels for vehicle and Dex-injected mice. The bars represent means ± SE for n = 4 vehicle-injected and n = 5 Dex-injected mice. *Significantly different from vehicle injected, P < 0.05. D: quantification of MSTN pre-mRNA levels relative to GAPDH levels for vehicle and Dex-injected mice. The bars represent means ± SE for n = 4 vehicle-injected and n = 5 Dex-injected mice. *Significantly different from vehicle injected, P < 0.05. E: quantification of MSTN mRNA levels relative to GAPDH levels for vehicle and Dex-treated C2C12 myotubes. The bars represent means ± SE for n = 4 different experiments. *Significantly different from vehicle treated, P < 0.05. F: quantification of MSTN pre-mRNA levels relative to GAPDH levels for vehicle and Dex-treated C2C12 myotubes. The bars represent means ± SE for n = 4 different experiments. *Significantly different from vehicle treated, P < 0.05.
MSTN promoter activity is minimally changed by Dex treatment in vitro.
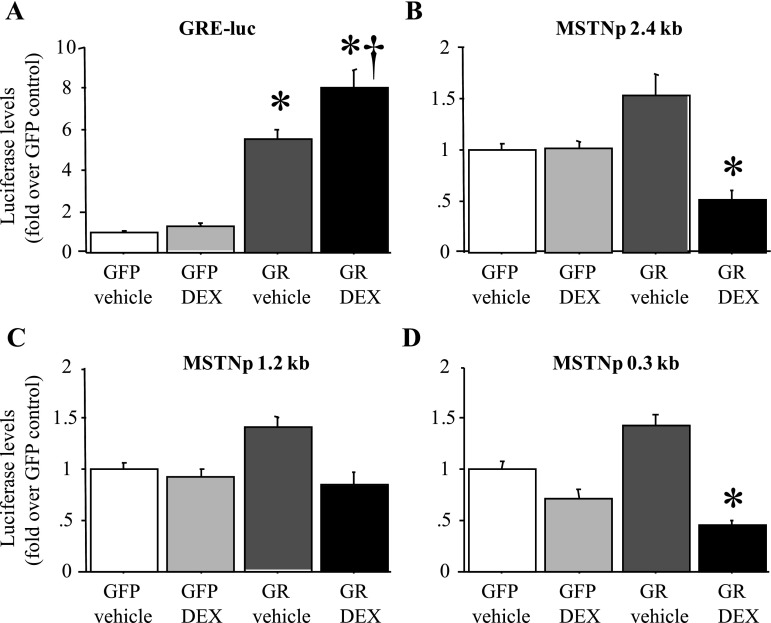
Previous data (20, 39) have suggested that the increase in MSTN mRNA levels with Dex treatment of C2C12 myotubes in vitro is due to an increase in MSTN promoter activity. Because GR levels can be low in C2C12 myotubes (57) and/or decreased by Dex treatment (44), we examined the effects of 2 days of Dex treatment on MSTN promoter activity of C2C12 myotubes with and without cotransfection with a GR expression construct. As a positive control, we examined the effects of Dex treatment with and without GR cotransfection on activity of a GRE-luciferase (GRE-luc) “sensor” construct in which luciferase activity is driven by tandem repeats of the GRE. Dex treatment alone had only minimal effects on activity of a GRE reporter construct, consistent with the interpretation that the C2C12 myotubes used in these studies expressed minimal amounts of GR (Fig. 3A). Cotransfection with the GR construct alone significantly increased activity of the GRE-luc reporter, presumably by allowing it to respond to endogenous glucocorticoids in FBS, and cotransfection with the GR construct and Dex treatment combined had the greatest effect on GRE-luc activity as expected (Fig. 3A).
Fig. 3.
The mouse MSTN promoter is minimally responsive to Dex treatment in vitro. A: activity of a glucocorticoid response element-luciferase (GRE-luc) “sensor” plasmid in response to green fluorescent protein (GFP) cotransfection and vehicle treatment, GFP cotransfection and Dex treatment, glucocorticoid receptor (GR) cotransfection and vehicle treatment, and GR cotransfection and Dex treatment. All values were normalized to the average of the GFP-cotransfected, vehicle-treated control. The GRE-luc construct responded to GR cotransfection alone and with Dex treatment. B–D: activity of the 2,400 bp (B), 1,200 bp (C), and 300 bp (D) mouse MSTN promoter-luciferase reporter plasmids in response to GFP cotransfection and vehicle treatment, GFP cotransfection and Dex treatment, GR cotransfection and vehicle treatment, and GR cotransfection and Dex treatment. All values were normalized to the average of the GFP-cotransfected, vehicle-treated control. The mouse MSTN promoter constructs were minimally responsive to GR cotransfection alone and were not increased by GR cotransfection with Dex treatment. Bars in all panels represent means ± SE for n = 3 separate experiments with 6–8 wells per experiment. *Statistically significantly different from GFP-cotransfected, vehicle-treated control, P < 0.05. †Statistically significantly different from GR-cotransfected, vehicle-treated control, P < 0.05.
In contrast, for the three different lengths of MSTN promoter, Dex treatment alone had no effect, GR cotransfection alone modestly and nonsignificantly increased MSTN promoter activity by ∼50%, and cotransfection with the GR construct and Dex treatment combined actually significantly decreased activity of the 2.4- and 0.3-kb length MSTN constructs (Fig. 3, B–D). Similar results were found with both shorter (6, 12, and 24 h) and longer (72 and 96 h) periods of Dex treatment, with lower (0.1 μM) and higher (10 μM) concentrations of Dex treatment, and with treatment with physiological levels of purified corticosterone for 2 days (data not shown). Thus we observed only modest effects of GR cotransfection on MSTN promoter activity, and Dex treatment with or without GR cotransfection did not increase MSTN promoter activity in C2C12 myotubes.
Examination of the sequence upstream of the MSTN coding region revealed no perfect matches to the GRE consensus sequence (GGTACANNNTGTTCT; 7) within ∼10,000 bp of the upstream promoter sequence (data not shown), but within the proximal 1,200 bp there were four partial matches to this sequence, though the best of these, GRE2, was only an 80% match (12/15 nucleotides) to the GRE consensus (Supplemental Fig. S1; Supplemental Material for this article is available online at the Journal website). GRE2 also showed the highest sequence conservation across species; it was 100% conserved between human and mouse while GREs 1, 3, and 4 showed just 86%, 66%, and 66% similarity, respectively, between the two species (Supplemental Fig. S1). We therefore mutated GRE2 so that it was further dissimilar to the GRE consensus (mouse GREmut sequence in Supplemental Fig. S1). Mutation of this element had no significant effect on either basal or GR-cotransfected (Supplemental Fig. S1) promoter activity. Thus our promoter data, consistent with the MSTN pre-mRNA data from Figs. 1 and 2 above, strongly suggest that glucocorticoids have minimal effects on MSTN transcription.
The MSTN 3′-UTR contains a well-conserved miR-27 recognition sequence.
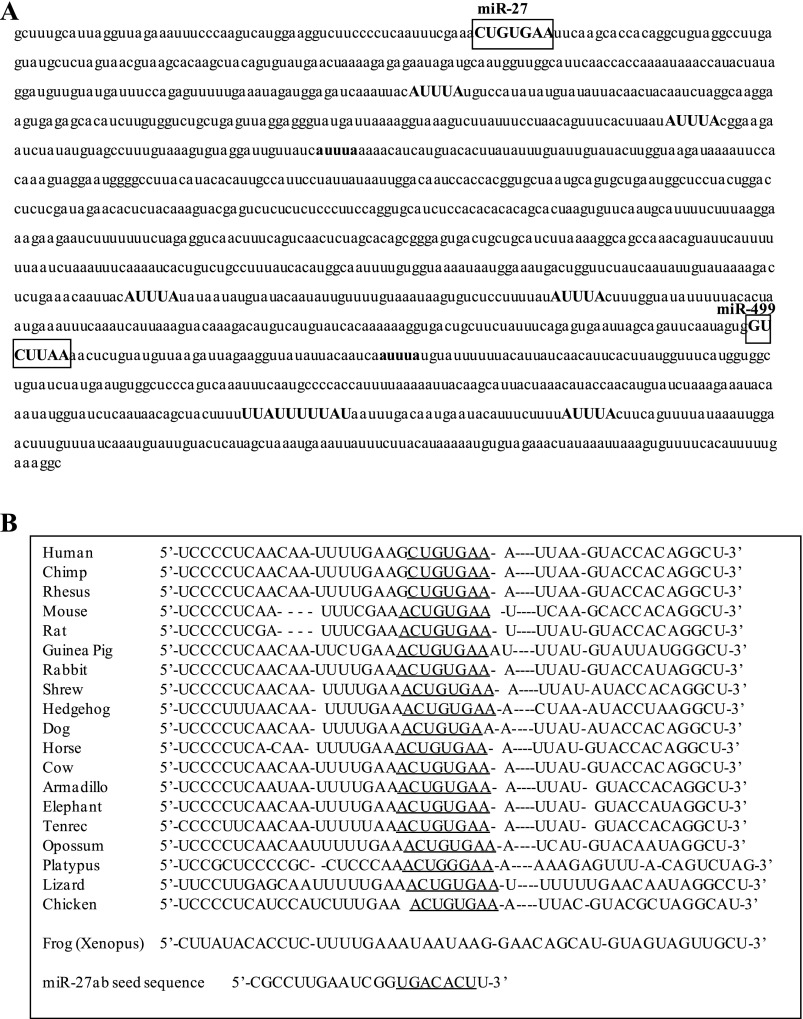
We then examined the MSTN 3′-UTR to identify potential element(s) that might mediate posttranscriptional mechanisms governing MSTN expression. The MSTN 3′-UTR contains several AUUUA elements (Fig. 4A) involved in deadenylase recruitment and 3′-5′ exonuclease degradation (25). Five of these (shown in all uppercase letters in Fig. 4A) are conserved between mouse and human. In addition, the microRNA prediction web site TargetScan predicted that the mouse MSTN 3′-UTR contains 70 putative microRNA recognition sequences. However, most of these are only found in mouse or a few other species and thus are not well conserved, and/or have low context scores predicting low strength/likelihood of microRNA binding. One site that was both highly conserved and had a high total context score was the one for miR-27ab (Fig. 4A). The miR-27ab recognition sequence is conserved as either an 8mer, exact match to nucleotides 2–8 of the microRNA seed sequence followed by an adenine, or as a 7mer-1A, an exact match to positions 2–7 of the mature microRNA (the seed) followed by an adenine, in 19 vertebrate species, including opossum, platypus, chicken, and lizard (Fig. 4B). Only in Xenopus was this sequence not conserved. The next highest conservation was for a 7mer-1A matching the miR-673 recognition sequence, which was conserved in 18 different species (all but platypus and Xenopus), but the context score for this site was low. In mouse, the miR-27ab sequence has a total context score of −0.42 (the combination of site-type distribution, 3′-pairing contribution, local adenylate/uridylate contribution, and position distribution, all variables affecting the strength of likely miR interaction; the larger the negative number, the stronger the site; 26); only 7 out of 70 predicted miR sites had a higher total context score than miR-27 based on the TargetScan algorithm, and of these only miR-499 has been shown to be expressed in detectable amounts in C2C12 myotubes (see below). Moreover, the TargetScan algorithm predicts 786 potential targets for miR-27a/b in the mouse, of which MSTN is ranked 154th on the basis of total context score. Thus the MSTN 3′-UTR contains several putative regulatory elements, but the miR-27 site in particular is the highest conserved across a wide range of species and shows one of the highest total context scores predicting microRNA binding.
Fig. 4.
The 3′-untranslated region (UTR) of the mouse MSTN gene contains several stability-regulating elements. A: sequence of the mouse MSTN 3′-UTR. AUUUA sites are shown in bold; ones that are conserved between mouse and human are shown in all uppercase letters. The putative recognition sequences for microRNA (miR)-27ab and for miR-499 are boxed. B: interspecies sequence alignment for the sequence around the miR-27ab recognition sequence. The recognition sequence for miR-27ab is underlined. The actual seed sequence of miR-27a is shown at bottom and is underlined.
MiR-27a and b are expressed in differentiated muscle cells.
To examine which, if any, of the microRNAs identified from the putative binding sites above were expressed in muscle cells, we used microRNA microarray technology to quantify levels of >700 microRNAs in differentiated C2C12 myotubes and from TA skeletal muscle. Three hundred thirty microRNAs were found to be within the detectable range of the chip's sensitivity in C2C12 myotubes. These are shown in Supplemental Table SI. Not surprisingly, the highest expression was that observed for the myomirs miRs-206 and 1, which are highly enriched in differentiated skeletal muscle (48). MiR-206 and miR-1-1 had levels of over 40,000 and 20,000 arbitrary units, respectively. MiR-21, -23a and 23-b, and -709 also showed levels close to or above 15,000 arbitrary units, while miR-499, which has been previously demonstrated to regulate MSTN expression (8), had values of ∼1,400 arbitrary units. MiR-27a and b demonstrated levels of ∼2,700 and 5,300 arbitrary units in C2C12 myotubes, which is considerably higher than the average of 1,147 arbitrary units across all detectable miR transcripts in C2C12 myotubes in this assay (Supplemental Table SI). MiR-27a and b were the 31st and 23rd most abundant miRs out of the 330 detectable miRs in this assay. Thus miR-27a and b are expressed in differentiated muscle cells at a level 2- to 5-fold higher than that of the average of all other detectable miRs.
In the adult mouse TA muscle, many fewer microRNAs were detectable; just 191 different microRNAs were above the limit of detection. The highest expressed microRNAs by this assay were miR-1, -709, -2,142, and -762 (Supplemental Table SII). MiR-27a and b were much less abundant in the adult fast-twitch TA relative to other microRNAs, representing the 72nd and 106th most abundant microRNAs, respectively, out of the 191 detectable microRNAs. This is similar to miR-206, which went from being the most abundant microRNA in C2C12 myotubes at over 40,000 arbitrary units to being the 21st most abundant microRNA in the adult TA with 974 arbitrary units (Supplemental Table SII). Thus expression of miR-27a and b is lower than other microRNAs but detectable in mature adult skeletal muscle.
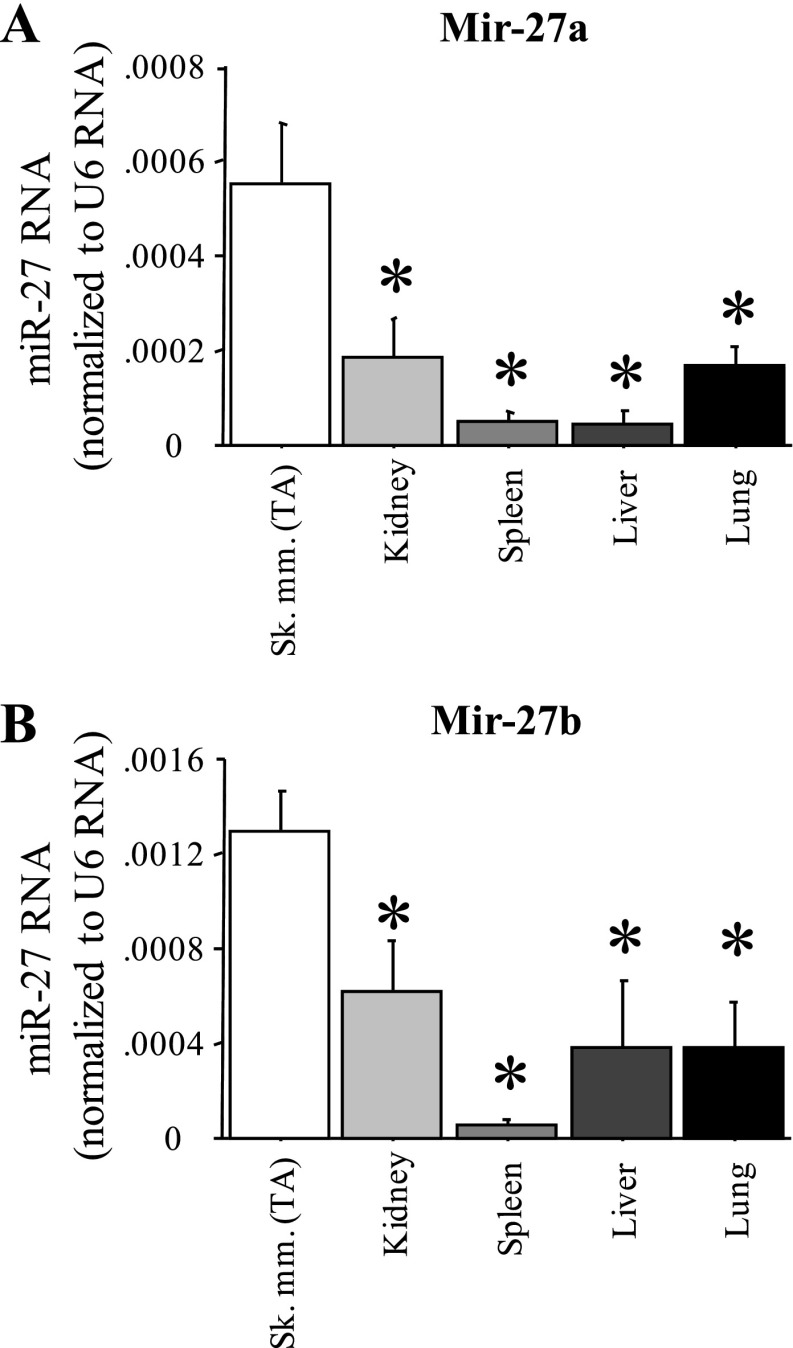
However, expression of miR-27a and b was greater in TA skeletal muscle than in several other tissues in the adult mouse. We used quantitative real-time PCR analysis of RNA isolated from the TA and from several other tissues from wild-type C57 mice to confirm expression of miR-27a and b in skeletal muscle and compared it to other adult mouse tissues. Both miR-27a and b were found in greater quantities in TA muscle than in kidney, spleen, liver, and lung (Fig. 5, A and B). Thus while miR-27a and b are not exclusively expressed in skeletal muscle and thus cannot be considered true myomirs, their expression is enriched in skeletal muscle relative to non-muscle tissues.
Fig. 5.
Muscle-enriched expression of miR-27a and b. Quantitative real-time PCR quantification of miR-27a (A) and miR-27b (B) levels relative to U6 RNA control for five different mouse tissues. Sk mm, skeletal muscle. Bars represent means ± SE for n = 4 tissues. *Significantly different from TA, P < 0.05.
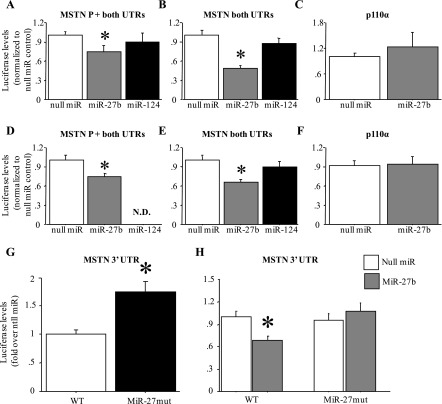
MiR-27b inhibits MSTN expression in vitro.
In both myoblasts (Fig. 6, A and B) and myotubes (Fig. 6, D and E), cotransfection of a miR-27b expression construct significantly attenuated activity of a MSTN 3′-UTR-luciferase reporter construct compared with cotransfection with a “null miR” control construct. Moreover, this inhibition was observed in two different constructs, one containing the MSTN promoter and 5′-UTR and one containing a minimal TATA box and the MSTN 5′-UTR along with the MSTN 3′-UTR (Fig. 6, A, B, D, and E). In addition, this effect was specific for miR-27b, since cotransfection with a miR-124 construct, which has no predicted binding site in the MSTN 3′-UTR, had no significant effect on activity of either MSTN 3′-UTR-luciferase reporter construct (Fig. 6, A, B, D, and E). Furthermore, this effect of miR-27b was specific for the MSTN 3′-UTR, since cotransfection of either myoblasts or myotubes with a p110α 3′-UTR-luciferase reporter did not affect activity of this construct (Fig. 6, C and F).
Fig. 6.
MiR-27b decreases activity of a mouse MSTN 3′-UTR-luciferase reporter construct in vitro. A and D: activity of a plasmid containing the mouse MSTN 1,200-bp promoter, 5′-UTR, and 3′-UTR. B and E: a plasmid containing a minimal MSTN promoter (20 bp upstream of the TATA box), 5′-UTR, and 3′-UTR. C and F: a plasmid containing the SV40 promoter and the p110α 3′-UTR. A–C: results for C2C12 myoblasts. D–F: results for C2C12 myotubes. All results were normalized to the null miR-cotransfected control. In each panel is shown the results of cotransfection with the null miR control, miR-27b, or miR-124. ND, no data. Cotransfection with miR-27b significantly attenuated activity of the MSTN but not p110α constructs, and miR-124 had no effect. Bars in all panels represent means ± SE for n = 3 different experiments with 6 wells per experiment. *Significantly different from null miR cotransfected, P < 0.05. G: effect of mutation of the miR-27 site of the mouse MSTN 3′-UTR on basal activity in C2C12 myotubes. Mutation of the miR-27 recognition sequence significantly increased activity relative to the wild-type (WT) construct. Bars represent means ± SE for n = 3 different experiments with 6 wells per experiment. *Significantly different from wild-type construct, P < 0.05. H: effects of mutation of the miR-27 site of the mouse MSTN 3′-UTR on miR-27b responsiveness. Mutation of the miR-27 recognition sequence abolished responsiveness to miR-27b cotransfection compared with the wild-type construct. Bars represent means ± SE for n = 3 different experiments with 6 wells per experiment. *Significantly different from wild-type construct cotransfected with null miR, P < 0.05.
Conversely, mutation of the miR-27 recognition sequence in the mouse MSTN 3′-UTR resulted in a significant increase in activity of this reporter construct relative to the wild-type construct under basal conditions (Fig. 6G). Moreover, mutation of this sequence also abolished responsiveness to miR-27b cotransfection (Fig. 6H). Together, these data strongly suggest that miR-27a/b directly regulate stability and/or translation of the MSTN mRNA through interaction with the predicted miR-27 recognition sequence.
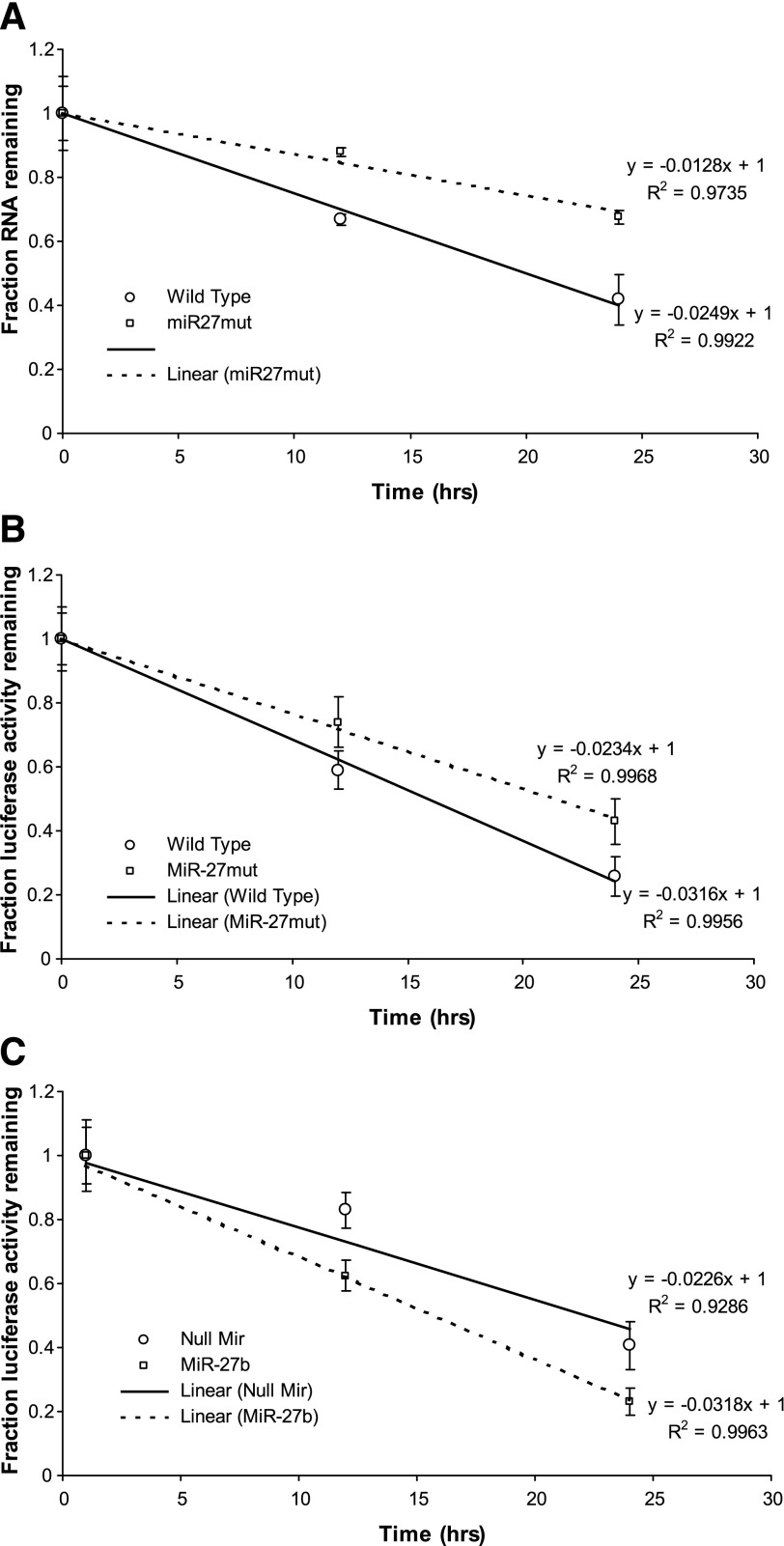
MiR-27 alters MSTN mRNA stability in vitro.
We next examined the effects of miR-27 on MSTN mRNA stability. Mutation of the miR-27 recognition sequence in the MSTN 3′-UTR conferred greater mRNA stability to a luciferase-MSTN 3′-UTR reporter construct relative to the wild-type MSTN 3′-UTR (Fig. 7A). In addition, we used luciferase activity as a readout of message decay with actinomycin D treatment as described by others (28, 55, 62, 65) to confirm the effects of miR-27 on MSTN 3′-UTR stability. Mutation of the miR-27 recognition sequence significantly increased luciferase activity of the MSTN 3′-UTR construct at 12 and 24 h of actinomycin D treatment compared with the wild-type construct (Fig. 7B). Conversely, cotransfection with the miR-27b expression construct significantly decreased luciferase activity of the MSTN 3′-UTR construct at 12 and 24 h of actinomycin D treatment compared with cotransfection with the null miR control (Fig. 7C).
Fig. 7.
MiR-27 alters MSTN mRNA stability in vitro. A: effect of cotransfection of the MSTN P + UTR construct with either the null miR control or a miR-27b expression construct on luciferase levels in the presence of actinomycin D. MiR-27b cotransfection results in a greater decrease in luciferase levels compared with cotransfection with the null miR construct. Points represent means ± SE for n = 3 different experiments. B: effect of mutation of the miR-27 recognition sequence on activity of the MSTN P + UTR construct in the presence of actinomycin D. Mutation of the miR-27 recognition sequence results in an attenuation of the decrease in luciferase levels compared with the wild-type construct. Points represent means ± SE for n = 3 different experiments with 6 wells per experiment. C: effect of cotransfection with either the null miR control or the miR-27b expression plasmid on activity of the MSTN P + UTR construct in the presence of actinomycin D. Cotransfection of the miR-27 recognition sequence results in a more rapid decrease in luciferase levels compared with the wild-type construct. Points represent means ± SE for n = 3 different experiments with 6 wells per experiment.
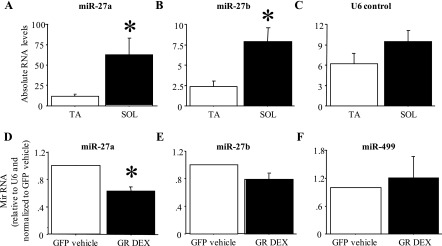
MiR-27a and b expression is greater in fast muscles and is decreased by Dex treatment.
Figure 8, A–C, shows the absolute levels of miR-27a, miR-27b, and a U6 control RNA in the fast-twitch TA and the slow-twitch Sol. MiR-27a and b RNA levels were significantly (approximately 5- and 3-fold) greater in Sol than in TA (Fig. 8, A and B); levels of the control RNA U6 were ∼50% greater in Sol than in TA but this was not significant (Fig. 8C). Conversely, 48 h of Dex treatment significantly decreased expression of miR-27a by 40–60% and decreased expression of miR-27b by ∼20%, though the latter was not significant (Fig. 8, D and E). Expression of miR-499 was not significantly affected by Dex treatment of C2C12 myotubes (Fig. 8F).
Fig. 8.
Differential expression of miR-27a and b in fast and slow muscles and in response to Dex treatment in vitro. A–C: expression of miR-27a (A), miR-27b (B), and U6 control RNA (C) in the TA and Sol muscle. Both miR-27a and b were expressed at a significantly higher level in Sol compared with TA. Bars represent means ± SE for n = 4 muscles each. *Significantly different from TA, P < 0.05. D–F: expression of miR-27a (D), miR-27b (E), and miR-499 (F) in GFP-cotransfected vehicle-treated and GR-cotransfected Dex-treated C2C12 myotubes. MiR-27a but not miR-27b was expressed at a significantly lower level in GR-cotransfected Dex-treated myotubes. Bars represent means ± SE for n = 3 different experiments. *Significantly different from GFP-cotransfected, vehicle-treated myotubes, P < 0.05.
MiR-27a and C9orf3 promoters are activated by calcineurin.
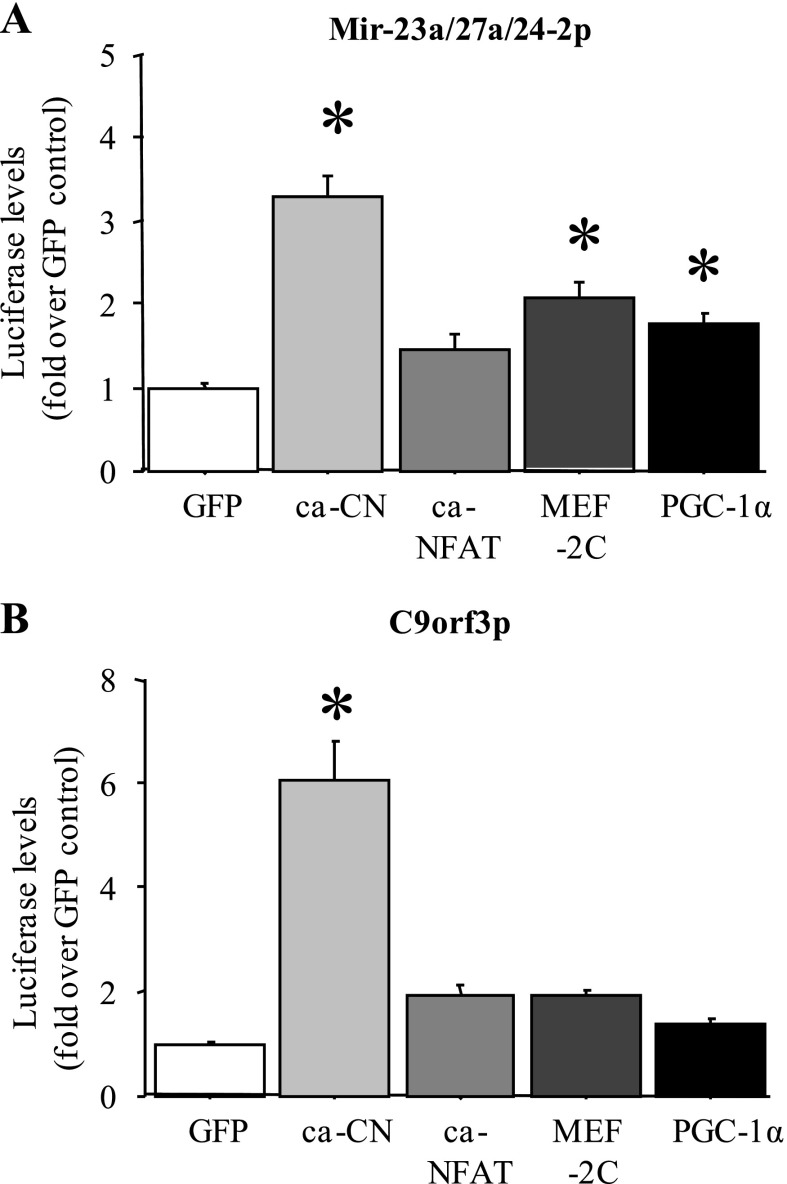
Because miR-27a and b RNA levels were significantly greater in slow vs. fast muscles, we examined whether transcription of these genes might be regulated by slow muscle-enriched signaling molecules and transcription factors. As shown in Fig. 9A, cotransfection of either the mouse miR-23a/27a/24–2 upstream region or the C9orf3 upstream promoter region, which governs expression of the C9orf3 gene within which the miR-23b/27b/24–1 cluster is located, with a constitutively active calcineurin construct resulted in a significant increase in luciferase activity relative to the cytomegalovirus (CMV)-GFP-transfected control in C2C12 myotubes in vitro (Fig. 9, A and B). In addition, cotransfection of the mouse miR-23a/27a/24–2 upstream region with either a CMV-myocyte enhancer-factor-2C (MEF-2C) or a peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) expression construct modestly but significantly increased luciferase activity in C2C12 myotubes, while a constitutively active NFATc3 construct had no significant effect. Cotransfection of C2C12 myotubes with the constitutively active NFATc3, wild-type MEF-2C, or PGC-1α construct had no significant effect on activity of the C9orf3 promoter (Fig. 9B).
Fig. 9.
MiR-27a and b promoters are responsive to slow-twitch-associated signaling factors. Activity of the mouse miR-27a (A) and mouse C9orf3 (B) upstream promoter regions in C2C12 myotubes with cotransfection with a GFP control plasmid, a constitutively active calcineurin (ca-CN) plasmid, a constitutively active nuclear factor of activated T cells (ca-NFAT3) plasmid, a wild-type myocyte enhancer-factor-2C (MEF-2C) plasmid, and a wild-type peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) plasmid. Cotransfection with the constitutively active calcineurin construct increased activity of both the miR-27a and the C9orf3 promoters, and MEF-2C and PGC-1α cotransfection increased activity of the miR-27a promoter construct. Bars represent means ± SE for n = 3 different experiments with 6 wells per experiment. *Significantly different from GFP-cotransfected control for each promoter, P < 0.05.
DISCUSSION
A key finding that led us to explore the contribution of posttranscriptional mechanisms to the regulation of MSTN expression was our inability to demonstrate a significant effect of glucocorticoids on MSTN promoter activity in vitro. As shown in Fig. 3, we observed only a modest increase in MSTN promoter activity with GR cotransfection alone, and a small but significant decrease in MSTN promoter activity in response to Dex treatment and cotransfection with a GR expression plasmid. This was true regardless of the length of the MSTN promoter construct used (ranging from >2,400 to ∼300 bp in length; Fig. 3, B–D) or the concentration or duration of Dex treatment used (data not shown) and is in direct contrast to the effects of Dex alone or with GR cotransfection on activity of a glucocorticoid “sensor” construct containing multimerized glucocorticoid response elements or GREs (Fig. 3A). It is not clear why our results differ from those of Ma et al. (39) and Du et al (20). However, the effects reported by both of these groups were also relatively modest, ranging from 1.4- to 2-fold (20, 39). The modest ∼1.5-fold increase in MSTN promoter activity with GR transfection alone for 48 h in the present study is therefore not at odds with the findings of these other studies, and while it is not inconceivable that transcriptional mechanisms contribute to differences in MSTN expression in this and other models, it seems likely that these modest effects are not of sufficient magnitude to completely explain the dramatic differences in MSTN expression in certain states. Moreover, it is not clear why cotransfection with a GR expression plasmid combined with Dex treatment actually decreased MSTN promoter activity, because this is completely at odds with the results of Du et al. and Ma et al. It is possible that species differences may account for this difference, since Du et al. examined the sheep MSTN promoter, Ma et al. examined the human MSTN promoter, and the present work examined the mouse MSTN promoter. Alternatively, it is possible that glucocorticoids have divergent effects on MSTN transcription and posttranscriptional regulation.
Moreover, we (1, 2) and others (10) have demonstrated that MSTN mRNA levels differ between fast-twitch and slow-twitch hindlimb muscles. The functional implications of this difference are not well characterized, but we recently demonstrated that the fast-twitch TA is more responsive to food deprivation-induced muscle atrophy (5) and thus differential expression and responsiveness of the MSTN gene in fast-twitch muscles may contribute to muscle-specific forms of muscle atrophy. In the present study, when MSTN pre-mRNA was quantified in fast vs. slow skeletal muscles and from control and Dex-treated skeletal muscle and C2C12 myotubes, only a modest and nonsignificant difference was observed across the different conditions, despite the fact that mature MSTN mRNA levels differed significantly by nearly 10-fold between fast and slow muscles and >2-fold between vehicle and Dex-treated myotubes in vitro (Figs. 1 and 2). Again, while this does not preclude transcriptional contributions to the regulation of MSTN expression, it does suggest that other mechanisms may also contribute to the differential expression of MSTN in these states. In contrast to the present results, we recently reported a significant increase in MSTN pre-mRNA that was nearly identical to the increase in mature MSTN mRNA in skeletal muscle in response to food deprivation (6), which demonstrates that transcription of the MSTN gene can be increased during certain physiological states. We further showed that this increase is glucocorticoid-dependent and appears to act via a C/EBP-dependent increase in MSTN transcription (6). This may seem at odds with the results of the present study, but it is likely that additional factors altered by food deprivation contribute to the transcriptional regulation of MSTN by glucocorticoids/C/EBPs during this state. Furthermore, we have not ruled out a contribution of miR-27-dependent posttranscriptional mechanisms during food deprivation, and it is thus possible that both transcriptional and posttranscriptional mechanisms contribute to the increase in MSTN expression during food deprivation.
Nevertheless, because we were unable to conclusively demonstrate that transcriptional changes were solely responsible for the differences in MSTN expression between fast and slow and between vehicle and Dex-treated muscle cells in the present study, we therefore explored the potential contribution of posttranscriptional mechanisms on the regulation of MSTN expression during these states. Examination of the MSTN 3′-UTR revealed a number of AUUUA elements, many of which were conserved across mice and humans (Fig. 4). AUUUA elements are commonly found in cytokine and other short-lived mRNAs and regulate 3′ to 5′ endonuclease degradation of mRNAs (25). We are aware of only one other example of posttranscriptional regulation of mRNA stability of a transforming growth family-β (TGF-β) family member through AUUUA elements, which is the regulation of BMP-2 mRNA stability by tumor necrosis factor-α (24). Members of the TGF-β family such as MSTN have rapid and potent effects on cell growth and differentiation similar to the effects cytokines have on their target tissues, and it is not too surprising that their expression may be regulated in a similar fashion.
We then examined the potential role of microRNAs in the regulation of MSTN expression for two reasons. First, microRNA regulation of skeletal muscle gene expression and cell proliferation, differentiation, and growth has been well established (63). Second, bioinformatic analysis revealed several putative microRNA-binding sites in the MSTN 3′-UTR, some of which showed great promise as candidates for further examination based on their conservation across a wide range of species and their predicted affinity for microRNA interaction. We chose to focus on miR-27a and b for several reasons. First, the putative miR-27ab site was identified using two separate microRNA prediction sites. In addition, the total context score of the miR-27ab site was one of the highest among the putative microRNA sites, while MSTN ranks highly as a putative target of miR-27a and b, meaning that the miR-27 recognition sequence is among the strongest predicted for MSTN while MSTN represents one of the most likely potential candidates of miR-27 regulation. Finally, expression of both miR-27a and b was higher than that of most miRs in C2C12 myotubes, and while not highly expressed relative to other microRNAs in adult TA muscle, was enriched in skeletal muscle relative to other tissues (Fig. 6).
MiR-27a and b are microRNAs associated with homologous microRNA clusters, the miR-23a/27a/24–2 and the 23b/27b/24–1 clusters found on human chromosomes 19 and 9, respectively. They both have the same “seed” sequence, UGACACU, which recognizes complementary sequences in the UTRs of specific genes, including MSTN. Previously published data support the hypothesis that miR-27a and b have profound effects on cell proliferation, differentiation, and/or survival. For example, both miR-27a and b appear to act as oncogenes that promote cell proliferation, and while other mRNA species, including those for FoxO1, prohibitin, and the tumor suppressor myt-1 (15, 29, 38, 66), have been implicated in this effect in non-muscle cells, miR-27-mediated downregulation of MSTN may also contribute to this as well, because MSTN functions as a “stasis factor” in skeletal muscle, inhibiting the proliferation, differentiation, and death of myogenic precursors (50, 51, 60) and protein accretion in differentiated muscle cells (60). In addition, members of the miR23a-27a-24–2 cluster can induce both caspase dependent and caspase-independent apoptosis in human embryonic kidney cells, at least in part by reducing expression of the prosurvival factor fas-associated death domain protein (FADD) (11). MSTN is a potent inhibitor of myogenic cell apoptosis (50), and again MSTN may be part of a suite of mRNAs downregulated by miR-27a and b to promote cell proliferation under growth-favorable conditions and cell differentiation and/or death under growth-unfavorable ones.
Recent evidence has suggested that members of the miR-23a and miR-23b microRNA clusters may inhibit TGF-β signaling on multiple levels. Upregulation of the miR-23a-27a-24–2 cluster blunts the tumor-suppressive activity of TGF-β in human hepatic carcinoma cells (31), while members of the miR-23b-37b-24–1 cluster appear to target SMADs for degradation in liver cells (52). Thus induction of either microRNA cluster may represent a common mechanism for attenuating signaling of TGF-β family members, including MSTN.
Our data confirm that MSTN is indeed a target of miR-27 signaling in vitro. Activity of a MSTN 3′-UTR-luciferase reporter construct was significantly attenuated in both myoblasts and myotubes by cotransfection with a miR-27b expression plasmid. This dampening effect was specific for MSTN, because cotransfection of a p110α 3′-UTR-luciferase reporter construct with a miR-27b expression plasmid had no significant effect in muscle cells, and it was also specific for miR-27b, because cotransfection of the MSTN 3′-UTR-luciferase reporter construct with an expression plasmid for miR-124, which is not predicted to have interactions with the MSTN 3′-UTR, had no significant effect on luciferase levels. We have not yet tested whether miR-27a cotransfection also affects MSTN 3′-UTR activity, since at present we do not have a mouse miR-27a expression plasmid. But given that both miR-27a and b recognize the same seed sequence, it is likely that it has qualitatively similar effects on MSTN expression, though these may be altered somewhat by differences in processing and/or seed affinity between the two miRs. In addition, our mutation data strongly suggest that the miR-27 recognition sequence binds to factors (presumably miR-27a or b) that depress MSTN 3′-UTR activity in normal C2C12 myotubes under basal states, and moreover that the inhibitory effects of miR-27b cotransfection on MSTN 3′-UTR activity depend critically on this sequence. Finally, our data suggest that miR-27 acts at least in part by decreasing MSTN mRNA stability in vitro. Together, these data strongly support a specific role for miR-27b in the posttranscriptional regulation of MSTN expression, and, in particular, our data suggest that miR-27a and b influence MSTN mRNA stability. However, miRs can also regulate gene expression via translational repression independent of their effects on mRNA stability (12), and it is therefore possible that miR-27a and b might also regulate expression of MSTN or other skeletal muscle genes in this manner as well.
The effect size of miR-27b cotransfection was relatively modest, decreasing MSTN UTR activity 30–60%, depending on the MSTN reporter construct and the cells (myoblasts vs. myotubes). However, this is identical to the effect sizes reported for miR transient cotransfection by others (17). Moreover, because regulation of mRNA stability is downstream of transcriptional regulation, it is possible that miR-27a and b, as well as other microRNA-dependent and -independent mechanisms regulating MSTN mRNA stability, may act as a “gatekeeper” on transcriptional changes, potentiating or attenuating their effects on overall MSTN mRNA levels. Future work will need to explore the interplay between transcriptional and posttranscriptional mechanisms in regulating MSTN expression in muscle cells to further define the relationship between these two processes.
Recently, evidence has accumulated supporting a role for other microRNAs in the regulation of MSTN expression. Transgenic overexpression of miR-208a in the heart suppresses the expression of several transcripts, including MSTN (9). Essential amino acids promote muscle hypertrophy and induce greater expression of miR-499, -208b, -23a, -1, and -206, while suppressing MSTN expression (19); more recent evidence has directly implicated miR-499 in downregulation of MSTN expression in vitro (8). Moreover, a naturally occurring mutation in the 3′-UTR of the sheep MSTN gene creates a miR-206 site causing translational inhibition of MSTN expression, resulting in more muscular sheep (16). Together with the present results, these data further support the contention that posttranscriptional regulation of MSTN by microRNA-dependent mechanisms is a major determinant of MSTN expression.
Several recent studies have also confirmed that microRNA expression can be regulated by glucocorticoids, consistent with the results of the present study. Expression of miR-132 and miR-17 is suppressed by glucocorticoids in neurons and in acute lymphoblastic leukemias, respectively (34, 47), similar to the suppression of miR-27 expression by glucocorticoid treatment in C2C12 myotubes reported here. Together, these data are consistent with previous work demonstrating that glucocorticoids can regulate gene expression via regulation of mRNA stability (33, 56) as well as through more traditional transcriptional mechanisms (7).
We report here that both miR-27a and b display slow muscle-specific expression complementary to that of MSTN which may explain at least in part the difference in MSTN expression between fast and slow skeletal muscles (1, 2, 10). MSTN promoter activity also shows fast-specific expression in vivo (53), and thus the fast-specific expression of MSTN may arise as a consequence of both transcriptional and posttranscriptional mechanisms. The relationship between MSTN and fiber type is complex, because MSTN is both expressed in a fiber-specific manner (53) and in turn influences fiber specificity (30), and MSTN regulation by miR-27a and b may therefore reflect a complex feedback circuit designed to establish and then maintain fiber specificity in adult muscle. A similar feed-forward circuit appears to exist between other microRNAs and fiber-specific gene products, most notably miR-208 and miR-499, which are also enriched in slow skeletal muscle relative to fast and which activate slow and repress fast myosin heavy chain isoform expression (61). MiR-206 also shows slow muscle enrichment (41), and we have shown that its expression is decreased during the slow-to-fast fiber type shift accompanying spaceflight-induced unloading (4).
Dex treatment also significantly decreased expression of miR-27a in C2C12 myotubes. MiR-27a and b RNA levels in the vehicle and Dex-treated TA muscle were not measured due to lack of RNA, and thus it is not possible to know whether they show a similar effect in vivo. However, this raises an interesting point as to whether changes in miR-27 expression in the TA might have biologically meaningful effects on MSTN (or other target gene) expression given its low expression in TA relative to Sol. But while miR-27a/b expression is lower in the TA relative to the Sol, they are still expressed at levels much higher than that observed in other tissues, including kidney, lung, and liver, as shown in Fig. 5C. Thus we feel that levels of miR-27a and/or b, while lower in the TA than in the Sol, are still sufficiently robust that changes in their expression are likely to have an effect on MSTN expression, though this still needs to be confirmed empirically. Moreover, changes in expression/activity of critical regulatory molecules can have profound effects even if their overall expression is not particularly high; indeed, most signal transduction proteins (kinases, transcription factors) are typically present in small quantities but changes in their expression/activity can still have profound effects on downstream functions. Thus we feel that the modulations in miR-27a/b expression reported here could be reasonably construed to have effects of biological significance in these models.
Finally, we demonstrate that promoters for the miR-23a-27a-24-2 cluster and C9orf3, the gene within which the miR-23b-27b-24-1 cluster resides, are responsive to calcineurin signaling in C2C12 myotubes, and activity of the miR-23a-27a-24-2 cluster promoter is also modestly but significantly upregulated by cotransfection with expression constructs for the PGC-1α and MEF-2 family of transcription factors (Fig. 9B). Calcineurin, PGC-1α, and MEF-2 have been strongly implicated in slow-muscle-specific gene expression (14, 37, 64). Thus at least part of the difference in expression of miR-27a and b between slow and fast muscles may be transcriptionally determined. However, one caveat needs to be mentioned with regard to the miR-23b-27b-24-1 cluster. First, while there has been some demonstration that miR-27b and C9orf3 RNA levels respond identically to some signals and thus may be regulated coordinately (22), other evidence suggests that each miR in this cluster may have its own separate upstream enhancer (58). We have not yet tested whether the miR-27b upstream regulatory region is responsive to slow-specific signals or glucocorticoid signaling but it may be a more proximal determinant of miR-27b expression. Lastly, preliminary tests of the effects of Dex treatment and GR cotransfection on activity of either the miR-27a or C9orf3 promoters were equivocal, with some experiments showing a significant reduction in activity of both promoters and others showing no effect (data not shown). It is therefore not clear at present whether glucocorticoids act via a direct transcriptional mechanism involving calcineurin or PGF-1α or via other posttranscriptional mechanisms (such as microRNA processing) to repress expression of miR-27a.
GRANTS
D. L. Allen was supported by National Institutes of Health Grant K01 AR-050505-01 for part of this work.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
REFERENCES
- 1. Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol 292: C188–C199, 2007. [DOI] [PubMed] [Google Scholar]
- 2. Allen DL, Cleary AS, Speaker KJ, Lindsay SF, Uyenishi J, Reed JM, Madden MC, Mehan RS. Myostatin, activin receptor IIb, and follistatin-like-3 gene expression are altered in adipose tissue and skeletal muscle of obese mice. Am J Physiol Endocrinol Metab 294: E918–E927, 2008. [DOI] [PubMed] [Google Scholar]
- 3. Allen DL, Du M. Comparative functional analysis of the cow and mouse myostatin genes reveals novel regulatory elements in their upstream promoter regions. Comp Biochem Physiol B Biochem Mol Biol 150: 432–439, 2008. [DOI] [PubMed] [Google Scholar]
- 4. Allen DL, Bandstra ER, Harrison BC, Thorng S, Stodieck LS, Kostenuik PJ, Morony S, Lacey DL, Hammond TG, Leinwand LL, Argraves WS, Bateman TA, Barth JL. Effects of spaceflight on murine skeletal muscle gene expression. J Appl Physiol 106: 582–595, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allen DL, Cleary AS, Lindsay SF, Loh AS, Reed JM. Myostatin expression is increased by food deprivation in a muscle-specific manner and contributes to muscle atrophy during prolonged food deprivation in mice. J Appl Physiol 109: 692–701, 2010. [DOI] [PubMed] [Google Scholar]
- 6. Allen DL, Cleary AS, Hanson AM, Lindsay SF, Reed JM. CCAAT/enhancer binding factor-δ expression is increased in fast skeletal muscle by food deprivation and regulates myostatin transcription in vitro. Am J Physiol Regul Integr Comp Physiol (September 15, 2010). doi:10.1152/ajpregu.00247.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beato M. Gene regulation by steroid hormones. Cell 56: 335–344, 1989. [DOI] [PubMed] [Google Scholar]
- 8. Bell ML, Buvoli M, Leinwand LA. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol Cell Biol 30: 1937–1945, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 119: 2772–2786, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlson CJ, Booth FW, Gordon SE. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases during hindlimb unloading. Am J Physiol Regul Integr Comp Physiol 277: R601–R606, 1999. [DOI] [PubMed] [Google Scholar]
- 11. Chhabra R, Adlakha YK, Hariharan M, Scaria V, Saini N. Upregulation of miR-23a-27a-24–2 cluster induces caspase-dependent and -independent apoptosis in human embryonic kidney cells. PLoS One 4: e5848, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 21: 452–460, 2009. [DOI] [PubMed] [Google Scholar]
- 13. Chen Y, Cao L, Ye J, Zhu D. Upregulation of myostatin gene expression in streptozotocin-induced type 1 diabetes mice is attenuated by insulin. Biochem Biophys Res Commun 388: 112–116, 2009. [DOI] [PubMed] [Google Scholar]
- 14. Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev 12: 2499–2509, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chintharlapalli S, Papineni S, Abdelrahim M, Abudayyeh A, Jutooru I, Chadalapaka G, Wu F, Mertens-Talcott S, Vanderlaag K, Cho SD, Smith R, 3rd, Safe S. Oncogenic microRNA-27a is a target for anticancer agent methyl 2-cyano-3,11-dioxo-18beta-olean-1,12-dien-30-oate in colon cancer cells. Int J Cancer 125: 1965–1974, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibé B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38: 813–818, 2006. [DOI] [PubMed] [Google Scholar]
- 17. Crist CG, Montarras D, Pallafacchina G, Rocancourt D, Cumano A, Conway SJ, Buckingham M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc Natl Acad Sci USA 106: 13383–13387, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dasarathy S, Muc S, Hisamuddin K, Edmison JM, Dodig M, McCullough AJ, Kalhan SC. Altered expression of genes regulating skeletal muscle mass in the portacaval anastomosis rat. Am J Physiol Gastrointest Liver Physiol 292: G1105–G1113, 2007. [DOI] [PubMed] [Google Scholar]
- 19. Drummond MJ, Glynn EL, Fry CS, Dhanani S, Volpi E, Rasmussen BB. Essential amino acids increase microRNA-499, -208b, and -23a and downregulate myostatin and myocyte enhancer factor 2C mRNA expression in human skeletal muscle. J Nutr 139: 2279–2284, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du R, An XR, Chen YF, Qin J. Some motifs were important for myostatin transcriptional regulation in sheep (Ovis aries). J Biochem Mol Biol 40: 547–553, 2007. [DOI] [PubMed] [Google Scholar]
- 21. Elferink CJ, Reiners JJ., Jr Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. Biotechniques 20: 470–477, 1996. [DOI] [PubMed] [Google Scholar]
- 22. Feng J, Iwama A, Satake M, Kohu K. MicroRNA-27 enhances differentiation of myeloblasts into granulocytes by post-transcriptionally downregulating Runx1. Br J Haematol 145: 412–423, 2009. [DOI] [PubMed] [Google Scholar]
- 23. Frost RA, Nystrom GJ, Jefferson LS, Lang CH. Hormone, cytokine, and nutritional regulation of sepsis-induced increases in atrogin-1 and MuRF1 in skeletal muscle. Am J Physiol Endocrinol Metab 292: E501–E512, 2007. [DOI] [PubMed] [Google Scholar]
- 24. Fukui N, Ikeda Y, Ohnuki T, Hikita A, Tanaka S, Yamane S, Suzuki R, Sandell LJ, Ochi T. Pro-inflammatory cytokine tumor necrosis factor-alpha induces bone morphogenetic protein-2 in chondrocytes via mRNA stabilization and transcriptional up-regulation. J Biol Chem 281: 27229–27241, 2006. [DOI] [PubMed] [Google Scholar]
- 25. Gingerich TJ, Feige JJ, LaMarre J. AU-rich elements and the control of gene expression through regulated mRNA stability. Anim Health Res Rev 5: 49–63, 2004. [DOI] [PubMed] [Google Scholar]
- 26. Grimson A, Farh KKH, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA Targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, Fries R, Hanset R, Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17: 71–74, 1997. [DOI] [PubMed] [Google Scholar]
- 28. Gou Q, Liu CH, Ben-Av P, Hla T. Dissociation of basal turnover and cytokine-induced transcript stabilization of the human cyclooxygenase-2 mRNA by mutagenesis of the 3′-untranslated region. Biochem Biophys Res Commun 242: 508–512, 1998. [DOI] [PubMed] [Google Scholar]
- 29. Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 284: 23204–23216, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hennebry A, Berry C, Siriett V, O'Callaghan P, Chau L, Watson T, Sharma M, Kambadur R. Myostatin regulates fiber-type composition of skeletal muscle by regulating MEF2 and MyoD gene expression. Am J Physiol Cell Physiol 296: C525–C534, 2009. [DOI] [PubMed] [Google Scholar]
- 31. Huang S, He X, Ding J, Liang L, Zhao Y, Zhang Z, Yao X, Pan Z, Zhang P, Li J, Wan D, Gu J. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer 123: 972–978, 2008. [DOI] [PubMed] [Google Scholar]
- 32. Huey KA, Haddad F, Qin AX, Baldwin KM. Transcriptional regulation of the type I myosin heavy chain gene in denervated rat soleus. Am J Physiol Cell Physiol 284: C738–C748, 2003. [DOI] [PubMed] [Google Scholar]
- 33. Ing NH. Steroid hormones regulate gene expression posttranscriptionally by altering the stabilities of messenger RNAs. Biol Reprod 72: 1290–1296, 2005. [DOI] [PubMed] [Google Scholar]
- 34. Kawashima H, Numakawa T, Kumamaru E, Adachi N, Mizuno H, Ninomiya M, Kunugi H, Hashido K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience 165: 1301–1311, 2010. [DOI] [PubMed] [Google Scholar]
- 35. Lang CH, Silvis C, Nystrom G, Frost RA. Regulation of myostatin by glucocorticoids after thermal injury. FASEB J 15: 1807–1809, 2001. [DOI] [PubMed] [Google Scholar]
- 36. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993. [DOI] [PubMed] [Google Scholar]
- 37. Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 418: 797–801, 2002. [DOI] [PubMed] [Google Scholar]
- 38. Liu T, Tang H, Lang Y, Liu M, Li X. MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett 273: 233–242, 2009. [DOI] [PubMed] [Google Scholar]
- 39. Ma K, Mallidis C, Artaza J, Taylor W, Gonzalez-Cadavid N, Bhasin S. Characterization of 5′-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. Am J Physiol Endocrinol Metab 281: E1128–E1136, 2001. [DOI] [PubMed] [Google Scholar]
- 40. Ma K, Mallidis C, Bhasin S, Mahabadi V, Artaza J, Gonzalez-Cadavid N, Arias J, Salehian B. Glucocorticoid-induced skeletal muscle atrophy is associated with upregulation of myostatin gene expression. Am J Physiol Endocrinol Metab 285: E363–E371, 2003. [DOI] [PubMed] [Google Scholar]
- 41. McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 102: 306–313, 2007. [DOI] [PubMed] [Google Scholar]
- 42. McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94: 12457–12461, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387: 83–90, 1997. [DOI] [PubMed] [Google Scholar]
- 44. Menconi M, Gonnella P, Petkova V, Lecker S, Hasselgren PO. Dexamethasone and corticosterone induce similar, but not identical, muscle wasting responses in cultured L6 and C2C12 myotubes. J Cell Biochem 105: 353–364, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mosher DS, Quignon P, Bustamante CD, Sutter NB, Mellersh CS, Parker HG, Ostrander EA. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 3: e79, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Plant PJ, Brooks D, Faughnan M, Bayley T, Bain J, Singer L, Correa J, Pearce D, Binnie M, Batt J. Cellular markers of muscle atrophy in chronic obstructive pulmonary disease (COPD). Am J Respir Cell Mol Biol 42: 461–471, 2009. [DOI] [PubMed] [Google Scholar]
- 47. Rainer J, Ploner C, Jesacher S, Ploner A, Eduardoff M, Mansha M, Wasim M, Panzer-Grümayer R, Trajanoski Z, Niederegger H, Kofler R. Glucocorticoid-regulated microRNAs and mirtrons in acute lymphoblastic leukemia. Leukemia 23: 746–752, 2009. [DOI] [PubMed] [Google Scholar]
- 48. Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA 103: 8721–8726, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reisz-Porszasz S, Bhasin S, Artaza JN, Shen R, Sinha-Hikim I, Hogue A, Fielder TJ, Gonzalez-Cadavid NF. Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of myostatin. Am J Physiol Endocrinol Metab 285: E8768–E8788, 2003. [DOI] [PubMed] [Google Scholar]
- 50. Ríos R, Carneiro I, Arce VM, Devesa J. Myostatin regulates cell survival during C2C12 myogenesis. Biochem Biophys Res Commun 280: 561–566, 2001. [DOI] [PubMed] [Google Scholar]
- 51. Ríos R, Carneiro I, Arce VM, Devesa J. Myostatin is an inhibitor of myogenic differentiation. Am J Physiol Cell Physiol 282: C993–C999, 2002. [DOI] [PubMed] [Google Scholar]
- 52. Rogler CE, Levoci L, Ader T, Massimi A, Tchaikovskaya T, Norel R, Rogler LE. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology 50: 575–584, 2009. [DOI] [PubMed] [Google Scholar]
- 53. Salerno MS, Thomas M, Forbes D, Watson T, Kambadur R, Sharma M. Molecular analysis of fiber type-specific expression of murine myostatin promoter. Am J Physiol Cell Physiol 287: C1031–C1040, 2004. [DOI] [PubMed] [Google Scholar]
- 54. Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688, 2004. [DOI] [PubMed] [Google Scholar]
- 55. Schwerin M, Maak S, Hagendorf A, von Lengerken G, Seyfert HM. A 3′-UTR variant of the inducible porcine hsp70.2 gene affects mRNA stability. Biochim Biophys Acta 1578: 90–94, 2002. [DOI] [PubMed] [Google Scholar]
- 56. Staples KJ, Bergmann MW, Barnes PJ, Newton R. Evidence for post-transcriptional regulation of interleukin-5 by dexamethasone. Immunology 109: 527–535, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sultan KR, Henkel B, Terlou M, Haagsman HP. Quantification of hormone-induced atrophy of large myotubes from C2C12 and L6 cells: atrophy-inducible and atrophy-resistant C2C12 myotubes. Am J Physiol Cell Physiol 290: C650–C659, 2006. [DOI] [PubMed] [Google Scholar]
- 58. Sun F, Wang J, Pan Q, Yu Y, Zhang Y, Wan Y, Wang J, Li X, Hong A. Characterization of function and regulation of miR-24–1 and miR-31. Biochem Biophys Res Commun 380: 660–665, 2009. [DOI] [PubMed] [Google Scholar]
- 59. Sunnerhagen P. Cytoplasmatic post-transcriptional regulation and intracellular signalling. Mol Genet Genomics 277: 341–355, 2007. [DOI] [PubMed] [Google Scholar]
- 60. Taylor WE, Bhasin S, Artaza J, Byhower F, Azam M, Willard DH, Jr, Kull FC, Jr, Gonzalez-Cadavid N. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am J Physiol Endocrinol Metab 280: E221–E228, 2001. [DOI] [PubMed] [Google Scholar]
- 61. Van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17: 662–673, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang J, Pitarque M, Ingelman-Sundberg M. 3′-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem Biophys Res Commun 340: 491–497, 2006. [DOI] [PubMed] [Google Scholar]
- 63. Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol 21: 461–469, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, Williams RS. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J 19: 1963–1973, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yasuda S, Wada S, Arao Y, Kogawa M, Kayama F, Katayama S. Interaction between 3′ untranslated region of calcitonin receptor messenger ribonucleic acid (RNA) and adenylate/uridylate (AU)-rich element binding proteins (AU-rich RNA-binding factor 1 and Hu antigen R). Endocrinology 145: 1730–1738, 2004. [DOI] [PubMed] [Google Scholar]
- 66. Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of microRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol 76: 582–588, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.