Abstract
Organotypic cultures of primary human airway epithelial cells have been used to investigate the morphology, ion and fluid transport, innate immunity, transcytosis, infection, inflammation, signaling, cilia, and repair functions of this complex tissue. However, we do not know how closely these cultures resemble the airway surface epithelium in vivo. In this study, we examined the genome-wide expression profile of tracheal and bronchial human airway epithelia in vivo and compared it with the expression profile of primary cultures of human airway epithelia grown at the air-liquid interface. For comparison, we also investigated the expression profile of Calu-3 cells grown at the air-liquid interface and primary cultures of human airway epithelia submerged in nutrient media. We found that the transcriptional profile of differentiated primary cultures grown at the air-liquid interface most closely resembles that of in vivo airway epithelia, suggesting that the use of primary cultures and the presence of an air-liquid interface are important to recapitulate airway epithelia biology. We describe a high level of similarity between cells of tracheal and bronchial origin within and between different human donors, which suggests a very robust expression profile that is specific to airway cells.
Keywords: tracheal and bronchial epithelia, cell culture models
the airway epithelium lines the luminal surface of an arrayed system of cylinders with fractal geometry designed to conduct gases from the environment into the alveoli. This epithelium is complex both in cell type and function. Cell types include ciliated and nonciliated columnar cells, secretory cells, and basal cells, although this nomenclature might represent a simplified classification (14). The cells are arranged in a pseudostratified pattern with all cells contacting the basement membrane but only some reaching the luminal surface. Moreover, the barrier formed by the tight junctions defines two distinct compartments, apical and basolateral. Many of the functions attributed to airway epithelia require a small volume of liquid at the apical surface acting to separate the cells from luminal gases. Furthermore, the barrier formed by the tight junctions separate the basolateral compartment from the apical compartment, which is lined by a thin layer of fluid and constitutes the air-liquid interface.
The study of biological processes in epithelial cells in vivo can be limited by both their residence within the lungs and, in some cases, their interactions with the environment, extracellular matrix, and other cell types. Cell culture methods allow a reduction in these confounding variables. We routinely use differentiated primary human airway epithelia cultures grown at the air-liquid interface (13) based on methods initially developed by Yamaya et al. (20) and Whitcutt et al. (18). Under these conditions, cells develop tight junctions and a differentiation state similar to that reached in vivo, with ciliated, basal, and secretory cells. We also use primary airway cultures grown on plastic surfaces (1) submerged in nutrient media and transformed cell lines that can be grown both submerged and at the air-liquid interface, although they fail to represent all the airway cell types (10).
Morphological analysis has suggested similarities between in vivo and differentiated primary human airway epithelia cultures grown at the air-liquid interface (13). Here, we asked how closely the genome-wide expression profile of these cells matches the in vivo profile. We compare these conditions to the frequently used Calu-3 cell line grown at the air-liquid interface and to primary cultures of human airway epithelia grown on a plastic surface and submerged in nutrient media.
METHODS
Study population.
For the in vivo airway epithelia samples, subjects were evaluated in the Weill Medical College of Cornell University National Institutes of Health General Clinical Research Center. After signing an informed consent, a Cornell University Institutional Review Board-approved protocol was followed to assess the clinical status of study subjects. Pure populations of in vivo airway epithelium were obtained from eight healthy individuals as previously described (11). From each individual, both tracheal and bronchial brushings were obtained (n = 16; Table 1). Differential cell counts in all samples were within expected values; therefore, all samples were included in subsequent steps of analysis. For the in vitro samples, human donor lungs were obtained from eight individuals without primary lung diseases whose lungs were determined to be unsuitable for organ transplantation. The University of Iowa Institutional Review Board approved the use of human tissues. Mean age of study subjects in years is 45.1 ± 5.6 (95% confidence interval) for in vivo and 49.0 ± 13.06 (95% confidence interval) for in vitro samples. There were no differences in age (P = 0.65), sex (P = 1.00), race (P = 0.07), or smoker status (P = 1.00) between the two groups.
Table 1.
Differential cell counts of in vitro differentiated airway epithelia vs. in vivo airway epithelia
| Cell Type | In Vitro Airway Epithelia | In Vivo Airway Epithelia | P |
|---|---|---|---|
| Inflammatory | 0 | 0.5 ± 0.2 | 0.007 |
| Ciliated | 39.6 ± 3.9 | 45.7 ± 2.7 | 0.26 |
| Secretory | 25.4 ± 2.5 | 7.7 ± 0.8 | <0.001 |
| Basal | 26.5 ± 1.3 | 17.2 ± 1 | <0.001 |
| Other | 9.1 ± 2.3 | 28.9 ± 2.8 | <0.001 |
Values are means ± SE, n = 16. Differential cell counts of primary cultures of human airway epithelia (in vitro) were performed as described in Ref. 17 and compared with airway epithelia in vivo.
Primary cultures of human airway epithelia.
Primary cultures of human airway epithelia were grown at the air-liquid interface as described previously (13). From each donor, tracheal and bronchial epithelial cells were cultured separately (n = 16). Cells were used after 2 wk in culture, when complete differentiation is usually reached (21) and the transepithelial resistance is 700–1,200 Ω·cm2. Average (± SD) time in culture for cells included in this study was 27 ± 10 days. Typical cell yield is 750,000–800,000 per culture in a 0.6-cm2 surface. Epithelia were maintained in DMEM/F-12 with 1% penicillin-streptomycin, 50 mg/ml gentamicin, and 2% Ultroser G (Pall BioSepra, Cergy, France) in a humidified incubator with 5% CO2 at 37°C. Differential cell counts were performed as described in Ref. 17 on randomly chosen hematoxylin and eosin-stained sections of cultured cells.
Cultures of Calu-3 cells.
Calu-3 cells [American Type Culture Collection (ATCC) cat. no. HTB-55] were prepared and grown at the air-liquid interface and maintained in the same nutrient media and conditions as described above for primary cultures of human airway epithelia.
Microarray hybridizations.
Total RNA was extracted from all samples using TRIzol reagent (Invitrogen, Carlsbad, CA). Five micrograms of total RNA was processed using the Affymetrix GeneChip One-Cycle Target Labeling Kit (Affymetrix, Santa Clara, CA) following the manufacturer's protocols. Biotinylated cRNA was hybridized to a custom GeneChip Human Airway Array (HsAirway; Affymetrix). The HsAirway was composed of ∼23,000 probe sets derived from sequencing of cDNA libraries prepared from human lung, primary airway epithelial cells, and human alveolar macrophages (16, 19). The arrays were washed, stained, and scanned using the Affymetrix Fluidics Station 450 and Affymetrix GeneChip Scanner 3000. Global target intensity was set to 1,500. Prechip quality control criteria for samples were: clear 18S and 28S RNA bands with A260/280 ratio > 1.8, RNA integrity number (Agilent Bioanalyzer RIN) > 6, and cRNA amplification of >4-fold. Postchip quality control criteria required a visually defect-free chip image, scaling factor ≤ 10, >40% present calls, 3′-5′ ratio of the GAPDH gene < 3, and RNA degradation plot without significant degradation.
Data analysis.
Samples that passed prechip and postchip quality control criteria were included in data analysis. Raw data .cel files were imported into Partek Genomics Suite v. 6.4 for computation of robust multiarray analysis (RMA) expression values (12). The data were deposited in the Gene Expression Omnibus web site (http://www.ncbi.nlm.nih.gov/geo/) under GEO acc. no. GSE20502. Hierarchical clustering analysis was performed in GenePattern (http://www.broadinstitute.org/cancer/software/genepattern/) (15). RMA values generated in Partek were used to generate a .gct format file. The .gct file was used as the input file for the “HierarchicalClustering” module in GenePattern with Pearson correlation used as distance measure and pairwise complete linkage as clustering method. The resulting output files were used in the “HierarchicalClusteringViewer” module of GenePattern, and a .txt sample information file was used to label each sample with sample category, donor number, and tissue site (bronchus or trachea) as necessary. The resulting file was edited in the GNU Image Manipulation Program (GIMP) v. 2.6 (http://www.gimp.org/) for further annotation.
ANOVA was performed in Partek Genomics Suite. Data in the resulting spreadsheet containing P values and log2-transformed fold changes were used in Prism v. 5.0 for Macintosh to generate volcano plots. To perform gene ontology (GO) analysis, Partek Genomics Suite was used to generate a list containing probes that pass a threshold of false discovery rate of 0.01 (1%). Probes in this spreadsheet were ranked according to fold change from highest to lowest, generating two lists, one for genes with higher expression in vivo and one for genes with higher expression in vitro. The list was used in GOrilla (http://cbl-gorilla.cs.technion.ac.il/) (8, 9) to generate directed acyclic graphs of the GO terms enriched in the submitted gene list. The generated GO term lists were filtered to include only those with ≥5 genes enriched in the GO term.
RESULTS
Unsupervised hierarchical clustering of airway epithelial cells.
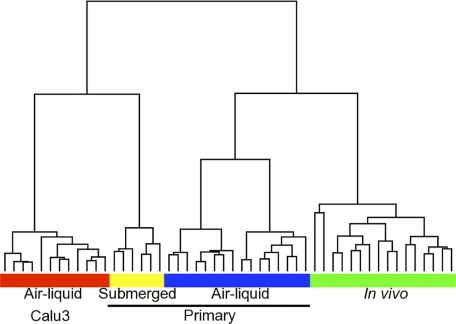
We obtained human in vivo airway epithelia from tracheal and bronchial brush biopsies (22). This technique has the advantage of being minimally invasive, safe, and well-tolerated by healthy volunteers. The cells obtained by this method are predominantly epithelial cells (Table 1). To compare the transcriptional profile of human in vivo airway epithelia from trachea and bronchus with differentiated primary human airway epithelia cultures, also from trachea and bronchus and grown at the air-liquid interface, we performed unsupervised hierarchical clustering. We also included the profile of Calu-3 cultures grown at the air-liquid interface and primary cultures submerged in nutrient media. We performed the clustering using Pearson dissimilarity as distance measure and complete linkage cluster analysis. Figure 1 shows that primary cultures grown at air-liquid interface cluster together with samples from in vivo airway epithelia. In contrast, samples from Calu-3 cells or primary submerged cultures are more dissimilar compared with in vivo airway epithelia.
Fig. 1.
Unsupervised hierarchical clustering of airway epithelial cells. RNA was extracted from samples of Calu-3 cells (n = 12; red box), in vitro airway epithelia submerged in nutrient media (n = 6; yellow box), in vitro differentiated airway epithelia (n = 16; blue box), and in vivo airway epithelia (n = 16; green box). Biotinylated cRNA was synthesized following the manufacturer's protocol and then hybridized to a custom GeneChip Human Airway Array (HsAirway; Affymetrix). Normalized data were analyzed in GenePattern (http://www.broadinstitute.org/cancer/software/genepattern/) using the “HierarchicalClustering” module with Pearson correlation as distance measure and pairwise complete linkage as clustering method.
The expression profile of tracheal and bronchial epithelia is similar both in vitro and in vivo.
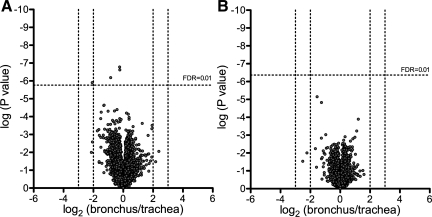
We asked whether the transcriptional profile of epithelia obtained from two distinct localizations in the airways (trachea or main bronchi) were different. We also investigated differentiated primary human airway cultures obtained from donor lungs, cells from either the trachea or 1st to 3rd generation bronchi. In vivo (Fig. 2A) and in vitro (Fig. 2B), tracheal and bronchial epithelia cultures were analyzed using a one-way ANOVA model. In both analyses, the volcano plots show similar transcriptional profiles. These data suggest that the transcriptional profile of tracheal and bronchial epithelia is similar and that this similarity is not explained by the culture conditions alone since we obtained comparable results in vivo.
Fig. 2.
One-way ANOVA comparing tissue from bronchus and trachea. Normalized microarray expression data from tracheal and bronchial samples of in vivo airway epithelia (A) and in vitro differentiated airway epithelia (B) were analyzed using 1-way ANOVA. Average probe fold change levels between bronchial and tracheal samples with their respective P values are displayed as volcano plots. FDR, false discovery rate.
Unsupervised hierarchical clustering of in vivo airway epithelia and in vitro differentiated airway epithelia.
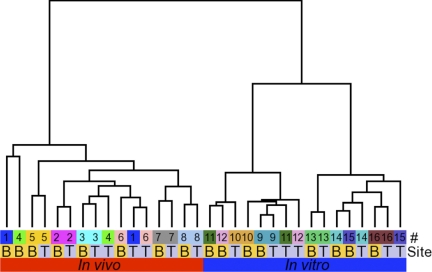
We hypothesized that interdonor variability in the transcriptional profile would result in grouping of samples based on donor in an unsupervised hierarchical clustering analysis of the samples. As a comparison, we contrasted the closeness of trachea and bronchus from the same donor. In vivo airway epithelia and in vitro differentiated airway epithelia were classified according to condition (in vitro or in vivo), tissue type (bronchus or trachea), and date of hybridization (to account for potential batch effects). We performed unsupervised hierarchical clustering using Pearson dissimilarity as distance measure and complete linkage cluster analysis. Contrary to our hypothesis, when we analyzed in vivo samples, three out of eight donor pairs (trachea-bronchus) did not cluster together (Fig. 3). Moreover, five out of eight pairs of in vitro donor bronchi-trachea did not cluster together. This suggests that the similarity between the transcriptional profiles of airway epithelia from different donors is very high. The hierarchical clustering also shows some batch effects in the in vitro samples that are dependent on hybridization date. These data suggest that normal human airway epithelia have an exceptionally constant transcriptional profile that sometimes prevails above interdonor and environmental variability.
Fig. 3.
Unsupervised hierarchical clustering of in vivo airway epithelia and in vitro differentiated airway epithelia. Normalized microarray expression data from in vitro differentiated airway epithelia (n = 16; blue box) and in vivo airway epithelia (n = 16; red box) were analyzed in GenePattern using the HierarchicalClustering module with Pearson correlation as distance measure and pairwise complete linkage as clustering method. Labels denote donor (#) and tracheal (T) or bronchial (B) source of tissue.
Differentially expressed genes between in vivo airway epithelia and in vitro differentiated airway epithelia.
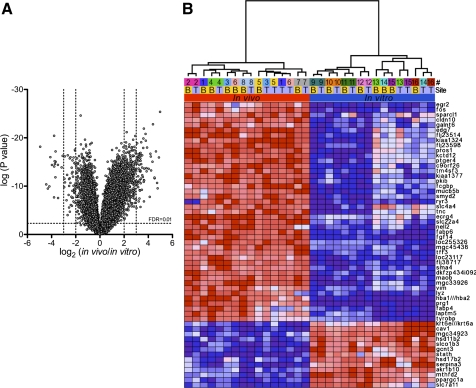
We speculated that the differences between the in vitro and in vivo conditions including available nutrients, different cell type composition, and exposure to the environment would result in differences in the transcriptional profile. In vivo airway epithelia and in vitro differentiated airway epithelia cultures were analyzed using a one-way ANOVA model. The volcano plot in Fig. 4A shows that 62 genes were expressed at lower levels and 391 genes at higher levels in vivo compared with in vitro when setting a threshold of >2-fold (log2) change and a false discovery rate (FDR) < 1%. We then plotted a heat map (Fig. 4B) comprising 56 genes that pass a threshold of >3-fold (log2) change and an FDR < 1% in the ANOVA comparison. This relatively small number of differentially expressed genes suggested that functional differences might be discerned from these data.
Fig. 4.
ANOVA for expression data from in vivo airway epithelia and in vitro differentiated airway epithelia. A: normalized microarray expression data from samples of in vivo airway epithelia and in vitro differentiated airway epithelia were analyzed using 1-way ANOVA. Average probe fold change levels between in vitro and in vivo samples with their respective P values are displayed as a volcano plot. Genes differentially expressed at a log2 fold change level > 3 with an FDR < 1% are displayed as a heat map in B. Labels denote donor (#) and tracheal (T) or bronchial (B) source of tissue.
GO analysis of differentially expressed genes between in vivo airway epithelia and in vitro differentiated airway epithelia.
GO is a collection of controlled terms describing the biology of a gene product in an organism. Analysis is based on the association of genes with GO terms related to selected functional processes, cell compartments, cell types, or user-defined categories. This facilitates interpretation of changes in a transcriptional profile in a biologically relevant context. GO analysis was performed using GOrilla (8, 9) as detailed in methods. Table 2 shows GO terms enriched in vivo, and Table 3 shows GO terms enriched in vitro. We found that in vivo epithelia had higher expression of genes related to cilia including multiple axonemal dynein (DNAH) genes bbs4, bbs5, alms, pkd1, ift172, lca5, and catsper2. This suggests that in vivo epithelia have more ciliated cells than differentiated primary airway epithelia cultures or that the expression levels of genes associated with cilia function and structure are upregulated in vivo (6, 7). Moreover, there were differences in genes involved in immune processes including: ptprc; complement-related components such as c1qa, c1qb, c6, c5ar1, and cfp; and cfd, mnda, tlr5, ccl4, alox5ap, nfkbiz, cxcl6, and cx3cr1. These genes are highly expressed in macrophages (3, 4), suggesting that as expected, the in vitro differentiated airway epithelia lack intraepithelial dendritic cells and macrophages. We also found that in vitro differentiated airway epithelia had higher expression of genes involved in metabolism and respiration such as ppargc1a, serpina3, acsl1, cav1, ppap2a, hsd17b2, g6pd, sdhd, far1, and multiple NADH dehydrogenase (ndufb) subcomplexes. These data suggest that either the nutrient media lacks or has too many of certain nutrients or growth factors. Finally, to our surprise, we found very few differences in genes related to ion transport, antimicrobial peptides (excluding lysozyme), surfactants, or cell adhesion molecules.
Table 2.
Gene ontology (GO) analysis of expression data from in vivo airway epithelia
| Biological Process | |||
|---|---|---|---|
| GO Term | Description | P | Enrichment (N, B, n, b) |
| GO:0006952 | Defense response | 1.60e−05 | 2.45 (2,804, 87, 394, 30) |
| GO:0002252 | Immune effector process | 7.92e−05 | 7.13 (2,804, 24, 131, 8) |
| GO:0048584 | Positive regulation of response to stimulus | 1.83e−04 | 7.49 (2,804, 20, 131, 7) |
| GO:0001539 | Ciliary or flagellar motility | 2.25e−04 | 4.16 (2,804, 7, 674, 7) |
| GO:0006470 | Protein amino acid dephosphorylation | 3.27e−04 | 4.19 (2,804, 21, 319, 10) |
| GO:0002253 | Activation of immune response | 4.25e−04 | 8.03 (2,804, 16, 131, 6) |
| GO:0009611 | Response to wounding | 4.45e−04 | 2.71 (2,804, 50, 372, 18) |
| GO:0007018 | Microtubule-based movement | 4.46e−04 | 2.91 (2,804, 25, 540, 14) |
| GO:0006469 | Negative regulation of protein kinase activity | 4.98e−04 | 6.17 (2,804, 17, 187, 7) |
| GO:0033673 | Negative regulation of kinase activity | 4.98e−04 | 6.17 (2,804, 17, 187, 7) |
| GO:0051348 | Negative regulation of transferase activity | 7.45e−04 | 5.83 (2,804, 18, 187, 7) |
| GO:0030705 | Cytoskeleton-dependent intracellular transport | 8.30e−04 | 2.69 (2,804, 29, 540, 15) |
| GO:0007266 | Rho protein signal transduction | 8.68e−04 | 3.43 (2,804, 7, 817, 7) |
| GO:0050778 | Positive regulation of immune response | 8.91e−04 | 7.13 (2,804, 18, 131, 6) |
| GO:0050896 | Response to stimulus | 8.96e−04 | 1.41 (2,804, 381, 501, 96) |
| Molecular Function | |||
|---|---|---|---|
| GO Term | Description | P | Enrichment (N, B, n, b) |
| GO:0003777 | Microtubule motor activity | 6.08e−05 | 3.30 (2,804, 22, 540, 14) |
| GO:0004871 | Signal transducer activity | 4.18e−04 | 1.48 (2,804, 265, 579, 81) |
| GO:0060089 | Molecular transducer activity | 4.18e−04 | 1.48 (2,804, 265, 579, 81) |
| GO:0017017 | MAPK tyrosine/serine/threonine phosphatase activity | 4.30e−04 | 7.32 (2,804, 6, 319, 5) |
| GO:0033549 | MAPK phosphatase activity | 4.30e−04 | 7.32 (2,804, 6, 319, 5) |
| GO:0022891 | Substrate-specific transmembrane transporter activity | 4.91e−04 | 1.56 (2,804, 100, 988, 55) |
| GO:0008201 | Heparin binding | 5.15e−04 | 4.32 (2,804, 8, 568, 7) |
| GO:0005539 | Glycosaminoglycan binding | 5.15e−04 | 4.32 (2,804, 8, 568, 7) |
| GO:0022857 | Transmembrane transporter activity | 5.90e−04 | 1.54 (2,804, 105, 988, 57) |
| GO:0043168 | Anion binding | 6.95e−04 | 5.21 (2,804, 12, 314, 7) |
| GO:0031404 | Chloride ion binding | 6.95e−04 | 5.21 (2,804, 12, 314, 7) |
| GO:0004888 | Transmembrane receptor activity | 7.50e−04 | 1.65 (2,804, 106, 770, 48) |
| GO:0016791 | Phosphatase activity | 7.71e−04 | 3.08 (2,804, 40, 319, 14) |
| GO:0004721 | Phosphoprotein phosphatase activity | 8.09e−04 | 3.58 (2,804, 27, 319, 11) |
| GO:0004725 | Protein tyrosine phosphatase activity | 8.19e−04 | 4.16 (2,804, 19, 319, 9) |
| GO:0042578 | Phosphoric ester hydrolase activity | 9.11e−04 | 2.61 (2,804, 58, 333, 18) |
| Cell Component | |||
|---|---|---|---|
| GO Term | Description | P | Enrichment (N, B, n, b) |
| GO:0005576 | Extracellular region | 1.06e−10 | 1.97 (2,804, 220, 550, 85) |
| GO:0030286 | Dynein complex | 5.33e−07 | 4.82 (2,804, 14, 499, 12) |
| GO:0005858 | Axonemal dynein complex | 5.13e−06 | 5.62 (2,804, 8, 499, 8) |
| GO:0044447 | Axoneme part | 5.13e−06 | 5.62 (2,804, 8, 499, 8) |
| GO:0005929 | Cilium | 4.29e−05 | 2.23 (2,804, 31, 935, 23) |
| GO:0005886 | Plasma membrane | 1.58e−04 | 1.30 (2,804, 339, 994, 156) |
| GO:0005615 | Extracellular space | 2.20e−04 | 2.01 (2,804, 62, 698, 31) |
| GO:0005875 | Microtubule-associated complex | 4.86e−04 | 2.78 (2,804, 28, 540, 15) |
| GO:0044421 | Extracellular region part | 6.00e−04 | 1.97 (2,804, 98, 480, 33) |
| GO:0044463 | Cell projection part | 9.47e−04 | 3.07 (2,804, 22, 499, 12) |
Data from 1-way ANOVA comparing expression data of in vivo airway epithelia were used to generate a list containing probes that pass a threshold of false discovery rate (FDR) of 0.01 (1%). Probes were ranked according to fold change from highest to lowest, generating 2 lists, 1 for genes with higher expression in vivo, and 1 for genes with higher expression in vitro. The lists were used in GOrilla for GO analysis, and the resulting GO term lists enriched in vivo (above) and in vitro (Table 3) are shown. Enrichment (N, B, n, b) is defined as follows: N, total number of genes; B, total number of genes associated with a specific GO term (“target set” and “background set”); n, number of genes in the target set; b, number of genes in the target set associated with a specific GO term; Enrichment = (b/n)/(B/N).
Table 3.
GO analysis of expression data from in vitro differentiated airway epithelia
| Biological Process | |||
|---|---|---|---|
| GO Term | Description | P | Enrichment (N, B, n, b) |
| GO:0055114 | Oxidation reduction | 1.56e−07 | 1.63 (2,816, 159, 979, 90) |
| GO:0019216 | Regulation of lipid metabolic process | 2.39e−06 | 30.09 (2,816, 12, 39, 5) |
| GO:0008544 | Epidermis development | 2.71e−06 | 6.12 (2,816, 21, 241, 11) |
| GO:0009888 | Tissue development | 6.11e−06 | 2.39 (2,816, 35, 843, 25) |
| GO:0031667 | Response to nutrient levels | 2.33e−05 | 33.13 (2,816, 10, 34, 4) |
| GO:0015849 | Organic acid transport | 3.03e−05 | 20.32 (2,816, 21, 33, 5) |
| GO:0046942 | Carboxylic acid transport | 3.03e−05 | 20.32 (2,816, 21, 33, 5) |
| GO:0006629 | Lipid metabolic process | 5.48e−05 | 5.42 (2,816, 136, 42, 11) |
| GO:0006091 | Generation of precursor metabolites and energy | 5.89e−05 | 1.67 (2,816, 101, 917, 55) |
| GO:0009991 | Response to extracellular stimulus | 6.07e−05 | 27.61 (2,816, 12, 34, 4) |
| GO:0022900 | Electron transport chain | 7.90e−05 | 1.86 (2,816, 54, 979, 35) |
| GO:0006461 | Protein complex assembly | 1.99e−04 | 2.16 (2,816, 55, 641, 27) |
| GO:0008202 | Steroid metabolic process | 2.94e−04 | 13.33 (2,816, 32, 33, 5) |
| GO:0048519 | Negative regulation of biological process | 3.54e−04 | 2.06 (2,816, 378, 116, 32) |
| GO:0008285 | Negative regulation of cell proliferation | 4.51e−04 | 4.59 (2,816, 42, 146, 10) |
| GO:0006099 | Tricarboxylic acid cycle | 4.57e−04 | 3.00 (2,816, 14, 737, 11) |
| GO:0051187 | Cofactor catabolic process | 4.57e−04 | 3.00 (2,816, 14, 737, 11) |
| GO:0046356 | Acetyl-CoA catabolic process | 4.57e−04 | 3.00 (2,816, 14, 737, 11) |
| GO:0009109 | Coenzyme catabolic process | 4.57e−04 | 3.00 (2,816, 14, 737, 11) |
| GO:0042127 | Regulation of cell proliferation | 5.97e−04 | 1.60 (2,816, 93, 945, 50) |
| GO:0048523 | Negative regulation of cellular process | 6.22e−04 | 2.03 (2,816, 370, 116, 31) |
| Molecular Function | |||
|---|---|---|---|
| GO Term | Description | P | Enrichment (N, B, n, b) |
| GO:0016491 | Oxidoreductase activity | 1.17e−07 | 1.59 (2,816, 183, 979, 101) |
| GO:0005342 | Organic acid transmembrane transporter activity | 5.46e−06 | 27.94 (2,816, 18, 28, 5) |
| GO:0046943 | Carboxylic acid transmembrane transporter activity | 5.46e−06 | 27.94 (2,816, 18, 28, 5) |
| GO:0004866 | Endopeptidase inhibitor activity | 1.01e−05 | 6.41 (2,816, 23, 191, 10) |
| GO:0030414 | Protease inhibitor activity | 1.01e−05 | 6.41 (2,816, 23, 191, 10) |
| GO:0022890 | Inorganic cation transmembrane transporter activity | 1.27e−05 | 2.19 (2,816, 52, 767, 31) |
| GO:0015078 | Hydrogen ion transmembrane transporter activity | 1.88e−05 | 2.02 (2,816, 44, 980, 31) |
| GO:0015077 | Monovalent inorganic cation transmembrane transporter activity | 3.98e−05 | 1.98 (2,816, 45, 980, 31) |
| GO:0004857 | Enzyme inhibitor activity | 1.56e−04 | 4.21 (2,816, 42, 191, 12) |
| GO:0004867 | Serine-type endopeptidase inhibitor activity | 1.90e−04 | 6.88 (2,816, 15, 191, 7) |
| GO:0016616 | Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | 2.02e−04 | 4.90 (2,816, 33, 174, 10) |
| GO:0009055 | Electron carrier activity | 3.24e−04 | 1.72 (2,816, 67, 979, 40) |
| GO:0016614 | Oxidoreductase activity, acting on CH-OH group of donors | 4.62e−04 | 4.50 (2,816, 36, 174, 10) |
| GO:0004129 | Cytochrome c oxidase activity | 4.91e−04 | 2.76 (2,816, 15, 817, 12) |
| GO:0016675 | Oxidoreductase activity, acting on heme group of donors | 4.91e−04 | 2.76 (2,816, 15, 817, 12) |
| GO:0016676 | Oxidoreductase activity, acting on heme group of donors, oxygen as acceptor | 4.91e−04 | 2.76 (2,816, 15, 817, 12) |
| GO:0015002 | Heme-copper terminal oxidase activity | 4.91e−04 | 2.76 (2,816, 15, 817, 12) |
| GO:0022891 | Substrate-specific transmembrane transporter activity | 8.65e−04 | 1.53 (2,816, 163, 734, 65) |
| GO:0022857 | Transmembrane transporter activity | 9.65e−04 | 1.50 (2,816, 179, 734, 70) |
| Cell Component | |||
|---|---|---|---|
| GO Term | Description | P | Enrichment (N, B, n, b) |
| GO:0044429 | Mitochondrial part | 7.34e−06 | 1.47 (2,816, 210, 984, 108) |
| GO:0005576 | Extracellular region | 1.37e−04 | 2.91 (2,816, 279, 66, 19) |
| GO:0005739 | Mitochondrion | 1.60e−04 | 1.31 (2,816, 326, 984, 149) |
| GO:0044455 | Mitochondrial membrane part | 3.66e−04 | 1.93 (2,816, 58, 781, 31) |
| GO:0001533 | Cornified envelope | 4.18e−04 | 8.35 (2,816, 7, 241, 5) |
Data from 1-way ANOVA comparing expression data of in vitro differentiated airway epithelia were used to generate a list containing probes that pass a threshold of FDR of 0.01 (1%). Probes were ranked according to fold change from highest to lowest, generating 2 lists, 1 for genes with higher expression in vivo, and 1 for genes with higher expression in vitro. The lists were used in GOrilla for GO analysis, and the resulting GO term lists enriched in vivo (Table 2) and in vitro (above) are shown. Enrichment (N, B, n, b) is defined as follows: N, total number of genes; B, total number of genes associated with a specific GO term (target set and background set); n, number of genes in the target set; b, number of genes in the target set associated with a specific GO term; Enrichment = (b/n)/(B/N).
DISCUSSION
The study of airway epithelia relies on human studies, animal models, organotypic culture models, and cell lines. However, as with any other model, fidelity, reproducibility, and simplicity have to be balanced with biological relevance. Here, we show that primary cultures of human airway epithelia share striking similarities in gene expression patterns with in vivo airway epithelia and that those similarities rely on both culture of primary nonimmortalized cells and the presence of an air-liquid interface.
GO analysis reveals that the differences between in vitro and in vivo airway epithelia are related to a few pathways that may represent differences in cell composition (Table 1). In particular, immune effector cells expected to be present in vivo and not in vitro resulted in marked differences in expression levels of genes related to inflammatory pathways. Similarly, a higher proportion of ciliated columnar cells in vivo might contribute to the increased expression levels of genes related to cilia in vivo. Of interest, mRNA of lysozyme, the first antimicrobial protein discovered in 1922, is highly expressed in human airway epithelia in vivo but is expressed at significantly lower levels in in vitro differentiated airway epithelia (5). Finally, the expression levels of genes related to some pathways appeared similar in vivo and in vitro. These included cell adhesion molecules, antimicrobial peptides, ion transport proteins, surfactant proteins, and others.
We found that airway epithelia from trachea and bronchi had very similar expression profiles. Since donors of in vivo airway epithelia were all nonsmoking normal volunteers, we speculated that differences in genetics would result in differentially expressed genes. A recent study compared the transcriptional profile of in vivo airway epithelia with in vitro airway epithelia from a single donor (7). Analysis of different donors for the in vitro airway epithelia in this study allowed us to compare the interdonor (samples from different donors) with intradonor variability (trachea vs. bronchus from the same donor). Moreover, the inclusion of Calu-3 cell lines and submerged airway epithelia allowed us to analyze interdonor differences in the context of more dramatic changes in genotype (Calu-3) and culture conditions (submerged airway epithelia). To our surprise, the transcriptional profiles failed to identify donors reproducibly. The localization of sample (tracheal vs. bronchial) and the donor seem to be of approximately equal importance as determinants of transcriptional profile variability. This suggests that in normal subjects, the airway epithelia are remarkably constant in their transcriptional profile. In this study, we speculated that genetic diversity within the in vitro airway epithelia group would best represent diversity within a population, but the high degree of similarity between samples suggests that using a small number of biological replicates to study the transcriptional profile of the airway is an acceptable approach. We predict that using the transcriptional profile to compare healthy subjects with subjects with disease should reliably identify differences between these two groups. However, as with any profiling study, caution must be exercised when using microarray analysis of tissue samples to study changes in transcriptional regulation in disease states. A change in the expression level of a gene or group of genes could represent either a change in regulation of a biological pathway or differences in cell type composition between the samples. We suggest an approach that could allow the detection of this bias in microarray analysis by generating GO terms for cell type-specific gene expression patterns. The overrepresentation of these terms in an analysis could alert the user toward cell type bias. This approach is already in partial implementation thanks to the availability of resources such as the Gene Expression Atlas and cell type-specific studies in organs such as the kidney (2).
In summary, airway epithelial cells grown in primary cultures closely recapitulate the transcriptional profile of in vivo airway epithelia. Both the use of primary cells for culture and the presence of an air-liquid interface play an important role in cell differentiation. These data also establish a baseline for comparing airway epithelia in health and disease.
GRANTS
B.-G. Harvey, A. E. Tilley, and R. G. Crystal are supported, in part, by National Institutes of Health (NIH) Grants RO1-HL-074326, P50-HL-084936, and UL1-RR-024996. P. B. McCray, Jr., is supported, in part, by NIH Grants P50-HL-61234 and N01-AI-30040, Cystic Fibrosis Foundation Grant MCCRAY00V0, and the Roy J. Carver Charitable Trust. J. Zabner is supported, in part, by NIH Grants P50-HL-61234 and P30-DK-54759 and Cystic Fibrosis Foundation Grant RDP R458-CR07.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
ACKNOWLEDGMENTS
We acknowledge the University of Iowa DNA Core, Cells and Tissue Core, and Cell Morphology Core. We thank Bart Brown and Tom Bair for technical support and Shyam Ramachandran, David A. Stoltz, and Michael J. Welsh for insightful discussions.
REFERENCES
- 1. Banerjee B, Kicic A, Musk M, Sutanto EN, Stick SM, Chambers DC. Successful establishment of primary small airway cell cultures in human lung transplantation. Respir Res 10: 99, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, McMahon AP, Patterson LT, Little MH, Potter SS. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell 15: 781–791, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carolan BJ, Harvey BG, Hackett NR, O'Connor TP, Cassano PA, Crystal RG. Disparate oxidant gene expression of airway epithelium compared to alveolar macrophages in smokers. Respir Res 10: 111, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clarke S, Gordon S. Myeloid-specific gene expression. J Leukoc Biol 63: 153–168, 1998 [DOI] [PubMed] [Google Scholar]
- 5. Dajani R, Zhang Y, Taft PJ, Travis SM, Starner TD, Olsen A, Zabner J, Welsh MJ, Engelhardt JF. Lysozyme secretion by submucosal glands protects the airway from bacterial infection. Am J Respir Cell Mol Biol 32: 548–552, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Jong PM, van Sterkenburg MA, Hesseling SC, Kempenaar JA, Mulder AA, Mommaas AM, Dijkman JH, Ponec M. Ciliogenesis in human bronchial epithelial cells cultured at the air-liquid interface. Am J Respir Cell Mol Biol 10: 271–277, 1994 [DOI] [PubMed] [Google Scholar]
- 7. Dvorak A, Tilley AE, Shaykhiev R, Wang R, Crystal RG. Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am J Respir Cell Mol Biol. doi: 10.1165/rcmb.2009-0453OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eden E, Lipson D, Yogev S, Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 3: e39, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forbes I. Human airway epithelial cell lines for in vitro drug transport and metabolism studies. Pharm Sci Technolo Today 3: 18–27, 2000 [DOI] [PubMed] [Google Scholar]
- 11. Harvey BG, Heguy A, Leopold PL, Carolan BJ, Ferris B, Crystal RG. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J Mol Med 85: 39–53, 2007 [DOI] [PubMed] [Google Scholar]
- 12. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karp PH, Moninger TO, Weber SP, Nesselhauf TS, Launspach JL, Zabner J, Welsh MJ. An in vitro model of differentiated human airway epithelia. Methods for establishing primary cultures. Methods Mol Biol 188: 115–137, 2002 [DOI] [PubMed] [Google Scholar]
- 14. Mercer RR, Russell ML, Roggli VL, Crapo JD. Cell number and distribution in human and rat airways. Am J Respir Cell Mol Biol 10: 613–624, 1994 [DOI] [PubMed] [Google Scholar]
- 15. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet 38: 500–501, 2006 [DOI] [PubMed] [Google Scholar]
- 16. Scheetz TE, Zabner J, Welsh MJ, Coco J, Eyestone M, Bonaldo MF, Kucaba T, Casavant TL, Soares MB, McCray PB., Jr Large-scale gene discovery in human airway epithelia reveals novel transcripts. Physiol Genomics 17: 69–77, 2004 [DOI] [PubMed] [Google Scholar]
- 17. Vermeer PD, Panko L, Karp P, Lee JH, Zabner J. Differentiation of human airway epithelia is dependent on erbB2. Am J Physiol Lung Cell Mol Physiol 291: L175–L180, 2006 [DOI] [PubMed] [Google Scholar]
- 18. Whitcutt MJ, Adler KB, Wu R. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell Dev Biol 24: 420–428, 1988 [DOI] [PubMed] [Google Scholar]
- 19. Winder AA, Wohlford-Lenane C, Scheetz TE, Nardy BN, Manzel LJ, Look DC, McCray PB., Jr Differential effects of cytokines and corticosteroids on toll-like receptor 2 expression and activity in human airway epithelia. Respir Res 10: 96, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamaya M, Finkbeiner WE, Chun SY, Widdicombe JH. Differentiated structure and function of cultures from human tracheal epithelium. Am J Physiol Lung Cell Mol Physiol 262: L713–L724, 1992 [DOI] [PubMed] [Google Scholar]
- 21. Zabner J, Zeiher BG, Friedman E, Welsh MJ. Adenovirus-mediated gene transfer to ciliated airway epithelia requires prolonged incubation time. J Virol 70: 6994–7003, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zavala DC, Rossi NP, Bedell GN. Bronchial brush biopsy. A valuable diagnostic technique in the presurgical evaluation of indeterminate lung densities. Ann Thorac Surg 13: 519–528, 1972 [DOI] [PubMed] [Google Scholar]