Abstract
Previous microarray analyses identified 22 microRNAs (miRNAs) differentially expressed in paired ectopic and eutopic endometrium of women with and without endometriosis. To investigate further the role of these miRNAs in women with endometriosis, we conducted an association study aiming to explore the relationship between endometriosis risk and single-nucleotide polymorphisms (SNPs) in miRNA target sites for these differentially expressed miRNAs. A panel of 102 SNPs in the predicted miRNA binding sites were evaluated for an endometriosis association study and an ingenuity pathway analysis was performed. Fourteen rare variants were identified in this study. We found SNP rs14647 in the Wolf–Hirschhorn syndrome candidate gene1 (WHSC1) 3′UTR (untranslated region) was associated with endometriosis-related infertility presenting an odds ratio of 12.2 (95% confidence interval = 2.4–60.7, P = 9.03 × 10−5). SNP haplotype AGG in the solute carrier family 22, member 23 (SLC22A23) 3′UTR was associated with endometriosis-related infertility and more severe disease. With the individual genotyping data, ingenuity pathways analysis identified the tumour necrosis factor and cyclin-dependant kinase inhibitor as major factors in the molecular pathways. Significant associations between WHSC1 alleles and endometriosis-related infertility and SLC22A23 haplotypes and the disease severe stage were identified. These findings may help focus future research on subphenotypes of this disease. Replication studies in independent large sample sets to confirm and characterize the involvement of the gene variation in the pathogenesis of endometriosis are needed.
Keywords: miRNA, endometriosis, single-nucleotide polymorphism, haplotype
Introduction
Endometriosis is a common disease causing pelvic pain and reduced fertility in 6–10% of reproductive aged women at considerable cost to health systems (Giudice and Kao, 2004; Berkley et al., 2005; Guo and Wang, 2006). Although the aetiology of endometriosis is still not well understood, there is wide support for Sampson's theory that endometrial fragments are carried in retrograde menstrual flow through the fallopian tubes and implant in the pelvic cavity (Sampson, 1927). Other aetiological factors have been proposed to influence establishment and progression of the disease, including hormonal (Nyholt et al., 2009), immune (Weiss et al., 2009) and environmental alterations (Guo, 2009). Genetic inheritance has been shown to be a major risk factor for endometriosis in studies that show the incidence of endometriosis is increased among the relatives of endometriosis patients compared with population controls (Zondervan et al., 2001).
Molecular studies have focused considerable effort on expression, regulation and inheritance of genes in endometriotic disease. In recent years, it has been shown that small non-coding microRNAs (miRNAs), are important components of complex gene regulatory networks (Reinhart and Bartel, 2002; Aravin et al., 2007). miRNAs are endogenous ∼19–22 nt RNAs that are a component of the miRNA-induced silencing effector complex (mRISC). The microRNA sequence directs the sequence-specific binding of the mRISC complex to one or more target mRNAs, repressing their translation to protein (Bartel, 2009). In this way, protein production can be inhibited and cellular functions altered without altering the target gene's sequence. However, a genetic aberration in miRNA binding could result in an inherited defect in protein translation of a target mRNA. We hypothesized that an inherited single-nucleotide polymorphism (SNP) and/or a mutation near a miRNA binding site may have potential impact in women with endometriosis and contribute to the development of endometeriotic disease. Recent research (Halvorsen et al., 2010) has shown that SNPs can affect the RNA structure of untranslated regions (UTRs), and these changes in conformation are known to affect the accessibility of miRNA target sites (Doench and Sharp, 2004). Furthermore, research (Wang et al., 2006) swapping miRNA target sites between UTRs-demonstrated miRNA target sites are repressionally active due to contextual signals surrounding target sites. Another study highlighted the importance of the sequence surrounding miRNA target sites for normal miRNA regulation by mutating the regions surrounding the target sites and showing loss of miRNA repression (Mencia et al., 2009).
Recent evidence has shown that miRNAs are involved in many biological processes, including cell differentiation, proliferation and apoptosis that commonly occur in endometriosis (Pan and Chegini, 2008; Glinskii et al., 2009). Compelling studies have implicated miRNAs in various human diseases, such as cancers (Kontorovich et al., 2009; Satzger et al., 2009), diabetes (Yang and Kaye, 2009), schizophrenia (Coyle, 2009), inflammation and viral infection (Hufner et al., 2009; Mattes et al., 2009), as well as in endometriosis (Pan et al., 2007; Burney et al., 2009; Ohlsson Teague et al., 2009b). Ye et al. (2008) found an association between oesophageal cancer risk and a functional SNP located in the pre-mir423 binding site. In addition, Chin et al. and Tian et al. provided evidence for association between lung cancer and the number of miRNA functional SNPs (Chin et al., 2008; Tian et al., 2009).
Previous microarray analyses identified 22 miRNAs differentially expressed in paired ectopic and eutopic endometrium of women with and without endometriosis (Ohlsson Teague et al., 2009b). To investigate the potential impact of these miRNAs in women with endometriosis, we conducted an association study aiming to explore the relationship between SNPs in target sites for these differentially expressed miRNA and endometriosis risk. We designed and evaluated a panel of 102 SNPs in predicted miRNA target sites and conducted an association study in endometriosis cases and controls. Given the key regulatory function of miRNAs in gene expression stability, understanding the mechanisms of how endometrial miRNAs are regulated and identifying their specific target genes might lead to the development of novel therapeutic treatments for endometriosis.
Materials and Methods
Participants
Participants were 958 endometriosis cases and 959 controls from an Australian study of endometriosis. The vast majority of cases (n=926) were drawn from families with affected sister pairs (ASPs) (one per family), and 32 were from non-ASP families. The cases with surgically confirmed endometriosis and with the most severe stage of disease were chosen from ASPs (Zhao et al., 2006). Disease severity was assessed using the revised American Fertility Society (rAFS) classification system (The American Fertility Society, 1985). However, the time from diagnosis to recruitment was variable and there was variable detail in the clinical records available. We therefore collapsed stage of disease into two classes for analysis (rAFS I/II and rAFS III/IV) (Zhao et al., 2007). Cases with rAFS Stages I and II were more common (59%) than the cases with rAFS Stages III and IV (41%). Twenty-five percent of cases (n=236) self-reported a history of ‘problems conceiving’, and were therefore considered to be a subfertility group of women diagnosed from endometriosis. The remainder reported no problems conceiving (44.2%) or had never tried to conceive (29.3%).
A similar number of 959 unrelated female controls were selected from a twin study of gynaecological health (Treloar et al., 1999). These controls had never been diagnosed with the disease on the basis of either self-report or medical records were available. No evidence of endometriosis was reported in the 27% of control women who reported having a hysterectomy and/or laparoscopy, therefore supporting a low risk of endometriosis in our control sample. The mean ages (±SD) of the cases and controls at the time of data collection were 35.82±8.87 and 45.60±11.98 years, respectively. Ethics approval was obtained from the Human Research Ethics Committee of the Queensland Institute of Medical Research and the Australian Twin Registry.
miRNA target SNP selection
Twenty-two miRNAs were previously identified by microarray analysis (Ohlsson Teague et al., 2009a). Potential miRNA target sites in genomic sequences for the list of miRNAs were assessed by using the Miranda algorithm (Betel et al., 2008). The binding energy of hybridization between the miRNA and target as ≤−30 kcal/mole and a cut-off score of 120 were considered to be efficient for functional binding specificity. The genome locations of these target sites were determined and searched against dbSNP (build 126) to identify genes within target sites. In this case if a gene was considered a target based on the criteria above, all target sites for that miRNA within that gene were searched for SNPs regardless of the binding energies of the individual sites. When searching for SNPs within the target sites, 2 nt were added to each side of all the target sites since SNPs which are close to the target site may also affect miRNA binding. In total, there were 2657 target sites across 145 genes included in the search of dbSNP database. A total of 243 SNPs were identified within target sites using the UC Santa Cruz genome browser (UCSC version 18, 2006).
We selected a subset of these SNPs for genotyping by first selecting any target SNPs located in regions of significant or suggestive linkage to endometriosis on chromosomes 10, 7 and 20 (Treloar et al., 2005; Zondervan et al., 2007). A total of 24 SNPs were identified including 6 SNPs on chromosome 10, 9 SNPs on chromosome 7 and 9 SNPs on chromosome 20. To supplement this list, we then screened the SNPs using ingenuity pathways analysis (IPA) software (http://www.ingenuity.com/) and the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway). Of the remaining 219 target SNPs, 78 SNPs were also selected into the study excluding 123 unmapped SNPs or the SNPs located in unknown genes. Eighteen SNPs failed in the assay design stage, as determined by Sequenom Assay Design software (Sequenom, version 3.1).
Individual genotyping
SNP sequences were downloaded from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and were cross checked in Sequenom databases (https://mysequenom.com) before assay design. Multiplex assays were designed for these 102 miRNA target SNPs using the Sequenom MassARRAY Assay Design software (version 3.1). SNPs were typed using iPLEXTM Gold chemistry and analysed using a Sequenom MassARRAY Compact Mass Spectrometer (Sequenom Inc., San Diego, CA, USA). The PCR and the iplex reactions were performed according to the manufacturer's instructions as described in our previous study (Zhao et al., 2006). Briefly, the 2.5 ml PCRs were performed using 12.5 ng genomic DNA, 1 unit of Taq polymerase (Sequenom Inc.), 500 µM of each dNTP, 2 mM of MgCl2 and 100 nM of each PCR primer (Integrated DNA Technologies Inc., Coralville, IA, USA). PCR thermal cycling program and a two-step 200 short cycles program for extension reaction were used as described in our previous study (Zhao et al., 2006). The post-PCR products were spotted on a SpectroChip (Sequenom Inc.), and data were processed and analysed by MassARRAY TYPER 4.0 software (Sequenom Inc.).
Data analysis
SNP genotypes were tested for departures from Hardy–Weinberg equilibrium (HWE). The PLINK genetic analysis package (http://pngu.mgh.harvard.edu/purcell/plink/) was used to calculate allele frequencies and to access the association of miRNA target SNPs with endometriosis. To obtain nominal P-values, 10 000 permutation tests were performed to obtain a region-wide empirical P-value for each SNP and SNP haplotype. To identify specific relationships between disease phenotype and SNP genotype, we conducted analyses stratifying cases by disease severity and phenotypic characteristics. Each subgroup was compared with the large Australian control sample. We assessed statistical significance at P<0.05 after correction for testing data for multiple SNPs, which for the endometriosis subgroup analyses became 0.01 (0.05/3) after permutation testing. Pairwise linkage disequilibrium (LD), haplotype frequencies and blocks were determined by Haploview version 4.1 using the default method of Gabriel et al. (2002). To test for non-independence of association signals, we analysed multiple SNPs jointly by logistic regression using the PLINK program (Arya et al., 2009).
Results
miRNA target SNPs
We searched for SNPs in target sites for published miRNAs differentially expressed in ectopic versus eutopic endometrial tissues and genotyped 102 miRNA target SNPs in an Australian sample comprising 958 endometriosis cases and 959 controls. The miRNA binding sites for each SNP are shown in the Supplementary data, table. Genotypes were obtained for most individuals with a mean completion rate of 99.4%. Of the 102 markers, 47 were not polymorphic (monomorphic) in our case–control sample. There were 41 polymorphic variants with allele minor allele frequencies >0.1%. Genotype frequencies for the 41 polymorphisms were in HWE in the control sample. The minor allele frequencies of the 41 miRNA target SNPs ranged from 0.16 to 46.4% in the cases and 0.05 to 46.8% in the controls (Table I). There was some evidence for allelic association between endometriosis and SNPs rs35091219 and rs1736215 with an asymptotic pointwise P<0.01 (Table I). The minor T allele of SNP rs35091219 was associated with endometriosis (case frequency=0.017; control frequency=0.007), whereas the minor C allele of SNP rs1736215 was more frequent in controls (case frequency=0.421; control frequency=0.469). However, the differences were not significant after correcting for multiple testing (P>0.1).
Table I.
Association analyses of miRNA target SNPs genotyped in endometriosis patients compared with 959 controls.
| Cases (n = 958) |
||||||
|---|---|---|---|---|---|---|
| #SNP | SNP position | Gene | Control (n = 959) | MAF | P | Odd ratio (95% CI) |
| rs12758341 | chr1:6084546 | KCNAB2 | 0.121 | 0.121 | 0.982 | 0.998 (0.821–1.213) |
| rs2092507 | chr1:6086995 | CHD5 | 0.123 | 0.119 | 0.646 | 0.955 (0.786–1.161) |
| rs12039801 | chr1:226436235 | C1orf69 | 0.039 | 0.045 | 0.417 | 1.142 (0.828–1.575) |
| rs14647 | chr4:1953633 | WHSC1 | 0.001 | 0.004 | 0.058 | 4.000 (0.848–18.86) |
| rs3813486 | chr6:3215104 | SLC22A23 | 0.128 | 0.135 | 0.558 | 1.058 (0.877–1.267) |
| rs1127473 | chr6:3215577 | SLC22A23 | 0.068 | 0.073 | 0.594 | 1.070 (0.835–1.371) |
| rs35091219 | chr6:3216786 | SLC22A23 | 0.007 | 0.017 | 0.005 | 2.476 (1.295–4.732) |
| rs3211066 | chr6:3216787 | SLC22A23 | 0.134 | 0.153 | 0.094 | 1.167 (0.974–1.399) |
| rs4718067 | chr7:63453831 | BC031335 | 0.305 | 0.296 | 0.548 | 0.958 (0.834–1.101) |
| rs1993894 | chr7:63454923 | BC031335 | 0.286 | 0.280 | 0.659 | 0.969 (0.841–1.115) |
| rs11762700 | chr7:63455322 | BC031335 | 0.171 | 0.159 | 0.309 | 0.915 (0.771–1.086) |
| rs621054 | chr7:63455611 | BC031335 | 0.357 | 0.360 | 0.817 | 1.016 (0.890–1.160) |
| rs1404679 | chr7:63456067 | BC031335 | 0.301 | 0.298 | 0.805 | 0.983 (0.855–1.129) |
| rs10274229 | chr7:149126986 | SSPO | 0.001 | 0.002 | 0.319 | 2.991 (0.318–28.78) |
| rs1122307 | chr7:157026137 | PTPRN2 | 0.076 | 0.079 | 0.798 | 1.032 (0.813–1.308) |
| rs1254967 | chr10:43010397 | GALNACT-2 | 0.163 | 0.183 | 0.095 | 1.154 (0.975–1.366) |
| rs7894262 | chr10:73765059 | DNAJB12 | 0.026 | 0.036 | 0.081 | 1.388 (0.959–2.009) |
| rs2227579 | chr10:75341295 | C10orf55 | 0.009 | 0.010 | 0.629 | 1.173 (0.613–2.247) |
| rs11156473 | chr10:133454858 | FLJ46300 | 0.163 | 0.148 | 0.206 | 0.893 (0.749–1.064) |
| rs45477500 | chr11:2826028 | KCNQ1 | 0.017 | 0.022 | 0.290 | 1.286 (0.806–2.051) |
| rs8813 | chr11:3065082 | OSBPL5 | 0.248 | 0.236 | 0.405 | 0.939 (0.810–1.089) |
| rs1053746 | chr11:3065261 | OSBPL5 | 0.031 | 0.027 | 0.439 | 0.861 (0.588–1.259) |
| rs17178177 | chr11:3065447 | OSBPL5 | 0.110 | 0.122 | 0.255 | 1.122 (0.920–1.368) |
| rs2285676 | chr11:17364601 | KCNJ11 | 0.381 | 0.400 | 0.234 | 1.082 (0.950–1.233) |
| rs1045721 | chr11:133754816 | B3GAT1 | 0.135 | 0.115 | 0.057 | 0.830 (0.684–1.006) |
| rs1459706 | chr14:69580639 | SLC8A3 | 0.091 | 0.083 | 0.368 | 0.902 (0.720–1.129) |
| rs8020841 | chr14:69581912 | SLC8A3 | 0.219 | 0.202 | 0.213 | 0.903 (0.770–1.060) |
| rs3204173 | chr14:104547703 | CDCA4 | 0.176 | 0.183 | 0.588 | 1.047 (0.887–1.235) |
| rs12890396 | chr14:104587271 | GPR132 | 0.324 | 0.328 | 0.807 | 1.017 (0.888–1.165) |
| rs3803359 | chr15:38450040 | DISP2 | 0.161 | 0.158 | 0.771 | 0.975 (0.820–1.159) |
| rs1736215 | chr17:17072927 | FLCN | 0.469 | 0.421 | 0.003 | 0.824 (0.725–0.936) |
| rs34304528 | chr19:5794576 | FUT3 | 0.128 | 0.139 | 0.336 | 1.096 (0.910–1.320) |
| rs874232 | chr19:5794609 | FUT3 | 0.429 | 0.448 | 0.226 | 1.082 (0.952–1.230) |
| rs268674 | chr19:45592705 | PRX | 0.059 | 0.056 | 0.658 | 0.940 (0.715–1.236) |
| rs268672 | chr19:45593444 | PRX | 0.442 | 0.464 | 0.165 | 1.094 (0.964–1.243) |
| rs3803947 | chr20:1236966 | SDCBP2 | 0.464 | 0.461 | 0.853 | 0.988 (0.870–1.122) |
| rs13163 | chr20:1237585 | SDCBP2 | 0.058 | 0.064 | 0.488 | 1.098 (0.843–1.432) |
| rs6121315 | chr20:30083007 | C20orf160 | 0.142 | 0.141 | 0.932 | 0.992 (0.827–1.190) |
| rs817321 | chr20:62060778 | ZNF512B | 0.013 | 0.010 | 0.437 | 0.787 (0.430–1.442) |
| rs3830028 | chrX:151978585 | PNMA3 | 0.049 | 0.056 | 0.362 | 1.141 (0.859–1.517) |
| rs1045069 | chrX:151979017 | PNMA3 | 0.432 | 0.432 | 0.963 | 1.003 (0.882–1.140) |
MAF, minor allele frequency.
Fourteen variants were rare in our sample with the variant allele seen in no more than three individuals and these were not included in the association analysis. Of these, six were found only in endometriosis cases, five were found only in unrelated controls and three were found in both cases and controls (Table II).
Table II.
Rare miRNA variants identified in this study.
| miRNA SNPa | Location | Gene | No case | Allele freq (%) | No control | Allele freq (%) |
|---|---|---|---|---|---|---|
| rs12082745 | chr1:6086867 | CHD5 | 1 | 0.11 | ||
| rs7542469 | chr1:201723721 | PRELP | 2 | 0.21 | ||
| rs11545893 | chr4:1952426 | WHSC1 | 1 | 0.11 | ||
| rs10034373 | chr4:1952453 | WHSC1 | 1 | 0.11 | 2 | 0.21 |
| rs17132112 | chr4:1954282 | WHSC2 | 1 | 0.11 | ||
| rs7742745 | chr6:3216228 | SLC22A23 | 3 | 0.31 | ||
| rs913981 | chr9:129688166 | ST6GALNAC6 | 1 | 0.11 | ||
| rs8073668 | chr17:3711258 | CAMKK1 | 1 | 0.11 | ||
| rs9652820 | chr17:17074576 | FLCN | 1 | 0.21 | ||
| rs3862149 | chr17:17075630 | FLCN | 2 | 0.21 | ||
| rs8068566 | chr17:44157228 | HOXB13 | 1 | 0.11 | 1 | 0.11 |
| rs374028 | chr19:5817549 | FUT5 | 1 | 0.11 | ||
| rs2233837 | chr20:29536061 | REM1 | 1 | 0.11 | 1 | 0.11 |
| rs10597644 | chr22:20131554 | HIC2 | 1 | 0.11 |
Numbers of cases and minor allele frequencies were determined by Sequenom SNP analysis of 958 endometriosis cases and 959 unrelated controls.
aSNPs in the miRNA target sites were assessed by using Miranda algorithm.
Association with disease characteristics
We further evaluated association results by stratifying cases according to disease severity and phenotypic characteristics. Differences were observed with nominal P-values<0.05 for several SNPs (rs3211066, rs7894262, rs1736215, rs874232 and rs3803947) and higher minor allele frequencies were observed for SNP rs35091219 in all different case groups compared with controls. These differences were not significant after correcting for multiple testing (data not shown). The smallest pointwise P-value was 9×10−5 [χ2=15.3, odd ratio (95% confidence interval)=12.2 (2.4–60.8)] for SNP rs14647, located in the Wolf–Hirschhorn syndrome candidate 1 gene (WHSC1) 3′UTR in 236 endometriosis subfertility cases compared with 959 unrelated controls (Table III). This difference remained significant after correction for multiple testing (P=0.005). It was noted, the minor A allele frequencies for this SNP were very low, at 0.013 in the 236 subfertility cases and 0.001 in the 959 controls (Table III).
Table III.
Association analyses of miRNA SNPs genotyped in phenotype subgroups of endometriosis patients compared with 959 controls.
| Diagnosed rAFS Stages I/II (559) |
Diagnosed rAFS Stages III/IV (394) |
Subfertility cases (236) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #SNP | Gene | Control F | MAF | P | Odd ratio (95% CI) | MAF | P | Odd ratio (95% CI) | MAF | P | Odd ratio (95% CI) |
| rs12758341 | KCNAB2 | 0.121 | 0.116 | 0.653 | 0.949 (0.753–1.194) | 0.129 | 0.593 | 1.071 (0.834–1.375) | 0.128 | 0.695 | 1.063 (0.784–1.440) |
| rs2092507 | CHD5 | 0.123 | 0.115 | 0.509 | 0.926 (0.736–1.164) | 0.123 | 0.999 | 1.000 (0.777–1.287) | 0.126 | 0.901 | 1.020 (0.751–1.383) |
| rs12039801 | C1orf69 | 0.039 | 0.050 | 0.182 | 1.277 (0.891–1.830) | 0.039 | 0.992 | 1.002 (0.650–1.546) | 0.035 | 0.641 | 0.877 (0.506–1.522) |
| rs14647 | WHSC1 | 0.001 | 0.002 | 0.585 | 1.716 (0.241–12.20) | 0.006 | 0.014 | 6.063 (1.174–31.32) | 0.013 | 9.03E–05 | 12.25 (2.464–60.87) |
| rs3813486 | SLC22A23 | 0.128 | 0.123 | 0.672 | 0.953 (0.762–1.192) | 0.149 | 0.148 | 1.192 (0.940–1.512) | 0.166 | 0.033 | 1.352 (1.024–1.785) |
| rs1127473 | SLC22A23 | 0.068 | 0.059 | 0.329 | 0.858 (0.632–1.166) | 0.089 | 0.063 | 1.332 (0.984–1.804) | 0.072 | 0.769 | 1.060 (0.717–1.569) |
| rs35091219 | SLC22A23 | 0.007 | 0.016 | 0.014 | 2.387 (1.165–4.891) | 0.018 | 0.009 | 2.634 (1.232–5.629) | 0.023 | 0.001 | 3.475 (1.547–7.806) |
| rs3211066 | SLC22A23 | 0.134 | 0.137 | 0.795 | 1.029 (0.830–1.276) | 0.173 | 0.008 | 1.355 (1.080–1.700) | 0.193 | 0.001 | 1.547 (1.188–2.014) |
| rs4718067 | BC031335 | 0.305 | 0.294 | 0.525 | 0.949 (0.807–1.116) | 0.300 | 0.806 | 0.978 (0.815–1.172) | 0.279 | 0.274 | 0.883 (0.705–1.104) |
| rs1993894 | BC031335 | 0.286 | 0.271 | 0.363 | 0.926 (0.785–1.093) | 0.293 | 0.702 | 1.036 (0.863–1.245) | 0.284 | 0.927 | 0.990 (0.792–1.237) |
| rs11762700 | BC031335 | 0.171 | 0.160 | 0.434 | 0.924 (0.757–1.127) | 0.159 | 0.451 | 0.917 (0.732–1.149) | 0.136 | 0.060 | 0.758 (0.568–1.013) |
| rs621054 | BC031335 | 0.357 | 0.363 | 0.732 | 1.027 (0.881–1.198) | 0.353 | 0.877 | 0.986 (0.829–1.174) | 0.368 | 0.638 | 1.052 (0.853–1.297) |
| rs1404679 | BC031335 | 0.301 | 0.297 | 0.806 | 0.980 (0.834–1.152) | 0.300 | 0.934 | 0.992 (0.828–1.190) | 0.281 | 0.384 | 0.905 (0.724–1.132) |
| rs10274229 | SSPO | 0.001 | 0.003 | 0.115 | 5.124 (0.532–49.32) | NA | NA | NA | NA | NA | NA |
| rs1122307 | PTPRN2 | 0.076 | 0.080 | 0.729 | 1.050 (0.797–1.382) | 0.077 | 0.991 | 1.002 (0.733–1.370) | 0.072 | 0.748 | 0.939 (0.637–1.383) |
| rs1254967 | GALNACT-2 | 0.163 | 0.188 | 0.077 | 1.191 (0.981–1.445) | 0.176 | 0.391 | 1.101 (0.883–1.373) | 0.153 | 0.624 | 0.933 (0.706–1.233) |
| rs7894262 | DNAJB12 | 0.026 | 0.033 | 0.277 | 1.270 (0.825–1.956) | 0.041 | 0.047 | 1.576 (1.003–2.476) | 0.055 | 0.001 | 2.159 (1.329–3.507) |
| rs2227579 | C10orf55 | 0.009 | 0.008 | 0.804 | 0.902 (0.401–2.031) | 0.014 | 0.239 | 1.575 (0.735–3.376) | 0.008 | 0.925 | 0.949 (0.318–2.833) |
| rs11156473 | FLJ46300 | 0.163 | 0.142 | 0.126 | 0.850 (0.691–1.047) | 0.157 | 0.721 | 0.959 (0.764–1.205) | 0.138 | 0.187 | 0.823 (0.617–1.099) |
| rs45477500 | KCNQ1 | 0.017 | 0.022 | 0.345 | 1.293 (0.758–2.206) | 0.022 | 0.395 | 1.293 (0.714–2.343) | 0.032 | 0.035 | 1.929 (1.036–3.592) |
| rs8813 | OSBPL5 | 0.248 | 0.255 | 0.686 | 1.036 (0.873–1.228) | 0.211 | 0.042 | 0.812 (0.665–0.993) | 0.230 | 0.413 | 0.905 (0.713–1.149) |
| rs1053746 | OSBPL5 | 0.031 | 0.027 | 0.541 | 0.870 (0.557–1.359) | 0.027 | 0.553 | 0.858 (0.518–1.423) | 0.032 | 0.918 | 1.031 (0.580–1.834) |
| rs17178177 | OSBPL5 | 0.110 | 0.141 | 0.013 | 1.325 (1.061–1.653) | 0.096 | 0.259 | 0.852 (0.646–1.125) | 0.109 | 0.929 | 0.985 (0.713–1.363) |
| rs2285676 | KCNJ11 | 0.381 | 0.392 | 0.549 | 1.048 (0.900–1.220) | 0.416 | 0.092 | 1.157 (0.976–1.370) | 0.409 | 0.276 | 1.121 (0.913–1.378) |
| rs1045721 | B3GAT1 | 0.135 | 0.123 | 0.333 | 0.896 (0.718–1.119) | 0.102 | 0.019 | 0.728 (0.558–0.950) | 0.091 | 0.011 | 0.644 (0.458–0.904) |
| rs1459706 | SLC8A3 | 0.091 | 0.091 | 0.928 | 0.988 (0.764–1.278) | 0.074 | 0.142 | 0.793 (0.582–1.081) | 0.081 | 0.454 | 0.870 (0.603–1.254) |
| rs8020841 | SLC8A3 | 0.219 | 0.203 | 0.307 | 0.907 (0.753–1.093) | 0.199 | 0.250 | 0.883 (0.714–1.092) | 0.213 | 0.777 | 0.964 (0.749–1.241) |
| rs3204173 | CDCA4 | 0.176 | 0.179 | 0.831 | 1.021 (0.842–1.239) | 0.189 | 0.426 | 1.091 (0.881–1.351) | 0.165 | 0.577 | 0.926 (0.707–1.213) |
| rs12890396 | GPR132 | 0.324 | 0.321 | 0.861 | 0.986 (0.842–1.155) | 0.339 | 0.452 | 1.070 (0.897–1.276) | 0.305 | 0.434 | 0.917 (0.737–1.140) |
| rs3803359 | DISP2 | 0.161 | 0.155 | 0.643 | 0.953 (0.778–1.168) | 0.159 | 0.879 | 0.983 (0.783–1.233) | 0.163 | 0.927 | 1.013 (0.771–1.331) |
| rs1736215 | FLCN | 0.469 | 0.413 | 0.003 | 0.799 (0.688–0.927) | 0.430 | 0.070 | 0.857 (0.725–1.013) | 0.422 | 0.067 | 0.827 (0.675–1.014) |
| rs34304528 | FUT3 | 0.128 | 0.139 | 0.411 | 1.095 (0.882–1.359) | 0.138 | 0.487 | 1.090 (0.855–1.389) | 0.167 | 0.027 | 1.365 (1.035–1.799) |
| rs874232 | FUT3 | 0.429 | 0.435 | 0.758 | 1.024 (0.882–1.188) | 0.468 | 0.061 | 1.173 (0.993–1.386) | 0.511 | 0.001 | 1.390 (1.136–1.702) |
| rs268674 | PRX | 0.059 | 0.055 | 0.643 | 0.927 (0.672–1.278) | 0.055 | 0.677 | 0.926 (0.645–1.330) | 0.066 | 0.561 | 1.130 (0.749–1.705) |
| rs268672 | PRX | 0.442 | 0.445 | 0.871 | 1.012 (0.872–1.175) | 0.491 | 0.020 | 1.218 (1.032–1.439) | 0.475 | 0.203 | 1.140 (0.932–1.395) |
| rs3803947 | SDCBP2 | 0.464 | 0.467 | 0.869 | 1.013 (0.873–1.174) | 0.449 | 0.487 | 0.942 (0.797–1.114) | 0.394 | 0.006 | 0.751 (0.611–0.922) |
| rs13163 | SDCBP2 | 0.058 | 0.059 | 0.930 | 1.014 (0.741–1.388) | 0.069 | 0.317 | 1.187 (0.848–1.662) | 0.072 | 0.267 | 1.252 (0.841–1.865) |
| rs6121315 | C20orf160 | 0.142 | 0.144 | 0.899 | 1.014 (0.821–1.252) | 0.138 | 0.776 | 0.966 (0.759–1.228) | 0.140 | 0.932 | 0.988 (0.739–1.320) |
| rs817321 | ZNF512B | 0.013 | 0.013 | 0.990 | 0.996 (0.513–1.933) | 0.005 | 0.081 | 0.401 (0.139–1.160) | 0.006 | 0.254 | 0.503 (0.151–1.676) |
| rs3830028 | PNMA3 | 0.049 | 0.058 | 0.289 | 1.192 (0.861–1.650) | 0.053 | 0.668 | 1.085 (0.747–1.577) | 0.059 | 0.378 | 1.216 (0.787–1.877) |
| rs1045069 | PNMA3 | 0.432 | 0.449 | 0.363 | 1.072 (0.923–1.244) | 0.410 | 0.298 | 0.914 (0.772–1.082) | 0.419 | 0.628 | 0.951 (0.775–1.166) |
MAF, minor allele frequency.
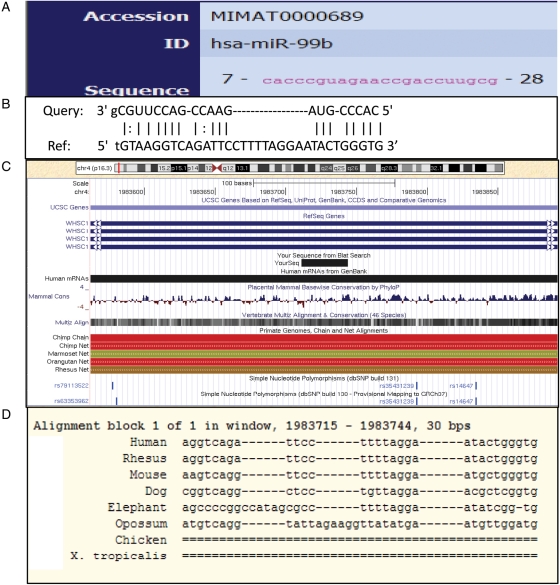
To identify a relationship between miR-99b and SNP rs14647 in the WHSC1 gene, a detailed search was conducted using the Miranda package (Enright et al., 2003) and the UCSC genome browser (hg18, www.genome.ucsc.edu). The mature miRNA sequence of has-miR-99b (MIMAT0000689) was obtained from mirBase (http://www.mirbase.org; Fig. 1A) and a miR-99b target site in the WHSC1 gene was identified using the Miranda algorithm. The target site in the WHSC1 3′UTR has a strong binding energy to miR-99b and a 3′ 8 nt seed region (Fig. 1B). SNP rs14647 is located at the extreme end of the WHSC1 3′UTR region and is within 50 nt of the miR-99b binding site (Fig. 1C). We used the UCSC ‘PhastCons'track (http://compgen.bscb.cornell.edu/phast/) to evaluate the conservation of the target binding region (Fig. 1D). Seed conservation was observed in some placental mammals which maintained a good binding energy to the WHSC1 3′UTR, indicating that a mutation in the surrounding regions of the target site could disrupt miRNA regulation of WHSC1 protein production and contribute to the development of the disease. Previous mutation studies have shown mutations near miRNA binding sites can affect mRNA stability/translation, presumably by altering the local RNA secondary structure in the usually highly folded 3′UTR region (Doench and Sharp, 2004; Didiano et al., 2010).
Figure 1.
Relationship between miR-99b and SNP rs14647 in the Wolf–Hirschhorn syndrome candidate gene 1 (WHSC1) gene. (A) The mature sequence of has-miR-99b (MIMAT0000689) was downloaded from mirBase (http://www.mirbase.org), (B) a miR-99b target site in the WHSC1 gene was identified using the Miranda algorithm. (C) SNP rs14647 is located at the extreme end of the WHSC1 3′-UTR region and within about 50 nt of the miR-99b binding site. (D) The “PhastCons” track (http://compgen.bscb.cornell.edu/phast/) at UCSC was used to evaluate the conservation of the target binding region of the WHSC1 3′-UTR.
Haplotype analysis on SLC22A23 variations
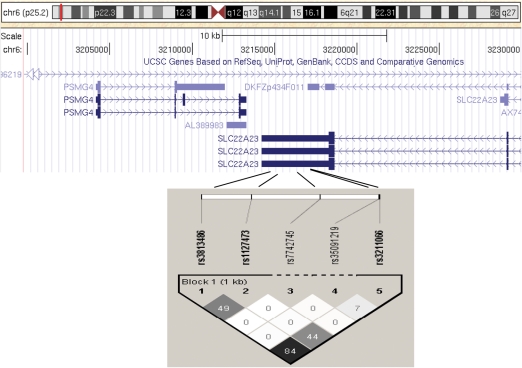
We performed haplotype analysis on genes that have more than two SNPs genotyped in our sample set. SNP haplotypes in solute carrier family 22, member 23 (SLC22A23) were shown to be associated with endometriosis subgroups after multiple testing. Therefore, we present detailed information on SLC22A23 only. We typed five miRNA target SNPs in the 3′UTR of SLC22A23 gene located on chromosome 6p25.2. Figure 2 shows the LD plot for the five miRNA SNPs from SLC22A23. The risk allele C of SNP rs7742745 was detected only in 3/958 cases (∼0.16%), whereas the observed minor allele frequency of SNP rs35091219 was very low (∼0.7%, see above), so both SNPs were excluded from further haplotype analysis. Therefore, we conducted haplotype analysis for the three SNPs: rs3813486, rs1127473 and rs3211066 (Fig. 2). There are four major haplotypes deduced, according to the criteria of Gabriel et al. (2002).
Figure 2.
Haplotype analyses for the contiguous solute carrier family 22 member 23 (SLC22A23) miRNA SNPs. One haplotype block was determined and linkage disequilibrium (LD) patterns from five miRNA SNPs in SLC22A23 were estimated utilizing Haploview program (white, r2 = 0; shades of grey, 0 < r2<1; black, r2 = 1).
One major haplotype AGC (haplotype 1 in Table IV) was predicted in our all samples, representing 85.2% of alleles, with three minor haplotypes being GAG (haplotype 2 in Table IV) at 7%, GGG (haplotype 3 in Table IV) at 5.8% and AGG (haplotype 4 in Table IV) at 1.6%. The frequencies of the predicted haplotypes for the three SLC22A23 miRNA SNPs listed from highest to lowest in disease groups are shown in Table IV. The major haplotype AGC represented 80.3% of alleles in subfertility cases and 86.1% of alleles in controls. Evidence for association was detected with a corrected P-value of 0.0064 (Table IV). The strongest associations were found for the fourth haplotype, with P-values of 5.0 × 10−4, 1 × 10−4 and 1.3 × 10−3 in the rAFS Stages III/IV group, subfertility and total endometriosis samples when compared with total controls, respectively. The P-values were significant after correcting for multiple testing (P=0.002, 0.002 and 0.005 for Stages III/IV group, subfertility and total cases, respectively), these measured haplotypes account for 2.2% of risk of endometriosis in our population.
Table IV.
Haplotype analysis on solute carrier family 22 member 23 (SLC22A23) miRNA SNPs (rs3813486/rs1127473/rs3211066) in phenotype subgroups of endometriosis patients compared with 959 controls.
| Haplotypea | Phonotype | No case | Frequency | Ratiosb | cCHISQ | P |
|---|---|---|---|---|---|---|
| 1 | rAFS Stages I/II | 559 | 0.860 | 0.858, 0.861 | 0.050 | 0.823 |
| 2 | 0.065 | 0.059, 0.068 | 0.864 | 0.353 | ||
| 3 | 0.058 | 0.059, 0.058 | 0.027 | 0.869 | ||
| 4 | 0.013 | 0.019, 0.009 | 5.392 | 0.020 | ||
| 1 | rAFS Stages III/IV | 394 | 0.850 | 0.824, 0.861 | 6.229 | 0.013 |
| 2 | 0.074 | 0.089, 0.068 | 3.618 | 0.057 | ||
| 3 | 0.058 | 0.058, 0.058 | 0.004 | 0.949 | ||
| 4 | 0.014 | 0.027, 0.009 | 12.302 | 5.00E–04 | ||
| 1 | Subfertility | 236 | 0.850 | 0.803, 0.861 | 10.091 | 0.002 |
| 2 | 0.069 | 0.072, 0.068 | 0.109 | 0.742 | ||
| 3 | 0.064 | 0.089, 0.058 | 6.191 | 0.013 | ||
| 4 | 0.014 | 0.032, 0.009 | 14.601 | 1.00E–04 | ||
| 1 | Total | 958 | 0.852 | 0.843, 0.861 | 2.477 | 0.116 |
| 2 | 0.070 | 0.073, 0.068 | 0.347 | 0.556 | ||
| 3 | 0.058 | 0.058, 0.058 | 0.012 | 0.914 | ||
| 4 | 0.016 | 0.022, 0.009 | 10.367 | 0.001 |
Haplotype frequencies and haplotype association analyses were estimated utilizing Haploview program and PLINK genetic analysis package.
aHaplotype 1 = AGC; 2 = GAG; 3 = GGG; 4 = AGG.
bCase, control frequency ratios.
cCases versus 959 controls.
Logistic regression analysis on multiple loci
The logistic regression analysis was performed based on four functional SNPs: rs14647, rs3813486, rs1127473 and rs3211066, because there was some evidence for association with endometriosis subfertility. SNP rs14647, located on chromosome 4p16.3 in the WHSC1 gene, is clearly not in LD (as it is on a different chromosome) with any of the chromosome 6 SNPs rs3813486, rs1127473 or rs3211066 (SLC22A23). Both SNPs rs14647 and rs3211066 gave the strongest evidence for association with unconditioned P-value of 0.002 and 0.001, respectively. The effects of SNPs rs14647 and rs3211066 on endometriosis-related infertility were completely independent from conditioned logistic regression analysis.
Among the three SNPs in close proximity to each other on chromosome 6 (rs3813486, rs1127473 and rs3211066), SNP rs3211066 showed the strongest evidence for association with endometriosis-related infertility for both unconditional and conditional models. After conditioning on rs3211066, evidence for association at rs3813486 and rs1127473 was no longer significant (P=0.1186, P=0.1178, respectively), indicating their association is not independent and may be due to them being in LD with rs3211066.
Pathway analysis of miRNA target SNPs
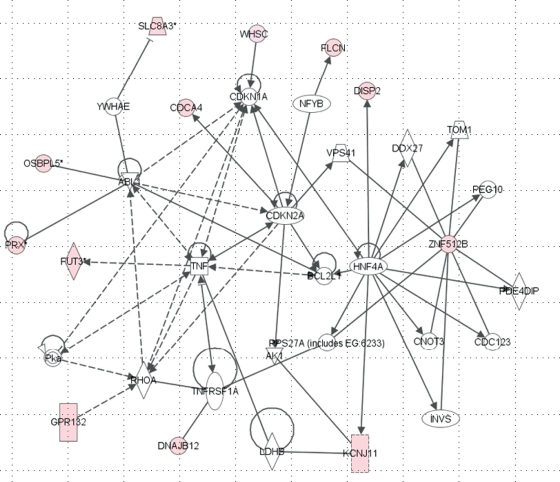
We have assessed 41 of the miRNA target SNPs with individual genotyping data, focusing on the molecular pathways they are potentially involved in using IPA. Several significant common pathways overlapping the data set were identified, mainly with TNF and cyclin-dependent kinase inhibitor (CDKN) centrally located in the molecular network (P=6.9E–04 to 3.6E–02). One of these, the cell cycle, cellular growth and proliferation pathway, was predicted to be involved in endometriosis development (Leyendecker et al., 1998) (Fig. 3). IPA identified 12 SNP-related genes in the pathway with molecular networks. The remaining other pathways were significantly enriched in the pathogenesis of endometriosis, including cell morphology, cellular assembly and organization; gene expression, nervous system development and function and carbohydrate metabolism.
Figure 3.
Cell cycle, cellular growth and proliferation pathways detected by ingenuity pathways analysis. Forty-one miRNA target SNPs with individual genotype data were assessed, 12 miRNA SNP-related genes were detected in the pathway with tumour necrosis factor (TNF) and cyclin-dependant kinase inhibitor (CDKN) centrally located in the molecular network.
Discussion
Studies in endometriosis have identified several miRNAs differentially expressed in ectopic versus eutopic endometrial tissues (Ohlsson Teague et al., 2009b), endometrial cells (Pan et al., 2007) and in women with endometriosis versus controls (Burney et al., 2009). In general, few miRNAs were detected in more than one study suggesting complex relationships of miRNAs in gene regulatory networks during development of endometrial lesions. Although the relationship between miRNA and mRNA in endometriosis has been evaluated in two studies (Pan et al., 2007; Ohlsson Teague et al., 2009b), a link between miRNAs and effects of DNA variation at their target sites has not been previously investigated. We hypothesized that mutations in target binding sites for the differentially expressed miRNAs could alter transcriptional regulation resulting in changes in expression of the target gene(s) and modifying endometriosis risk.
We used stringent criteria to identify target genes for 22 miRNAs differentially expressed (Ohlsson Teague et al., 2009b) in ectopic and eutopic endometrium. Once target genes were detected, we searched for SNPs in all corresponding sites for each miRNA within the target gene. This search identified 243 SNPs and we genotyped a subset of 102 SNPs in an Australian sample comprising 958 endometriosis cases and 959 controls. Forty-six percent of SNPs were not polymorphic in our sample. Fourteen SNPs showed evidence for rare minor alleles present in no more than three individuals. These were distributed roughly equally between being present in cases only, controls only or in both cases and controls.
Infertility is a major symptom associated with endometriosis and almost half of women with endometriosis are infertile (American Fertility Society, 1985; Guo and Wang, 2006). There is growing evidence that molecular dysfunction in eutopic endometrium contributes to endometriosis susceptibility and may also contribute to endometriosis-related infertility. A number of genes that code for proteins such as integrin (alpha V, beta 3), nitric oxide synthase, heat shock protein and interleukin-11, are reported to have an important role in endometrial receptivity (Agarwal et al., 2005; Bondza et al., 2009). The WHSC1 gene is located on chromosome 4p16.3 and is associated with Wolf–Hirschhorn syndrome. Patients with this gene defect suffer from craniofacial defects, intellectual disability, growth delay and 30% have genital anomalies including anomalies of the Mullerian system (female reproductive system). In a Whsc1-deficient mouse model reported that surviving Whsc1 heterozygotes were mostly fertile, but homozygous embryos showed growth retardation and died within 10 days of birth (Nimura et al., 2009). SNP rs14647 is located near a miRNA target site, between WHSC1 and WHSC2 and in the 3′UTR region of both genes (the two genes are transcribed in opposite directions). UTRs are the regulatory elements of genes, and play important roles in RNA's translation and decay. The WHSC1 and WHSC2 3′UTR region may be involved in dsRNA degradation when both genes are transcribed with overlapping UTRs. The current study is a genetic study in a large case–control sample set aiming to investigate the potential impact of 102 miRNA SNPs in women with endometriosis. Until now, most SNPs identified in genetic studies are not fully confirmed by functional study. However, it is well established that genetic studies not only attempt to identify functional SNPs but also ‘tag’ the approximate location of disease variants (Wang et al., 2010). Another unknown SNP in high LD with rs14647 might either increase or decrease the accessibility of the miRNA target site, leading to inactivation of the target site by this or other unknown mechanisms. A recent study by Halvorsen et al. showed that multiple specific SNPs in UTRs have a structural consequence and can result in a disease phenotype (Halvorsen et al., 2010). We found the rs14647A allele in the WHSC1 gene was most strongly associated with risk of endometriosis-related infertility with an increased frequency up to 13-fold when compared with our controls, suggesting that the SNP may have local effects on the RNA structural consequence of the associated genes.
We also found significant evidence for association between SLC22A23 haplotypes with SNPs in miR-125a-3p binding sites and predisposition to endometriosis. SLC22A23 belongs to a large family of transmembrane proteins that function as uniporters, symporters and antiporters to transport organic ions across cell membranes (Jacobsson et al., 2007). The SLC22A23 target site harbours five miRNA target SNPs: rs3813486, rs1127473, rs7742745, rs35091219 and rs3211066. Mutation in the functional SNP rs7742745 was only detected in three cases and not in 959 controls, suggesting a possible role in risk for endometriosis in these patients. Haplotype analysis for three SNPs (rs3813486, rs1127473 and rs3211066), excluding the rare variants, supports a role for variation in SLC22A23 in endometriosis risk. Haplotypes in SLC22A23 were strongly associated with infertility and more severe stage of the disease. The three SNPs are in one major haplotype block, with the common haplotype AGC representing 80.3% of alleles in subfertility cases and 86.1% of alleles in controls. The minor haplotype AGG accounted for 2.2% of alleles in all cases and 0.9% of alleles in all controls. The association detected with these haplotypes suggests that these variants may play a role in expression of the SLC22A23 transcript levels. The association results remained significant after correction of multiple testing, but need to be replicated in independent samples. Comprehensive genotyping in large, powerful, samples with appropriate data on disease subphenotypes will be required to ultimately confirm and characterise the involvement of the gene variation in the pathogenesis of endometriosis.
To evaluate the miRNA target SNPs that potentially contribute to molecular pathways in endometriosis, we performed pathway analysis using IPA software. By assessing 41 miRNA SNPs with the genotype data, IPA identified several overlapping common pathways constituted by two major molecular networks: TNF and CDKN, both are centrally located in the cellular growth and proliferation pathways. TNF is a pro-inflammatory cytokine and is known to play important roles in the pathological processes involved in endometriosis (Barcz et al., 2000; Braun et al., 2002). CDKN2A is an important tumour suppressor gene and was found to be less expressed in endometriosis lesions when compared with normal controls from immunohistochemical studies (Cameron et al., 2002; Goumenou et al., 2006). The functional relationship between TNF/NF-κB (nuclear factor of kappa light polypeptide gene enhancer in B-cells) signalling and CDKN2A in tumorigenesis has been explored in a mouse model (Altomare et al., 2009), but their functions and relationships in the pathogenesis of endometriosis are unclear. In women with endometriosis, activated macrophages secrete TNF-α, which then activates NF-κB and other inflammatory mediators to promote epithelial cell proliferation and in turn contributes to the disease progress. Two reports from same group provided evidence that inhibition of TNF reduces endometriosis. Treatment with recombinant human TNFRSF1A (tumour necrosis factor receptor superfamily, member 1A) or anti-TNF antibody in baboons, effectively inhibited the development of endometriosis (D'Hooghe et al., 2006; Falconer et al., 2006). TNF-α has been shown to inhibit asbestos-induced cytotoxicity via a NF-κB-dependent pathway (Yang et al., 2006) and NF-κB inhibition reduces cell proliferation, but increases apoptosis of endometriotic lesions (Gonzalez-Ramos et al., 2008). A previous study provided the information that supports the gene-to-function relationship between binding of promoter fragment from the human CDKN1A gene and human WHSC2 protein occurs in Hct 116 cells (Gomes et al., 2006). We found evidence for association with functional miRNA SNPs/SNP haplotypes in WHSC1 and SLC22A23 loci. Both loci have not previously been reported to be associated with endometriosis. Given that these miRNA SNPs impact molecular networks and their potential targets in the pathogenesis of endometriosis, our findings may provide valuable insights for the future investigations.
Although the present work supports our hypothesis that miRNA target SNPs might be involved in the development of endometriosis, it remains to be determined whether the association between these SNPs and endometriosis risk is due to an effect on miRNA binding. Functional studies in ectopic and eutopic endometrial tissues would be required, but the current results need first to be replicated in independent large sample sets to confirm and characterise the involvement of the gene variation in the pathogenesis of endometriosis.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors’ roles
The principal authors’ roles were as follows: Z.Z.: design, acquisition of data, analysis and interpretation, drafting manuscript and final approval. L.C.: conception and design, interpretation, revising for critical content and final approval. D.R.N.: analysis and interpretation of data, revising for critical content and final approval. B.C.: acquisition of data, revising for critical content and final approval. S.A.T.: conception and design, interpretation, revising for critical content and final approval. M.L.H.: conception and design, interpretation, revising for critical content and final approval. G.W.M.: conception and design, interpretation, revising for critical content and final approval.
Funding
This study was supported by grants to G.W.M. from the National Institute of Child Health and Human Development (HD050537) and National Health and Medical Research Council of Australia (339430 and 339446).
Acknowledgements
We thank Dr Daniel T. O'Connor for confirmation of diagnosis and staging of disease from clinical records of 295 of the 958 cases; Barbara Haddon for co-ordination of family recruitment, blood and phenotype collection; Genetic and Molecular Epidemiology Laboratories staff for sample processing and DNA preparation.
References
- Agarwal A, Gupta S, Sharma R. Oxidative stress and its implications in female infertility—a clinician's perspective. Reprod Biomed Online. 2005;11:641–650. doi: 10.1016/s1472-6483(10)61174-1. [DOI] [PubMed] [Google Scholar]
- Altomare DA, Menges CW, Pei J, Zhang L, Skele-Stump KL, Carbone M, Kane AB, Testa JR. Activated TNF-alpha/NF-kappaB signaling via down-regulation of Fas-associated factor 1 in asbestos-induced mesotheliomas from Arf knockout mice. Proc Natl Acad Sci USA. 2009;106:3420–3425. doi: 10.1073/pnas.0808816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Fertility Society. Revised American Fertility Society classification of endometriosis. Fertil Steril. 1985;43:351–352. doi: 10.1016/s0015-0282(16)48430-x. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Arya R, Hare E, Del Rincon I, Jenkinson CP, Duggirala R, Almasy L, Escalante A. Effects of covariates and interactions on a genome-wide association analysis of rheumatoid arthritis. BMC Proc. 2009;3(Suppl 7):S84. doi: 10.1186/1753-6561-3-s7-s84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcz E, Kaminski P, Marianowski L. Role of cytokines in pathogenesis of endometriosis. Med Sci Monit. 2000;6:1042–1046. [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkley KJ, Rapkin AJ, Papka RE. The pains of endometriosis. Science. 2005;308:1587–1589. doi: 10.1126/science.1111445. [DOI] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondza PK, Maheux R, Akoum A. Insights into endometriosis-associated endometrial dysfunctions: a review. Front Biosci (Elite Ed) 2009;1:415–428. doi: 10.2741/E38. [DOI] [PubMed] [Google Scholar]
- Braun DP, Ding J, Dmowski WP. Peritoneal fluid-mediated enhancement of eutopic and ectopic endometrial cell proliferation is dependent on tumor necrosis factor-alpha in women with endometriosis. Fertil Steril. 2002;78:727–732. doi: 10.1016/s0015-0282(02)03318-6. [DOI] [PubMed] [Google Scholar]
- Burney RO, Hamilton AE, Aghajanova L, Vo KC, Nezhat CN, Lessey BA, Giudice LC. MicroRNA expression profiling of eutopic secretory endometrium in women with versus without endometriosis. Mol Hum Reprod. 2009;15:625–631. doi: 10.1093/molehr/gap068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RI, Maxwell P, Jenkins D, McCluggage WG. Immunohistochemical staining with MIB1, bcl2 and p16 assists in the distinction of cervical glandular intraepithelial neoplasia from tubo-endometrial metaplasia, endometriosis and microglandular hyperplasia. Histopathology. 2002;41:313–321. doi: 10.1046/j.1365-2559.2002.01465.x. [DOI] [PubMed] [Google Scholar]
- Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al. A SNP in a let-7 microRNA complementary site in the KRAS 3' untranslated region increases non-small cell lung cancer risk. Cancer Res. 2008;68:8535–8540. doi: 10.1158/0008-5472.CAN-08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle JT. MicroRNAs suggest a new mechanism for altered brain gene expression in schizophrenia. Proc Natl Acad Sci USA. 2009;106:2975–2976. doi: 10.1073/pnas.0813321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Hooghe TM, Nugent NP, Cuneo S, Chai DC, Deer F, Debrock S, Kyama CM, Mihalyi A, Mwenda JM. Recombinant human TNFRSF1A (r-hTBP1) inhibits the development of endometriosis in baboons: a prospective, randomized, placebo- and drug-controlled study. Biol Reprod. 2006;74:131–136. doi: 10.1095/biolreprod.105.043349. [DOI] [PubMed] [Google Scholar]
- Didiano D, Cochella L, Tursun B, Hobert O. Neuron-type specific regulation of a 3'UTR through redundant and combinatorially acting cis-regulatory elements. RNA. 2010;16:349–363. doi: 10.1261/rna.1931510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer H, Mwenda JM, Chai DC, Wagner C, Song XY, Mihalyi A, Simsa P, Kyama C, Cornillie FJ, Bergqvist A, et al. Treatment with anti-TNF monoclonal antibody (c5N) reduces the extent of induced endometriosis in the baboon. Hum Reprod. 2006;21:1856–1862. doi: 10.1093/humrep/del044. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–1799. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- Glinskii AB, Ma J, Ma S, Grant D, Lim CU, Sell S, Glinsky GV. Identification of intergenic trans-regulatory RNAs containing a disease-linked SNP sequence and targeting cell cycle progression/differentiation pathways in multiple common human disorders. Cell Cycle. 2009;8:3925–3942. doi: 10.4161/cc.8.23.10113. [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ramos R, Van Langendonckt A, Defrere S, Lousse JC, Mettlen M, Guillet A, Donnez J. Agents blocking the nuclear factor-kappaB pathway are effective inhibitors of endometriosis in an in vivo experimental model. Gynecol Obstet Invest. 2008;65:174–186. doi: 10.1159/000111148. [DOI] [PubMed] [Google Scholar]
- Goumenou AG, Matalliotakis IM, Tzardi M, Fragouli IG, Mahutte NG, Arici A. p16, retinoblastoma (pRb), and cyclin D1 protein expression in human endometriotic and adenomyotic lesions. Fertil Steril. 2006;85(Suppl 1):1204–1207. doi: 10.1016/j.fertnstert.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Guo SW. Epigenetics of endometriosis. Mol Hum Reprod. 2009;15:587–607. doi: 10.1093/molehr/gap064. [DOI] [PubMed] [Google Scholar]
- Guo SW, Wang Y. Sources of heterogeneities in estimating the prevalence of endometriosis in infertile and previously fertile women. Fertil Steril. 2006;86:1584–1595. doi: 10.1016/j.fertnstert.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufner K, Horn A, Derfuss T, Glon C, Sinicina I, Arbusow V, Strupp M, Brandt T, Theil D. Fewer latent herpes simplex virus type 1 and cytotoxic T cells occur in the ophthalmic division than in the maxillary and mandibular divisions of the human trigeminal ganglion and nerve. J Virol. 2009;83:3696–3703. doi: 10.1128/JVI.02464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson JA, Haitina T, Lindblom J, Fredriksson R. Identification of six putative human transporters with structural similarity to the drug transporter SLC22 family. Genomics. 2007;90:595–609. doi: 10.1016/j.ygeno.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Kontorovich T, Levy A, Korostishevsky M, Nir U, Friedman E. SNPs in miRNA binding sites and miRNA genes as breast/ovarian cancer risk modifiers in Jewish high risk women. Int J Cancer. 2009 doi: 10.1002/ijc.25065. [DOI] [PubMed] [Google Scholar]
- Leyendecker G, Kunz G, Noe M, Herbertz M, Mall G. Endometriosis: a dysfunction and disease of the archimetra. Hum Reprod Update. 1998;4:752–762. doi: 10.1093/humupd/4.5.752. [DOI] [PubMed] [Google Scholar]
- Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-e>5 to Wolf–Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- Nyholt DR, Gillespie NG, Merikangas KR, Treloar SA, Martin NG, Montgomery GW. Common genetic influences underlie comorbidity of migraine and endometriosis. Genet Epidemiol. 2009;33:105–113. doi: 10.1002/gepi.20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2009a doi: 10.1093/humupd/dmp034. [Epub to heads of print] [DOI] [PubMed] [Google Scholar]
- Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, Perry N, Wagaarachchi P, Robertson SA, Print CG, Hull LM. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009b;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Chegini N. MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 2008;26:479–493. doi: 10.1055/s-0028-1096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Luo X, Toloubeydokhti T, Chegini N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol Hum Reprod. 2007;13:797–806. doi: 10.1093/molehr/gam063. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Sampson JA. Metastatic or embolic endometriosis, due to the menstrual dissemination of endometrial tissue into the venous circulation. Am J Pathol. 1927;3:93–110.. [PMC free article] [PubMed] [Google Scholar]
- Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2009 doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- Tian T, Shu Y, Chen J, Hu Z, Xu L, Jin G, Liang J, Liu P, Zhou X, Miao R, et al. A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev. 2009;18:1183–1187. doi: 10.1158/1055-9965.EPI-08-0814. [DOI] [PubMed] [Google Scholar]
- Treloar SA, O'Connor DT, O'Connor VM, Martin NG. Genetic influences on endometriosis in an Australian twin sample . Fertil Steril. 1999;71:701–710. doi: 10.1016/s0015-0282(98)00540-8. [DOI] [PubMed] [Google Scholar]
- Treloar SA, Wicks J, Nyholt DR, Montgomery GW, Bahlo M, Smith V, Dawson G, Mackay IJ, Weeks DE, Bennett ST, et al. Genomewide linkage study in 1176 affected sister pair families identifies a significant susceptibility locus for endometriosis on chromosome 10q26. Am J Hum Genet. 2005;77:365–376. doi: 10.1086/432960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Wang K, Bucan M, Grant SF, Schellenberg G, Hakonarson H. Strategies for genetic studies of complex diseases. Cell. 2010;142:351–353. doi: 10.1016/j.cell.2010.07.025. author reply 353–355. [DOI] [PubMed] [Google Scholar]
- Weiss G, Goldsmith LT, Taylor RN, Bellet D, Taylor HS. Inflammation in reproductive disorders. Reprod Sci. 2009;16:216–229. doi: 10.1177/1933719108330087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Kaye DM. Mechanistic insights into the link between a polymorphism of the 3'UTR of the SLC7A1 gene and hypertension. Hum Mutat. 2009;30:328–333. doi: 10.1002/humu.20891. [DOI] [PubMed] [Google Scholar]
- Yang H, Bocchetta M, Kroczynska B, Elmishad AG, Chen Y, Liu Z, Bubici C, Mossman BT, Pass HI, Testa JR, et al. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-kappaB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci USA. 2006;103:10397–10402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila Pa) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZZ, Nyholt DR, Le L, Martin NG, James MR, Treloar SA, Montgomery GW. KRAS variation and risk of endometriosis. Mol Hum Reprod. 2006;12:671–676. doi: 10.1093/molehr/gal078. [DOI] [PubMed] [Google Scholar]
- Zhao ZZ, Nyholt DR, Le L, Thomas S, Engwerda C, Randall L, Treloar SA, Montgomery GW. Genetic variation in tumour necrosis factor and lymphotoxin is not associated with endometriosis in an Australian sample. Hum Reprod. 2007;22:2389–2397. doi: 10.1093/humrep/dem182. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR, Kennedy SH. The genetic basis of endometriosis. Curr Opin Obstet Gynecol. 2001;13:309–314. doi: 10.1097/00001703-200106000-00011. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Treloar SA, Lin J, Weeks DE, Nyholt DR, Mangion J, MacKay IJ, Cardon LR, Martin NG, Kennedy SH, et al. Significant evidence of one or more susceptibility loci for endometriosis with near-Mendelian inheritance on chromosome 7p13–15. Hum Reprod. 2007;22:717–728. doi: 10.1093/humrep/del446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.