Abstract
Background
Comparative genomics resources, such as ortholog detection tools and repositories are rapidly increasing in scale and complexity. Cloud computing is an emerging technological paradigm that enables researchers to dynamically build a dedicated virtual cluster and may represent a valuable alternative for large computational tools in bioinformatics. In the present manuscript, we optimize the computation of a large-scale comparative genomics resource—Roundup—using cloud computing, describe the proper operating principles required to achieve computational efficiency on the cloud, and detail important procedures for improving cost-effectiveness to ensure maximal computation at minimal costs.
Methods
Utilizing the comparative genomics tool, Roundup, as a case study, we computed orthologs among 902 fully sequenced genomes on Amazon’s Elastic Compute Cloud. For managing the ortholog processes, we designed a strategy to deploy the web service, Elastic MapReduce, and maximize the use of the cloud while simultaneously minimizing costs. Specifically, we created a model to estimate cloud runtime based on the size and complexity of the genomes being compared that determines in advance the optimal order of the jobs to be submitted.
Results
We computed orthologous relationships for 245,323 genome-to-genome comparisons on Amazon’s computing cloud, a computation that required just over 200 hours and cost $8,000 USD, at least 40% less than expected under a strategy in which genome comparisons were submitted to the cloud randomly with respect to runtime. Our cost savings projections were based on a model that not only demonstrates the optimal strategy for deploying RSD to the cloud, but also finds the optimal cluster size to minimize waste and maximize usage. Our cost-reduction model is readily adaptable for other comparative genomics tools and potentially of significant benefit to labs seeking to take advantage of the cloud as an alternative to local computing infrastructure.
Keywords: cloud computing, elastic computing cloud, Roundup, comparative genomics, high performance computing, Amazon, orthologs
Introduction
Roundup1 is one of many bioinformatics applications that computationally compares hundreds, and soon to be thousands, of genomes to predict the evolutionary relationship between genes, organisms, and biological functions.2 At the core of Roundup is the reciprocal smallest distance algorithm (RSD),3,4 which uses global sequence alignment and estimates of evolutionary distance to detect orthologous genes between pairs of organisms. The RSD algorithm employs a computational pipeline that includes local homology (BLAST),5 global alignment (clustalw),6 and maximum likelihood estimates of evolutionary distances (paml)7 to find sequences in a pair of genomes that have reciprocally close evolutionary distances. The use of evolutionary distance and reciprocity in RSD can more effectively recover putative orthologs than other methods, such as reciprocal BLAST procedures. 4 However, this improvement in quality comes at the cost of increased computation. For comparative genomics resources like Roundup, the demand for computational power scales with the square of the number of genomes. A survey of UniProtKB,8 a comprehensive resource of protein sequences, shows that the number of fully sequenced genomes has doubled almost every year since 2003 while the increase in processing speed has lagged behind, continuing to grow under Moore’s Law of doubling every 18–24 months. As a consequence, Roundup’s computational demand has grown as much as eight times faster than processor capacity. This disparity is consistent with other complex bioinformatics computational tasks and underscores the necessity of building optimized code even in cases where massive compute capacity is available, and of seeking alternative strategies for computational processing in general.
In designing computational genomics tools that keep pace with the rate of genome sequencing, the challenge is to optimize the computational task while simultaneously optimizing the cost associated with expansion of local compute clusters. Issues that add to cost or complexity of expanding local compute clusters include: funding (up front cost, grant funding), efficiency (variable usage patterns, long queues), maintenance costs (datacenters, system administrators, shared usage), and the gap between demand and scaling the local cluster (demand can fluctuate hourly and daily while renting new space, hiring administrators, buying and installing new computers can take months and years).9 An emerging solution to these challenges is cloud computing.
Cloud computing offers rapid scaling, reduced management, pay-as-you-go pricing, code reproducibility and the potential for 100% utilization.10,11 The demand pricing model of cloud services is especially attractive when compared to local clusters that have highly variable usage patterns and the standard management and logistical issues—(eg, hiring systems administrators, acquiring space, installation, configuration, maintenance and full costs including electricity, etc.)—typical of local compute farms.12 However, the value of the cloud is vastly diminished if its ‘elastic’ (scaled up or down depending on demand) nature is not fully exploited.
In the present manuscript, we demonstrate fundamental principles to optimize large-scale bioinformatic operations using the comparative genomics tool Roundup and the cloud services offered by Amazon. We highlight the importance of optimizing code in order to reduce costs, and also describe the steps necessary to estimate costs on standard cloud infrastructures prior to launch. We show how our procedures can substantially reduce cloud- computing costs and discuss how the principles we have developed can be generalized to other types of bioinformatic tasks.
Methods
Cloud implementation
We previously published a series of methods to port the reciprocal smallest distance algorithm of Roundup to the Amazon cloud.13 Briefly, the cloud-based pipeline uses a combination of Simple Storage Service (S3), Elastic Compute Cloud (EC2), and Elastic MapReduce (EMR) provided by Amazon Web Services.14–16 The services are all based on a pay-per-use model (Cost Breakdown). We used the persistent storage provided by S3 to store Roundup’s code base, all genomes (Genome Data), executable programs linked to Roundup (BLAST, ClustalW, and codeml), and all computed results. For computations, we provisioned 8-core High-CPU instances with 7 GB of memory and 1.7 TB of local storage from EC2. For workflow management, we used EMR, Amazon’s implementation of Hadoop17 on EC2. By default, Hadoop is configured to run Java programs. Thus, we devised an approach to use the streaming mode of EMR to allow any program to be executed from the command line and still take advantage of MapReduce {Wall, 2010 #205}. This approach enabled us to run our existing codebase with almost no changes and demonstrated that the method could be easily adapted to any complex task and codebase.
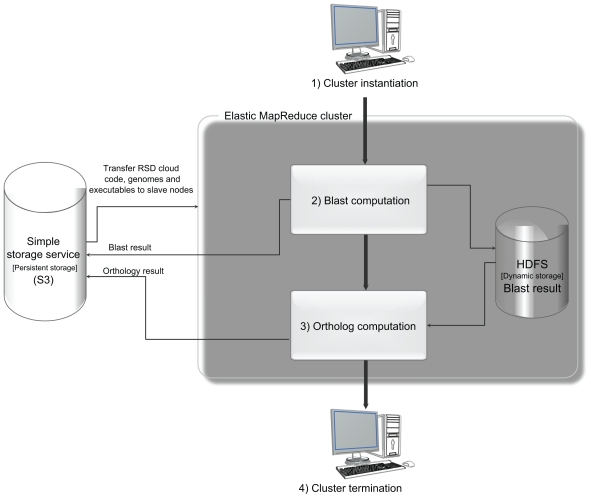
We used the Ruby command line interface18 to launch a cluster and configure jobs on EMR. The RSD pipeline was divided into BLAST computation followed by ortholog computation. Figure 1 demonstrates the cloud-computing infrastructure for Roundup. We monitored job completion status on the Hadoop user interface by establishing a SOCKS server on the local machine and an SSH tunnel between the local machine and master node.
Figure 1.
Cloud computing setup for executing the reciprocal smallest distance algorithm (RSD) used within the Roundup resource. Thin lines represent components of RSD for cloud computing infrastructure and connections among them. Thick lines emphasize the multi-step execution of the cloud- computing pipeline for execution of the RSD package. Steps 2 and 3 are the major stages of RSD, the BLAST computation stage, and ortholog detection stage. Further description of the workflow is provided in the text and in Wall et al.13
Code optimization
For the purposes of the present case study, and to ensure maximal cost reduction on the cloud, we optimized the speed and stability of RSD. Specific improvements included reducing disk I/O, increasing in-memory caching, and eliminating excessive invocations of the BLAST executable. In addition, we rewrote the RSD algorithm to simultaneously find orthologs for any number of parameter combinations, a feature particularly useful for Roundup because it contains orthologs for 12 combinations of E-values (1e-20, 1e-15, 1e-10, 1e-5) and divergence values (0.2, 0.5, 0.8). This optimized cloud-ready RSD may be found at http://wall.hms.harvard.edu/software.
Genome data
We tested the optimized configuration of RSD on a selection of 334 new genomes available in the Roundup database, which contained 568 genomes in total at the time of writing. Our genome selection contained a wide array of genomes from across the tree of life, and consequently a wide variety of sizes and sequence complexity sufficient to test the speed and cost of the cloud. The number of amino acid sequences per genome varied by over 2 orders of magnitude, with an average of 4,415 sequences. The total storage requirement to host all 334 genomes was 4.3 gigabytes (GB).
Cost breakdown
Following from our previous experiences with Amazon cloud services,13 we used 8-core High-CPU instances listed at a price of $0.68/hr at the time of writing. In addition, we used the Elastic MapReduce (EMR) processing service at $0.12 and S3 storage at $0.15 per gigabyte.
“Bin” model of computation time
We developed a model to simulate the computation time of executing RSD on a set of genomes on cluster of a given size. Given that RSD is a two-stage computational process—first BLAST is run for all pairs of genomes and then ortholog detection is run using pre-computed BLAST results to calculate orthologs over a range of parameters—we fit two linear models to account for computational complexity of both stages. For the BLAST stage of the RSD pipeline, we first fit a linear model between BLAST computational time and the product of the number of sequences in a pair of genomes. This model was based on the work of Altschul et al,19 who show that the expected time complexity of BLAST is O(WN), where W is linear in the length of a query sequence and N is the number of residues in the subject genome. By extension, running BLAST on all the sequences in a query genome is O(QN), where Q is the number of residues in the query genome. We used the number of sequences in a genome as an proxy for the number of residues, a proxy that, based on fit of the linear model (r2 = 0.8; P = 2.2e-16), yielded time estimations accurate enough for the purpose of ordering genome computations on a cluster in order to approximately minimize cluster idle time. For the ortholog detection stage, we fit a linear relationship between time and the minimum number of sequences in a pair of genomes (r2 = 0.3; P = 2.2e-16). This was based on a time complexity analysis of the ortholog detection stage, which internally performs O(1) work for each sequence in the genome with the fewest sequences (unpublished results). We fit these models using a preselected set of 30 genomes ranging in size from 75 to 66,710 sequences that were adequately representative of the phylogenetic diversity present in our larger sample.
We elected to estimate the overall runtime of an EMR cluster for both the BLAST and ortholog detection stages of the RSD algorithm, utilizing two different strategies of ordering the jobs for submission: 1) lexicographically by genome name and 2) descending order of estimated runtime. The first ordering was designed to be essentially random with respect to runtime, while the second was designed to avoid mixtures of long and short jobs that would tend to increase the number of idle nodes in a cluster at the end of a computation. In our simulations, we grouped jobs into “bins” equal to the number of nodes in a cluster, for a wide range of cluster sizes. We continually repopulated bins as computations on any given node in the cluster completed, with repopulation dictated by the style of ordering described above. We used the time estimates from these simulations to compare the alternative ordering strategies in terms of their predicted utilization of the cloud and percentage of idle nodes for a wide range of cloud cluster sizes in an effort to determine the optimal balance between speed of computation and cost.
Results
For the purposes of testing RSD on the cloud, we added 334 genomes to the 568 genomes currently in the Roundup database. This addition amounted in a total of 245,323 genome-to-genome pairs, following the formula (N*O + N*(N – 1)/2), where N was the number of new genomes and O was the number of pre-existing genomes in Roundup. The genomes varied considerably in size, and because RSD scales with the product of the sizes of the input genome pair, the time to process individual pairs of genomes also varied considerably, from 35 seconds to 71 hours.
Roundup optimization and computation
Reduction in disk I/O, in-memory caching, and the single invocation of the BLAST executable for a given genome-to-genome comparison lead to a two-fold increase in the speed of the BLAST stage of RSD in comparison to our un-optimized RSD code. In addition, simultaneous detection of orthologs for all 12 RSD parameter combinations reduced the total number of calculations by 12 and yielded a 7-fold increase in the speed of the orthology detection stage. These optimizations totaled to a 4.9 fold increase in speed over the earlier version of RSD across the set of genomes in Roundup. An additional benefit of the code optimization was increased stability, leading to fewer failed jobs and machines, an important benefit when running more jobs per machine and more machines per cluster.
Using the optimized code, we initiated computation of our 245,323 genome-to-genome comparisons on a 50-node cluster. In the testing phase of this run, we determined that the variation in genome size and con-comitant variation in runtime left a large percent-age of the cluster idle while long-running jobs finished computations. Because “out-of-the-box”, EMR did not have the capacity to reduce the size of the cluster without also terminating active processes, the variation in job size translated directly into wasted expense. To avoid this undesirable property of EMR on EC2, we elected to group genomes by size in terms of numbers of sequences (and presumably by runtime), reasoning that this would yield more effective utilization of nodes within a cloud cluster. To test this reasoning, we devised a strategy to predict computation time, percentage of cluster usage, and total costs that would model the optimal conditions for running RSD on the cloud.
Deployment of a strategy for optimal use of a cloud cluster
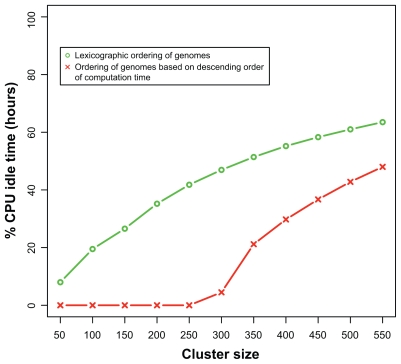
We designed a model to simulate the computational time over a range of different cluster sizes and orderings of jobs. Based on historical records of runtimes, we were able to determine that our model could predict actual run performance with greater than 95% accuracy. Figure 2 depicts a computational simulation for the BLAST stage of Roundup, and demonstrates the differences in efficiency between ordering jobs by genome name and ordering jobs in descending rank of estimated runtime. The simulation yielded the percentage of cluster idle time for the genomes currently in Roundup and provided a direct measurement of wasted expense, as idle nodes contributed to the total cost at the same rate as operational nodes. The simulation demonstrated that increasing the size of a cluster results in a tradeoff between reduction of over-all computation time (via a larger cluster that can run more jobs simultaneously) and percentage of cluster idle time. For example, Figure 2 illustrates the inflection point at which the percentage of idle time across the cluster outweighs the overall speed of computation for the BLAST stage of the RSD algorithm. Figure 2 also highlights the fact that when jobs were ordered by estimated runtime rather than lexicographically by genome name, a cluster size of 250 minimized the total runtime while also minimizing the percentage of idle processors. In general, our simulations demonstrated that ordering jobs by descending order of runtime rather than by genome name yielded the most efficient cloud costs for a wide range of cluster sizes.
Figure 2.
Graphical model to predict % CPU idle time for the BLAST stage of the RSD cloud-computing pipeline. The plot estimates CPU idle time for computation of 902 genomes on clusters of varying size ranging from 50 to 550 instances. The green line represents idle time when genomes are ordered lexicographically by genome name. The red line represents idle time when genomes are ordered in decreasing order of runtime. Under this simulation, use of 250-node cluster with jobs ordered by runtime rather than by genome name yielded the optimal cost-to-computation ratio.
By taking advantage of these cost estimation models and our cloud-optimized RSD code, we successfully completed 245,323 genome comparisons in just over 200 hours for a total cost of $8,000 USD. Plugging the same computation into our cost estimation model with order determined lexicographically by genome name, the total cost would have exceeded $11,000 USD, at least 40% more than our observed costs.
Discussion
The pay-per-use model of cloud computing provides an attractive alternative for high performance computing algorithms in bioinformatics. However, few case studies have been published to demonstrate how and when such infrastructure can be of benefit. In the present manuscript, we use a common ortholog detection tool, the reciprocal smallest distance algorithm (RSD) to demonstrate best usage practices of the Amazon Elastic Computing Cloud (EC2). We detail how RSD can be run on EC2 via the web service Elastic MapReduce (EMR), a service that facilitates spawning and management of large numbers of jobs over a cloud-based cluster of predefined size.
Amazon’s EMR is a Hadoop-based implementation of MapReduce on Amazon’s cloud. Hadoop was originally developed with the intent of crawling, indexing and processing huge amounts of data for Internet search engines. Several companies like Yahoo!, Facebook, and New York Times have taken advantage of Hadoop with great success. In general, the style of cluster computing embodied by EMR offers considerable advantages including job monitoring and flow management. However, EMR and similar Hadoop-based computing frameworks do not adapt well to batch-based algorithms like RSD. One significant disadvantage identified through our case study was the inability to identify and systematically terminate idle processors without impacting active jobs running on other nodes within the same cluster. Given the variance in genome size and sequence complexity among fully sequenced genomes, the time required to compute orthologs can vary widely, from minutes to days. We determined that such variation in runtime translates directly into substantial cloud waste, in terms of idle processors and unnecessary costs, if jobs run using EMR on EC2 are not managed appropriately a priori.
To account for this issue, we devised a model to consider the relationship between cluster size, job order, and idle time, as a means to predict and avoid unnecessary costs. We demonstrate how this model can be used to design an appropriate strategy for submitting jobs to a cloud-computing cluster and to determine the ideal size of the cloud cluster that maximizes both computational speed and cluster occupancy while minimizing costs. Applying our model to the RSD ortholog detection algorithm, we were able to estimate runtime for a wide array of genometo- genome comparisons, and test the performance of two potential means of queuing the jobs for submission to EC2 via EMR, either lexicographically by genome name, or in descending order of runtime. Our tests demonstrated that the latter approach significantly improved performance and maximized utilization of the cloud cluster. By grouping our jobs by runtime, we were able to run 245,323 RSD ortholog detection processes in 200 hours for a total cost of $8,000 USD. Had we ordered by genome name, or randomly with respect to runtime, our observed costs would have been at least 40% higher, stressing the importance of job management prior to launch.
Our case study highlights the importance of using services like EMR with some caution, and provides a valuable model of how to do so. Given the relative simplicity of our model, we expect it to be adaptable to other bioinformatics programs seeking to invest in cloud services for either supplanting or augmenting existing local computing infrastructure.
Conclusions
When faced with computational bottlenecks imposed by availability of local resources, cloud-computing services like Amazon represent potential alternatives. In the present manuscript, we used a common ortholog detection tool (the reciprocal smallest distance algorithm, RSD) to directly test the efficacy of the cloud offered by Amazon. Our work was motivated by our own local computing bottlenecks, and by the need to keep the coverage of genomes in Roundup1 up-to-date with the explosive rate of genome sequencing. We demonstrated how best to utilize the EMR webservice offered by Amazon for optimal job management and minimal costs. Specifically, we designed a model to predict job runtime and costs for an array of cloud cluster sizes, and showed how this model can be used to identify the optimal cluster size as well as the best strategy for ordering jobs prior to submission to the cloud. Most importantly, we show how our model can be used to achieve at least a 40% reduction in overall cloud computing costs. Our effective use of the cloud enabled a dramatic expansion of the Roundup database20 to over 900 genomes, making it among the largest publicly available orthology databases. In summary, our case study indicates that the cloud is a viable solution for boosting large-scale projects like Roundup, and provides a best-practice model for cloud computing that can be adapted to similar comparative genomics algorithms.
Acknowledgements
We would like to thank the Amazon Web Services for providing access to computational resources throughout the duration of the project. We are grateful to Prasad Patil, Rimma Pivovarov, Kristian Che St-Gabriel, and Jike Cui for feedback and advice during the project. Our work was funded by grants from the National Science Foundation (0543480 and 0640809) and National Institutes of Health (LM009261 and LM009261-02S1) awarded to Dennis P. Wall.
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Deluca TF, Wu IH, Pu J, et al. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006 Jun 15; doi: 10.1093/bioinformatics/btl286. [DOI] [PubMed] [Google Scholar]
- 2.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 2009 Jan;5(1):e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wall DP, Deluca T. Ortholog detection using the reciprocal smallest distance algorithm. Methods Mol Biol. 2007;396:95–110. doi: 10.1007/978-1-59745-515-2_7. [DOI] [PubMed] [Google Scholar]
- 4.Wall DP, Fraser HB, Hirsh AE. Detecting putative orthologs. Bioinformatics. 2003 Sep 1;19(13:):1710–11. doi: 10.1093/bioinformatics/btg213. [DOI] [PubMed] [Google Scholar]
- 5.Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A. 1990 Jul;87(14:):5509–13. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003 Jul 1;31(13:):3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997 Oct;13(5:):555–6. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 8.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 9.Rosenthal A, Mork P, Li MH, Stanford J, Koester D, Reynolds P. Cloud computing: a new business paradigm for biomedical information sharing. J Biomed Inform. 2009 Apr;43(2:):342–53. doi: 10.1016/j.jbi.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Dudley JT, Pouliot Y, Chen R, Morgan AA, Butte AJ. Translational bioinformatics in the cloud: an affordable alternative. Genome Med. 2010;2(8):51. doi: 10.1186/gm172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudley JT, Butte AJ. In silico research in the era of cloud computing. Nat Biotechnol. 2010 Nov;28(11:):1181–5. doi: 10.1038/nbt1110-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armbrust M, Fox A, Griffith R, et al. Above the clouds: a Berkeley view of cloud computing. EECS Department, University of California; Berkeley: 2009. Feb 10 [Google Scholar]
- 13.Wall DP, Kudtarkar P, Fusaro VA, Pivovarov R, Patil P, Tonellato PJ. Cloud computing for comparative genomics. BMC Bioinformatics. 2010;11:259. doi: 10.1186/1471-2105-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean J, Ghemawat S. MapReduce: Simplified Data Processing on Large Clusters. OSDI’04: Sixth Symposium on Operating System Design and Implementation; San Francisco, CA. 2004. [Google Scholar]
- 15.http://developer.amazonwebservices.com/connect/entry.jspa?externalID=2264&categoryID=266
- 16.http://aws.amazon.com/console/
- 17.http://hadoop.apache.org/core/
- 18.Elastic Map Reduce Ruby Client. [Accessed on 2010 Nov 10]. Available at: http://developer.amazonwebservices.com/connect/entry.jspa?externalID=2264&categoryID=266.
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3:):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.http://roundup.hms.harvard.edu