Summary
Background
Aging is under genetic control in C. elegans but the mechanisms of lifespan regulation are not completely known. MicroRNAs (miRNAs) regulate various aspects of development and metabolism and one miRNA has been previously implicated in lifespan.
Results
Here we show that multiple miRNAs change expression in C. elegans aging, including novel miRNAs, and that mutations in several of the most up-regulated miRNAs lead to lifespan defects. Some act to promote normal lifespan and stress resistance while others inhibit these phenomena. We find that these miRNAs genetically interact with genes in the DNA damage checkpoint response pathway and in the insulin signaling pathway.
Conclusions
Our findings reveal that miRNAs both positively and negatively influence lifespan. Since several miRNAs up-regulated during aging regulate genes in conserved pathways of aging and thereby influence lifespan in C. elegans, we propose that miRNAs may play important roles in stress response and aging of more complex organisms.
Keywords: microRNAs, longevity, lifespan, aging, stress, C. elegans
Introduction
In nature, longevity is characterized by a remarkable degree of variability, but the factors that dictate this plasticity of aging are largely unknown. Research pioneered in C. elegans has identified multiple genetic mechanisms that play a significant role during organismal aging [1]. The accumulated evidence demonstrates that aging is influenced by a complex interplay of regulatory mechanisms that respond to stress, nutrient availability and environment. Mutations in the daf-2 insulin/IGF receptor gene cause an extension of lifespan and stress resistance by deregulating the function of the DAF-16/FOXO transcription factor [1]. Similarly, mutations in genes that modulate the response to caloric restriction and DNA repair have significant effects on longevity, through mechanisms independent of the insulin pathway [1]. Moreover, these pathways are evolutionarily conserved in higher organisms. Nevertheless, the complete genetic circuits by which these genes modulate aging are not fully elucidated.
MicroRNAs (miRNAs), discovered in C. elegans [2], are endogenous, noncoding RNAs with important roles in controlling gene expression and development in plants and animals. Diverse functions have been attributed to miRNAs - including roles in developmental timing, neuronal development, metabolism, immunity, lifespan and cancer [3] - and target predictions suggest that they may regulate thousands of human genes [4]. Still, only a small number of the thousands of known miRNAs have been implicated in a specific biological function, while the role of the vast majority of miRNAs remains unknown. Mutations in the developmental timing genes, lin-4 (a founding member of the miRNA family) and its target lin-14 significantly alter the normal aging process of C. elegans, thus revealing a degree of miRNA control of aging in C. elegans [5]. In addition, microarray analysis in C. elegans revealed distinct miRNA expression changes during aging [6]. These observations show that miRNAs may function in pathways that impact lifespan but little is known about the extent of their roles and their mechanisms.
Results
Deep-Sequencing of small RNAs from aging C. elegans
To identify miRNAs with functions in lifespan, we undertook a deep sequencing survey of miRNAs from aged tissue in wild-type (N2) and in a long-lived daf-2(e1370) mutant C. elegans. We hypothesized that aging-specific miRNAs might exhibit differential expression in older animals or in mutants with aberrant lifespan, such as daf-2(e1370), which live twice as long as wild-type C. elegans [7]. Total RNA was collected from N2 and daf-2 animals when they reached young adulthood (Day 0) and when animals reached Day 10, an inflection point when wild-type animals begin dying. We obtained a total of 889,762 sequences of small RNAs (15-25nt) from the 4 samples of C. elegans examined, of which 626,616 sequences (70.4%) aligned perfectly to the C. elegans genome (Table S1). Overall, our survey detected perfect matches to the mature forms of 120 out of the 155 miRNAs annotated in miRBase 13.0, which indicates the significant sequencing depth of our study. This allowed us to identify miRNAs, including novel species, that are enriched in aging animals.
Novel microRNAs in aged C. elegans
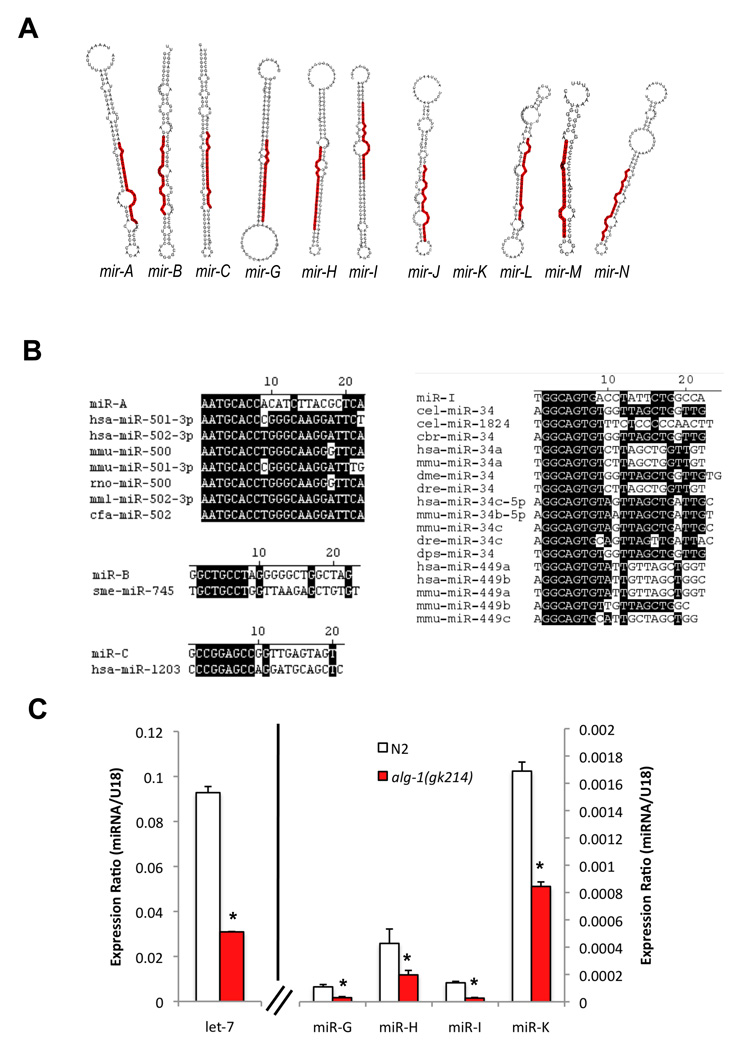
We hypothesized that yet-unidentified miRNAs may contribute to gene regulatory networks that affect lifespan. After discarding previously annotated ncRNAs and mRNAs from the 626,616 C. elegans aligned reads, we obtained 83,439 non-redundant, unannotated sequences from C. elegans. These 83,439 sequences, representing 142,689 total reads, were tested to determine if they represent novel C. elegans miRNAs. To identify likely miRNA candidates we used the miRNA-discovery software package, miRDeep, which identifies novel miRNAs based on the stability of their putative pre-miRNA hairpins in addition to "signature reads" - sequences found with the deep sequencing data that are consistent with miRNA biogenesis [8]. Our analysis revealed a set of 11 sequences that are predicted to fold into hairpin structures characteristic of bona-fide miRNAs (Fig. 1a) and represent likely novel miRNAs. Moreover, several of these novel miRNA candidates (mir-A, mir-B, mir-C and mir-I) share homology over their "seed" sequence (nucleotides 2–8 of the mature miRNA) with miRNAs in higher eukaryotes, including 3 human miRNAs, suggesting that they may be family members of these miRNAs (Fig. 1b). Interestingly, miR-I appears to be a new member of the miR-34 miRNA family, and miR-34 is one of the miRNAs that exhibited the greatest increase in expression with aging (see below) (Fig. S2 and Table 1).
Figure 1. Novel miRNAs identified in aged C. elegans.
(A) Secondary structures of putative precursor hairpins corresponding to miRNAs identified in a pool of RNA enriched for aged C. elegans. The predicted mature sequence is highlighted in red.
(B) Conservation of novel miRNAs miR-A, miR-B, miR-C and miR-I to known miRNAs. Identical nucleotides are highlighted in black.
C) Validation of expression of candidate novel miRNAs. The expression of novel miRNAs mir-G, mir-H, mir-I and mir-K was confirmed by TaqMan qRT-PCR in wild-type (N2) C. elegans. Consistent with their classification as miRNAs, their expression was significantly reduced in alg-1(gk214) mutant animals. The expression of the miRNA let-7 is shown as a positive control. The expression of a non-miRNA small RNA species (snoRNA sn2429) does not change in alg-1(gk214) background (DNS). *, p < 0.05, alg-1(gk215) compared to wild-type (N2) (Student's t-test).
Table 1.
Summary of largest expression changes of miRNAs with aging
| Most Upregulated with aging |
Most Downregulated with aging |
||||||
|---|---|---|---|---|---|---|---|
| miRNA | N2 | daf2 | Homology | miRNA | N2 | daf2 | Homology |
| miR-246 | 5.90 | 9.46 | let-7 | −7.38 | −5.39 | let-7 | |
| miR-71 | 4.04 | 4.21 | miR-41 | −6.78 | −2.34 | ||
| miR-34 | 3.93 | 3.28 | miR-34 | miR-70 | −5.70 | −3.80 | |
| miR-253 | 3.31 | 1.44 | miR-220 | miR-252 | −5.02 | −2.22 | |
| miR-238 | 2.88 | 2.43 | miR-12 | miR-62 | −4.30 | −2.91 | |
| miR-239b | 2.85 | 6.37 | miR-12 | miR-36 | −4.10 | −2.62 | |
| miR-239a | 2.15 | 1.91 | miR-12 | miR-59 | −3.56 | −1.41 | |
| miR-1829b | −3.44 | −2.31 | |||||
| miR-236 | −3.20 | −3.69 | miR-200b | ||||
| miR-37 | −3.20 | −2.15 | |||||
| miR-237 | −3.14 | −5.70 | lin-4, miR-125 | ||||
| miR-793 | −2.95 | −2.13 | |||||
| miR-43 | −2.72 | −2.10 | |||||
| miR-65 | −2.65 | −1.76 | |||||
| miR-40 | −2.59 | −2.50 | |||||
| miR-77 | −2.27 | −1.48 | |||||
| miR-79 | −2.22 | −4.93 | miR-9 | ||||
| miR-81 | −2.18 | −1.88 | |||||
| miR-82 | −2.17 | −1.82 | |||||
| miR-259 | −2.16 | −1.37 | |||||
| miR-38 | −2.14 | −1.84 | |||||
| miR-64 | −2.11 | −1.61 | |||||
| miR-35 | −2.06 | −1.81 | |||||
|
Most Upregulated in daf-2 |
Most Downregulated in daf-2 |
||||||
| miRNA | Day 0 | miRNA | Day 10 | miRNA | Day 0 | miRNA | Day 10 |
| miR-237 | 2.94 | miR-62 | 3.00 | miR-1829c | −4.49 | miR-242 | −2.24 |
| miR-49 | 2.26 | miR-49 | 2.93 | miR-239b | −3.40 | ||
| miR-253 | 2.21 | miR-252 | 2.22 | miR-259 | −2.80 | ||
| miR-44 | 2.05 | miR-231 | 2.19 | miR-246 | −2.04 | ||
| miR-45 | 2.05 | miR-85 | −2.00 | ||||
| miR-62 | 2.03 | ||||||
Top. Summary of miRNAs with >2-fold change in expression from day 0 to day 10 of adulthood in wild-type C. elegans (N2) in our cloning survey, after normalizing for total reads that match the C. elegans genome (values that represent >2-fold change are indicated in bold). Also shown are those miRNAs that belong to miRNA families conserved in other phyla (miR-12 is a Drosophila homolog). Bottom. List of miRNAs with >2-fold expression changes in daf-2(e1370) mutants as compared to wild-type (N2) animals on day 0 and day 10 of adulthood.
Several of these novel candidates – such as miR-N, miR-L, miR-M, miR-G, miR-H, miR-I and miR-K - are expressed preferentially in aged C. elegans (Fig. S1). In addition, these miRNAs show reduced expression in daf-2(lf) worms in both young and old animals (Fig. S1). Thus, these novel miRNAs seem to be preferentially expressed in normal, aged worms – a result that is consistent with the rationale of the experiment. Finally, four miRNAs - miR-A, miR-B, miR-C and miR-I are expressed only in the daf-2 samples, and may represent miRNAs that are normally under the regulation of the IIS pathway.
In order to validate the expression of our candidate novel miRNAs, we designed custom TaqMan assays specific to the putative mature sequence of the four candidate miRNAs for which we had the fewest reads - miR-G, miR-H, miR-I, and miR-K. Our qRT-PCR results showed that indeed all four candidates were expressed in aged C. elegans, although very much less frequently, as expected, than previously characterized miRNAs, such as let-7 (Fig. 1c). Importantly, the expression of these tested candidate miRNAs was significantly reduced in mutant C. elegans animals with attenuated expression of the miRNA-associated factor ALG-1 (Fig. 1c). As alg-1 has been shown to be important for mature miRNA accumulation [9], our results are consistent with the classification of mir-G, mir-H, mir-I and mir-K as novel miRNAs.
Changes in expression of miRNAs with aging
In an effort to discover miRNAs with functions specific to post-developmental processes, we measured the expression changes of all miRNAs in our sequencing data over the course of aging and in long-lived mutants. We observed a relative overall reduction in the expression of known miRNAs between days 0 and 10 of adulthood in both wild-type and daf-2 animals. Whereas nearly half of all sequencing reads in Day 0 wild-type adult animals represent known miRNAs (47.9%), by Day 10 the frequency of known miRNAs dropped to 33.6% of all genome-matching reads (Table S1). This pattern was similar in the long-lived daf-2 mutants (45.1% at Day 0 to 38.1% by Day 10). We found that although the expression levels of many miRNAs remain unchanged, a subset of miRNAs exhibit considerable changes in expression with aging (Fig. S2). For purposes of comparing expression changes of individual miRNAs, here we considered only those 81 miRNAs for which we obtained more than 10 sequences reads between the 2 samples being compared. Using this criteria, 8.6% miRNAs increased and 28.4% decreased in expression by more than 2-fold from Day 0 to Day 10 of adulthood in wild-type N2 animals (Fig. S2, Table 1). daf-2 animals exhibited a nearly identical pattern: 9.8% increased while 25.6% miRNAs decreased (Fig. S2, Table 1).
miR-246 was the most up-regulated miRNA during aging, showing nearly a 6-fold increase in expression from Day 0 to Day 10 in wild-type. Other miRNAs that exhibited strong increases in expression during aging in wild-type include miR-71, which became the second most abundant miRNA by Day 10 of aging (>4-fold up-regulation), miR-34 (4-fold up-regulation), miR-253 (3.3-fold up-regulation), and the homologous miRNAs miR-238 (2.9-fold up-regulation) and miR-239a/b (>2-fold up-regulation) (Fig. S2 and Table 1). Conversely, let-7 was the most down-regulated miRNA in aging - exhibiting a 7.4-fold decrease in expression from day 0 to day 10 of adulthood in N2 animals (Table 1). Other miRNAs that decreased in expression with aging include miR-41 (>6-fold down-regulation), miR-70 (>5-fold down-regulation) and miR-252 (>5-fold down-regulation). Notably, two miRNAs with dynamic changes in expression with aging, let-7 and mir-34, are known to be involved in cancer [10, 11]. In a further link between miRNAs and lifespan, a recent report has implicated let-7 in aging-induced senescence in mouse neuronal stem cells [12].
We validated our measurements of the expression of several miRNAs by using several established measures of RNA expression. We tested five miRNAs that we cloned more frequently in old animals (miR-246, miR-238, miR-71, miR-240 and miR-67) and three that we cloned less frequently in old animals (let-7, miR-70, miR-237), and found that the changes in expression with aging detected by TaqMan qRT-PCR correlated with the expression changes we observed by cloning for both N2 (r=0.90) and daf-2 animals (r=0.90) (Fig. S3). We also used northern blot to compare the expression changes with aging of representative miRNAs, such as miR-246, the most highly up-regulated miRNA in our cloning survey, and, as a control, miR-66, whose expression remains constant. Consistent with our cloning results, we found that in both wild-type and daf-2 mutants that miR-66's expression remains unchanged while miR-246's expression increases significantly with aging (Fig. S3b).
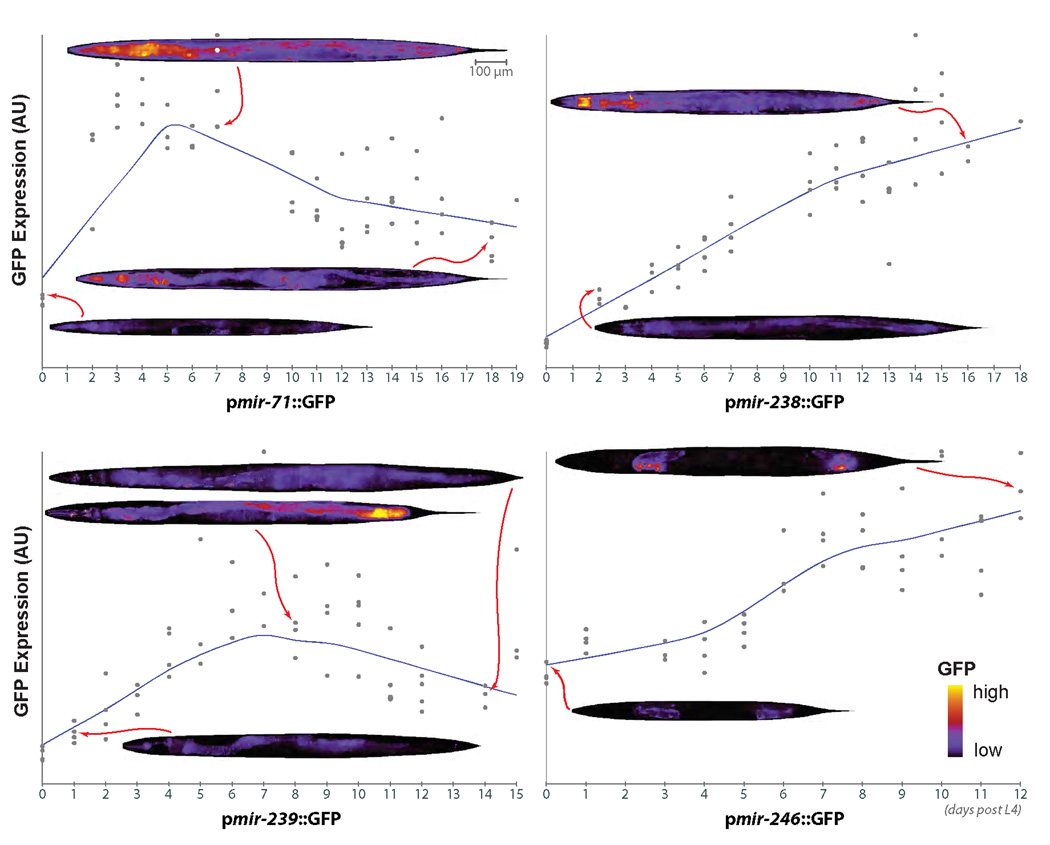
In order to characterize the spatio-temporal expression pattern of these miRNAs during aging, we examined GFP expression driven by promoters of aging-associated miRNAs (Fig. 2). In confirmation of our deep-sequencing and qRT-PCR findings, we observe a significant increase in expression of GFP driven by the promoters of mir-71, mir-238, mir-239 and mir-246 during aging (Fig. 2). Interestingly, both pmir-71::GFP and pmir-239::GFP reach maximum GFP expression between days 5–7 of adulthood, which subsequently wanes, whereas pmir-238::GFP and pmir-246::GFP expression increases steadily until the end of life. As reported before, miR-71 and miR-238 are expressed ubiquitously and miR-246 is expressed in gonadal sheath cells [13]. During aging, pmir-71::GFP is noticeably increased throughout the intestine and in the pharynx (Fig. 2). By contrast, pmir-239::GFP expression is seen primarily in intestine and in neurons (Fig. 2). Although the GFP expression patterns in transgenic animals may not fully reflect the endogenous expression patterns of these miRNAs (particularly as expression of genes introduced by microinjection is often silenced in the germline), the observation of increased expression of these miRNAs in intestine, neurons and somatic gonad is particularly interesting as signaling in these tissues has been previously shown to regulate lifespan in C. elegans [1].
Figure 2. Spatio-temporal expression changes in miRNA expression during aging.
The expression of GFP in C. elegans reporter strains for 4 aging-associated miRNAs was surveyed during adulthood. Consistent with deep-sequencing and qRT-PCR results, the expression of GFP driven by the promoters of mir-71, mir-238, mir-239 and mir-246 increased during aging. miR-71 and miR-238 are expressed ubiquitously and miR-246 is expressed in gonadal sheath cells, as reported before [13]. During aging, pmir-71::GFP expression increases in the intestine and in the pharynx, while pmir-239::GFP expression is seen primarily in intestine and in neurons, tissues previously implicated in regulation of lifespan in C. elegans [1]. Representative images showing changes in GFP intensity are shown (warmer colors indicate higher GFP expression).
Changes in expression of miRNAs in daf-2 animals
The insulin/insulin-like growth factor-1 (IGF-1) receptor, DAF-2, is a key component of the insulin/insulin-like IGF-1 signaling (IIS) pathway in C. elegans that responds to environmental signals to regulate stress response and lifespan. Previous studies have identified a wide variety of mRNAs whose expression is altered in daf-2 mutant animals and found that a significant number of these genes could affect lifespan when mutated [14]. Similarly, we hypothesized that miRNAs that exhibit significantly altered expression in daf-2 animals may be good candidates for novel lifespan determining genes. In an effort to identify miRNAs that depend on the IGF-1/daf-2 pathway, we compared the miRNA profiles of wild-type animals with those in daf-2(e1370) mutants.
We observe that the vast majority of miRNAs exhibit very similar changes in expression with aging between N2 and daf-2 animals (r=0.83) suggesting that the changes in miRNA expression that we observed are reproducible across strains and that the expression of the majority of miRNAs is not affected by a loss of DAF-2 (Fig. S2). Although the profile of miRNA expression in N2 and daf-2 animals was similar, we found a few specific miRNAs that exhibit altered expression in daf-2 mutant animals (Fig. S2, Table 1). Notably, several of these miRNAs also exhibited altered expression with aging (see below), suggesting that they may be particularly good candidates for genes that influence lifespan. Interestingly, the miRNA with the greatest increase in expression in young daf-2 animals, miR-237, is one of the most down-regulated miRNAs in older animals (Table 1). In fact, other miRNAs exhibit this pattern - miR-62 and miR-252 are among the most up-regulated miRNAs in daf-2 mutant animals, when compared to wild-type of the same age (Day 10), in addition to being among the most down-regulated miRNAs with aging. Conversely, miR-239b which appears to be down-regulated in young daf-2 animal is also among the most up-regulated microRNAs with aging (Table 1). Collectively, these daf-2-associated miRNAs reveal the intersection of the daf-2 pathway with miRNA-regulated genes that impact lifespan (Table 1).
Certain aging-enriched miRNAs function in lifespan control
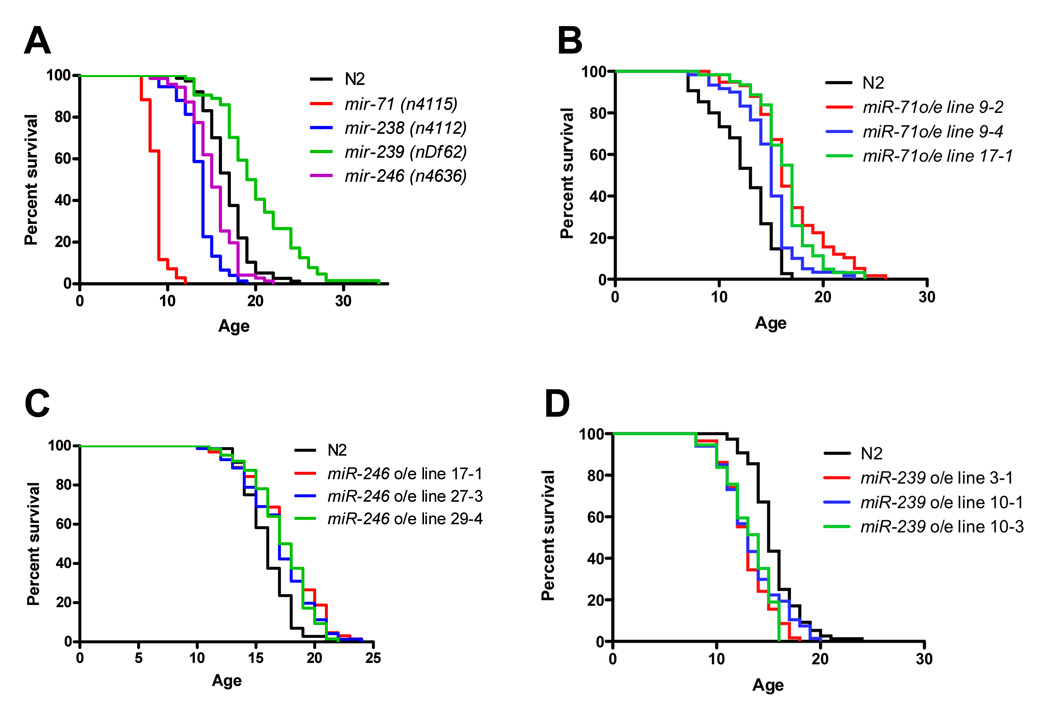
We considered the hypothesis that miRNAs displaying dramatic changes in expression across aging and in daf-2 mutants may function in lifespan control. Specifically, we focused on miRNAs that are over-expressed with aging since the up-regulation of specific miRNAs appears to be a common response of various organisms to stress conditions such as DNA damage, hypoxia and nutrient deprivation [10, 15–18]. In order to uncover putative functions for these miRNAs in lifespan, we examined mutants that contain deletions for the miRNAs of interest (Table 1) from the C. elegans Genome Consortium (CGC). These mutant C. elegans were backcrossed 3–6 times to the control laboratory wild-type C. elegans strain (N2) before assaying lifespan. Knock-out mutants for all miRNAs that exhibit greater than 2-fold increases in expression with aging were tested: mir-246(n4636), mir-71(n4115), mir-34(gk437), mir-253(nDf64), mir-238(n4112), and mir-239(nDf62). In addition, we also tested the lifespans of mir-240(n4541) and mir-67(n4899) - knock-out mutants of miRNAs that exhibited more modest increases of expression with aging (>1.5 fold). Lifespan assays on these mutants revealed that four of the miRNAs that exhibit the largest expression changes with aging - miR-71, miR-238, miR-239, and miR-246 - also influence lifespan (Fig. 3). Specifically, deletions of miR-71, miR-238 and miR-246 decreased lifespan, while deletion of miR-239 lead to reproducible and significant lifespan extension as compared to wild-type animals (Fig. 3a). The mean lifespan of these mutants was altered by ~10–50% in relation to wild-type N2 animals (Table S3). Other miRNA mutants tested - mir-34(gk437 and n4276), mir-253(nDf64), mir-240(n4541) and mir-67(n4899) - did not exhibit significantly altered lifespan (data not shown). Although the effects observed with our aging-associated miRNA mutants are not as dramatic as the aberrant lifespan observed in such mutants as daf-2 and age-1 (which typically double the lifespan of C. elegans [1]), they are within the range of other well characterized effectors of C. elegans lifespan such as the 5–20% extension induced by a clk-1 mutation [19, 20], the 12–20% extension caused by sir-2.1 overexpression [21], the 14% lifespan extension induced by resveratrol treatment [22], and the significance cut-offs (>5–10% extension) used in two large-scale RNAi screens [23, 24].
Figure 3. Deletion or over-expression of aging-associated miRNAs affects lifespan.
(A) Mutations in differentially expressed miRNAs affect lifespan. Four miRNA genes that are among the most over-expressed in aged animals - mir-71, mir-238, mir-239, and mir-246 - exhibit significant changes in lifespan when mutated. C. elegans mutants that contain deletions of miR-71, miR-238 and miR-246 - mir-71(n4115) (red), mir-238(n4112) (blue), and mir-246(n4636) (green) exhibit significantly reduced lifespan compared to wild-type (N2) animals (black), whereas deletion of miR-239 – mir-239(nDf62) – extends lifespan.
(B–C) miR-71 and miR-246 overexpression extends life. Three C. elegans lines that overexpress (o/e) miR-71 or miR-246 exhibit longer lifespans compared to wild-type N2 animals.
(D) miR-239 overexpression reduces lifespan. Three lines overexpressing (o/e) miR-239 (red, blue and green) exhibit significantly shorter lifespan as compared to wild-type N2 animals (black).
All lifespan assays conducted at 20°C. Statistics are shown in Table 3.
Since miR-71, miR-238 and miR-246 were highly up-regulated during aging (Table 1), these results suggest that these miRNAs increase during aging and promote longevity. An alternative explanation for the shorter lifespans of mir-71, mir-238 and mir-246 mutants might lie in non-specific sickness caused by deletion of these genes. However, none of these mutants exhibited any physiological or behavioral abnormalities during growth and development [25][26]; Fig. S4 and Table S4). In assays that tested morphology, growth, development, locomotion, pharyngeal pumping, defecation, egg-laying, and dauer formation these mutants all exhibited wild-type behavior [25, 26]. Nevertheless, and in order to test directly for a role in lifespan, we generated, via microinjection, transgenic C. elegans that over-express the aging-associated miRNAs miR-71, miR-238, miR-239 and miR-246 via extrachromosomal arrays [27]. Strikingly, we observe that over-expression of both miR-71 and miR-246 significantly and reproducibly increase longevity while miR-239 overexpression reduces lifespan (Fig. 3 b–d). Overexpression of miR-238 did not affect lifespan, suggesting that either this miRNA is not a bona fide aging gene or that its over-expression may cause confounding effects such as developmental abnormalities (Fig. S4a). Importantly, the lifespan phenotypes caused by miR-71, miR-239 or miR-246 overexpression are the opposite of what we observe when these miRNAs are deleted (Fig. 3a). Together, these observations strongly suggest a direct role for miR-71 and miR-246 to promote longevity and a function for miR-239 in pathways that antagonize longevity in C. elegans.
Stress Resistance of aging miRNAs
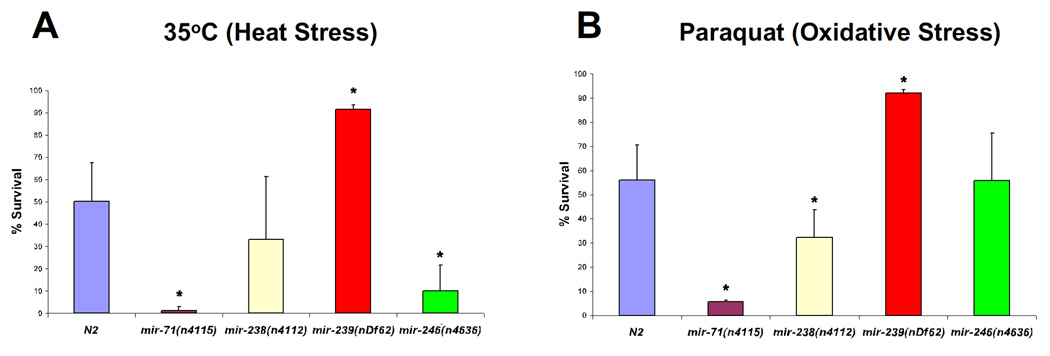
Since stress resistance is associated with lifespan [28], we considered the possibility that deletions of aging-associated miRNAs may also impact the response of C. elegans to stress. To that end, we tested the heat stress and oxidative stress response of C. elegans mutant adults bearing deletions of mir-71, mir-238, mir-239 and mir-246. Compared to the wild-type N2 strain, we observed that the long-lived mutant mir-239(nDf62) exhibited significant resistance to both heat stress (35°C) and oxidative stress (paraquat) (Fig. 4). Conversely, the short-lived mutant mir-71(n4115 exhibited a significantly increased sensitivity to both heat and oxidative stress while mir-238(n4112) and mir-246(n4636) caused increased sensitivity to oxidative and heat stress, respectively (Fig. 4). These results demonstrate that miR-71, miR-238, miR-239 and miR-246 function in pathways that regulate the response of C. elegans to conditions of environmental stress, in support of our observations that these miRNAs are necessary for normal lifespan in C. elegans.
Figure 4. Stress resistance in miRNA mutants correlates with their lifespan phenotypes.
(A) In response to heat shock (12h at 35°C), the short-lived miRNA mutants mir-71(n4115) and mir-246(n4636) exhibit increased thermo-sensitivity while the long-lived mir-239(nDf62) mutant exhibited significantly increased thermo-resistance as compared to wild-type animals (N2).
(B) Similarly, in response to oxidative stress (Paraquat, 6 hours), the short-lived mutants mir-71(n4115) and mir-238(n4112) exhibited decreased survival while the long-lived mir-239(nDf62) mutant exhibited significantly increased survival compared to wild-type animals.
Each strain (n>30) was examined in triplicate. Survival (%) and error bars represent the average survival and standard deviation between triplicate samples, respectively. p-values were calculated using Student's t-test: p=0.42 for mir-238(n4112) (heat stress); p=0.99 for mir-246(n4636) (paraquot); *, p < 0.05, compared to wild-type (N2).
Aging-associated miRNAs function through the IGF-1/insulin-like pathway
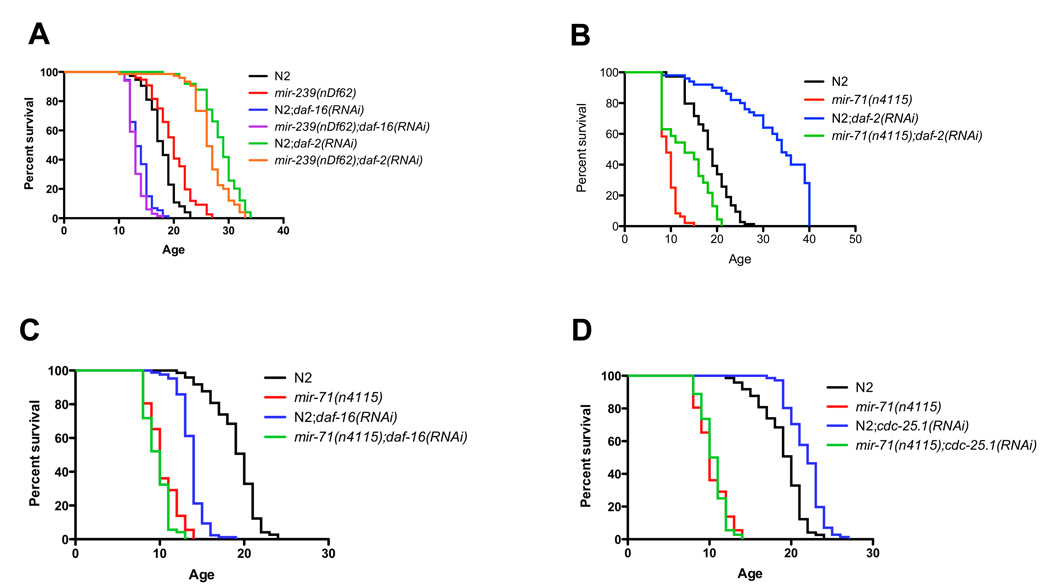
In order to understand the biological pathways that mediate the function of aging-associated miRNAs, we tested their function in the IGF-1/insulin-like pathway, which is known to mediate longevity in C. elegans, Drosophila and mouse. We tested if the aging phenotypes of mir-71(n4115), mir-238(n4112), mir-239(nDf62) and mir-246(n4636) would be affected in daf-2 and daf-16 loss-of-function environments. We found that RNAi directed against daf-2 and daf-16 modified the lifespan phenotypes of mir-71(n4115) and mir-239(nDf62), suggesting that these miRNAs function through the insulin-like pathway in C. elegans. Indeed, daf-16(RNAi) completely abolished the long lifespan of mir-239(nDf62) mutants, showing that the long-lived phenotype of mir-239 mutants depends on the presence of daf-16 (Fig. 5a). In addition, mir-239(nDf62) mutant animals did not further enhance lifespan in a daf-2(RNAi) background, suggesting that mir-239 and daf-2 function in the same genetic pathway (Fig. 5a). Similarly, we observed that mir-71(n4115) animals grown on daf-2(RNAi) were shorter lived than wild-type animals grown on daf-2(RNAi), which demonstrates that loss of miR-71 partially suppresses the long lifespan induced by daf-2(RNAi) (Fig. 5b). In addition, daf-16(RNAi) did not further decrease the lifespan of mir-71(n4115) animals, suggesting that mir-71 and daf-16 function in the same pathway (Fig. 5c). By contrast, we found that the reduction in lifespan of mir-246(n4636) mutants in daf-2(RNAi) was comparable to the effect of loss of miR-246 in an empty vector RNAi (~15% reduction in lifespan), suggesting that miR-246 does not function through daf-2 (Fig. S6a). In addition, we found that miR-238 is not necessary for longevity induced by loss of DAF-2 as mir-238(n4112) did not affect the lifespan of daf-2(RNAi) animals suggesting that miR-238 functions upstream of daf-2 or through an independent pathway (Fig. S6b). Together, these observations strongly suggest that miR-71 and miR-239 function through the IGF-1/insulin-like pathway to mediate lifespan in C. elegans.
Figure 5. miR-239 and miR-71 function through the IGF-1/insulin-like and the DNA Damage Checkpoint pathways.
(A) The long lifespan of mir-239(nDf62) is suppressed by daf-16(RNAi) but is unaffected by control (empty vector) RNAi. mir-239(nDf62) does not further enhance the lifespan on daf-2(RNAi), suggesting that they function in the same pathway.
(B–C) The long lifespan of daf-2(RNAi) is partially suppressed by mir-71(n4115) and daf-16(RNAi) does not further decrease the lifespan of mir-71(n4115), suggesting miR-71 functions in the same pathway as DAF-2 and DAF-16.
(D) miR-71 additionally functions in DNA damage checkpoint pathway. mir-71(n4115) fully suppresses the longevity induced by RNAi to CDC-25.1, a factor of the DNA damage checkpoint pathway.
All lifespan assays conducted at 20°C. Statistics are shown in Table S5.
miRNA function in DNA damage response
Since mir-71(n4115) did not fully suppress the long lifespan of animals on daf-2(RNAi), we considered whether miR-71 might also function in other pathways of aging. To that end, we tested whether the absence of a functional copy of miR-71 might affect the lifespan of mutants in the DNA damage checkpoint pathway, which has been shown to affect lifespan in C. elegans [29], We found that the increased lifespan observed when RNAi is directed against two checkpoint proteins – CDC-25.1 and CHK-1 – was completely suppressed in a mir-71(n4115) genetic background, suggesting that the function of CDC-25.1 and CHK-1 on lifespan depends on miR-71 (Fig. 5d and Fig. S6c).
miRNAs affect expression of aging pathway genes
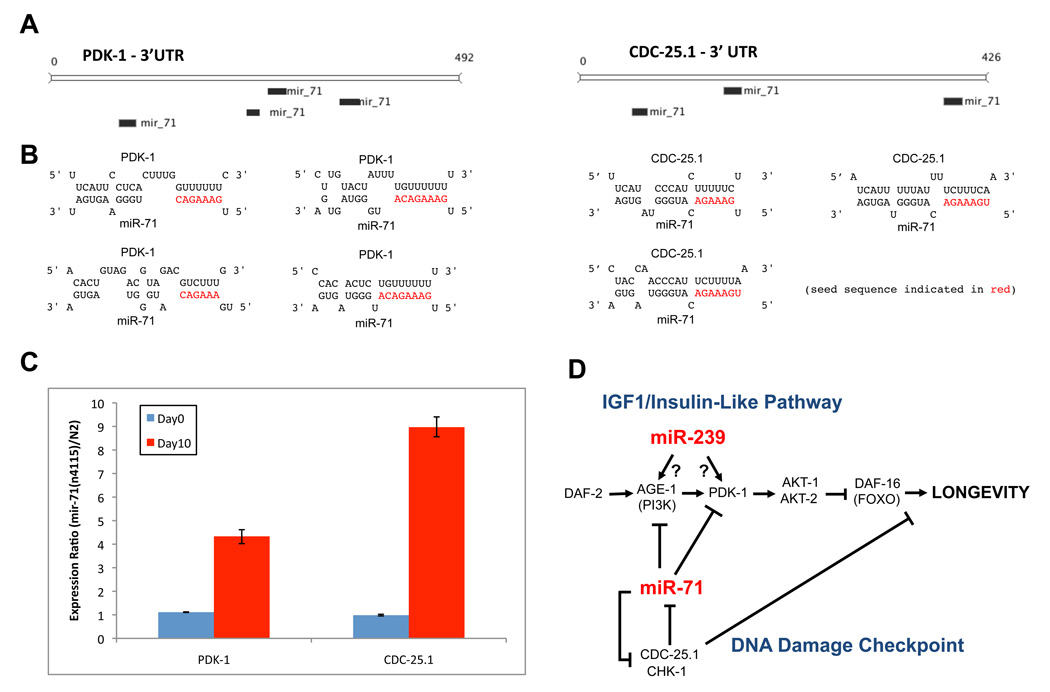
In order to understand the molecular basis of miRNA function in IIS and in the DNA damage checkpoint pathways, we examined the 3′ UTRs of genes in these pathways and searched for putative binding sites to the aging-associated miRNAs. We found that several genes in the IGF-1/insulin like pathway such as AGE-1, PDK-1, AKT-1 and DAF-16 have putative binding sites for miR-71 (Table S6) as predicted by mirWIP [30]. In addition, CHK-1, CDC-25.1 and CDC-25.2 are also predicted to be targets of miR-71 (Table S6). Indeed, miR-71 is predicted to have 4 binding sites in the 3’UTR of PDK-1 and 3 binding sites in CDC-25.1 (Fig. 6a). Since CDC-25.1 and PDK-1 antagonize lifespan, we reasoned that miR-71 may promote longevity by down-regulating these genes during adulthood. In order to test this hypothesis, we assayed the expression levels of these genes by qRT-PCR in a mir-71(n4115) background and found that the mRNA levels of both PDK-1 and CDC-25.1 in Day 10 adults were significantly elevated in mutants that lack miR-71, as compared to wild-type N2 animals (Fig. 6c). Interestingly, in young adults (Day 0), the levels of PDK-1 and CDC-25.1 were unchanged in a mir-71(n4115) background as compared to N2 animals suggesting that miR-71 functions to down-regulate these genes in older animals specifically (Fig. 6c). We also examined the expression levels of other predicted targets of miR-71 – AGE-1, DAF-16 and CHK-1 – in a mir-71(n4115) background and we found that the levels of these mRNAs were unchanged in Day 0 adults and modestly increased (< 2-fold) in Day 10 mir-71(n4115) adults, compared to wild-type worms.
Figure 6. Targets of miR-71 in IIS and DNA Damage Checkpoint pathways.
(A–B) Schematic representation and secondary structures of miR-71 binding sites on the 3’UTR of PDK-1, a component of IIS pathway, and on the 3’UTR of CDC-25.1, a component of the DNA Damage Checkpoint pathway.
(C) In a miR-71 deletion genetic background (mir-71(n4115)), the mRNA levels of PDK-1 and CDC-25.1 are significantly up-regulated specifically in older adult animals (Day 10) as compared to wild-type N2 animals, as measured by qRT-PCR, and after normalization by endogenous control ACT-1.
(D) Model for molecular and genetic interactions of age-related miRNAs with known aging pathways. Arrows denote positive interaction, -| denotes negative interactions.
Since we found that miR-239 function depends on DAF-16, we also searched for potential targets of miR-239 in the IIS pathway. Interestingly, however, none of the core genes in the IIS pathway are predicted to be targeted by miR-239. When we examined the expression of genes in the IIS pathway by qRT-PCR, we found a decrease in the mRNA levels of AGE-1 and PDK-1 in Day 10 mir-239(nDf62) animals (2.2-fold and 2.1-fold, respectively) (Fig. S6d), suggesting that miR-239 may act upstream of AGE-1/PDK-1 through a factor that is yet to be identified (Fig. 6d).
CONCLUSIONS
Our results reveal a greater involvement of miRNAs in C. elegans lifespan than previously appreciated, with some promoting and others suppressing longevity. We observed significant expression changes of multiple miRNAs during aging and also identified novel miRNAs in aged C. elegans. Analysis of the lifespan of strains of C. elegans containing mutations in miRNAs that exhibit the most significant changes of expression during aging demonstrated that three of the miRNAs that are most over-expressed in aged animals - miR-71, miR-238 and miR-246 - act to increase longevity in C. elegans, while a fourth miRNA - miR-239 - normally limits lifespan. We found, by genetic criteria, that both miR-71 and miR-239 function through the insulin-signaling pathway. In addition, we observed that miR-71 also interacts with the DNA damage response pathway, as it is required for the longevity extension caused by knock-down of CDC-25.1 and CHK-1. Given that miR-71 is predicted to target CDC-25.1 and CHK-1, this suggests a possible negative feedback loop, whereupon miR-71’s ability to promote longevity is antagonized by CDC-25.1, but miR-71, in turn, negatively regulates these genes (Fig. 6d). Consistent with this idea, we observed that in mutant animals that lack miR-71, the expression of CDC-25.1 in older animals is increased 8-fold. The fact that miR-71 functions in both the IIS and the DNA checkpoint pathways suggests that miR-71 is a possible link between these pathways (Fig. 6d).
Notably, the lifespan abnormalities reported here are the first observable phenotypes reported for these miRNAs. Indeed, an earlier survey failed to identify any phenotypes during growth and development for the vast majority of miRNA knock-out mutants in C. elegans [25]. Given that the majority of miRNAs individually seem to not be essential for C. elegans development, our observation of dramatic effects on adult lifespan for four independent loss-of-function mutants is striking. These results suggest that miR-71, miR-239 and miR-246 may be aging-specific genes - functioning specifically during adulthood to regulate genetic pathways that promote lifespan. Consistent with this idea, we observed that the up-regulation in mRNA levels of two predicted targets of miR-71 was specific to older animals. The absence of any phenotypes beside abnormal lifespans for these miRNA mutants further suggests that they might not be genes with antagonistic pleiotrophic function - i.e. beneficial early in life but detrimental post-reproduction. However, we cannot rule out a subtle function for these miRNAs in development - indeed, these miRNAs are expressed dynamically during development, especially miR-71 [31, 32]. It is possible that the trade-offs inherent to the roles of these miRNAs may only be apparent in specific environmental conditions, such as nutrient limitation and non-optimal temperature. In this case, these miRNAs may be used to fine-tune the response of C. elegans to environmental stress. In support of this idea, we observe that the miRNA mutants that we found to exhibit abnormal lifespan also show correspondingly abnormal responses to stress. These results are in agreement with previous studies that demonstrate that miRNAs may be particularly well suited to modulate organismal response to stress [15]. Since miRNAs have the potential to regulate multiple targets, increasing the level of one miRNA could potentially affect the action of a vast number of target genes. Indeed, the up-regulation of specific miRNAs has been noted in response to a variety of sources of stress in different organisms - DNA damage and oncogenic stress to mammalian cell lines, [10, 16], hypoxia in neoplastic cells [17], and nutrient deprivation in plants [18]. In that context, it is particularly interesting that we observe aberrant lifespan and stress responses in C. elegans mutants of miRNAs that are up-regulated with aging. These observations lend credence to the idea that miRNAs may be particularly useful modulators of gene regulatory pathways that respond to stress to affect an organism's lifespan. Since miRNAs exhibit high degrees of conservation of sequence and function it is likely that miRNAs also play extensive roles in aging in more complex animals.
MATERIALS AND METHODS
Strains and culture methods
The wild-type C. elegans strain N2 (Bristol) was cultured using standard protocols [33]. Mutant strains were obtained from the Caenorhabditis Genetics Center or generated in house and are listed in Supplemental Experimental Methods.
Lifespan assays
Lifespan assays were performed at 20°C as previously described [5]. Statistical analysis was done using Graphpad Prism 5.0 and JMP software to determine survival difference (log-rank Mantel-Cox test). Summary of lifespan statistics is shown in Tables S3–S4.
Stress Assays
Heat stress and oxidative stress (Paraquat) experiments were performed as previously described [34]. At least 30 animals were tested for each strain. Mean and standard deviation were determined from experiments done in triplicate. p-values were calculated using Student's t-test.
RNA isolation and sequencing
Total RNA was harvested from worm pellets, size-selected (15–25nt) and miRNAs were cloned as described before [35, 31]. The resulting cDNAs were then concatemerized, as described [36], and sequenced by 454 deep sequencing (454 Lifesciences / Roche) [37].
Computational data analysis
A total of 889,762 reads corresponding to small RNAs (15–30nt) were obtained by 454 deep sequencing. The deep-sequencing reads were aligned to the C. elegans genome (version WS190, obtained from Wormbase) and the number of sequence reads that correspond to known miRNAs was assessed using perfect sequence matching to a database of known miRNAs (miRBase, v.13.0). To compare the differential expression of miRNAs across samples, the number of miRNA reads in each sample was normalized to the total number of reads in each sample that matched the C. elegans genome (Table S1).
Novel miRNA Discovery
To identify novel miRNAs we used the software package miRDeep [8]. Beginning with all sequence reads that match the C. elegans genome perfectly, we discarded sequences that match previously annotated ncRNAs and mRNAs, using NCBI blastall. In addition, matches to known C. elegans miRNAs (miRBase v.13) and 21U-RNAs [37, 38, 32] were removed by perfect sequence matching. The remaining reads were then mined for putative novel miRNAs using miRDeep, using default settings [8], and secondary structures of putative pre-miRNA hairpins were generated using RNAFold [39].
Computational target prediction
We used the program mirWIP to identify targets of aging-associated miRNAs [30]. mirWIP targets are enriched for characteristics of bona-fide miRNA targets, including the structural accessibility of target sequences, the total free energy of miRNA-target hybridization and the topology of base-pairing to the 5' seed region of the miRNA [30]. As mirWIP’s target predictions originate from immuno-precipitation of the RISC factors AIN-1 and AIN-2 in C. elegans, they are particularly appropriate to our study, and are reported to have a lower false-positive rate than other methods [30]. mirWIP predictions were obtained from http://146.189.76.171/query.php; (predictions for mir-238 were not available for mirWIP).
Raw sequencing reads from 454 deep sequencing and processed small RNA sequences were deposited in NCBI's Gene Expression Omnibus [GSE24510]. Additional experimental details available in Supplemental Experimental Methods.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. elegans Genetic Center for supplying strains. ADL was supported by a National Research Service Award Post-Doctoral Fellowship from the NIH (1F32AG030851). ZP was supported by a Jane Coffin Childs Post-Doctoral Fellowship. FJS was supported by a Breakthrough in Gerontology grant from the American Federation for Aging Research, the Ellison Medical Foundation and the NIH (AG033921).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 6.Ibáñez-Ventoso C, Yang M, Guo S, Robins H, Padgett RW, Driscoll M. Modulated microRNA expression during adult lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:235–246. doi: 10.1111/j.1474-9726.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 7.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 8.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 9.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 10.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJM. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 15.Leung AKL, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol. Cell. Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 19.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272:1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 20.Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 21.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 22.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 23.Lee SS, Lee RYN, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 24.Hansen M, Hsu A, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Olsen A, Vantipalli MC, Lithgow GJ. Checkpoint proteins control survival of the postmitotic cells in Caenorhabditis elegans. Science. 2006;312:1381–1385. doi: 10.1126/science.1124981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M, Ding Y, Ambros V. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nat. Methods. 2008;5:813–819. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato M, de Lencastre A, Pincus Z, Slack F. Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol. 2009;10:R54. doi: 10.1186/gb-2009-10-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 36.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 37.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 38.Batista PJ, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. Chapter 12. Curr Protoc Bioinformatics. 2009 doi: 10.1002/0471250953.bi1202s26. Unit12.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.