Abstract
The 5′-untranslated regions (5′-UTRs) of all gammaretroviruses contain a conserved “double hairpin motif” (ΨCD) that is required for genome packaging. Both hairpins (SL-C and SL-D) contain GACG tetraloops that, in isolated RNAs, are capable of forming “kissing” interactions stabilized by two intermolecular G-C base pairs. We have determined the three-dimensional structure of the double hairpin from the Moloney Murine Leukemia Virus (MoMuLV) ([ΨCD]2, 132-nucleotides, 42.8 kDaltons) using a 2H-edited NMR spectroscopy-based approach. This approach enabled the detection of 1H-1H dipolar interactions that were not observed in previous studies of isolated SL-C and SL-D hairpin RNAs using traditional 1H-1H correlated and 1H-13C-edited NMR methods. The hairpins participate in intermolecular cross-kissing interactions (SL-C to SL-D’ and SLC’ to SL-D), and stack in an end-to-end manner (SL-C to SL-D and SL-C’ to SL-D’) that gives rise to an elongated overall shape (ca. 95 Å by 45 Å by 25 Å). The global structure was confirmed by cryo-electron tomography (cryo-ET), making [ΨCD]2 simultaneously the smallest RNA to be structurally characterized to date by cryo-ET and among the largest to be determined by NMR. Our findings suggest that, in addition to promoting dimerization, [ΨCD]2 functions as a scaffold that helps initiate virus assembly by exposing a cluster of conserved UCUG elements for binding to the cognate nucleocapsid domains of assembling viral Gag proteins.
Keywords: retrovirus, RNA structure, NMR spectroscopy, cryo-electron tomography, genome packaging
Considerable effort has been made over the past three decades to understand the mechanisms that retroviruses use to selectively package their RNA genomes.1-8 All retroviruses contain two positive-strand RNA genomes that are encapsidated within the central core of the virus.9 Both copies of the genome are required for replication, allowing strand-transfer to occur at strand breaks during reverse transcription and promoting genetic diversity through recombination in heterozygous particles.10-13 Genome selection is mediated by packaging elements, called Ψ-sites, which are typically located within the 5′-untranslated region (5′-UTR) of the RNA.1,2,9,14,15 Elements that promote RNA packaging generally overlap with those that promote dimerization,4,16 and both processes are mediated by the nucleocapsid domain (NC) of the retroviral Gag protein. The RNA exists as a non-covalently linked dimer in mature particles, and although small amounts of monomeric genomes can be isolated under mildly denaturing conditions from young virions,17 there is now considerable genetic evidence that genomes are selected for packaging as dimers, supporting early proposals that dimerization and packaging are mechanistically coupled.5,7,18-24
Much of what is currently known about genome packaging has been derived from studies of the Moloney Murine Leukemia Virus (MoMuLV). MoMuLV is a gammaretrovirus that has been widely used as a vector for gene delivery2 and is closely related to Xenotropic murine leukemia virus (MLV)-related virus (XMRV), a potential human pathogen25 with links to aggressive forms of prostate cancer26 and Chronic Fatigue Syndrome.27 Although both disease links have been questioned,28,29 very recent studies suggest the CFS may be caused by a genetically diverse group of MLV-related viruses30. Early nucleotide accessibility mapping, phylogenetic analyses and free energy calculations indicated that the MoMuLV 5′-UTR consists of a series of closely spaced hairpins, and that the RNA undergoes changes in secondary structure upon dimerization.31-34 Mutagenesis studies revealed that dimerization and packaging are promoted by four stem loops, two that are capable of forming intermolecular duplexes (DIS1 and DIS2)16,31,33,35-38 and two that can form “kissing interactions” mediated by base pairing between residues within their conserved GACG tetraloops (SLC and SLD).39 The latter hairpins are essential for genome packaging and have been proposed to function as “double hairpin motif” (ΨCD).40
We recently presented evidence that register shifts in the base pairing of DIS-1 and DIS-2 that occur upon dimerization expose conserved UCUG elements that are capable of binding NC with high affinity, Figure 1.41 The UCUG elements are sequestered by base pairing in the monomeric RNAs and unable to bind NC. Although these studies suggest well-defined roles of the DIS-1 and DIS-2 hairpins in regulating exposure of NC binding sites, the mechanistic role of the ΨCD double hairpin remains less well understood. Deletion of ΨCD can reduce vector RNA packaging to ~1% of wild-type levels, and introduction of a segment containing ΨCD and some adjacent residues (A285-C419) into a non-packaged vector RNA can promote packaging to nearly wild-type levels.42 However, isolated SL-C, SL-D, and ΨCD RNAs do not exhibit significant affinity for NC.43 In addition, although the GACG tetraloops are capable of forming kissing dimer contacts in small RNAs, they do not appear to contribute significantly to the monomer-dimer equilibrium measured for the intact 5′-UTR (although they enhance the rate of equilibration in vitro).31 Structures have been determined by NMR for the kissing dimer form of the isolated SL-D hairpin,39 as well as for larger RNAs that contain loop mutations that prevent kissing interactions,41,44 and site-directed hydroxyl radical cleavage and ribose reactivity (SHAPE) experiments have been used to derive a model for the tandem hairpin of the closely related Moloney Murine Sarcoma Virus (MoMuSV, which differs from the MoMuLV tandem hairpin by a single G338/U substitution).45 The latter studies suggested that the MuSV [ΨCD]2 RNA forms a highly stable, globular structure, in which the GACG tetraloops of SL-C and SL-D participate in cross-kissing (SL-C to SL-D’ and SL-C’ to SL-D) interactions.46
Figure 1.

Conformational states detected by NMR for the MoMuLV ΨCES site. (a) Unspliced MoMuLV genome showing the location of ΨCES. (b) Secondary structures of the monomeric ΨCES site. DIS-2 and SL-C exist in two alternate conformations (the palindromic sequence is shown in green, and conserved GACG sequences required for kissing interactions are shown in blue). (c) Secondary structure of the dimeric core encapsidation signal ([ΨCES]2). A frame shift upon dimerization exposes UCUG elements (red) that can function as potential NC binding sites.
To better understand the role of ΨCD in dimerization and packaging, we have characterized the solution-state equilibrium properties of the tandem hairpin in the context of the intact core encapsidation signal (ΨCES), which includes DIS-2, SL-C and SL-D, and have determined the structure of the dimeric MoMuLV tandem hairpin ([ΨCD]2) using a 2H-edited NMR approach. The NMR data confirmed the cross-kissing interactions predicted by chemical probing and mutagenesis.46 However, local and global structural features, and particularly the elongated overall shape of the NMR structure, differ significantly from the more globular SHAPE-derived model. We therefore independently assessed the overall shape of [ΨCD]2 by cryo-electron tomography (cryo-ET). Both the individual RNA densities in the unfiltered tomograms and the final cryo-ET average density map were consistent with the elongated structure determined by NMR. The combination of high-resolution local structural interactions derived by 2H-edited 2D NOESY NMR with global structural shape information derived by cryo-ET could serve as a general approach for structural studies of larger RNAs, particularly those that adopt multiple equilibrium conformations in solution. Our findings are consistent with a packaging model in which the tandem hairpin functions as a scaffold that, upon dimerization, helps expose of a cluster of high-affinity Gag binding sites.
RESULTS
The double hairpin exists in solution as interconverting conformers
The conformational behavior of the double hairpin motif was initially characterized in the context of a larger ¬CES construct that included residues of DIS-2, Fig. 1. Previous studies showed that ΨCES exists predominantly as a monomer at low RNA concentrations and low ionic strength, and that the DIS-2 hairpin adopts two slowly inter-converting conformers,41 Fig. 1b. We now show that the non-kissing form of SL-C also exists in two slowly-interconverting conformations, one in which conserved residues G229-G332 form a tetraloop and G338 to A341 form a bulge, an another in which several of these residues form Watson-Crick base pairs, Fig. 1c. Both of these structures (and only these structures) are predicted to occur based on free energy calculations with MFOLD.47 Analysis of 2D NOESY spectra obtained for an isolated SL-C RNA confirmed the presence of the predicted structures (Supplementary Figure S1), and NMR spectra obtained for native ΨCES exhibited exchange cross peaks consistent with those observed for SL-C and indicative of the conformational equilibrium shown in Fig. 1. The exchange peaks persisted in NMR spectra obtained for ¬CES under physiological concentrations of salts that favor dimerization (140 mM KCl, 10 mM NaCl, 1 mM MgCl2), even though the intensities of the resolved diagonal peaks indicated that the population of non-kissing conformer is low (< 5%).
MFOLD calculations indicated that substitution of the U328-A333 base pair preceding the SL-C GACG tetraloop by an A-U base pair would prevent formation of the minor, non-kissing conformer. Interestingly, a comparison of the nucleotide sequences of the gammaretroviruses revealed that this substitution occurs naturally. We therefore prepared RNA constructs in which these residues were swapped. As predicted, these conservative substitutions (U328A/A333U) precluded formation of the alternate, non-kissing SL-C conformer and gave rise to NMR spectra that lacked the associated conformational exchange peaks (Figure 2B and Supplementary Figure S1). Therefore, in order to reduce crowding in the NMR spectra and simplify analyses, subsequent NMR studies were conducted with RNAs containing the U328A/A333U substitutions, Figure 2c.
Figure 2.

Effects of a U328-A333 to A328-U333 base pair substitution on the RNA conformational equilibrium and gRNA packaging into the virus. (a) A portion of 2D NOESY spectrum obtained for wild type [ΨCES]2. Exchange peaks associated with the alternate conformations of SL-C shown in Figure 1 are outlined by red (A/G-H8) and blue (A-H2) boxes. (b) Portion of 2D NOESY spectrum obtained for the U328A/A333U mutant form of [ΨCES]2. Exchange peaks observed in the wild type spectrum are no longer observed, consistent with MFOLD calculations indicating that the substitution favors the SL-C secondary structure shown in (c). (c) Secondary structure and dimer interface consistent with the NMR data. (d) Riboprobe used for quantification of genomic RNA packaging, which includes sequences complementary to portions of the MoMuLV 5′ untranslated region as well as to host 7SL RNA. Thin line: flanking vector-derived sequences; boldface lines: MoMuLV- and 7SL-derived sequences. Numbers indicate RNA map positions. (e) RNase protection assay of RNA extracted from virions produced by transient transfection of 293T cells. Migration of probe fragments protected by intact MoMuLV genomic RNA, Ψ deletion variant, and 7SL are indicated on the right. Lane 1: undigested probe (P); 2: RNA size markers (M); 3: RNA from wild type leader virus; 4: “classic Ψ” deletion virus (Δ215-568)8; 5: virus produced by leader-deleted GagPol expression plasmid (helper); 6: SL-C mutant virus (U328A/A333U); 7: RNA extracted from mock transfected cell media (mock); 8: digested probe alone control (C). Genomic RNA packaging values determined by normalizing gRNA to 7SL: Lane 3 (WT): 100%; 2 (“classic Ψ” deletion): 0.4%; 3 (leader-deficient helper): ≤ 0.04%; 5 (U328A/A333U): 98%.
To ensure that the conservative U328A/A333U substitution did not have unexpected effects on RNA packaging in vivo, RNA encapsidation efficiencies were measured for RNAs containing mutant and wild type leader sequences. Virions were produced by transient transfection of human 293T cells, and quantified with values for wild type particles set to 100%. Previous work has demonstrated that virion quantification by virion protein (eg: reverse transcriptase activity48) or by host 7SL RNA yields indistinguishable values.49,50 7SL is a host RNA that is incorporated into retroviruses in proportion to virion proteins, in a manner independent of viral genomic RNA.49-51 Therefore, the amount of 7SL RNA in a cell-free virus sample is directly proportional to virion proteins and can be used to normalize for the amount of virus in that sample. The RNAse protection assay that was used to quantify viral RNA packaging is shown in Figure 2d,e. Figure 2d shows the riboprobe used, which allowed simultaneous quantification of encapsidated 7SL and virion genomic RNA. RNase digestion products are shown in Figure 2e. Analysis of RNA isolated from virions produced by transient transfection revealed that the U328A/A333U mutant RNA was packaged at levels comparable to wild type genomic RNA, while a mutant from which regions classically defined as sufficient to promote packaging of a heterologous RNA8 were deleted was packaged more than 200-fold less well than either wild type or the U328A/A333U mutant.
Confirmation of cross-kissing interactions by segmental labeling
NMR spectra obtained for ¬CES under conditions that favor dimerization exhibit signals diagnostic of kissing interactions involving the GACG tetraloops of SL-C and SL-D. Unfortunately, the chemical shift differences between SL-C and SL-D loop residues are very small, and it was not possible to unambiguously differentiate between SL-C to SL-C, SL-D to SL-D, and SL-C to SL-D kissing modes on the basis of the 2D NOESY spectra obtained for fully protonated, or nucleotide-specifically protonated/perdeuterated [¬CES]2 RNAs. We therefore obtained 2D NOESY data for a ¬CES sample prepared by segmentally ligating differentially deuterated fragments using T4 RNA ligase. Specifically, we ligated an SL-BC fragment, in which the guanosines were fully protonated and all other nucleotides were perdeuterated (GH-SL-BC), with an SL-D fragment that contained protonated adenosines and deuterated G, C and U nucleotides (AH-SL-D), Figure 3a,b. The AH-SL-D fragment was prepared using a plasmid that encoded the hammerhead and HDV ribozymes at the 5′- and 3′-ends of the RNA, respectively, in order to obtain homogeneous products with adenosine and 2′-3′-cyclic phosphate groups at the 5′- and 3′-termini, respectively.52 After ribozyme cleavage, the SL-D fragment was treated with polynucleotide kinase to produce the desired 5′-monophosphorylated (donor) terminus, and was used in the ligation reaction with excess amounts of the SL-BC fragment. Efficient ligation was achieved without the use of stints, with typical yields (based on the limiting SL-D fragment) of ~85%, Fig. 3b.
Figure 3.

Evidence for inter-molecular cross-kissing interaction between SL-C and SL-D’. (a) Cartoon representation of the ΨCES sample employed, prepared by ligating G-protonated, A,C,U-perdeuterated SL-BC with A-protonated, G,C,U-perdeuterated SL-D (the ligation point is indicated by an asterisk). Protonated guanosine and adenosine residues are colored by green and red, respectively, and perdeuterated nucleotides are colored black. Blue arrows denote long range NOEs observed in the 2D NOESY spectrum obtained for this sample. (b) Denaturing PAGE data obtained for the transcribed HH-SL-D-HDV RNA and the cleavage products (left) and solutions prior and subsequent to ligation (right). SL-Dp indicates SL-D harboring the 3′-2′,3′-cyclic phosphate, and SL-pDp denotes SL-D harboring a 5′-monophosphate and 3′-2′,3′-cyclic phosphate. SL-BC and SL-D were used as markers. (c) Portion of 2D NOESY spectrum obtained for the dimeric, kissing form of [SL-Cm]2 (nt 320-345). NOE cross-peaks involving the G334-H4′ and A330-H2 protons, and the unusual upfield chemical shift of the G334-H4′ proton, are characteristic of the kissing interface. (d) A portion of 2D NOESY spectrum obtained for the segmentally 1H/2H labeled RNA sample shown in (a). The detection of NOE cross-peaks between the G334-H4′ and A364-H2 protons provides unambiguous evidence for an SL-C:SL-D type of kissing interface (and not SL-C:SL-C or SL-D:SL-D).
2D NOESY spectra obtained for the [SL-C]2 kissing dimer exhibited spectral features similar to those reported previously for [SL-D]2. In particular, the H4′ proton of G334 (corresponding to C368 of SL-C) gave rise to an unusual upfield-shifted NMR signal (2.8 ppm) and exhibited an inter-molecular NOE with the aromatic H2 proton of A330, Fig. 3c. A similar pattern of NOEs was observed in the 2D NOESY spectrum obtained for the fully protonated [ΨCES]2 sample (data not shown). Although these data confirmed that SL-C and SL-D both participate in kissing interactions, they did not allow unambiguous differentiation between SL-C:SL-C/SL-D:SL-D and SL-C:SL-D kissing modes. However, the observation of a A364-H2 to G334-H4′ NOE cross peak in the spectrum obtained for the segmentally labeled [ΨCES]2 sample (Fig. 3d) provides clear evidence that the kissing interface is formed by the loop residues of SL-C and SL-D.
NMR assignment of [ΨCD]2 by 2H-editing
To date, only a small handful of structures have been reported for RNAs comprising more than 50 nucleotides.53 We therefore focused our initial structural studies on the 132-nucleotide [ΨCD]2 dimer. A major impediment to NMR studies of larger RNAs is severe signal degeneracy resulting from the presence of only four different common nucleotides. Although spectral resolution of smaller RNAs can be increased through the use of multi-dimensional 13C-edited NMR experiments, this approach is problematic when applied to larger RNAs due to signal losses and broadening resulting from strong 1H-13C dipolar coupling of aromatic C-H groups that are critical for assignment and structure determination.54 We therefore employed a strategy that involves the collection and analysis of 2D NOESY spectra for RNA samples prepared by in vitro transcription using different combinations of fully protonated, fully deuterated, and partially deuterated nucleotides. This approach was intended to avoid sensitivity and resolution problems associated with 13C labeling. Throughout this paper, we use a nomenclature in which only the proton-containing nucleotides are denoted, with superscripts indicating the positions of protons in partially deuterated nucleotides (8, 2, and R = protonated on the C-8, C-2, and ribose carbons, respectively) and a lack of superscripts denoting full protonation. For example, a sample of [ΨCD]2 that contains fully protonated guanosines, perdeuterated cytidines and uridines, and adenosines with deuterons on the ribose and aromatic C-2 carbons and a proton on the aromatic C-8 carbon, is denoted G,A8-[ΨCD]2. Differentially deuterated samples utilized in these studies included: A,G8-, G,A8-, U,G8,A8-, C,G8,A8-, G,U6-, and G,A2,R-[ΨCD]2.
As expected, the 2D NOESY spectrum obtained for fully protonated [ΨCD]2 exhibited broad, overlapping cross peaks and was not assignable, Supplementary Figure S2. However, spectra exhibiting good resolution and sensitivity were obtained for several selectively deuterated [ΨCD]2 RNAs. Portions of the 2D NOESY spectrum obtained for G,A8-[ΨCD]2 are shown in Figure 4a. In this spectrum, intra-residue NOE cross peaks for all guanosines, G-H1′ to A-H8 NOEs for all sequential G(i)-A(i+1) pairs, and most of the expected H8-to-H8 NOEs involving sequential purines were readily detected. Because all of the adenosine ribose protons were substituted by deuterium, inter-residue A(i+1)-H8 to G(i)-H2′/3′ NOEs were also readily identified. Assignments of partially overlapping H2′ and H3′ signals were confirmed by comparison with spectra obtained for the isolated RNA stem loops. Note that some of the H8-to-H8 NOEs could not be unambiguously assigned due to small chemical shift differences between the neighboring aromatic protons and/or the proximity of the associated NOE cross peaks to the intense diagonal. The spectrum also exhibited exchange cross peaks corresponding to non-kissing SL-C and SL-D species. Although the intensities of the exchange peaks were significant, intensities of the diagonal and NOE cross peaks associated with the non-kissing species were small, when measurable, and no intra- or inter-residue NOEs were detected for the non-kissing species. Based on a qualitative assessment of the signal intensities, we estimate that, under the conditions of the NMR experiments, the population of the non-kissing species is ca. 5 ± 3% of that of the kissing species.
Figure 4.


Portions of 2D NOESY spectra obtained for partially deuterated [ΨCD]2 samples. Sequential aromatic to ribose (and H5) and aromatic to aromatic proton connectivities are shown in the upper and lower panels, respectively. (a) Data obtained for G,A8-[ΨCD]2 (fully protonated G, C8-protonated A (with C2 and ribose carbons deuterated), and fully deuterated C and U residues) showing intraresidue G-H8 to H1′ and sequential G(i)-H8/H1′ to G(i+1)-H8 and A(i+1)-H8 protons. (b) Data obtained for G,A2,R-[ΨCD]2 (fully protonated G, C8-deuterated A (with C2 and ribose carbons protonated), fully deuterated C and U) showing intraresidue A-H2 to –H1′ and sequential and long-range A-H2 to G- and A-H1′ protons (top) and A-H2 to G- and A-H8 protons (bottom). (c) U,G8,A8-[ΨCD]2 contains fully protonated U, fully deuterated C, and C-8-protonated (C2 and ribose carbons deuterated) A and G. Signals associated with the U-H5/6 protons are significantly broader than those of the G- and A-H8 protons, apparently due to H5-H6 dipolar interactions. (d) G,U6,A8-[ΨCD]2 contains fully protonated G, C6-protonated U (with C5 and ribose carbons deuterated), C8-protonated A (with C2 and ribose carbons deuterated), and fully deuterated C. Signals associated with the U-H6 protons are narrower than those shown in panel (a) due to the deuteration of the C5 and ribose carbons.
Sequential and long-range NOEs involving the adenosine-H2 protons were assigned primarily from spectra obtained for G,A2,R-[ΨCD]2, Figure 4b. Intense, well resolved cross peaks were observed for all of the expected sequential and cross-strand A-H2 to G/A-H1′ proton pairs, in addition to the expected A(i)-H1′ to G(i+1)-H8 NOEs, Figure 4b. Notably, residues A353 and A354 of the linker that connects SL-C and SL-D exhibited sequential inter-residue aromatic-to-ribose proton NOEs consistent with A-form helical stacking, with A353-H2 exhibiting intense NOEs with the H1′ and H8 protons of A354 and G310, and with A354-H2 exhibiting NOEs with the G309-H1′ and H8 and C355-H6 protons (observed in the A2R,G8C6-[ΨCD]2 spectra). These data indicate that SL-C and SL-D are arranged in an end-to-end manner, in which the 3′-residue of SL-C, the linker adenosines, and the 5′-residues of SL-D are stacked in an extended, A-like geometry.
NMR signals observed for pyrimidine H6 and H5 protons in spectra obtained for [ΨCD]2 samples containing fully protonated cytosine or uracil residues were significantly broader than those associated with the purine H8 protons. For example, as shown in Figure 4c, 1H NMR linewidths associated with U-H6 protons in 2D NOESY spectra obtained for U,A8,G8-[ΨCD]2 were typically more than three-times greater than those associated with the G-H8 and A-H8 protons (the exception being U319, which forms an unstructured bulge; see below). Several purine H8-to-uracil proton NOEs could be assigned from these spectra, mainly because ΨCD contains only seven uracil residues. Analogous spectra obtained for ΨCD RNAs containing fully protonated cytosines were largely not assignable due to severe signal overlap (data not shown). To eliminate the relatively strong H5-to-H6 dipolar coupling that appears to be primarily responsible for the broader NMR signals, ΨCD samples were prepared using pyrimidine nucleotides containing a proton on the C6 carbon and deuterons on all other carbons. The quality of the NMR spectra obtained for these samples was significantly improved, enabling assignment of all expected purine-H1′ to pyrimidine-H6′ proton connectivities and most purine-H8 to pyrimidine-H6 connectivities. Representative spectra and assignments made for G,U6,A8-[ΨCD]2 are shown in Figure 4d.
The NMR spectra obtained for the highly deuterated samples also exhibited cross-strand NOEs (i.e., between protons on two different strands of a given helix) for protons that are likely to be separated by more than 5.0 Å. For example, in the NMR spectrum obtained for A,G8-[ΨCD]2, relatively intense G359-H8 to A371-H2, G323-H8 to A343-H2, and G334-H8 to A328-H2 NOEs were readily detected (Supplementary Figure S3). Analogous cross peaks observed in spectra obtained for G,A2,R-[ΨCES]2 (Figure 4b) were originally attributed to spin diffusion, due to the close proximity of the A-H2 and G-H1′ protons. Since the G-H1′ protons are substituted by deuterium in the A,G8-labeled sample, cross-strand A-H2 to G-H8 NOEs observed in these spectra can only be attributed to direct, long-range dipolar interactions.
Using the above approach, 100% of the non-exchangeable aromatic and H1′ signals were assigned. Signal overlap precluded independent assignment of a majority of the H2′ and H3′ signals, but by comparisons with higher resolution 2D NOESY and 2D 1H-13C correlated NMR spectra obtained for isolated SL-C and SL-D hairpins, signals for > 90% of the purine H2′ and H3′ protons and > 70% of the pyrimidine H2′ and H3′ protons of [ΨCD]2 could be identified.
The improved resolution provided by the partially deuterated samples enabled us to correct a previously misassigned NOE associated with A340. Specifically, NMR spectra obtained for C,G8,A8-[ΨCD]2 revealed that a A340-H8 to C342-H1′ cross peak, which was previously attributed to spin diffusion via the intervening A341-H1′ and/or –H2 protons,44 was actually due to a direct dipole-dipole interaction. The A340-H8 proton also exhibited an intense intra-residue H8-to-H1′ NOE that was obscured by signal overlap in previously obtained NMR spectra, indicating that this residue adopts a syn conformation. As described below, these assignment corrections led to a change in the orientation of A340, but did not significantly affect other residues of the GGAA bulge in structures calculated for [ΨCD]2.
The 1H NMR chemical shifts measured for the stem loops of [ΨCD]2 were nearly indistinguishable from those observed for the isolated SL-C and SL-D hairpin RNAs, indicating that the structures should also be similar. The 1H NMR chemical shifts of the adenosine residues that link SL-C with SL-D differed by 0.02-0.83 ppm (Δδ) relative to the shifts observed previously for a 101-nucleotide RNA containing mutations in the loops of SL-B, SL-C and SL-D that prevented dimerization (Δδ for the H8, H2 and H1′ protons of 0.052, 0.271, 0.019 ppm for A353 and 0.325, 0.831 and 0.455 ppm for A354). This construct (SL-BmCmDm-UU) contained two additional non-native 3′-uridines that formed base pairs with A353 and A354 of the linker, and this is likely responsible for the observed chemical shift differences. Interestingly, the NOE cross peak patterns observed for A353 and A354 of both [ΨCD]2 and SL-BmCmDm-UU are consistent with A-form helical stacking. Thus, relatively intense NOEs were observed between A353-H2 and protons on the following (A354-H1′, -H2, and –H8) and cross-strand (G310-H1′) residues, and between A354-H2 and the H1′ proton on the following residue (C355). In addition, residues C352 through C355 exhibited sequential ribose(i) to H8/6(i+1) and H8/6(i) to H5(i+1) NOEs consistent with a structure in which residues C352 through C355 stack in a continuous, A-form like manner.
Structure calculations
A total of 1248 unique and functionally non-redundant NOE-derived 1H-1H distance restraints were obtained from the 2H-edited NOESY spectra, Table 1. Hydrogen bond restraints and torsion angle restraints were employed as flat-well potentials with values centered at those observed in idealized A-form helices for segments exhibiting NOE cross peak patterns and intensities consistent with A-form helical conformations (residues G310-G313, C316-G318, G320-G323, G324-A328, U333-C337, C342-C352 of SL-C and C355-A362, U367-G374 of SL-D). Major groove inter-phosphate distances were loosely restrained for these residues using database potentials derived from high-resolution X-ray crystal structures.53 These restraints were required to avoid collapse of the major groove that can result from the asymmetric distribution of distance restraints in A-form helical segments.44,53,55,56 Inter-molecular cross-kissing H-bond restraints consistent with the NOE results obtained for the segmentally-labeled sample were employed (see above), but no intra-molecular H-bond or torsion angle restraints were employed for the residues of any of the loops or bulges. Because the NOEs associated with A353 and A354 are consistent with an A-form helical stacking, loose inter-phosphate distance and torsion angle restraints (ideal ± 50°) were employed for these residues as well. In addition, weak restraints were employed for the phosphorus, ribose and aromatic carbon atoms to enforce symmetry between the two molecules of the dimer.
Table 1.
NMR Restraint and Structure Statisticsa
| NOE-Derived Distance restraints | |
| Intraresidue | 268 |
| Sequential | 440 |
| Long Range (|i-j| > 1) | 54 |
| Total | 1248 |
| Restraints per restrained residue | 9.7 |
| H-bond Restraintsb | 576 |
| Base Pair Planarity Restraints | 100 |
| P-P Distance Restraints | 298 |
| Backbone Torsion Angle Restraints | 624 |
| Total Restraints per Restrained Residue | 29.4 |
| Target Function (Å2) | 4.11 ± 0.05 |
| Upper Distance Violations (Å) | 0.03 ± 0.00 |
| Maximum Upper Distance Violation (Å) | 0.22 ± 0.01 |
| Lower Distance Violations (Å) | 0.02 ± 0.00 |
| Maximum Lower Distance Violation (Å) | 0.15 ± 0.01 |
| Sum of van der Waals Violations (Å) | 15.9 ± 0.2 |
| Maximum van der Waals Violation (Å) | 0.13 ± 0.01 |
| Torsion Angle Violations (°) | 0.21 ± 0.01 |
| Maximum Torsion Angle Violation (°) | 1.46 ± 0.12 |
| Molprobity120 Clash Scored | 0.72 (99th percentile) |
| RMSD (All heavy atoms, Å) | 0.40 ± 0.12 |
Results for an ensemble of 20 refined [ΨCD]2 structures generated with Cyana.113
A total of four restraints per hydrogen bond were employed.
A total of two restraints per base pair were employed.
Calculated on-line at: http://molprobity.biochem.duke.edu/
An ensemble of 20 structures with lowest target function (4.11 ± 0.05 Å2) was obtained from an initial pool of 160 structures generated using Cyana.57 The structure is well defined by the NMR data, with best-fit superpositions of all heavy atoms affording pairwise RMS deviations (relative to mean atomic coordinates) of 0.40 ± 0.12 Å, Table 1 and Figure 5a,b. Residues G309 and U319 (and the corresponding residues in the symmetrical dimer) were not experimentally restrained and therefore exhibit poorer convergence. These residues give rise to intense and relatively narrow 1H NMR signals and are therefore likely to be disordered. Statistical information regarding the restraints employed, restraint violations, and the structure convergence is provided in Table 1.
Figure 5.

Structure of [ΨCD]2 determined by NMR. (a,b) Orthogonal views of the ensemble of 20 structures calculated with Cyana showing the degree of convergence and overall shape and asymmetry of the dimer. The 5′- and 3′-terminal residues are labeled, along with the position of the experimentally unrestrained U319 residue. Carbon atoms of the two molecules of the dimer are colored gray and green. (c,d) Stereoviews showing (c) the structure of the A-minor K-turn and it’s proximity to the kissing interface, and (d) stacking of the A353 and A354 residues between the stems of SL-C and SL-D. (e) View showing one molecule of the symmetrical dimer. Adenosines that bridge SL-C and SL-D (blue) and residues that make intermolecular kissing contacts (red) are highlighted.
Description of the [ΨCD]2 NMR structure
Many features of the [ΨCD]2 structure are consistent with those observed in NMR structures calculated previously for native and mutant fragments of ΨCES.39,43,44 Thus, residues G310-G323 adopt an A-form helical “lower stem” in which unpaired residues A314 and C315 are internally stacked, U319 forms an unstructured extra-helical bulge, and residues G338-A341 form an A-minor K-turn type structure that connects the lower stem with the upper stem (G324-C337). As observed previously for a mutant ΨCES RNA (in which residues of the GACG tetraloop were mutated to prevent dimerization44), the nucleobase of G338 stacks against C337 in an A-like manner, G339 adopts a syn conformation and packs against the nucleobase of A340, and A341 packs against the nucleobase of C342 and forms an A-minor like interaction with A324 and A325 of the upper stem, Figure 5c. In addition, we now have evidence that A340 adopts a syn conformation, and as indicated above, NOE spectra obtained for the C,G8,A8-[ΨCD]2 sample provides clear evidence for direct A340-H8 to C342-H1′/–H4′ dipolar interactions (which were originally erroneously attributed to spin diffusion via A341). As such, the orientation of A340 in the [ΨCD]2 structure differs from that reported earlier, and is now shown to pack against the nucleobase of G339 and the nucleobase and ribose moieties of A341 and C342, respectively (Fig. 5c). The structures and interactions of the two symmetrical kissing interfaces (SL-C to SL-D’ and SL-C’ to SL-D) are essentially indistinguishable from those observed previously for a dimeric [SL-D]2 kissing hairpin.39
A surprising feature of the [ΨCD]2 structure is that the lower stems of SL-C and SL-D are stacked in an “end-to-end” orientation, in which the linker residues A353 and A354 are stacked in an A- helical manner between the G310-C352 base pair of SL-C and the C355-G374 base pair of SL-D, Figure 5d. Although similar end-to-end stacking interactions were observed in the monomeric, mutant ΨCES RNA, those interactions were attributed to non-native base pairing between the linker adenosines and two 3′-uridines that were included to accommodate an HpaI restriction site used for sub-cloning.44 The [ΨCD]2 RNA used in the present studies was transcribed from a template that encoded a 3′-hammerhead ribozyme which, after cleavage, afforded a product RNA that terminated at G374 and did not contain additional native or non-native residues. Thus, in the [ΨCD]2 structure, residues C352-C355 are stacked in a continuous, A-form like helical conformation as observed previously for ΨCES, even though that A354 and A355 do not form Watson-Crick base pairs. As such, the structure of [ΨCD]2 is both elongated and flat, with overall dimensions of ca. 95 Å by 45 Å by 25 Å.
The 5′-guanosine residues do not form base pair or stacking interactions and appear disordered in the structure. This is consistent with earlier NMR studies showing that this guanosine (G309) interacts directly with the zinc finger domain of the MoMuLV NC protein. The two guanosines of the symmetrical dimer (G309 and G309′) are separated by ~20 Å in the NMR structure, Figure 5, a finding that has important implications regarding the likely structure of the preceding DIS-2 duplex (see below).
Cryo-electron tomography of [ΨCD]2
Because of the inherent paucity of long-range structural information available in the NOE data, attempts were made to establish relative helix orientations and overall molecular shape using NMR-derived residual dipolar couplings (RDCs) and small angle X-ray scattering (SAXS), respectively. NMR-derived RDCs and RCSAs have been used previously to establish global structural features,44,53,58-85 including inter-helical orientations.44,63,64,68,86-91 Unfortunately, neither of these approaches was successful. As indicated above, ΨCD exists as a monomer at low ionic strength, and under the conditions of the NMR studies, a small amount of the monomer species (or extended dimer species, in which one of the hairpins does not participate in kissing interactions) persists and is detectable in the NMR spectra. Despite the low abundance of these species, their higher apparent rotational mobilities lead to significant contributions to the RDC spectra obtained, precluding quantitative analysis of the RDC data. In addition, dynamic light scattering (DLS) and SAXS data obtained for ΨCD under conditions of the NMR experiments were consistent with the presence of a small amount of one or more higher-MW species. Unfortunately, sample heterogeneity, and particularly the presence of even small amounts of high MW contaminants, can complicate quantitative analysis of the SAXS data. We therefore carried out structural studies by cryo-electron tomography (cryo-ET).
Cryo-ET allows for the direct detection of molecular assemblies within a given sample, and for heterogeneous samples, species of similar size and/or topological properties can be classified for further analysis. A representative projection of 15 slices of unfiltered cryo-ET data obtained for [ΨCD]2 is shown in Figure 6a. Numerous punctate and elongated dark densities are clearly visible in the tomogram. In contrast, cryo-ET data obtained for a sample containing only buffer (the same buffer used to prepare the [ΨCD]2 sample) and processed under identical conditions lacked these features, Figure 6b, indicating that the distinct densities observed using the [ΨCD]2 samples are due to electron scattering by the RNA. As an additional control, cryo-ET data were also obtained for [ΨCES]2. The tomograms obtained for this larger RNA exhibited densities with volumes and topological features consistent with the side-by-side packing of the DIS-2 helix and the [ΨCD]2 tandem hairpin (Supplementary Figure S9). These findings collectively indicate that RNAs as small as [ΨCD]2 (132 residues) are amenable to structural studies by cryo-ET. Previously, the smallest reported molecule studied by cryo-EM was a 78 kDA, 252 residue DNA tetrahedron. The structure of this highly symmetric and rigid molecule was determined by single particle methods with symmetry enforced.92
Figure 6.
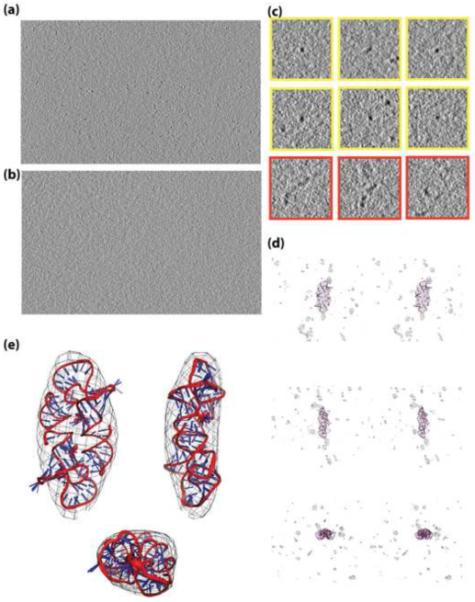
(a) A projection though 15 slices of a tomogram of [ΨCD]2. The dark densities are proposed to correspond to molecules of [ΨCD]2. They consist of individual compact point-like densities, multiple adjacent densities, and extended multimers when viewed in projection. Only subvolumes corresponding to well separated, compact densities were chosen for further processing. (b) A projection though 15 slices of a tomogram of buffer (the same buffer used for NMR studies). Wavy texture is most likely due to irregularities in the surface, since this texture is more prominent at the air-ice interface. There are no distinguishable compact densities present. Some ice contamination is also visible. (c) Two dimensional views of representative densities that were utilized for further analysis (yellow); densities attributed to higher molecular weight multimers, which were not utilized for further analysis, are also shown (red). Point-like densities in this projection were then examined in 3D to verify that they were also compact in the third dimension. (d) Stereo views of one representative subvolume out of those chosen for 3D alignment and averaging, shown from three different angles. A representative [ΨCD]2 NMR structure is fitted into the density for comparison. The surrounding noise is representative of the noise around all chosen particles. (e) Orthogonal views of the averaged cryo-ET subvolumes fitted to the ensemble of [ΨCD]2 NMR structures. The overall shape and dimensions of the averaged cryo-ET density is in good agreement with the structure calculated by NMR. Although U319 resides outside of the averaged cryo-ET density, this residue is disordered in the NMR structure and appears to undergo rapid conformational averaging.
In addition to the compact densities discussed above, densities consistent with larger, elongated structures were observed in the [ΨCD]2 tomograms (red boxes in Figure 6c). These more extended densities adopted random overall shapes. In view of the facts that (i) the NMR spectra obtained for the liquid state samples exhibited exchange cross peaks indicative of a minor population of monomeric, non-kissing species, and (ii) the DLS and SAXS data suggest the presence of a minor population of a higher molecular weight species, we attribute the smaller and extended densities in the cryo-ET tomograms to monomers, partly dissociated dimers (i.e., dimers linked by only a single kissing structure) and higher-order multimers. The [ΨCD]2 concentration used for cryo-ET was ~ 40-fold lower than that used for NMR measurements, which may explain why the non-kissing species were more abundant in the cryo-ET data than in the NMR data.
Stereo views of an unfiltered subvolume of a representative cryo-ET tomogram showing a typical compact density, along with the relative noise level, are shown in Figure 6d (movies of the raw data are provided in Supplementary Figures S5 and S6). For comparison, a representative [ΨCD]2 NMR structure has been fitted into the density. The signal-to-noise ratio of the unfiltered cryo-ET data is clearly sufficient for alignment and averaging, and the overall dimensions of the compact densities are consistent with the dimensions of the [ΨCD]2 NMR structure, Figure 6d.
A total of 38 such subvolumes with compact densities were computationally extracted, classified, aligned and averaged (see Methods), affording an averaged cryo-ET density with approximate length of 95 Å, a width of 45 Å and a thickness of 25 Å at its narrowest point, Figure 6e. The averaged cryo-ET density map exhibits two-fold symmetry, even though no symmetry was assumed or applied in the reconstruction, alignment and averaging process, and appears to be fully consistent with the NMR ensemble, Figure 6e. Although U319 resides outside of the averaged cryo-ET density (Fig. 6e), this residue is disordered in the NMR structure and appears to undergo rapid conformational averaging (based on the unusually narrow 1H NMR lineshapes and lack of significant inter-residue NOEs; see above). Interestingly, the cryo-ET average density exhibits a slightly concave shape about the approximate two-fold axis, which also appears consistent with the NMR structures, Fig. 6e.
DISCUSSION
Approach for RNA Structure Determination
The application of 2D NOESY NMR spectroscopy to partially deuterated [ΨCD]2 samples served as an effective approach for assignment of the non-exchangeable aromatic and H1′ protons. Assignment of the H2′ and H3′ signals was generally more challenging due to signal overlap, and in many cases it was only possible to make assignments based on comparisons with higher resolution 2D NOESY, TOCSY, and 1H-13C correlated NMR spectra obtained for isolated SL-C and SL-D hairpins. Except for the two adenosines that link SL-C and SL-D, 1H NMR frequencies and NOE cross peak patterns and intensities observed for residues of [ΨCD]2 were nearly indistinguishable from those observed for the isolated RNA hairpins, providing confidence in the resonance assignments and indicating that the internal structures of the hairpins in [ΨCD]2 are similar to those observed for the monomeric and kissing dimer forms of the isolated hairpins.
The NMR data obtained for one of the partially deuterated [ΨCD]2 samples (C,A8,G8-[ΨCD]2) led to the identification of inter-residue dipolar interactions that had previously been attributed to spin diffusion. As such, the orientation of A340 of the GGAA bulge of the [ΨCD]2 structure reported here differs from its orientation in previously reported structures from our laboratory.41,44 Although this correction did not lead to significant changes in the structure of the SL-C hairpin or the GGAA bulge (all of the A-minor93 and K-turn94-like interactions observed in the previous studies44 were also observed here), it does illustrate the power of 2H-editing for differentiating between spin diffusion and direct dipolar interactions. The structures calculated for [ΨCD]2 exhibited remarkably good convergence, despite the paucity of long-range NOE-derived restraints (the only intermoleculear NOEs detected were for protons at the kissing interfaces). This can be attributed to the fact that the internal structures of the hairpins, bulges, linkers, and kissing interfaces were all well defined by the local structural restraints, and as such, there was only one general intermolecular arrangement that satisfied both the internal NOE data and the intermolecular cross-kissing interactions.
Attempts to confirm the overall topological features of the [ΨCD]2 NMR structure using NMR-derived residual dipolar coupling (RDC) and residual chemical shift anisotropy (RCSA) information were unsuccessful due to the fact that, under all experimental conditions employed, the [ΨCD]2 samples contained a small residual population of monomeric and/or extended multimeric RNAs with non-kissing hairpins. The favorable NMR relaxation properties of these species (due to more rapid rotational diffusion), combined with the significantly less favorable relaxation properties of the fully folded dimeric RNA, precluded quantitative measurement of RDC and RCSA values for the dimer. Attempts to obtain information on the overall shape of [ΨCD]2 by SAXS, which has been used by others for RNA structure refinement,81,95,96 were also unsuccessful due conformational heterogeneity and the presence of multimers. Fortunately, we were able to obtain global shape information for [ΨCD]2 by cryo-ET. Although the resolution of the cryo-ET data did not allow detection of individual base pairs or the precise separation of the helices, the overall shape and dimensions of the individual and averaged cryo-ET subvolume densities, as well as the symmetry of the density average, was consistent with the NMR-derived structure of [ΨCD]2. The cryo-ET data also confirmed the presence of multimers, as well as extended species (monomers and dimers) inferred from the chemical exchange cross peaks in the 2D NOESY spectra.
The MoMuLV [ΨCD]2 NMR structure differs considerably from the MuSV [ΨCD]2 structure proposed on the basis of SHAPE and site-directed hydroxyl radical cleavage analyses.46 In the NMR structure, the 5′-guanosine of a given RNA strand resides near the kissing interface formed by the SL-C loop of the same molecule, whereas in the SHAPE structure, the 5′-guanosine resides near the SL-C’ loop of the neighboring strand. Also, the stems of SL-C and SL-D of a given strand are packed in a nearly side-by-side arrangement in the SHAPE structure, but are arranged end-to-end in the NMR structure. As a consequence, the NMR structure has an elongated overall shape, consistent with the cryo-ET data, whereas the SHAPE model is more compact and has a more isotropic, globular shape. These disparities could conceivably be due to the presence of alternate RNA conformers in solution (detected by both NMR and cryo-ET), which could potentially complicate interpretation of bulk chemical reactivity results. However, we cannot rule out the possibility that differences in nucleotide sequence (the MoMuSV and MoMuLV tandem hairpins differ by a G338U substitution) or sample conditions may be responsible for the observed differences.
Cryo-ET with subvolume classification, alignment and averaging is emerging as a useful technique for studying heterogeneous samples at low resolution. Compared with spectroscopic methods that rely on bulk sample measurements for determining molecular shape, cryo-ET appears superior for RNAs like ΨCD that adopt multiple equilibrium conformations. The low resolution global structural information provided by cryo-ET is complementary to the high resolution local structural information obtainable by NMR, making this an attractive combination for structural studies of modest sized (~150 nucleotide) RNAs.
Biological Implications
Mutagenesis and packaging studies have provided compelling evidence that ΨCES is both critical for genome packaging40,42,97-99 and capable of functioning as an autonomous packaging signal.42 The DIS-2 hairpin of ΨCES appears to contribute to packaging by regulating exposure of one or two UCUG elements located between DIS-2 and SL-C.41,100 These elements are sequestered by base pairing within the lower stem of the monomeric DIS-2, and a register shift in base pairing that occurs upon dimerization exposes one or both of the UCUG elements, thereby enabling high affinity binding to NC.41 An upstream element that also promotes dimerization and packaging (DIS-1) was shown to behave similarly.41 In addition, a large fragment of the 5′-UTR that spans from the primer binding site (PBS) through the gag start codon and includes all residues necessary for efficient RNA packaging also exhibits dimerization-dependent NC binding, with the monomeric RNA binding 1 or 2 NC proteins with high affinity and dimer binding tightly to 12 NC molecules.100 Mutations in this large RNA that inhibited dimerization in vitro led to dramatic reductions in RNA packaging in vivo.100 These findings collectively suggest that diploid genome selection is regulated by an RNA structural switch mechanism in which ~12 high affinity NC binding sites with the MoMuLV 5′-UTR become exposed upon dimerization. A resulting Gag12:RNA2 complex could then serve to nucleate virus assembly, Figure 7. Such a mechanism is consistent with recent findings for HIV-1 assembly, which appears to be initiated by a complex comprising two RNA genomes and fewer than ~12 Gag proteins.101
Figure 7.

Potential mechanism for the early stages of MoMuLV virus assembly. Initially synthesized genomes (yellow and gray cylinders) are monomeric and adopt structures that sequester the high affinity NC (Gag) binding sites (red lines). Upon dimerization, the tandem hairpin element functions as a scaffold that helps expose a cluster of guanosine-containing segments that would normally be sequestered by base pairing and stacking interactions and unable to bind the NC. The ribonucleoprotein complex, formed by assembly of a critical number of Gag molecules (possibly twelve100) onto the dimeric 5′-UTR, then becomes anchored at the plasma membrane where it functions as a nucleation site for the non-specific binding of additional Gag molecules.
The double hairpin is immediately preceded by two UCUG sequences (U301CUG304 and U306CUG309), and is immediately followed by a closely related U377UCG380 sequence, all of which are potentially capable of binding NC with high affinity if maintained in an exposed, unstructured-like state.102 We propose that the tandem hairpin functions not only to promote dimerization, but also as a scaffold that helps maintain exposure of a clustered group of NC binding sites. Although U301-G304 were shown to participate in non-canonical base pairs that extended the length of the DIS-2 helix in isolated ΨCES fragments,43,44 modeling studies indicate that residues U301 through G309 would need to exist in a relatively extended conformation in order to bridge between the well-separated ends of the DIS-2 duplex and the relatively proximal 5′-ends of the [ΨCES]2 dimer. Thus, in the context of the native [ΨCES]2 dimer, residues U301CUG304 and U306CUG309 should both exist in extended, unstructured conformations, affording a total of six high affinity NC binding sites per dimer (including the 3′-proximal U377UCG380 element). The stage is now set for structural and NC (and Gag) binding studies of a MoMuLV ΨCES construct that contains all six predicted NC binding sites (underway).
MATERIALS AND METHODS
Plasmids for RNA synthesis
pMoMuLV-SL-BCD-HDV
pMoMuLV-SL-BCD-HDV was generated by polymerase chain reaction (PCR) on pNCA, which contains the proviral DNA of MoMuLV. DNA fragments were amplified with oligonucleotide MoMuLV-276f (5′-CCAGTGAATTCTAATACGACTCACTATAGGCGGTACTAGTTAGCTAACTAGCTCTGTATCTGGCGG-3′), carrying an Eco RI site and a T7 RNA polymerase promoter, and oligonucleotide MoMuLV-374r (5′-CCAGTGCTAGCCCCTGGGACGTCTCCCAGGGTTGCGGCCGGGTGTT-3′), carrying an Nhe I site. The amplified product was inserted in the pHDV4 plasmid (kind gift from Dr. Conn) after EcoR I and Nhe I digestions (New England Biolabs).
pMoMuLV-SL-BCswapD-HDV
pMoMuLV-SL-BCcwapD-HDV was generated by PCR based mutagenesis. Oligonucleotide MoMuLV-Cswapf (5′-GCGGACCCGTGGTGGAACAGACGTGTTCGGAACACCCGGCCGC-3′) and MoMuLV-Cswapr (5′-GCGGCCGGGTGTTCCGAACACGTCTGTTCCACCACGGGTCCGC-3′) were used to insert mutations (nt 328 T to A and nt 333 T to A). Two DNA fragments were amplified on pMoMuLV-SL-BCD-HDV with oligonucleotide M13f (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′), annealing to pUC19 at the 5′ end of the primer lies 47 nucleotides upstream of the EcoRI restriction site, and oligonucleotide MoMuLV-Cswapr, and with oligonucleotide MoMuLV-Cswapf and oligonucleotide M13r (5′-AGCGGATAACAATTTCACACAGGA-3′), annealing to the pUC19 vector at the 5′ end of the primer lies 48 nucleotides downstream of the HindIII restriction site, respectively. Another PCR was performed on the two PCR products with oligonucleotide M13f and oligonucleotide M13r. The amplified product was inserted in the pUC19 plasmid after Eco RI and Hind III digestions (New England Biolabs).
pMoMuLV-SL-CD-HDV
pMoMuLV-SL-CD-HDV was generated by PCR on pMoMuLV-SL-BCD-HDV, which contains the proviral DNA of MoMuLV. DNA fragments were amplified with oligonucleotide MoMuLV-309f (5′-CCAGTGAATTCTAATACGACTCACTATAGGCGGACCCGTGGTGGAACTGACGAGTTCGGAACAC-3′), carrying an Eco RI site and a T7 RNA polymerase promoter, and oligonucleotide M13r. The amplified product was inserted in the pUC19 plasmid after EcoR I and Hind III digestions (New England Biolabs).
pMoMuLV-SL-CcwapD-HDV
pMoMuLV-SL-CcwapD-HDV was generated by PCR. Two DNA fragments were amplified on pMoMuLV-SL-CD-HDV oligonucleotide MoMuLV-309mf (5′-CCAGTGAATTCTAATACGACTCACTATAGGCGGACCCGTGGTGGAACAGACGTGTTCGGAACAC -3′), carrying an Eco RI site, a T7 RNA polymerase promoter and two mutations in SL-C sequence, and oligonucleotide M13r. The amplified product was inserted in the pUC19 plasmid after Eco RI and Hind III digestions (New England Biolabs).
pMoMuLV-HH-SL-D-HDV
PCR was performed on pMoMuLV-SL-CcswapD-HDV with oligonucleotide HH-354in-f (5′-GGTCTGATGAGAGCGAAAGCTCGAAACAGCTGTGAAGCTGTCACCCTGGGAGACGTCCCAGGG-3′), carrying a part of the hammer-head sequence and oligonucleotide M13r. The resulting PCR product was used in a second PCR reaction performed with oligonucleotide HH-354out-f (5′-CCAGTGAATTCTAATACGACTCACTATAGGGAATCCAGGGTCTGATGAGAGCGAAAGCTCGAAACAGC-3′), carrying an Eco RI site, a T7 RNA polymerase promoter, and a part of HH sequence, and M13r. The amplified product was inserted in the pUC19 plasmid after Eco RI and Hind III digestions (New England Biolabs).
Selective C8 protonation and deuteration of ATP and GTP
The partially deuterated rUTP reagents used for in vitro transcription was obtained from Cambridge Isotope Laboratories (CIL, Massachusetts) and ProSpect Pharma (Columbia, MD), and perdeuterated rNTPs were from CIL and Silantes (GmbH, Germany). Protonation at the C8 position of perdeuterated rGTP and rATP was achieved by incubation with ~5 equivalents of Triethylamine (TEA) in 1H2O at 60 °C for 24 hours and for 5 days, respectively.103,104 Deuteration of the C8 carbon of fully-protonated GTP and ATP was achieved by incubation in 2H2O (99.8 % deuteration; CIL). The TEA was subsequently removed by lyophylization.
RNA synthesis by in vitro transcription
All plasmids were amplified using E. coli DH5α. The pMoMuLV-SL-CD-HDV, pMoMuLV-SL-CcwapD-HDV and pMoMuLV-HH-SL-D-HDV plasmids were linearized by Eco RV (New England Biolabs) at 1 μg/unit for 16 hours at 37 °C. The linearized DNA was then extracted twice with phenol-chloroform and precipitated with ethanol. The pellet was washed with 70 % (v/v) ethanol, and the DNA was dissolved in sterile distilled water. Synthetic oligo (5′-TGCGGCCGGGTGTTCCGAACTCGTCAGTTCCACCACGGGTCCGCCAGATACAGAGCTAGTTAGCTAACTAGTACCGCCTATAGTGAGTCGTATTA-3′) was used as the template for SL-BCm, wherein the first and second nucleotides contained 2′-O-CH3 methyl groups to prevent the addition of non-templated nucleotides. This template was mixed with DNA containing the positive-sense strand of the T7 promoter (5′-TAATACGACTCACTATA-3′), heated and then cooled to anneal the two strands so as to form the double-stranded promoter.
RNAs were synthesized by in vitro transcription105 using T7 RNA polymerase in 30 ml reactions, each containing 2.5 mg of template, 20 mM MgCl2, 2 mM spermidine, 80 mM Tris-HCl (pH 8.1), 4 mM of each NTP, 2 mM DTT, and 0.3 mg T7 RNA polymerase. To cleave ribozyme, further 10 mM MgCl2 were added to transcription mixtures and were incubated for 16 hours and quenched by 35 mM EDTA. Selectively protonated RNA samples were prepared using appropriate combinations of deuterated and fully or partially protonated NTPs. After denaturation at 96 °C for five minutes, the RNA was purified by electrophoresis on urea-containing polyacrylamide denaturing gels. The concentration of each sample was determined by measuring the optical absorbance at 260 nm.
Enzymatic RNA ligation
The donor SL-D RNA synthesized using the pHH-SLD-HDV plasmid (a kind gift from Graeme L. Conn) 52 contains a 2′,3′-cyclic-phosphate at the 3′ end and a hydroxyl group on the 5′ end after ribozyme cleavage. The presence of a 2′,3′-cyclic-phosphaphate is advantageous since it prevents undesired self ligation products. However, ligation of two RNA fragments requires the donor RNA to be 5′ mono-phosphorylated. In order to preserve the 3′ end, this was achieved by treating SL-D with polynucleotide kinase that is defective for 3′ phosphatase activity (New England Biolabs). The acceptor SL-BC RNA was synthesized using an oligonucleotide template and hence contained the desired 5′ triphospate terminal and 3′ hydroxyl terminal and could be used without further treatment. The 5′ triphosphate terminal is not capable of self-ligation and the 3′ hydroxyl terminal acts as an acceptor during the ligation reaction. The two fragments (SL-BCm: SL-D = 1.5 : 1.0) were mixed in 50 mM Tris-HCl (pH. 7.8), 10 mM MgCl2, 1 mM ATP, 10 mM Dithiothreitol, 15 % of PEG8000 (w/v) and RNA ligase (New England Biolabs).
Plasmids for virology
MoMuLV RNA packaging was assessed using derivatives of MoMuLV-gag-pol-puro: an intact proviral construct in which the env open reading frame has been replaced by the puromycin N-acetyltransferase gene (puroR) driven by a simian virus 40 (SV40) promoter 106. The “classic Ψ” deletion (Δ215-568) and the SL-C mutant (U328A/A333U) were generated by overlap PCR, sequenced, and used to replace the corresponding portions of the parental MoMuLV-gag-pol-puro vector. The helper plasmid pNGVL-3′-gag-pol, which expresses MoMuLV Gag and Gag-Pol from a 5′ leader-deleted transcript driven by the CMV promoter, has been described previously 107.
Cells and virus
293T cells (human embryonic kidney cells expressing SV40 T antigen) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum and 1% penicillin-streptomycin. Virus was produced by transient transfection of the MoMuLV-gag-pol-puro or pNGVL-3′-gag-pol plasmids into 293T cells by calcium phosphate precipitation 108. Virus-containing media were harvested and pooled at 24, 36, and 48 hours post transfection, filtered through a 0.2-μm filter, and stored at −70° prior to use.
Ribonuclease protection assays
RNA was isolated from pelleted virus using a previously described proteinase K-based extraction protocol 22 and quantified by RNase protection using a chimeric MoMuLV-7SL riboprobe complementary to both portions of the MoMuLV 5′ untranslated region (nts. 55-255) and part of 7SL (Figure 2d) 49. Previously described RNAse protection assay approaches 49 were modified by extending hybridization times to 16h and digesting using only RNase T1. The riboprobe protected 201nt of the wild type and the SL-C mutant (U328A/A333U) RNAs, 159nt of the “classic Ψ” deletion (Δ215-568) RNA, and 100nt of 7SL RNA. Bands were quantified by PhosphorImager analysis and adjusted for the number of radiolabelled Cs incorporated. RNA packaging was quantified by normalizing the amount of MoMuLV RNA in each lane to the amount of co-packaged 7SL.
NMR Spectroscopy
Samples for NMR studies (250 μL of 800 μM RNA in D2O (99.8 %; Cambridge Isotope Laboratories, Massachussetts) in a Shigemi sample tube (Japan)) were prepared in TRIS-d11 buffer (pD = 7.0) containing NaCl (80 mM). NMR data were collected with a Bruker AVANCE spectrometer (800 MHz, 1H; sample temperature = 37 °C), and were processed with NMRPipe/NMRDraw109 and analyzed with NMRView110. Non-exchangeable 1H assignments were obtained from 2D NOESY111,112 (τm=100 ms) data collected at 37 °C, with real and imaginary components of indirect dimension collected in an interleaved manner to minimize t1 artifacts.
NMR Structure Calculations
Structures were calculated and refined with CYANA113 using the AMBER114 residue library. Upper-limit distance restraints of 2.7 Å, 3.3 Å and 5.0 Å were employed for direct NOE cross peaks of strong, medium and weak intensities, respectively, for all cross peaks except those associated with the intraresidue H8/6-to-H2′ and –H3′ interactions. For these proton pairs, upper distance limits of 4.2 Å and 3.2 Å were therefore employed for NOEs of medium and strong intensity, respectively.44 Cross-helix P-P distance restraints (with 20% weighting coefficient) were employed for A-form helical segments to prevent the generation of structures with collapsed major grooves:44,53,56 P(i)-P(i+2) (cross-helix phosphorus of the i+2 base pair) = 16.1 - 17.1 Å, P(i)-P(i+3) = 14.2 - 15.2 Å; P(i)-P(i+4) = 11.7 -12.7 Å; P(i)-P(i+5) = 9.4 -10.4 Å; P(i)-P(i+6) = 9.0 -10.0 Å. Torsion angle restraints for A helical stem residues were centered around published A-form RNA values (α=-62°, β=180°, γ=48°, δ=83°, ε=−152°, ζ=−73°)115 with allowed deviations of ± 50°. Four restraints per hydrogen bond were employed to enforce approximately linear NH-N and NH-O bond distances of 1.85 ± 0.05 Å, and two lower limit restraints per base pair were employed to weakly enforce base pair planarity (20% weighting coefficient) (G-C base pairs: G-C4 to C-C6 ≥ 8.3 Å and G-N9 to C-H6 ≥ 10.75 Å. A-U base pairs: A-C4 to U-C6 ≥ 8.3 Å and A-N9 to U-H6 ≥ 10.75 Å).
Cryo-Electron Tomography
The RNA sample (24 μM [ΨCD]2 in TRIS buffer, pH 7.0, containing 140 mM KCl and 2 mM MgCl2) was applied onto glow-discharged 200-mesh Quantifoil R 1.2/1.3 copper holey grids (Quantifoil Micro Tools GmBH, Germany) pre-treated with 15nm gold particles. The gold particles were used as fiducial markers during the tomographic reconstruction. The grid with the sample was blotted and vitrified using Mark III Vitrobot (FEI, USA) and stored in liquid nitrogen. Data collection was performed on a JEM2200FS electron microscope with a field emission gun and an in-column energy filter (energy slit of 10eV) at 200kV and ~23,123x detector magnification from −60 to +60° in 2° increments. The intended defocus was 8 μm. The cumulative specimen dose was ~70-90 electrons/Å2. Tilt series were collected on a Gatan 4k × 4k CCD camera using SerialEM 116. Final sampling was 6.49Å/pixel. Tomograms were reconstructed using IMOD and visualized by 3dmod.117
Densities visually examined with Chimera118 could be grouped qualitatively by volume. Two general groups were observed, one in which the densities had volumes approximately consistent with a 132 nucleotide RNA, and another in which densities had considerably larger volumes. In some cases the density was found to be extended in the third dimension, forming elongated, non-symmetric assemblies (i.e linear chains rather than the expected dimers), which was not obvious from the two dimensional projection. A total of 47 subvolumes from the group with smaller densities was subjected to an all-vs-all orientation search, which was performed independent of the NMR-derived [ΨCD]2 structure.119 In order to reduce the noise, the raw subvolumes were low-pass filtered to 40Å and spherically masked to a size slightly larger than a single particle. To speed the computational analysis, the data were shrunk in volume by a factor of two. Resulting rotations and translations were applied back to the original “raw” (i.e. not low pass filtered, masked or shrunk) subvolumes before averaging. It should be noted that because of the limited tilt range (−60 to +60°), it was easier to identify [ΨCD]2 structures that were oriented with their long axes more or less along the “z”, simply because this orientation produces the highest density in projection.
The averaging method used is an iterative procedure to align subvolumes to each other. The initial averages were obtained from the pairs of particles having the highest cross-correlation (excluding pairs where either member appears higher in the list).119 This procedure eventually generated two similar 3D class averages, with 33 and 14 subvolumes, respectively. In the next round of refining the classification, the 33-subvolume cryo-ET average was used as a template to which all initial subvolumes were compared and translation and rotation operations were applied. The number of subvolumes that were included in the final average was determined by the quality of their cross-correlation to the initial average and included a final total of 38 (Supplemental Figure S4). Additional control specimens (buffer, [ΨCES]2), see Results section) were prepared and analyzed as described above.
Supplementary Material
Acknowledgements
Support from the NIH (GM42561 to M.F.S., CA069300 to A.T., P41RR02250 to W.C.) is gratefully acknowledged. A.S.-M. was supported by NIH MARC U*Star (GM08663) and HHMI education grants, and R.I. was supported by NIH Training Grant No 5T90DK070121-05 through the Gulf Coast Consortia. We are grateful to Alexander Grishaev (NIAID, NIH) for collecting SAXS data and assisting in its interpretation.
Abbreviations used
- cryo-ET
cryo-electron tomography
- DLS
dynamic light scattering
- MoMuLV
Moloney murine leukemia virus
- MW
molecular weight
- NC
nucleocapsid
- NMR
nuclear magnetic resonance
- UTR
untranslated region
- HIV-1
human immunodeficiency virus 1
- ITC
isothermal titration calorimetry
- gRNA
genomic RNA
- RPA
RNase protection assay
- RT
reverse transcriptase
- SAXS
small angle X-ray scattering
- SV40
simian virus 40
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers. Coordinates have been deposited in the Protein Data Bank with accession number 2l1f. 1H NMR chemical shift assignments have been deposited in the BMRB with accession number 17083.
REFERENCES
- 1.Rein A. Retroviral RNA packaging: A review. Arch. Virology. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr. Top. Microbiol. Immun. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Jewell NA, Mansky LM. In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 2000;81:1889–1899. doi: 10.1099/0022-1317-81-8-1889. [DOI] [PubMed] [Google Scholar]
- 4.Paillart J-C, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nature Reviews Microbiology. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 5.Russell RS, Liang C, Wainberg MA. Retrovirology. 2 September edition. Vol. 1. 2004. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes,no, probably? [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greatorex J. Retrovirology. 18 August edition. Vol. 1. 2004. The retroviral RNA dimer linkage: different structures may reflect different roles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Souza V, Summers MF. How retroviruses select their genomes. Nature Reviews Microbiology. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 8.Mann R, Mulligan RC, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 9.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor Laboratory Press; Plainview, N.Y.: 1997. [PubMed] [Google Scholar]
- 10.Varmus HE. Form and function of retroviral proviruses. Science. 1982;216:812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- 11.Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 12.Onafuwa A, An W, Robson ND, Telesnitsky A. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J. Virol. 2003;77:4577–4587. doi: 10.1128/JVI.77.8.4577-4587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73:451–80. doi: 10.1128/MMBR.00012-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swanstrom R, Wills JW. Synthesis, assembly and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Press; Plainview, N.Y.: 1997. pp. 263–334. [PubMed] [Google Scholar]
- 15.Berkhout B. Prog. Nucl. Acid Res. and Mol. Biol. Vol. 54. Academic Press, Inc.; 1996. Structure and function of the human immunodeficiency virus leader RNA; pp. 1–34. [DOI] [PubMed] [Google Scholar]
- 16.Prats A-C, Roy C, Wang P, Erard M, Housset V, Gabus C, Paoletti C, Darlix J-L. Cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. Journal of Virology. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song R, Kafaie J, Yang L, Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007;371:1084–1096. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Levin JG, Grimley PM, Ramseur JM, Berezesky IK. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J. Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. Journal of Virology. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Housset V, De Rocquigny H, Roques BP, Darlix J-L. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. Journal of Virology. 1993;67:2537–2545. doi: 10.1128/jvi.67.5.2537-2545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hibbert CS, Mirro J, Rein A. mRNA molecules containing MLV packaging signals are encapsidated as dimers. J. Virol. 2004;78:10927–10938. doi: 10.1128/JVI.78.20.10927-10938.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flynn JA, King SR, Telesnitsky A. Nonrandom dimerization of murine leukemia virus genomic RNAs. J. Virol. 2004;78:12129–12139. doi: 10.1128/JVI.78.22.12129-12139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flynn JA, Telesnitsky A. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology. 2006;344:391–400. doi: 10.1016/j.virol.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu W-S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urisman A, Molinaro RJ, Fischer N, Plummer SJ, Casey G, Klein EA, Malathi K, Magi-Galluzzi C, Tubbs RR, Ganem D, Silverman RH, DeRisi JL. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci U S A. 2009;106:16351–6. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lombardi VC, Ruscetti FW, Gupta JD, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, Dean M, Silverman RH, Mikovits JA. Detection of an infectious retrovirus, XMRV, in blood cells of patients with Chronic Fatigue Syndrome. Science. 2009;326:585–589. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 28.Hohn O, Krause H, Barbarotto P, Niederstadt L, Beimforde N, Denner J, Miller K, Kurth R, Bannert N. Lack of evidence for xenotropic murine leukemia virus-related virus(XMRV) in German prostate cancer patients. Retrovirology. 2009;6:92. doi: 10.1186/1742-4690-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erlwein O, Kaye S, McClure MO, Weber J, Wills G, Collier D, Wessely S, Cleare A. Failure to detect the novel retrovirus XMRV in chronic fatigue syndrome. PLoS One. 2010;5:e8519. doi: 10.1371/journal.pone.0008519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo SC, Pripuzova N, Li B, Komaroff AL, Hung GC, Wang R, Alter HJ. Detection of MLV-related virus gene sequences in blood of patients with chronic fatigue syndrome and healthy blood donors. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1006901107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.De Tapia M, Metzler V, Mougel M, Ehresmann B, Ehresmann C. Dimerization of MoMuLV genomic RNA: Redefinition of the role of the palindromic stem-loop H1 (278-303) and new roles for stem-loops H2 (310-352) and H3 (355-374) Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- 32.Mougel M, Tounekti N, Darlix J-L, Paoletti J, Ehresmann B, Ehresmann C. Conformational analysis of the 5′ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Research. 1993;21:4677–4684. doi: 10.1093/nar/21.20.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy C, Tounekti N, Mougel M, Darlix J-L, Paoletti C, Ehresmann C, Ehresmann B, Paoletti J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Research. 1990;18:7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tounekti N, Mougel M, Roy C, Marquet R, Darlix J-L, Paoletti J, Ehresmann B, Ehresmann C. Effect of dimerization on the conformation of the encapsidation psi domain of Moloney murine leukemia virus RNA. Journal of Molecular Biology. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- 35.Girard P-M, Bonnet-Mathoniere B, Mauriaux D, Paoletti J. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 36.Ly H, Nierlich DP, Olsen JC, Kaplan AH. Moloney murine sarcoma virus genomic RNAs dimerize via a two-step process: a concentration-dependent kissing-loop interaction is driven by initial contact between consecutive guanines. J. Virol. 1999;73:7255–7261. doi: 10.1128/jvi.73.9.7255-7261.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oroudjev EM, Kang PCE, Kohlstaedt LA. An additional dimer linkage structure in Moloney Murine Leukemia Virus RNA. J. Mol. Biol. 1999;291:603–613. doi: 10.1006/jmbi.1999.2984. [DOI] [PubMed] [Google Scholar]
- 38.Ly H, Parslow TG. Bipartite signal for genomic RNA dimerization in the Moloney Murine Leukemia Virus. J. Virol. 2002;76:3135–3144. doi: 10.1128/JVI.76.7.3135-3144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim C-H, Tinoco I., Jr. A retroviral RNA kissing complex containing only two G-C base pairs. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konings DAM, Nash MA, Maizel JV, Arlinghaus RB. Novel GACG-hairpin pair motif in the 5′ untranslated region of type C retrovirus related to murine leukemia virus. Journal of Virology. 1992;66:632–640. doi: 10.1128/jvi.66.2.632-640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney Murine Leukaemia Virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 42.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J. Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Souza V, Melamed J, Habib D, Pullen K, Wallace K, Summers MF. Identification of a high-affinity nucleocapsid protein binding site within the Moloney Murine Leukemia Virus Ψ-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 2001;314:217–232. doi: 10.1006/jmbi.2001.5139. [DOI] [PubMed] [Google Scholar]
- 44.D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101 nucleotide core encapsidation signal of the Moloney Murine Leukemia Virus. J. Mol. Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 45.Badorrek CS, Gherghe CM, Weeks KM. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization. Proc. Natl. Acad. Sci. USA. 2006;103:13640–13645. doi: 10.1073/pnas.0606156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badorrek CS, Weeks KM. Architecture of a gamma retroviral genomic RNA dimer. Biochemistry. 2006;45:12664–12672. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- 47.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robson ND, Telesnitsky A. Effects of 3′ untranslated region mutations on plus-strand priming during moloney murine leukemia virus replication. J Virol. 1999;73:948–57. doi: 10.1128/jvi.73.2.948-957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Onafuwa-Nuga AA, King SR, Telesnitsky A. Nonrandom packaging of host RNAs in Moloney murine leukemia virus. J. Virol. 2005;79:13528–13537. doi: 10.1128/JVI.79.21.13528-13537.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Onafuwa-Nuga AA, Telesnitsky A, King SR. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA. 2006;12:542–6. doi: 10.1261/rna.2306306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rulli J,SJ, Hibbert CS, Mirro J, Pederson T, Biswal S, Rein A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007;81:6623–6631. doi: 10.1128/JVI.02833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucl. Acids Res. 2003;31:e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolbert BS, Miyazaki Y, Barton S, Kinde B, Starck P, Singh R, Bax A, Case DA, Summers MF. Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement. J. Biomol. NMR. 2010;47:205–219. doi: 10.1007/s10858-010-9424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J. Biomol. NMR. 2009 doi: 10.1007/s10858-009-9375-2. DOI 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clore GM, Kuszewski J. Improving the accuracy of NMR structures of RNA by means of conformational database potentials of mean force as assessed by complete dipplar coupling cross-validation. J. Am. Chem. Soc. 2003;125:1518–1525. doi: 10.1021/ja028383j. [DOI] [PubMed] [Google Scholar]
- 56.Rife JP, Stallings SC, Correll CC, Dallas A, Steitz TA, Moore PB. Comparison of the crystal and solution structures of two RNA oligonucleotides. Biophys. J. 1999;76:66–75. doi: 10.1016/S0006-3495(99)77178-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Güntert P, Wüthrich K. Improved Efficiency of Protein Structure Calculations from NMR Data Using the Program DIANA with Redundant Dihedral Angle Constraints. Journal of Biomolecular NMR. 1991;1:447–456. doi: 10.1007/BF02192866. [DOI] [PubMed] [Google Scholar]
- 58.Trantirek L, Urbasek M, Stefl R, Feigon J, Sklenar V. A method for direct determination of helical parameters in nucleic acids using residual dipolar couplings. J. Am. Chem. Soc. 2000;122:10454–10455. [Google Scholar]
- 59.Bayer P, Varani L, Varani G. Refinement of the structure of protein-RNA complexes by residual dipolar coupling analysis. J Biomol NMR. 1999;14:149–55. doi: 10.1023/a:1008360331296. [DOI] [PubMed] [Google Scholar]
- 60.Warren JJ, Moore PB. Application of dipolar coupling data to the refinement of the solution structure of the sarcin-ricin loop RNA. J Biomol NMR. 2001;20:311–23. doi: 10.1023/a:1011214214552. [DOI] [PubMed] [Google Scholar]
- 61.Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc Natl Acad Sci U S A. 2004;101:1177–82. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tjandra N, Tate S, Ono A, Kainosho M, Bax A. The NMR structure of a DNA dodecamer in an aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 63.Mollova ET, Hansen MR, Pardi A. Global structure of RNA determined with residual dipolar couplings. J. Am. Chem. Soc. 2000;122:11561–11562. [Google Scholar]
- 64.Ying J, Grishaev A, Latham MP, Pardi A, Bax A. Magnetic field induced residual dipolar couplings of imino groups in nucleci acids from measurements at a single magnetic field. J. Biomol. NMR. 2007;39:91–96. doi: 10.1007/s10858-007-9181-7. [DOI] [PubMed] [Google Scholar]
- 65.Latham MP, Hanson P, Brown DJ, Pardi A. Comparison of alignment tensors generated for native tRNAVal using magnetic fields and liquid crystalline media. J. Biomol. NMR. 2008;40:83–94. doi: 10.1007/s10858-007-9212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bax A, Grishaev A. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr. Opin. Struct. Biol. 2005;15:563–570. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 67.Lukavsky PJ, Puglisi JD. Structure determination of large biological RNAs. Methods in Enzymology. 2005;394:399–415. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 68.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nature Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 69.Al-Hashimi HM, Walter NG. RNA dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–9. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Al-Hashimi HM, Tolman JR, Majumdar A, Gorin A, Patel DJ. Determining stoichiometry in homomultimeric nucleic acid complexes using magnetic field induced residual dipolar couplings. J Am Chem Soc. 2001;123:5806–7. doi: 10.1021/ja0105865. [DOI] [PubMed] [Google Scholar]
- 71.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. Mg2+-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J Mol Biol. 2003;329:867–73. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Al-Hashimi HM, Patel DJ. Residual dipolar couplings: synergy between NMR and structural genomics. J Biomol NMR. 2002;22:1–8. doi: 10.1023/a:1013801714041. [DOI] [PubMed] [Google Scholar]
- 73.Al-Hashimi HM, Majumdar A, Gorin A, Kettani A, Skripkin E, Patel DJ. Field- and phage-induced dipolar couplings in a homodimeric DNA quadruplex: relative orientation of G.(C-A) triad and G-tetrad motifs and direct determination of C2 symmetry axis orientation. J Am Chem Soc. 2001;123:633–40. doi: 10.1021/ja003379y. [DOI] [PubMed] [Google Scholar]
- 74.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 75.Al-Hashimi HM, Gorin A, Majumdar A, Patel DJ. Alignment of the HTLV-I Rex peptide bound to its target RNA aptamer from magnetic field-induced residual dipolar couplings and intermolecular hydrogen bonds. J Am Chem Soc. 2001;123:3179–80. doi: 10.1021/ja004133w. [DOI] [PubMed] [Google Scholar]
- 76.Al-Hashimi HM. Beyond static structures of RNA by NMR: folding, refolding, and dynamics at atomic resolution. Biopolymers. 2007;86:345–7. doi: 10.1002/bip.20754. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Q, Al-Hashimi HM. Extending the NMR spatial resolution limit for RNA by motional couplings. Nat Methods. 2008;5:243–5. doi: 10.1038/nmeth.1180. [DOI] [PubMed] [Google Scholar]
- 78.Frank AT, Stelzer AC, Al-Hashimi HM, Andricioaei I. Constructing RNA dynamical ensembles by combining MD and motionally decoupled NMR RDCs: new insights into RNA dynamics and adaptive ligand recognition. Nucleic Acids Res. 2009;37:3670–9. doi: 10.1093/nar/gkp156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fisher CK, Al-Hashimi HM. Approximate reconstruction of continuous spatially complex domain motions by multialignment NMR residual dipolar couplings. J Phys Chem B. 2009;113:6173–6. doi: 10.1021/jp900411z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lawrence DC, Stover CC, Noznitsky J, Wu Z-R, Summers MF. Structure of the intact stem and bulge of HIV-1 Ψ-RNA stem loop SL1. J. Mol. Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 81.Zuo X, Wang J, Foster TR, Schwieters CD, Tiede DM, Butcher SE, Wang Y-X. Global molecular structure and interfaces: Refining an RNA:RNA complex structure using solution X-ray scattering data. J. Am. Chem. Soc. 2008;130:3292–3293. doi: 10.1021/ja7114508. [DOI] [PubMed] [Google Scholar]
- 82.Walsh JD, Wang YX. Periodicity, planarity, residual dipolar coupling, and structures. J Magn Reson. 2005;174:152–62. doi: 10.1016/j.jmr.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 83.Walsh JD, Cabello-Villegas J, Wang YX. Periodicity in residual dipolar couplings and nucleic acid structures. J Am Chem Soc. 2004;126:1938–9. doi: 10.1021/ja039446g. [DOI] [PubMed] [Google Scholar]
- 84.Wang J, Walsh JD, Kuszewski J, Wang YX. Periodicity, planarity, and pixel (3P): a program using the intrinsic residual dipolar coupling periodicity-to-peptide plane correlation and phi/psi angles to derive protein backbone structures. J Magn Reson. 2007;189:90–103. doi: 10.1016/j.jmr.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 85.Grishaev A, Ying J, Bax A. Pseudo-CSA restraints for NMR refinement of nucleic acid structures. J. Am. Chem. Soc. 2006;128:10010–10011. doi: 10.1021/ja0633058. [DOI] [PubMed] [Google Scholar]
- 86.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 87.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 88.Kim NK, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol. 2008;384:1249–61. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Skrisovska L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, Elliott DJ, Stevenin J, Allain FH. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007;8:372–9. doi: 10.1038/sj.embor.7400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 92.Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. High-resolution structural analysis of a DNA nanostructure by cryoEM. Nano Lett. 2009;9:2747–50. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]
- 93.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang J, Zuo X, Yu P, Xu H, Starich MR, Tiede DM, Shapiro BA, Schwieters CD, Wang YX. A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J Mol Biol. 2009;393:717–34. doi: 10.1016/j.jmb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang YX, Zuo X, Wang J, Yu P, Butcher SE. Rapid global structure determination of large RNA and RNA complexes using NMR and small-angle X-ray scattering. Methods. 2010 doi: 10.1016/j.ymeth.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang S, Temin HM. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mougel M, Zhang Y, Barklis E. cis-active structural motifs involved in specific encapsidation of Moloney Murine leukemia virus RNA. J. Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fisher J, Goff SP. Mutational analysis of stem-loops in the RNA packaging signal of the moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- 100.Miyazaki Y, Garcia E, King SR, Iyalla K, Loeliger K, Starck P, Syed S, Telesnitsky A, Summers MF. An RNA structural switch regulates diploid genome packaging by Moloney Murine Leukemia Virus. J. Mol. Biol. 2010;396:141–152. doi: 10.1016/j.jmb.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106:19114–9. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dey A, York D, Smalls-Mantey A, Summers MF. Composition and sequence dependent binding of RNA to the nucleocapsid protein of Moloney Murine Leukemia Virus. Biochemistry. 2005;44:3735–3744. doi: 10.1021/bi047639q. [DOI] [PubMed] [Google Scholar]
- 103.Rabi JA, Fox JJ. Nucleosides. LXXIX. Facile base-catalyzed hydrogen isotope labeling at position 6 of pyrimidine nucleosides. J Am Chem Soc. 1973;95:1628–32. doi: 10.1021/ja00786a044. [DOI] [PubMed] [Google Scholar]
- 104.Goodman L. Chemical synthesis and transformations of nucleosides. In: Ts’o, editor. Basic Principles in Nucleic Acid Chemistry. Vol. 1. Academic Press; New York: 1974. pp. 170–208. [Google Scholar]
- 105.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Meth. Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 106.Pfeiffer JK, Topping RS, Shin NH, Telesnitsky A. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J Virol. 1999;73:8441–7. doi: 10.1128/jvi.73.10.8441-8447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yang S, Delgado R, King SR, Woffendin C, Barker CS, Yang ZY, Xu L, Nolan GP, Nabel GJ. Generation of retroviral vector for clinical studies using transient transfection. Hum Gene Ther. 1999;10:123–32. doi: 10.1089/10430349950019255. [DOI] [PubMed] [Google Scholar]
- 108.Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Struhl K. Current protocols in molecular biology. John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 109.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 110.Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 111.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. Journal of Chemical Physics. 1979;71:4546–4553. [Google Scholar]
- 112.Macura S, Ernst RR. Elucidation of cross relaxation in liquids by two-dimensional NMR spectroscopy. Mol. Phys. 1980;41:95–117. [Google Scholar]
- 113.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for protein structure calculations with a new program, DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 114.Case DA. AMBER 7 Users Manual. University of California San Francisco; 2002. [Google Scholar]
- 115.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York, NY: 1984. [Google Scholar]
- 116.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 117.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 118.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 119.Schmid MF, Booth CR. Methods for aligning and for averaging 3D volumes with missing data. J Struct Biol. 2008;161:243–8. doi: 10.1016/j.jsb.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


