Abstract
The potent lipid mediator sphingosine-1-phosphate (S1P) regulates diverse physiological processes by binding to 5 specific GPCRs, although it also has intracellular targets. Here, we demonstrate that S1P, produced in the mitochondria mainly by sphingosine kinase 2 (SphK2), binds with high affinity and specificity to prohibitin 2 (PHB2), a highly conserved protein that regulates mitochondrial assembly and function. In contrast, S1P did not bind to the closely related protein PHB1, which forms large, multimeric complexes with PHB2. In mitochondria from SphK2-null mice, a new aberrant band of cytochrome-c oxidase was detected by blue native PAGE, and interaction between subunit IV of cytochrome-c oxidase and PHB2 was greatly reduced. Moreover, depletion of SphK2 or PHB2 led to a dysfunction in mitochondrial respiration through cytochrome-c oxidase. Our data point to a new action of S1P in mitochondria and suggest that interaction of S1P with homomeric PHB2 is important for cytochrome-c oxidase assembly and mitochondrial respiration.—Strub, G. M., Paillard, M., Liang, J., Gomez, L., Allegood, J. C., Hait, N. C., Maceyka, M., Price, M. M., Chen, Q., Simpson, D. C., Kordula, T., Milstien, S., Lesnefsky, E. J., Spiegel, S. Sphingosine-1-phosphate produced by sphingosine kinase 2 in mitochondria interacts with prohibitin 2 to regulate complex IV assembly and respiration.
Keywords: SphK2, oxidative phosphorylation, cytochrome-c oxidase
Sphingosine-1-phosphate (S1P) is a potent lipid mediator produced by two sphingosine kinase isoenzymes, SphK1 and SphK2. It regulates diverse physiological and pathological processes mainly by binding to 5 specific G-protein-coupled cell surface receptors (GPCR), termed S1P1–5. The majority of research to date has focused on signaling through these S1P receptors, but compelling evidence is emerging to support S1P having multiple intracellular functions independent of S1P receptors in organisms as diverse as yeast, plants, and mammals (1–3). Although yeast do not have S1P receptors, accumulation of phosphorylated sphingoid bases confers resistance to heat shock (4, 5). In plants, intracellular phosphorylated sphingoid bases regulate stomatal apertures and drought responses, likely independently of their two GPCR-like proteins, GCR1 and GCR2 (6, 7).
Numerous studies in mammalian cells have implicated intracellular actions of S1P in regulation of important biological functions, beginning with the demonstration that intracellular S1P may regulate calcium release independently of inositol trisphosphate receptors (8, 9). Indeed, caged S1P elicits calcium release in cells that do not respond to exogenous S1P (10). Consistent with these results, mast cells from sphk2−/− mice with reduced intracellular S1P have a defect in calcium mobilization in response to IgE receptor crosslinking that cannot be reversed with exogenous S1P (11). Moreover, overexpression of SphK1, which increases intracellular S1P, stimulated growth and survival in cells devoid of S1P receptors (12). In agreement, S1P produced intracellularly by SphK1 enhanced intestinal adenoma cell proliferation and tumor size in APCmin/+ mice independently of S1P receptors (13). Hence, intracellular actions of S1P appear to be biologically important but remain relatively unexplored.
SphK2, which is localized in the nucleus of many types of cells (14–18), was recently shown to produce S1P that binds to and inhibits histone deacetylases, HDAC1 and HDAC2, resulting in enhanced local histone acetylation and increased transcription of specific genes (18). To identify other intracellular targets, we utilized S1P affinity matrices to pull down proteins that bind S1P with high affinity (18). Here, we report that S1P binds with high affinity and specificity to prohibitin 2 (PHB2), a highly conserved protein predominantly localized to the inner mitochondrial membrane that has been implicated in regulating mitochondrial function (19–21), and demonstrate that this interaction in the mitochondria is important for the assembly and function of respiratory complex IV (cytochrome-c oxidase, COX) in the electron transport chain.
MATERIALS AND METHODS
Materials
S1P was obtained from Enzo Life Sciences International (Plymouth Meeting, PA, USA), and other lipids were from Avanti Polar Lipids (Alabaster, AL, USA). S1P, LPA, and control lipid conjugated beads were obtained from Echelon Biosciences (Salt Lake City, UT, USA). Antibodies against the following were used: PHB1 (Neomarkers/Lab Vision, Fremont, CA, USA); PHB2 (Millipore, Billerica, MA, USA); β-tubulin and lamin A/C (Cell Signaling Technology, Danvers, MA, USA); V5 and TOM20 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); complex I subunit NDuFA9, 70-kDa complex II subunit, core 2 subunit of complex III, subunit I of cytochrome-c oxidase, and subunit IV of cytochrome-c oxidase (Mitosciences, Boston, MA, USA); cyclophilin D, ANT, and VDAC (Calbiochem, San Diego, CA, USA); murine C-terminal SphK2 [kindly provided by Dr. Richard Proia, National Institutes of Health (NIH), Bethesda, MD, USA]; secondary antibodies (Jackson ImmunoResearch, West Grove, PA, USA). Rabbit polyclonal antiserum raised against a unique SphK2 peptide sequence (QALHIQRLRPKPEARPR) was purified as described previously (22). Primer probes for qPCR were obtained from Applied Biosystems (Carlsbad, CA, USA). Protein A/G beads were from Santa Cruz Biotechnology.
Pulldowns with lipid affinity matrices
Control, S1P, or LPA-coated agarose beads (Echelon Biosciences, Salt Lake City, UT, USA) equilibrated with binding buffer containing 10 mM HEPES (pH 7.8), 150 mM NaCl, and 0.5% Igepal were incubated with extracts (10 μg protein/1 μl beads) for 2 h at 4°C. Beads were washed, and bound proteins were analyzed by SDS-PAGE. Gels were stained with SyproRuby (Invitrogen, Carlsbad, CA, USA), washed, and exposed to UV light to visualize protein bands. Bands were excised, and polypeptides were sequenced by MALDI-TOF mass spectrometry (Center for Proteomics, University of Medicine and Dentistry of New Jersey, New Brunswick, NJ, USA).
[32P]S1P binding assay
Cell extracts were incubated with or without unlabeled lipids in the presence of [32P]S1P (0.1 nM, 6.8 μCi/pmol) in buffer containing 50 mM Tris (pH 7.5), 137 mM NaCl, 1 mM MgCl2, 2.7 mM KCl, 15 mM NaF, and 0.5 mM NaV3O4 for 60 min at 4°C. Samples were precleared with Protein A/G agarose beads and immunoprecipitated with anti-PHB1, anti-PHB2, or isotype matched IgG. Beads were collected, washed extensively, and bound [32P]S1P was quantified with a scintillation counter (18).
Mass spectrometric analysis of lipids
Sphingolipids were quantified by liquid chromatography, electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) with a Shimadzu LC-20AD binary pump HPLC system (Shimadzu, Kyoto, Japan) and an Applied Biosystems 4000 QTRAP operating in a triple quadrupole mode, as described previously (18). In some experiments, cell extracts were immunoprecipitated with anti-PHB1 or anti-PHB2 antibody or with control IgG, as described above, and bound sphingolipids were determined by LC-ESI-MS/MS.
Cell culture and transfection
MCF7 and HeLa cells were transfected with OnTargetPlus SmartPool siRNA against SphK2 (5′-CAAGGCAGCUCUACACUCA-3′; 5′-GAGACGGGCUGCUCCAUGA-3′; 5′-GCUCCUCCAUGGCGAGUUU-3′; 5′-CCACUGCCCUCACCUGUCU-3′), PHB2 (5′-GUGAAUAUCUCCCUGCGAG-3′; 5′-CCACAUCACAGAAUCGUAU-3′; 5′-CAUCAAACUUCGCAAGAUU-3′; 5′-CAGAAUAUCUCCAAGACGA-3′); and control siRNA from Dharmacon (Lafayette, CO, USA), as described previously (18).
Preparation of HeLa cell mitochondria and nuclear fractions
Mitochondria, cytoplasmic fractions, and nuclear fractions were isolated from 2 × 107 HeLa cells with the mitochondria isolation kit for cultured cells and the NePER kit (Pierce Chemical, Rockford, IL, USA), respectively, according to the manufacturer's instructions.
Mice and preparation of cardiac mitochondria
Protocols and studies involving animals were performed in accordance with the Virginia Commonwealth University Institutional Animal Care and Use Committee guidelines. C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA), and SphK2-knockout mice were obtained from Dr. Richard Proia (NIH). Twelve- to 16-wk-old male mice were used for preparation of heart mitochondria, as described previously (23). Briefly, hearts were harvested from mice while still beating and immediately placed in cold buffer containing 100 mM KCl, 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS; pH 7.4), 1 mM EGTA, 5 mM MgSO4, 1 mM ATP, and 0.2% BSA, as described previously (23). Cardiac tissue was homogenized with a glass tissue grinder, and the homogenate was centrifuged at 500 g for 10 min. The supernatant was then centrifuged at 3000 g for 10 min, and the mitochondrial pellet was washed and suspended in 100 mM KCl, 50 mM MOPS, and 0.5 mM EGTA. Soluble and membrane fractions were prepared from purified mitochondria exactly as described previously (24).
Western blot analysis
Cells were sonicated in buffer containing 50 mM Tris, 100 mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, 1 mM DTT, 1 mM PMSF, and 1:500 protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA). Equal amounts of proteins were separated by SDS-PAGE, and immunopositive bands were visualized by enhanced chemiluminescence, as described previously (18). Where indicated, immunopositive bands were detected with IRDye 800 anti-rabbit (Rockland, Gilbertsville, PA, USA) and AlexaFluor 680 anti-mouse (Invitrogen) antibodies and visualized with an Odyssey infrared imaging system (LiCor, Lincoln, NE, USA).
Mitochondrial oxidative phosphorylation
Oxygen consumption in mitochondria was measured using a Clark-type oxygen electrode at 30°C. Mitochondria were incubated in buffer containing 80 mM KCl, 50 mM MOPS (pH 7.4), 1 mM EGTA, 5 mM KH2PO4, and 1 mg/ml BSA. Glutamate/malate (complex I substrate; 20/5 mM), succinate (complex II substrate, 20 mM) plus rotenone (5 μM), and N,N,N,N-tetramethyl-p-phenylenediamine (TMPD)/ascorbate (complex IV substrate; 1–10 mM) plus rotenone were used as electron donors. State 3 (0.2 mM ADP-stimulated) and state 4 (ADP-limited) respiration rates, respiratory control ratio (RCR), rate of uncoupled respiration, and maximal rate of state 3 respiration (2 mM ADP) were measured as described previously (25).
Isolation of cardiomyocytes and measurement of oxygen uptake
Cardiomyocytes (∼15,000 cells), isolated as described previously (26), were suspended in MiR05 mitochondrial respiration medium containing 0.5 mM EGTA, 110 mM sucrose, 3 mM MgCl2, 20 mM taurine, 10 mM KH2PO4, 20 mM HEPES (pH 7.1), and 1 g/L BSA. A multiple substrate-uncoupler-inhibitor titration protocol was optimized with a high-resolution oxygraph (Oxygraph-2k; Oroboros, Innsbruck, Austria) to determine oxidative phosphorylation. Pyruvate/malate (5 mM and 2 mM, respectively) were used as complex I substrates. After permeabilization of cardiomyocytes with digitonin (10 μg/ml), consumption of oxygen was strongly activated by 2 mM ADP. Successive additions of rotenone (0.5 μM), succinate (complex II substrate; 10 mM), TTFA (40 μM), TMPD/ascorbate (complex IV substrates; 0.3 and 3 mM respectively), and then azide (15 mM), allowed determination of sensitive rates of oxidative phosphorylation using complex I, II, and IV substrates, respectively. Oxygen consumption was evaluated by the Oroboros DatLab4 software and expressed as nanomoles of oxygen per minute per milligram.
Measurement of oxidative phosphorylation in HeLa cells
HeLa cells (2.5×106/ml) were permeabilized at 30°C with digitonin (10 μg/ml) in medium containing 80 mM KCl, 10 mM Tris (pH 7.4), 3 mM MgCl2, 1 mM EDTA, and 5 mM KH2PO4. After injection of 2 mM of ADP, sensitive rates for complexes I, II, and IV were measured as described for cardiomyocytes.
Citrate synthase and electron transport chain enzyme activities
Citrate synthase activity was measured in cholate-solubilized, frozen-thawed mitochondria (27). NADH-cytochrome-c reductase, rotenone-sensitive (NCR; complexes I and III); ubiquinol:ferricytochrome-c oxido-reductase (complex III), and cytochrome-c oxidase (complex IV) were measured in detergent-solubilized, frozen-thawed mitochondria at 37°C, as described previously (25).
Immunocapture of PHB2 and COX
Mitochondrial proteins (300 μg) were precleared with magnetic protein A Dynabeads (Invitrogen) and then mixed overnight at 4°C with 0.6 μg of anti-PHB2 antibody (Bethyl, Montgomery, TX, USA) in buffer containing 50 mM Tris, 150 mM NaCl, 1% Triton X-100, and 1 mM EDTA, supplemented with protease and phosphatase inhibitors (Roche, Nutley, NJ, USA). The mixture was then incubated for 2 h at 4°C with protein A Dynabeads, which were captured with a Dynal magnet (Invitrogen), and bound proteins were analyzed by Western blot analysis. After solubilization of the mitochondrial protein (300 μg) in n-dodecyl-β-d-maltopyranoside, native COX was isolated with the complex IV rodent immunocapture kit (Mitosciences), according to the manufacturer's instructions.
Blue native PAGE (BN-PAGE)
Mitochondrial proteins were solubilized with n-dodecyl-β-d-maltopyranoside (detergent:protein ratio of 1.6:1, g/g), combined with Coomassie blue G-250 dye (detergent/dye ratio of 4:1, g/g), and separated on 4–16% Bis-Tris native PAGE gels (Invitrogen) (28). Proteins were visualized with Coomassie blue or analyzed by immunoblotting with mitochondria respiratory complex antibodies (Mitosciences), including complex I subunit NDuFA9, complex II subunit 70 kDa, complex III subunit core 2, complex IV subunit 1, and holo cytochrome-c oxidase.
In-gel COX activity
Gel strips were incubated in buffer containing 5 mg diaminobenzidine dissolved in 10 ml 50 mM sodium phosphate (pH 7.2) and 50 μM oxidized horse heart cytochrome c as described previously (29). After 30 min, reactions were stopped by addition of 50% methanol/10% acetic acid. The specificity for COX was assessed with antimycin A and azide.
Sphingosine kinase activity
SphK1 and SphK2 activities were determined by isotype-specific assays that have 20–30% overlap, exactly as described previously (30). Briefly, SphK1 activity was measured in the presence of 0.25% Triton X-100, which inhibits SphK2. SphK2 activity was determined in the presence of 4 mg/ml BSA and 1 M KCl, conditions in which SphK1 is inhibited. SphK-specific activity is expressed as picomoles S1P formed per minute per milligram protein (30).
Confocal microscopy
Cells grown on glass coverslips were mounted on glass slides and examined with a Zeiss LSM 510 Meta confocal microscope (software V3.25.0; Carl Zeiss, Thornwood, NY, USA) (18). To analyze mitochondrial inner membrane potential, cells were incubated for 30 min at 37° with 50 nM tetramethylrodamine methyl ester (TMRM; Molecular Probes, Carlsbad, CA, USA), and fluorescence measurements were made using 543 nm for excitation and a 560- to 615-nm bandpass emission filter. ImageJ 1.6.0_12 image analysis software (NIH) was used for computerized quantification of fluorescence intensity.
Statistical analysis
Data are expressed as means ± se. For oxidative phosphorylation of HeLa cells, differences between groups were compared by 1-way ANOVA. When a significant F value was obtained, means were compared using Tukey's test. For other analyses, statistical significance was determined with Student's t test for unpaired samples, and values of P < 0.05 were considered significant.
RESULTS
PHB2 is a bona fide S1P-binding protein
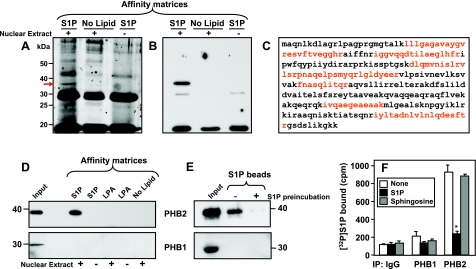
Recently, we have shown that HDAC1 and HDAC2 specifically bind S1P produced in the nucleus (18). Utilizing the rapid and sensitive method that we developed to pull down S1P-binding proteins with S1P immobilized on agarose beads (18), we sought to identify other proteins that specifically interact with S1P. A distinct protein band with an apparent molecular mass of ∼34 kDa was evident in the pulldown of nuclear proteins with S1P-conjugated beads that was not present in the absence of nuclear extract or when control unconjugated agarose beads were used (Fig. 1A). This band was excised and sequenced by MALDI-TOF mass spectrometry. Nine peptide sequences were obtained, spanning 40% of the amino acid sequence of prohibitin 2 (PHB2; Fig. 1C and Supplemental Table S1), an evolutionarily conserved and ubiquitously expressed membrane protein that is essential for cell growth and development (19–21). Western blot analysis with a specific PHB2 antibody confirmed the identity of this S1P-binding protein (Fig. 1B). Moreover, PHB2 was associated only with matrices carrying S1P but not with matrices without lipids or matrices carrying LPA, another bioactive lysophospholipid structurally similar to S1P (Fig. 1D). Preincubation of nuclear extracts with exogenous S1P abolished binding of PHB2 to the S1P affinity beads (Fig. 1E), further confirming the specificity of the association between PHB2 and S1P. In contrast, the closely related protein PHB1, which shares >50% identical amino acid residues with PHB2 (19–21), did not bind to S1P agarose beads (Fig. 1D, E). Next, direct binding of 32P-labeled S1P to endogenous PHB2 and PHB1 was evaluated, as described previously (18). 32P-labeled S1P was incubated with nuclear extracts from MCF7 cells, and PHB2 and PHB1 were then immunoprecipitated with specific antibodies. A significant amount of [32P]S1P was present in PHB2 immunoprecipitates that was markedly reduced by the addition of unlabeled S1P but not sphingosine (Fig. 1F). In contrast, very little radioactivity was detectable in the PHB1 immunoprecipitate, similar to that associated with isotype-matched IgG control antibody (Fig. 1F).
Figure 1.
A–C) Identification of PHB2 as an S1P binding protein. A) Control (no lipid) or S1P affinity beads were incubated without or with MCF7 nuclear extracts (NE) and extensively washed; bound proteins were resolved by SDS-PAGE and were visualized with SYPRO Ruby protein gel stain. Arrow indicates the band that was excised and sequenced by mass spectrometry. Dark stained bands that appear in the no extract lane are from the affinity matrix. B) Bound proteins from duplicate nuclear extracts were resolved by SDS-PAGE and analyzed by Western blotting with anti-PHB2 antibody. C) Amino acid sequence of PHB2. Red indicates peptide sequences identified by mass spectrometry. D–F) PHB2 specifically binds S1P. D) Nuclear extracts from MCF7 cells were incubated with control (no lipid), LPA, or S1P affinity matrices, as indicated, and washed extensively; bound proteins were separated by SDS-PAGE and then analyzed by Western blotting with anti-PHB2 or anti-PHB1 antibodies. Inputs are shown in the leftmost lanes. E) Nuclear extracts were preincubated without or with 10 μM S1P for 2 h, followed by pulldown with S1P beads. Beads were washed, and bound proteins were analyzed by Western blotting with anti-PHB2 or anti-PHB1 antibodies. F) Nuclear extracts from MCF7 cells were incubated for 30 min with [32P]S1P (0.1 nM) in the absence or presence of 10 μM unlabeled S1P or sphingosine. Cell lysates were then immunoprecipitated with antibodies against PHB1 or PHB2 or with control IgG, and radioactivity in the immunocomplexes was determined by scintillation counting. Data are expressed as means ± sd. *P < 0.05.
Because PHB2 is ubiquitously expressed, it was important to determine whether binding of S1P to PHB2 was a general phenomenon. PHB2 present in HeLa cell lysates also bound [32P]S1P in a specific manner, as binding was abolished by the addition of excess unlabeled S1P but not by sphingosine or LPA (Fig. 2A). Displacement of [32P]S1P bound to PHB2 by increasing concentrations of S1P indicated a Kd value of ∼1 μM (Supplemental Fig. S1). Dihydro-S1P, which lacks the trans double bond and is a ligand for all of the S1P receptors, also effectively competed with labeled S1P for binding to PHB2 (Fig. 2A).
Figure 2.
Endogenous S1P is bound to PHB2 in cells. A) HeLa cell lysates were incubated for 30 min with [32P]S1P (0.1 nM) in the absence or presence of 10 μM unlabeled S1P, dihydro-S1P (DHS1P), sphingosine, or LPA, as indicated. Cell lysates were then immunoprecipitated with antibodies against PHB2 or with isotype-matched IgG antibody, and radioactivity in the immunocomplexes was determined by scintillation counting. Data are expressed as means ± sd. *P < 0.05. B–D) HeLa cell lysates were immunoprecipitated with antibodies against PHB1 or PHB2 or with control IgG, as indicated. Immunocomplexes were captured with protein A/G agarose beads and washed extensively. B) Equal amounts of proteins were analyzed by Western blotting with specific antibodies for PHB1 and PHB2. C, D) Lipids were extracted from duplicate samples, and sphingolipids were analyzed by LC-ESI-MS/MS. C) Extracted ion chromatogram for LC-MS/MS reverse-phase separation of PHB2-specific antibody (red), and control IgG immunoprecipitates (blue). D) Quantification of S1P bound to IgG, PHB1, and PHB2 immunocomplexes. Data are means ± sd. E–G) Interaction of S1P and PHB2 in mitochondria. Mitochondria (mito), cytosol (cyto), and nuclear (nuc) fractions were isolated from HeLa cell lysates by subcellular fractionation. E) Proteins were analyzed by immunoblotting with anti-PHB1 and anti-PHB2 antibodies. Antibodies against subunit IV of cytochrome-c oxidase, COX (IV), lamin, and tubulin were used as specific organelle markers for mitochondria, nuclei, and cytosol, respectively. F) Proteins immunoprecipitated with antibodies against PHB2 or with control IgG were analyzed by immunoblotting with specific antibodies for PHB1 and PHB2. Similar results were obtained in two additional experiments. G) Mitochondria extracts were incubated with control (no lipid), sphingosine (Sph), or S1P affinity matrices, and washed extensively; bound proteins were separated by SDS-PAGE and then analyzed by Western blotting with anti-PHB2 or anti-PHB1 antibodies. Inputs are shown in the leftmost lanes.
We then sought to examine whether endogenous S1P is bound to PHB2 in vivo. To this end, we measured the abundance of sphingolipids in PHB2 and PHB1 immunoprecipitates from HeLa cell lysates by LC-ESI-MS/MS. In agreement with previous studies demonstrating that PHB2 and PHB1 interact with each other to form heteromers (31), PHB1 was effectively immunoprecipitated by anti-PHB2 and vice versa (Fig. 2B). Of all the sphingolipids present in these cells, only S1P was bound to immunoprecipitated PHB2 (Fig. 2C, D). Although both PHB1 and PHB2 were also effectively immunoprecipitated with anti-PHB1 antibody (Fig. 2B), endogenous S1P was not detected in this immunoprecipitate (Fig. 2D).
Mitochondrial homomeric PHB2 specifically binds S1P
Although PHB2 was initially named repressor of estrogen receptor activity (REA) because of its putative role in transcription regulation (32), more recently, it has become apparent that PHB2 functions predominantly within the mitochondria (reviewed in refs. 19, 20). Indeed, immunoblotting analysis of subcellular fractions of HeLa cells revealed that both PHB2 and PHB1 are present in the nucleus and in the mitochondria (Fig. 2E, F), in agreement with previous studies (33, 34). Moreover, as in the nuclear fraction, mitochondrial PHB1 was coimmunoprecipitated with anti-PHB2 antibody (Fig. 2F). Yet, only mitochondrial PHB2, and not PHB1, was bound to S1P affinity matrices (Fig. 2G). Once again, the binding was specific, as shown by the absence of significant association of PHB2 with sphingosine, or control affinity beads (Fig. 2G). Taken together, these data demonstrate that S1P only binds to PHB2, and not PHB1, and implies that S1P binds to monomeric or homomeric PHB2.
S1P is produced in the mitochondria by SphK2
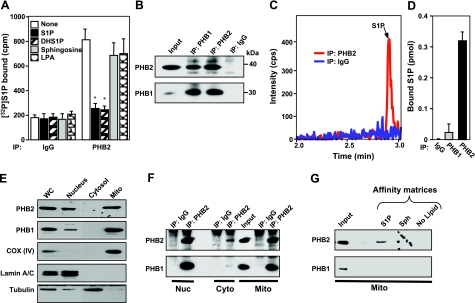
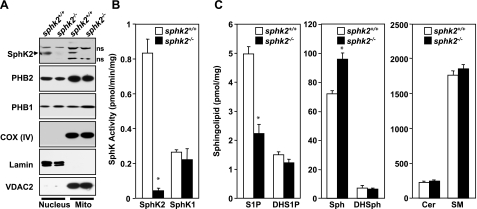
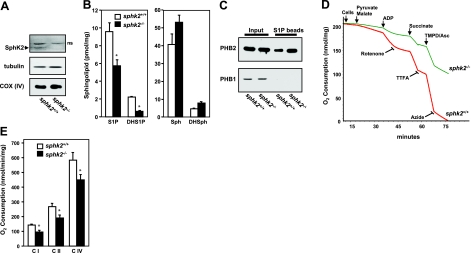
To examine production of S1P in the mitochondria, murine cardiac mitochondria were prepared by subcellular fractionation, and expression of SphK1 and SphK2 was determined by Western blot analysis and enzymatic activity. Purity of the cardiac mitochondria was demonstrated by the presence of the mitochondrial markers cytochrome-c oxidase subunit IV [COX (IV)], voltage-dependent anion channel (VDAC), cyclophilin D (CypD), and adenine nucleotide translocator (ANT), whereas little or no tubulin, or lamin, cytosol, and nuclear markers, respectively, were detected (Fig. 3A and data not shown). As expected (33, 34), both PHB1 and PHB2 were enriched in the nucleus and mitochondria (Fig. 3A). SphK2 protein with a molecular mass of ∼70 kDa was detected in the nuclear fraction, in agreement with previous studies (14, 15, 18). Notably, there was also significant expression of SphK2 in highly purified mitochondria (Fig. 3A), which was absent in mitochondrial and nuclear fractions from SphK2-null mice. Isotype-specific enzymatic assays further confirmed that SphK2 activity was present in the mitochondria (Fig. 3B). In contrast, the apparent enzymatic activity of SphK1 was much lower and the same in both wild-type and SphK2-null mitochondria, which was probably due to some overlap (∼25%) of SphK2 activity, as previously reported for this assay (35). In agreement, there was no detectable SphK1 protein in highly purified mitochondria (Supplemental Fig. S2), consistent with its predominant cytosolic localization (18). LC-ESI-MS/MS analyses revealed that S1P levels were much lower in cardiac mitochondria isolated from SphK2-null mice, whereas levels of sphingosine, the substrate of SphK2, were elevated (Fig. 3C). Dihydrosphingosine, ceramide, and sphingomyelin (Fig. 3C), as well as their different chain-length species (data not shown), were not significantly different. These results support the notion that SphK2 is the main isoform responsible for production of S1P in the mitochondria, although some of the S1P must come from extramitochondrial sources.
Figure 3.
SphK2 and S1P in cardiac mitochondria. A) Mitochondria (mito), and nuclear fractions were prepared from wild-type and sphk2−/− hearts. Equal amounts of proteins were separated by SDS-PAGE and immunoblotted with anti-SphK2, anti-PHB2, and anti-PHB1 antibodies. Antibodies against subunit IV of cytochrome-c oxidase, COX (IV), and VDAC2 or lamin were used as specific organelle markers for mitochondria and nuclei, respectively, and to show equal loading and transfer. ns, nonspecific bands. B) SphK1 and SphK2 activities were determined by isotype-specific assays (30) in extracts of mitochondria isolated from hearts of wild-type and SphK2-knockout mice. C) Lipids were extracted from mitochondria isolated from sphk2+/+ and sphk2−/− hearts and mass levels of sphingoid bases sphingosine (Sph) and dihydrosphingosine (DHS), and their phosphorylated products, S1P and dihydro-S1P (DHS1P), ceramide (Cer), and sphingomyelin (SM) were determined by LC-ESI-MS/MS. Data are expressed as means ± sd. *P < 0.01 vs. wild type.
Localization of SphK2 in the mitochondria
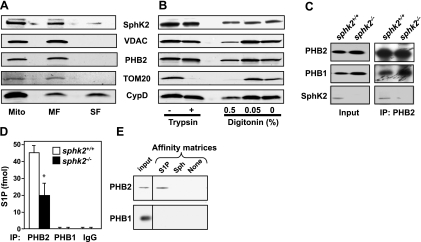
To clarify the submitochondrial localization of SphK2, soluble and membrane fractions prepared from purified mitochondria (24) were analyzed by Western blot analysis. Most of the SphK2 protein remained in the mitochondrial membrane pellets, as did the integral membrane proteins PHB2 and VDAC, whereas CypD, a matrix protein that is also linked to the inner membrane, was detected in both membrane and soluble fractions (Fig. 4A), as previously reported (24). Limited trypsin treatment, which removes the cytosol-exposed outer mitochondrial membrane proteins, as well as contaminating proteins nonspecifically adhering to the mitochondria outer membrane, had no effect on levels of SphK2, PHB2, or CypD, but it did remove the cytosolic-exposed outer membrane protein TOM20 (Fig. 4B). We further examined whether SphK2 is in the outer or inner mitochondrial membrane by selective disruption of the outer membrane with digitonin, which removed TOM20 but had no effect on levels of SphK2 or the inner membrane protein PHB2 (Fig. 4B). These results suggest that SphK2 does not adhere to the outer membrane of the mitochondria and is most likely located in the inner mitochondrial membrane.
Figure 4.
SphK2 localized in the mitochondrial inner membrane produces S1P that binds to endogenous PHB2. A) Membrane (MF) and soluble (matrix) fractions (SF) were prepared from wild-type heart mitochondria (mito). B) Mitochondria were treated with trypsin (50 μg/ml) or digitonin (0.05 and 0.5%) for 15 min at 4°C. Equal amounts of proteins were analyzed by Western blotting with the indicated antibodies. C) Heart mitochondria extracts from wild-type and sphk2−/− mice were immunoprecipitated with anti-PHB2 antibody, and immunocomplexes were analyzed by Western blotting with the indicated antibodies. D) Heart mitochondria extracts from wild-type and sphk2−/− mice were immunoprecipitated with anti-PHB1 or anti-PHB2 antibodies or control IgG as indicated; lipids were extracted from immunoprecipitates, and S1P was determined by LC-ESI-MS/MS. Data are means ± sd. *P < 0.05. E) Wild-type mitochondria extracts were incubated with S1P, sphingosine (Sph), or control (no lipid) affinity matrices and washed extensively, and bound proteins were separated by SDS-PAGE and then analyzed by Western blotting with anti-PHB2 or anti-PHB1 antibodies. Inputs are shown in the leftmost lanes.
It is well known that PHB2 is localized in the mitochondrial inner membrane, where it forms a large complex with PHB1 (20). In agreement, PHB1 was coimmunprecipitated from mitochondria with PHB2. SphK2 was also coimmunoprecipitated with PHB2 (Fig. 4C), bringing the enzyme that produces S1P in proximity to its mitochondrial target. Indeed, endogenous S1P, the product of SphK2, was tightly bound to mitochondrial PHB2 (Fig. 4D). Reduced levels of S1P were associated with endogenous PHB2 in mitochondria of SphK2-null mice (Fig. 4D), consistent with their lower content of S1P (Fig. 3C). As with HeLa mitochondria, S1P was not present in PHB1 immunoprecipitates (Fig. 4D) nor did PHB1 bind to S1P affinity matrices, in contrast to mitochondrial PHB2 (Fig. 4E).
Involvement of the interaction between PHB2 and S1P in respiratory chain complex assembly
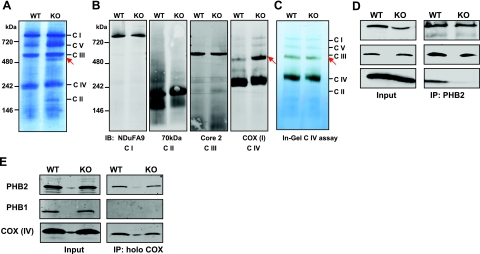
It was next important to determine whether depletion of SphK2 and S1P in the mitochondria affected the functions of PHB2 there. Because PHB2 has been demonstrated to assist in the assembly of the electron transport chain in mitochondria by binding subunits of respiratory complexes until they are fully assembled (36, 37), we next sought to evaluate the potential role of mitochondrial S1P generated by SphK2 in complex assembly. To this end, BN-PAGE was employed to analyze intact respiratory chain complexes. No differences were noted in the 5 electron transport chain complexes (complexes I–V), identified by molecular mass markers, between wild-type and SphK2-null mice (Fig. 5A). Identification of the complexes was confirmed by Western blot analyses after transfer to nitrocellulose with specific antibodies directed against component subunits (Fig. 5B). However, an additional stained band was present in the mitochondria from SphK2-null mice at ∼480 kDa. The additional band in the SphK2-null mitochondria was visualized with antibody against subunit I of complex IV, which also recognized the apparent 240-kDa monomeric form of complex IV (COX), composed of 13 subunits. In agreement, when this band was excised from the native blue gel and sequenced by mass spectrometry, 7 polypeptide sequences were identified that were subunits of COX (Supplemental Table S2). Consistent with previous results (28), specific in-gel staining for complex IV revealed that the majority of the enzymatic activity resided in the monomeric form and much less in the additional form. Yet, surprisingly, nearly the same activity was detected in the higher molecular mass complex present in both wild-type and SphK2-null mitochondria (Fig. 5C), despite the large differences in protein levels. These results suggest that the additional band detected in sphk2−/− mitochondria is a larger form of COX with decreased enzymatic activity.
Figure 5.
Aberrant interaction between PHB2 and COX in SphK2-null mitochondria. A–C) Analysis of mitochondrial electron transport chain complexes by BN-PAGE. A, B) Equal amounts of heart mitochondrial proteins from wild-type and sphk2−/− mice were separated by BN-PAGE (A) or transferred to nitrocellulose and immunoblotted with the indicated specific complex antibodies (B). C) In-gel catalytic activity assay for COX using the same gel as in A. C I–V, mitochondrial complexes I–V, respectively. Arrows indicate the additional band present only in sphk2−/− mitochondria. D, E) Interaction between PHB2 and COX. Mitochondria extracts from wild-type and sphk2−/− hearts were immunoprecipitated with anti-PHB2 antibody (D) or with an antibody against the holocytochrome-c oxidase complex (E) and then analyzed by Western blotting with the indicated antibodies. Inputs are shown at left. Results are representative of 3 experiments.
PHB2 in the mitochondrial inner membrane has been proposed to be a chaperone for the assembly of COX, associating and stabilizing its subunits (37–39). Therefore, we examined the possibility that this interaction may be impaired in sphk2−/− mitochondria with depleted levels of S1P. In agreement with a recent study (39), subunit IV of COX, a subunit necessary for COX assembly (40), was coimmunoprecipitated with PHB2 (Fig. 5D). Interestingly, this interaction was markedly reduced in sphk2−/− mitochondria (Fig. 5D). Assembly of the 13 subunits of the COX complex starts with the association of subunit I with subunit IV (41). Similarly, interaction of PHB2 with subunit I of COX was greatly reduced in mitochondria devoid of SphK2 (Supplemental Fig. S3). Moreover, only PHB2 but not PHB1 was pulled down when the native 13 subunit COX complex was immunocaptured (Fig. 5E), suggesting that there are specific associations between COX and PHB2 that do not include PHB1.
Involvement of S1P formed by SphK2 in mitochondrial respiration
As complex IV (COX) is the terminal complex in the respiratory chain, we wondered whether the association of PHB2 with S1P produced in the mitochondria by SphK2 also modulates mitochondrial respiration. To this end, oxidative phosphorylation was measured in intact heart mitochondria isolated from wild-type and sphk2−/− mice by polarography with a Clark-type electrode to evaluate the integrated function of the electron transport chain coupled to ATP synthesis. With glutamate plus malate as substrates to assess complex I-dependent electron transport, maximum state 3 respiratory rates measured in the presence of 2 mM ADP were 23% lower in heart mitochondria isolated from sphk2−/− mice than from wild-type mice, with preserved coupling of respiration (Table 1). Similar decreases in oxygen consumption were found when succinate was used as the electron donor for complex II (Table 1). Decreased respiration was also observed in the presence of the uncoupler of oxidative phosphorylation, 2,4-dinitrophenol, indicating that the respiratory defect is in the electron transport chain itself and not at the level of the phosphorylation apparatus that includes ATP-synthase (complex V). Moreover, complex IV-stimulated respiration measured with TMPD and ascorbate as substrate was also 25% lower in the knockout mice (Table 1), suggesting that the defect in respiration resides in complex IV.
Table 1.
Oxidative phosphorylation in heart mitochondria of wild-type and sphk2−/− mice
| Parameter | Wild type | sphk2−/− |
|---|---|---|
| Glutamate and malate | ||
| State 3 | 251 ± 6 | 195 ± 18* |
| State 4 | 29 ± 3 | 19 ± 5 |
| RCR | 9.0 ± 0.7 | 13.5 ± 3.0 |
| 2 mM ADP | 282 ± 6 | 211 ± 22* |
| DNP | 291 ± 11 | 198 ± 23* |
| Succinate | ||
| State 3 | 355 ± 18 | 274 ± 18* |
| State 4 | 100 ± 7 | 84 ± 9 |
| RCR | 3.6 ± 0.1 | 3.3 ± 0.2 |
| 2 mM ADP | 329 ± 18 | 253 ± 15* |
| DNP | 309 ± 25 | 236 ± 16* |
| TMPD and ascorbate | ||
| 2 mM ADP (azide sensitive) | 897 ± 32 | 710 ± 49* |
States 3 and 4 were measured in isolated heart mitochondria from wild-type (n=5) and sphk2−/− mice (n=6) with glutamate and malate as substrates of respiratory complex I, succinate as substrate of respiratory complex II, and TMPD and ascorbate as substrates of respiratory complex IV. Data are expressed as nanoatoms of oxygen per minute per milligram of protein. Values are expressed as means ± se.
P < 0.05 vs. wild type.
Enzymatic activities of electron transport chain complexes were investigated in solubilized mitochondria to determine whether defects in enzymatic activities, changes in composition of the inner mitochondrial membrane and/or transport defects of substrates might account for the decreased oxidation rates in intact SphK2-null mitochondria. However, there were no significant differences in activities of citrate synthase, an enzyme expressed exclusively in the mitochondrial matrix, rotenone-sensitive NADH-cytochrome-c reductase, which requires complexes I and III, ubiquitonol-ferricytochrome-c oxido-reductase (respiratory complex III), or cytochrome-c oxidase (respiratory complex IV) (Table 2). These results suggest that decreases in maximally expressed enzyme activities in detergent-solubilized mitochondria do not account for the reduced rates of integrated respiration detected in intact sphk2−/− mitochondria.
Table 2.
Electron transport chain subunit enzyme activities
| Genotype | CS (mU · mg−1) | NCR (mU · mg−1) | Complex III (mU · mg−1) | Complex IV (s−1 · mg−1) |
|---|---|---|---|---|
| Wild type | 2052 ± 178 | 2169 ± 98 | 5222 ± 264 | 576 ± 80 |
| Sphk2−/− | 1836 ± 134 | 2212 ± 295 | 5667 ± 512 | 669 ± 33 |
Citrate synthase (CS), NCR (complexes I and III), ubiquinol:ferricytochrome-c oxido-reductase (complex III), and COX (complex IV) activities were measured in frozen-thawed, cholate-solubilized heart mitochondria at 37°C. Values are expressed as means ± se; n = 3/group.
Oxidative phosphorylation of SphK2-deficient cardiomyocytes
Mitochondrial function may be regulated differently in situ compared to isolated organelles, when intracellular interactions with other structures, including sarcomeres and sarcolemma, are lost. Therefore, it was of interest to examine whether similar respiration defects were observed in cells that are deficient in SphK2. SphK2 is expressed in cardiomyocytes isolated from wild-type mice (Fig. 6A), and levels of S1P and dihydro-S1P were significantly lower in sphk2−/− cardiomyocytes (Fig. 6B), whereas their sphingoid base precursors were slightly elevated (Fig. 6B). Moreover, no significant differences were detected in levels of ceramide and sphingomyelin or their various acyl chain species (data not shown). Similar to isolated mitochondria, only cardiomyocyte PHB2, and not PHB1, bound to S1P affinity beads (Fig. 6C). A typical oxygraph experiment for isolated permeabilized wild-type and SphK2-null cardiomyocytes is shown in Fig. 6D. Rates of oxygen consumption measured in intact, digitonin-permeabilized cardiomyocytes were decreased in cardiomyocytes from sphk2−/− mice using substrates for complex I (pyruvate and malate), as well as with the substrate for complex II (succinate), by 34 and 29%, respectively (Fig. 6E). The site of the defect in the electron transport chain was localized by the use of TMPD and ascorbate as a substrate of complex IV. Oxidative phosphorylation solely through complex IV was significantly decreased (Fig. 6E), in line with the findings with isolated cardiac mitochondria. Thus, decreases in integrated respiration observed in isolated, intact mitochondria and in mitochondrial respiration in situ were localized to complex IV.
Figure 6.
SphK2, S1P, and respiration in cardiomyocytes. Cardiomyocytes were prepared from wild-type and sphk2−/− hearts. A) Equal amounts of lysate proteins were separated by SDS-PAGE and immunoblotted with the indicated antibodies. ns, nonspecific band. B) Lipids were extracted from sphk2+/+ and sphk2−/− cardiomyocytes, and mass levels of S1P, dihydro-S1P (DHS1P), sphingosine (Sph), and dihydrosphingosine (DHSph) were determined by LC-ESI-MS/MS. *P < 0.05 vs. wild type. C) Duplicate lysates were incubated with S1P affinity matrices and washed extensively, and bound proteins were separated by SDS-PAGE, then analyzed by Western blotting with anti-PHB2 or anti-PHB1 antibodies. Inputs are shown in the leftmost lanes. D, E) Respiratory complexes I, II, and IV in SphK2-null cardiomyocytes display a decreased rate of oxygen consumption. D) Representative respiration traces of digitonin-permeabilized wild-type (red) and sphk2−/− (green) cardiomyocytes. Sensitive rates were measured by successively adding substrates and specific inhibitors for complexes I, II, and IV. After stimulation with 2 mM ADP, each complex of sphk2−/− cardiomyocytes displayed a decreased rate of O2 consumption. E) Maximum rates of respiration of wild-type and sphk2−/− cardiomyocytes (n=4) in the presence of 2 mM ADP were measured with respiratory complex I, II, and IV substrates. Data are expressed as mean ± se respiration (nmol O2 · min−1 · mg protein−1). Similar results were obtained in 3 additional experiments. *P < 0.05 vs. wild type.
Depletion of SphK2 and PHB2 alters HeLa cell mitochondrial function
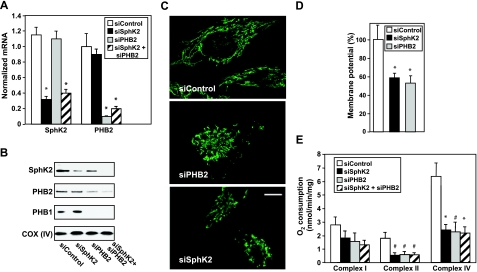
To confirm that the interaction between PHB2 and S1P generated by SphK2 regulates respiration in cells and to exclude the possibility that the respiratory defects observed in sphk2−/− heart mitochondria and cardiomyocytes were due to developmental differences, levels of SphK2 and PHB2 were acutely down-regulated in HeLa cells, and mitochondrial function was examined. Specific siRNAs targeted to SphK2 and PHB2 markedly decreased expression of the corresponding mRNAs (Fig. 7A). As expected (20), depletion of PHB2 was accompanied by the loss of its partner PHB1 (Fig. 7B). Depletion of PHB2 also reduced levels of SphK2 (Fig. 7B). Similarly, levels of Hax-1 (33) and subunits I, II, and IV of COX (39) are reduced by down-regulation of PHB2, supporting the notion that PHB2 is important for stabilization of several mitochondrial proteins, including SphK2. In contrast, levels of either PHB2 or PHB1 were not significantly affected by down-regulation of SphK2 (Fig. 7B). The loss of PHB2 in HeLa cells or mouse embryonic fibroblasts affects mitochondrial cristae and leads to the accumulation of fragmented mitochondria (33, 42). In agreement, mitochondria appeared fragmented and disorganized on PHB2 depletion, although this was less pronounced after SphK2 depletion (Fig. 7C). Similar to depolarization of the mitochondrial membrane due to PHB2 depletion, as expected from earlier work (33, 43, 44), SphK2 depletion also reduced membrane potential (Fig. 7D).
Figure 7.
SphK2 and PHB2 regulate HeLa cell mitochondrial function. HeLa cells were transfected with siControl, siSphK2, siPHB2, or both, as indicated. A, B) mRNA levels (A) and protein (B) were determined by quantitative PCR and Western blotting, respectively. C) Mitochondria were stained with anti-cyclophilin D antibody and visualized by confocal microscopy with a ×63 objective. Scale bar = 10 μm. D) Transfected HeLa cells were stained with TMRM, and mitochondrial membrane potential was determined by confocal microscopy with a ×40 objective. Data are expressed as mean ± se percentage of siControl. E) Maximum rates of respiration in the presence of 2 mM ADP were measured polarographically with respiratory complex I, II, and IV substrates. Data are expressed as mean ± se respiration (nmol O2 · min−1 · mg protein−1). Similar results were obtained in 3 additional experiments. *P < 0.05 vs. siControl.
The relative contributions of respiratory complexes I, II, and IV to overall mitochondrial respiration were next investigated. Depletion of either SphK2 or PHB2 reduced complex II and IV respiration rates by ∼65%, whereas the effect on complex I activity was less pronounced (Fig. 7E). Concurrent down-regulation of both SphK2 and PHB2 had the same inhibitory effects on complex II and IV activities as the individual siRNAs, suggesting that SphK2 and PHB2 actions may converge on the same rate-limiting step.
DISCUSSION
S1P has long been suspected to have intracellular targets and functions (1–3). Recently, we discovered that S1P in the nucleus of mammalian cells inhibits histone deacetylation, and we identified HDAC1 and HDAC2 as the first bona fide intracellular targets of S1P (18). Moreover, S1P binds to tumor necrosis factor receptor-associated factor 2 and is a cofactor for its E3 ubiquitin ligase activity (45). Utilizing S1P affinity beads, binding of labeled S1P, and mass spectrometry measurements of endogenously associated S1P, we have now unequivocally identified the mitochondrial protein PHB2 as yet another novel intracellular target of S1P. Our data suggest that SphK2 is localized to the inner mitochondrial membrane, where it can regulate levels of S1P, and we have demonstrated the importance of the interaction between mitochondrial S1P and PHB2 for proper assembly of COX, an electron transport chain complex, and mitochondrial respiration. Hence, our results point to a unique intracellular localization and function of S1P in the mitochondria important for oxidative phosphorylation and suggest that the intracellular location of S1P production dictates its function (18, 45).
PHB2 is essential for proper development, morphogenesis, and function of mitochondria, where it forms large, multimeric ring complexes with PHB1 in the inner mitochondria membrane (19, 20, 36). Remarkably, we found that only PHB2, and not PHB1, specifically binds S1P in MCF7 and HeLa cells, cardiomyocytes, and even in highly purified mitochondria. This raised the intriguing question: what is the physiological function of the interaction between S1P and PHB2?
In yeast, as well as in mammalian cells, PHB2 has been shown to directly associate with subunits of COX in mitochondria and stabilize them during their assembly (36–38). Interestingly, S1P is involved in the interaction of PHB2 with COX, since the interactions of subunits I and IV of COX with PHB2 were reduced in SphK2-null mitochondria with reduced S1P levels, whereas capture of holo COX yielded similar amounts. These findings suggest that PHB2 with bound S1P interacts with COX only during its assembly. Indeed, an aberrant, inactive form of COX was detected in mitochondria from SphK2-knockout mice. COX is assembled from preformed intermediates in a stepwise process. Its assembly starts with the association of subunit I, a mitochondrially encoded catalytic subunit, with subunit IV (41). Of the 10 nuclear DNA-encoded subunits, only subunits IV and Va are integrated at the early stages of assembly that involve formation of the catalytic core (46). Thus, S1P may be an important component for the chaperone function of PHB2 and proper assembly of COX.
Aberrant assembly of COX is often associated with respiratory chain defects in humans and may lead to deficiencies in oxidative phosphorylation (46). Indeed, depletion of SphK2 in mitochondria and reduction of S1P levels resulted in a significant decrease in integrated respiration observed in isolated intact mitochondria and in mitochondrial respiration in situ and in cardiomyocytes devoid of SphK2. The inability of exogenous S1P to restore respiration when added to SphK2-null cardiomyocytes (Supplemental Fig. S4) further supports the notion that the deficit in respiration is likely due to a defect in assembly of the COX complex. Acute down-regulation of SphK2 or PHB2 in HeLa cells also reduced oxidative phosphorylation through COX and decreased mitochondrial membrane potential. Thus, the interaction of S1P produced by SphK2 with PHB2 may play a role in the regulation of mitochondrial functions. This is consistent with several previous reports suggesting a role for prohibitins in the biogenesis of respiratory chains. Low prohibitin levels in plants result in reduced membrane potential and oxygen consumption (47). Similarly, knockdown of both phb-1 and phb-2 in Caenorhabditis elegans causes a slight reduction in oxygen consumption (48) and more robust reduction in mitochondrial membrane potential (49). Yeast cells depleted of prohibitins also have reduced mitochondrial membrane potential (43). Moreover, depletion of PHB1 in endothelial cells depolarized mitochondria and reduced complex I activity (50). In contrast, however, membrane potential, ATP levels, oxygen consumption, and electron transport chain activities were normal in PHB2-null mouse embryonic fibroblasts (42). It is not clear whether these variable effects on respiratory activity and mitochondrial membrane potential caused by depletion of prohibitins are due to cell type-specific differences, compensatory mechanisms in PHB2-null fibroblasts, different methods used to knock down expression of prohibitins, or different methods of measurements of mitochondrial function.
Abundant evidence indicates that large multimeric ring complexes of PHB1 and PHB2 in the inner membrane of mitochondria are the physiologically active structures (21). Nevertheless, our study points to a unique function of mono- or homomeric PHB2. Only PHB2, and not PHB1, binds to S1P and to subunit IV of COX. Our data also suggest that the association of PHB2 with S1P produced in the mitochondria by SphK2 is involved in proper assembly of COX and modulates mitochondrial respiration. A few previous studies also support the notion that PHB2 can function as a homomeric protein. PHB2, but not PHB1, interacts with the antiapoptotic protein Hax-1 to protect against apoptosis (33) and maintains the mitochondrial morphology via processing of the dynamin-like GTPase OPA1, a core component of the mitochondrial fusion machinery. Moreover, in the presence of estradiol, only mitochondrial PHB2 translocates to the nucleus where it represses ERα-dependent transcription (33). It has been proposed that there is an equilibrium between homomer and heteromer forms of PHB1 and PHB2, and only when they are not paired do they function as transcriptional repressors (31). Finally, capsaicin, the active component of chili peppers, binds to PHB2 but not to PHB1, induces its dissociation from ANT2, inhibits ADP uptake via ANT, and displaces PHB2 from the mitochondria to the nucleus (51). However, many other mitochondrial functions of PHB2 have been attributed to macromolecular complexes of PHB2 with PHB1 (21).
In summary, this work identifies another cellular location and function for SphK2 and S1P, provides new insight into the functions of PHB2 as a mitochondrial chaperone protein, and suggests that the interaction between S1P regulates the proper assembly of the electron transport chain and mitochondrial respiration.
Supplementary Material
Acknowledgments
This work was supported by U.S. National Institutes of Health (NIH) grant R37GM043880 (S.S.) and, in part, by P01AG15885 and a Medical Research Service Department of Veterans Affairs Merit Review grant (E.J.L.). G.S. was supported by National Research Service Award F30NS058008. M.P. was supported by French grant EXPLORA'DOC 2009. The authors thank Dr. Richard Proia (NIH, Bethesda, MD, USA) for a generous gift of SphK antibodies and the Lipidomics/Metabolomics facility for lipid analyses. Microscopy was performed at the Virginia Commonwealth University Microscopy Facility, supported, in part, by grant 5P30 NS047463 and NIH–National Cancer Institute Cancer Center Core grant 5P30CA016059.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Spiegel S., Milstien S. (2003) Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell. Biol. 4, 397–407 [DOI] [PubMed] [Google Scholar]
- 2. Saba J. D., Hla T. (2004) Point-counterpoint of sphingosine 1-phosphate metabolism. Circ. Res. 94, 724–734 [DOI] [PubMed] [Google Scholar]
- 3. Hannun Y. A., Obeid L. M. (2008) Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell. Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 4. Skrzypek M. S., Nagiec M. M., Lester R. L., Dickson R. C. (1999) Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J. Bacteriol. 181, 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oskouian B., Saba J. D. (2004) Death and taxis: what non-mammalian models tell us about sphingosine-1-phosphate. Semin. Cell. Dev. Biol. 15, 529–540 [DOI] [PubMed] [Google Scholar]
- 6. Coursol S., Fan L. M., Le Stunff H., Spiegel S., Gilroy S., Assmann S. M. (2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423, 651–654 [DOI] [PubMed] [Google Scholar]
- 7. Pandey S., Assmann S. M. (2004) The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16, 1616–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mattie M., Brooker G., Spiegel S. (1994) Sphingosine-1-phosphate, a putative second messenger, mobilizes calcium from internal stores via an inositol trisphosphate-independent pathway. J. Biol. Chem. 269, 3181–3188 [PubMed] [Google Scholar]
- 9. Ghosh T. K., Bian J., Gill D. L. (1994) Sphingosine 1-phosphate generated in the endoplasmic reticulum membrane activates release of stored calcium. J. Biol. Chem. 269, 22628–22635 [PubMed] [Google Scholar]
- 10. Meyer zu Heringdorf D., Liliom K., Schaefer M., Danneberg K., Jaggar J. H., Tigyi G., Jakobs K. H. (2003) Photolysis of intracellular caged sphingosine-1-phosphate causes Ca2+ mobilization independently of G-protein-coupled receptors. FEBS Lett. 554, 443–449 [DOI] [PubMed] [Google Scholar]
- 11. Olivera A., Mizugishi K., Tikhonova A., Ciaccia L., Odom S., Proia R. L., Rivera J. (2007) The sphingosine kinase-sphingosine-1-phosphate axis is a determinant of mast cell function and anaphylaxis. Immunity 26, 287–297 [DOI] [PubMed] [Google Scholar]
- 12. Olivera A., Rosenfeldt H. M., Bektas M., Wang F., Ishii I., Chun J., Milstien S., Spiegel S. (2003) Sphingosine kinase type 1 induces G12/13-mediated stress fiber formation yet promotes growth and survival independent of G protein-coupled receptors. J. Biol. Chem. 278, 46452–46460 [DOI] [PubMed] [Google Scholar]
- 13. Kohno M., Momoi M., Oo M. L., Paik J. H., Lee Y. M., Venkataraman K., Ai Y., Ristimaki A. P., Fyrst H., Sano H., Rosenberg D., Saba J. D., Proia R. L., Hla T. (2006) Intracellular role for sphingosine kinase 1 in intestinal adenoma cell proliferation. Mol. Cell. Biol. 26, 7211–7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Igarashi N., Okada T., Hayashi S., Fujita T., Jahangeer S., Nakamura S. I. (2003) Sphingosine kinase 2 is a nuclear protein and inhibits DNA synthesis. J. Biol. Chem. 278, 46832–46839 [DOI] [PubMed] [Google Scholar]
- 15. Okada T., Ding G., Sonoda H., Kajimoto T., Haga Y., Khosrowbeygi A., Gao S., Miwa N., Jahangeer S., Nakamura S. (2005) Involvement of N-terminal-extended form of sphingosine kinase 2 in serum-dependent regulation of cell proliferation and apoptosis. J. Biol. Chem. 280, 36318–36325 [DOI] [PubMed] [Google Scholar]
- 16. Sankala H. M., Hait N. C., Paugh S. W., Shida D., Lepine S., Elmore L. W., Dent P., Milstien S., Spiegel S. (2007) Involvement of sphingosine kinase 2 in p53-independent induction of p21 by the chemotherapeutic drug doxorubicin. Cancer Res. 67, 10466–10474 [DOI] [PubMed] [Google Scholar]
- 17. Ding G., Sonoda H., Yu H., Kajimoto T., Goparaju S. K., Jahangeer S., Okada T., Nakamura S. (2007) Protein kinase D-mediated phosphorylation and nuclear export of sphingosine kinase 2. J. Biol. Chem. 282, 27493–27502 [DOI] [PubMed] [Google Scholar]
- 18. Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., Spiegel S. (2009) Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 325, 1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Artal-Sanz M., Tavernarakis N. (2009) Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 20, 394–401 [DOI] [PubMed] [Google Scholar]
- 20. Merkwirth C., Langer T. (2009) Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta 1793, 27–32 [DOI] [PubMed] [Google Scholar]
- 21. Osman C., Merkwirth C., Langer T. (2009) Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 122, 3823–3830 [DOI] [PubMed] [Google Scholar]
- 22. Hait N. C., Sarkar S., Le Stunff H., Mikami A., Maceyka M., Milstien S., Spiegel S. (2005) Role of sphingosine kinase 2 in cell migration towards epidermal growth factor. J. Biol. Chem. 280, 29462–29469 [DOI] [PubMed] [Google Scholar]
- 23. Lesnefsky E. J., Chen Q., Moghaddas S., Hassan M. O., Tandler B., Hoppel C. L. (2004) Blockade of electron transport during ischemia protects cardiac mitochondria. J. Biol. Chem. 279, 47961–47967 [DOI] [PubMed] [Google Scholar]
- 24. Matas J., Young N. T., Bourcier-Lucas C., Ascah A., Marcil M., Deschepper C. F., Burelle Y. (2009) Increased expression and intramitochondrial translocation of cyclophilin-D associates with increased vulnerability of the permeability transition pore to stress-induced opening during compensated ventricular hypertrophy. J. Mol. Cell. Cardiol. 46, 420–430 [DOI] [PubMed] [Google Scholar]
- 25. Chen Q., Moghaddas S., Hoppel C. L., Lesnefsky E. J. (2006) Reversible blockade of electron transport during ischemia protects mitochondria and decreases myocardial injury following reperfusion. J. Pharmacol. Exp. Ther. 319, 1405–1412 [DOI] [PubMed] [Google Scholar]
- 26. Fisher P. W., Salloum F., Das A., Hyder H., Kukreja R. C. (2005) Phosphodiesterase-5 inhibition with sildenafil attenuates cardiomyocyte apoptosis and left ventricular dysfunction in a chronic model of doxorubicin cardiotoxicity. Circulation. 111, 1601–1610 [DOI] [PubMed] [Google Scholar]
- 27. Paillard M., Gomez L., Augeul L., Loufouat J., Lesnefsky E. J., Ovize M. (2009) Postconditioning inhibits mPTP opening independent of oxidative phosphorylation and membrane potential. J. Mol. Cell. Cardiol. 46, 902–909 [DOI] [PubMed] [Google Scholar]
- 28. Wittig I., Braun H. P., Schagger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 29. Wittig I., Karas M., Schagger H. (2007) High-resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 30. Liu H., Sugiura M., Nava V. E., Edsall L. C., Kono K., Poulton S., Milstien S., Kohama T., Spiegel S. (2000) Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J. Biol. Chem. 275, 19513–19520 [DOI] [PubMed] [Google Scholar]
- 31. He B., Feng Q., Mukherjee A., Lonard D. M., DeMayo F. J., Katzenellenbogen B. S., Lydon J. P., O'Malley B. W. (2008) A repressive role for prohibitin in estrogen signaling. Mol. Endocrinol. 22, 344–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montano M. M., Ekena K., Delage-Mourroux R., Chang W., Martini P., Katzenellenbogen B. S. (1999) An estrogen receptor-selective coregulator that potentiates the effectiveness of antiestrogens and represses the activity of estrogens. Proc. Natl. Acad. Sci. U. S. A. 96, 6947–6952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kasashima K., Ohta E., Kagawa Y., Endo H. (2006) Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2. J. Biol. Chem. 281, 36401–36410 [DOI] [PubMed] [Google Scholar]
- 34. Da Cruz S., Parone P. A., Gonzalo P., Bienvenut W. V., Tondera D., Jourdain A., Quadroni M., Martinou J. C. (2008) SLP-2 interacts with prohibitins in the mitochondrial inner membrane and contributes to their stability. Biochim. Biophys. Acta 1783, 904–911 [DOI] [PubMed] [Google Scholar]
- 35. Olivera A., Urtz N., Mizugishi K., Yamashita Y., Gilfillan A. M., Furumoto Y., Gu H., Proia R. L., Baumruker T., Rivera J. (2006) IgE-dependent activation of sphingosine kinases 1 and 2 and secretion of sphingosine 1-phosphate requires Fyn kinase and contributes to mast cell responses. J. Biol. Chem. 281, 2515–2525 [DOI] [PubMed] [Google Scholar]
- 36. Berger K. H., Yaffe M. P. (1998) Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nijtmans L. G., de Jong L., Artal Sanz M., Coates P. J., Berden J. A., Back J. W., Muijsers A. O., van der Spek H., Grivell L. A. (2000) Prohibitins act as a membrane-bound chaperone for the stabilization of mitochondrial proteins. EMBO J. 19, 2444–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nijtmans L. G., Artal S. M., Grivell L. A., Coates P. J. (2002) The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell. Mol. Life Sci. 59, 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsutsumi T., Matsuda M., Aizaki H., Moriya K., Miyoshi H., Fujie H., Shintani Y., Yotsuyanagi H., Miyamura T., Suzuki T., Koike K. (2009) Proteomics analysis of mitochondrial proteins reveals overexpression of a mitochondrial protein chaperon, prohibitin, in cells expressing hepatitis C virus core protein. Hepatology 50, 378–386 [DOI] [PubMed] [Google Scholar]
- 40. Dowhan W., Bibus C. R., Schatz G. (1985) The cytoplasmically-made subunit IV is necessary for assembly of cytochrome-c oxidase in yeast. EMBO J. 4, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nijtmans L. G., Taanman J. W., Muijsers A. O., Speijer D., Van den Bogert C. (1998) Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 254, 389–394 [DOI] [PubMed] [Google Scholar]
- 42. Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Lower B., Wunderlich F. T., von Kleist-Retzow J. C., Waisman A., Westermann B., Langer T. (2008) Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coates P. J., Jamieson D. J., Smart K., Prescott A. R., Hall P. A. (1997) The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr. Biol. 7, 607–610 [DOI] [PubMed] [Google Scholar]
- 44. Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brugger B., Westermann B., Langer T. (2009) The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordula T., Milstien S., Spiegel S. (2010) Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465, 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fernandez-Vizarra E., Tiranti V., Zeviani M. (2009) Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta 1793, 200–211 [DOI] [PubMed] [Google Scholar]
- 47. Ahn C. S., Lee J. H., Reum Hwang A., Kim W. T., Pai H. S. (2006) Prohibitin is involved in mitochondrial biogenesis in plants. Plant J. 46, 658–667 [DOI] [PubMed] [Google Scholar]
- 48. Artal-Sanz M., Tsang W. Y., Willems E. M., Grivell L. A., Lemire B. D., van der Spek H., Nijtmans L. G. (2003) The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans. J. Biol. Chem. 278, 32091–32099 [DOI] [PubMed] [Google Scholar]
- 49. Artal-Sanz M., Tavernarakis N. (2009) Prohibitin couples diapause signalling to mitochondrial metabolism during ageing in C. elegans. Nature 461, 793–797 [DOI] [PubMed] [Google Scholar]
- 50. Schleicher M., Shepherd B. R., Suarez Y., Fernandez-Hernando C., Yu J., Pan Y., Acevedo L. M., Shadel G. S., Sessa W. C. (2008) Prohibitin-1 maintains the angiogenic capacity of endothelial cells by regulating mitochondrial function and senescence. J. Cell Biol. 180, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuramori C., Azuma M., Kume K., Kaneko Y., Inoue A., Yamaguchi Y., Kabe Y., Hosoya T., Kizaki M., Suematsu M., Handa H. (2009) Capsaicin binds to prohibitin 2 and displaces it from the mitochondria to the nucleus. Biochem. Biophys. Res. Commun. 379, 519–525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.