Abstract
Targeted deletion of the Kcne2 potassium channel β subunit gene ablates gastric acid secretion and predisposes to gastric neoplasia in mice. Here, we discovered that Kcne2 deletion basolaterally reroutes the Kcnq1 α subunit in vivo in parietal cells (PCs), in which the normally apical location of the Kcnq1–Kcne2 channel facilitates its essential role in gastric acid secretion. Quantitative RT-PCR and Western blotting revealed that Kcne2 deletion remodeled fundic Kcne3 (2.9±0.8-fold mRNA increase, n=10; 5.3±0.4-fold protein increase, n=7) but not Kcne1, 4, or 5, and resulted in basolateral Kcnq1–Kcne3 complex formation in Kcne2−/− PCs. Concomitant targeted deletion of Kcne3 (creating Kcne2−/−Kcne3−/− mice) restored PC apical Kcnq1 localization without Kcne1, 4, or 5 remodeling (assessed by quantitative RT-PCR; n=5–10), indicating Kcne3 actively, basolaterally rerouted Kcnq1 in Kcne2−/− PCs. Despite this, Kcne3 deletion exacerbated gastric hyperplasia in Kcne2−/− mice, and both hypochlorhydria and hyperplasia in Kcne2+/− mice, suggesting that Kcne3 up-regulation was beneficial in Kcne2-depleted PCs. The findings reveal, in vivo, Kcne-dependent α subunit polarized trafficking and the existence and consequences of potassium channel β subunit remodeling.—Roepke, T. K., King, E. C., Purtell, K., Kanda, V. A., Lerner, D. J., Abbott, G. W. Genetic dissection reveals unexpected influence of β subunits on KCNQ1 K+ channel polarized trafficking in vivo.
Keywords: gastric acid, MiRP1, potassium channel
Parietal cells (PCs) achieve gastric acidification by virtue of an apical H+/K+ATPase (HKA) that pumps protons into the stomach lumen in exchange for K+ ions. To maintain this activity, K+ ions that enter the PC through the HKA must travel back into the stomach lumen across the apical membrane. This K+ ion efflux occurs primarily through the heteromeric KCNQ1–KCNE2 K+ channel (1, 2), with other K+ channels also possibly contributing (3, 4). KCNQ1 is a 6-transmembrane segment (TMS) α subunit from the S4 superfamily that forms functional, voltage-gated, homotetrameric, K+-selective channels in heterologous expression studies (5, 6). Originally named MinK-related peptide 1 (MiRP1), KCNE2 is a 1-TMS ancillary subunit from the KCNE gene family (7) (Fig. 1A). Here, for simplicity, we will use the KCNE nomenclature to refer to both genes and proteins; as per convention, human protein names are written in uppercase, mouse in lowercase; genes are written the same but in italics; where no specific species is implied, we will use uppercase. All five known KCNE gene products have been shown to regulate KCNQ1 function in heterologous expression studies (8). Two of these—KCNE2 and KCNE3, originally named MiRP2 (7)—endow KCNQ1 with constitutive activation, probably by favoring the activated conformation of the KCNQ1 voltage sensor (9–11). While KCNE2 and KCNQ1 colocalize in the PC apical membrane (Fig. 1B), KCNQ1–KCNE3 channels target to the basolateral membrane of colonic epithelial cells, where they regulate cAMP-stimulated chloride secretion (10, 12, 13).
Figure 1.
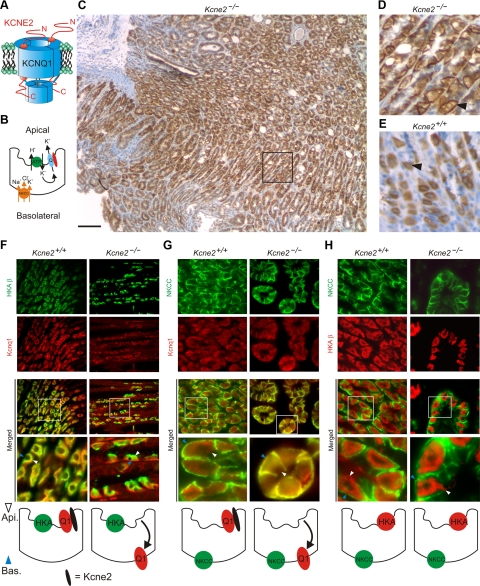
Reversed Kcnq1 trafficking in PCs of Kcne2−/− mice. A) Cartoon of a KCNQ1–KCNE2 complex. B) Cartoon of a PC showing location of HKA, NKCC1, and the KCNQ1–KCNE2 channel. C) KCNQ1 immunostaining (IS) in Kcne2−/− gastric mucosa. Scale bar = 100 μm. D) KCNQ1 IS in Kcne2−/− gastric mucosa (black box from panel C). Arrowhead indicates basolateral KCNQ1 staining. E) KCNQ1 IS in Kcne2+/+ gastric mucosa (same scale as panel D). Arrowhead indicates diffuse KCNQ1 staining due to localization at the invaginated apical membrane. F–H) Top: exemplar IF colabeling of Kcne2+/+ and Kcne2−/− gastric glands as indicated. “Merged” indicates merged view of the 2 panels above; bottom merged panel shows expanded view of the boxed region in the top merged panel. Yellow indicates colocalization. Blue arrowheads, PC basolateral side; white arrowheads, PC apical side. Representative of results from ≥2 mice, 3–5 sections/mouse/genotype. Bottom: cartoons summarizing IF data. F) Kcnq1 (red) and HKA β subunit (green). G) Kcnq1 (red) and NKCC (green). H) HKA β subunit (red) and NKCC (green). Width of view (except bottom merge): 100 μm (F); 75 μm (G, H).
Kcnq1−/− mice and Kcne2−/− mice show similar gastric phenotypes, characterized by achlorhydria, hypergastrinemia, and gastric glandular hyperplasia (1, 2, 14). PCs from either null show ∼10-fold reduced capacity to recover from proton loading, suggesting a primary defect in gastric acid secretion. The achlorhydria we previously observed in Kcne2−/− mice was striking given that Kcnq1, the pore-forming subunit of the complex, was still present, and in fact was strongly expressed in double the number of cells per gastric gland in Kcne2−/− mice compared to Kcne2+/+ mice (2). PCs are nonexcitable, and their membrane potential reportedly varies from −20 to −40 mV, with stimulation by secretagogues such as gastrin, histamine, or carbachol causing a shift to the hyperpolarized end of this spectrum (15). Current-voltage relationships measured using patch clamp of transfected KCNQ1 alone or with KCNE2 in mammalian nonpolarized cell lines indicate that KCNE2 reduces the voltage dependence of KCNQ1 activation (11); however, in the crucial −20- to −40-mV range, homomeric KCNQ1 channels pass more current (in e.g., 3-s pulses) at neutral pH than KCNE2–KCNQ1 complexes. While KCNQ1 channels are partially inhibited at low extracellular pH, KCNE2–KCNQ1 channel currents are increased; the former, however, still pass current even at pH 3, and low pH reduces homomeric KCNQ1 inactivation (16). The polarity of KCNQ1 trafficking would be expected to be fundamental to its role in PCs, and disruption of this trafficking an interesting candidate mechanism for the profound gastric effects produced by Kcne2 deletion. However, previous studies failed to find any effects of KCNE subunits on KCNQ1 localization in vitro in Madin-Darby canine kidney (MDCK) cells (in which KCNQ1 remained basolateral regardless of coexpression with each of KCNE1–5) or in vivo in the colonic epithelium (in which KCNQ1 was basolateral in both wild-type and Kcne3−/− mice) (13, 17). Further, we recently discovered that KCNQ1–KCNE2 plays a crucial role in thyroid hormone biosynthesis, and that this channel appears to be basolaterally located in thyrocytes (18), contrasting with its apical localization in PCs. These apparent paradoxes, and the relative lack of understanding of the mechanisms underlying polarized trafficking of ion channels in general, prompted us to determine the effects of Kcne gene deletion on KCNQ1 trafficking in mouse PCs in vivo.
MATERIALS AND METHODS
Generation of gene-targeted mice
All mice used were housed and utilized according to the NIH Guide for the Care and Use of Laboratory Animals and Weill Medical College of Cornell University animal care and use policies. Kcne2−/− mice were generated as described previously from Kcne2+/− × Kcne2+/− crosses (2). Kcne3 was disrupted through homologous recombination, using a targeting vector to replace the entire coding region, contained within the fourth exon, of the Kcne3 gene. Two homologous arms, a 3.9-kb sequence homologous to the 5′ region upstream from exon 4 and a 3.0-kb sequence homologous to the 3′ downstream region, were subcloned into a pVBTk-loxP-knockout backbone vector. The vector contained a neomycin resistance (Neor) cassette flanked by LoxP sites, allowing for the removal of the cassette on expression of Cre-recombinase, and a TK− selection marker (see Fig. 4A). The targeting vector was linearized at a unique I-Ceu1 restriction enzyme site outside of the homologous region and electroporated into Albino C57BL/6 (C2J) ES cells. Clones were positively selected for Neor, and integration of the null vector was confirmed through Southern blot analysis with a 5′ probe directed at a 126-bp sequence located outside the recombined region. The probe was amplified by PCR using the following primers: forward 5′-GCAGAAGGTAGGCACTTGGG-3′ and reverse 5′-ACTGGGGGAGACAATAGGCG-3′. Correctly targeted ES cells were injected into C57BL/6 blastocysts and implanted into female mice, which were bred with C57BL/6 males to generate chimeric progeny of a 50:50 C57BL/6:Albino B6 (C2J) genetic background. Chimeras were interbred to produce Kcne3+/− mice. The Kcne3−/− mice used in this study were bred from Kcne3+/− × Kcne3+/− crosses.
Figure 4.
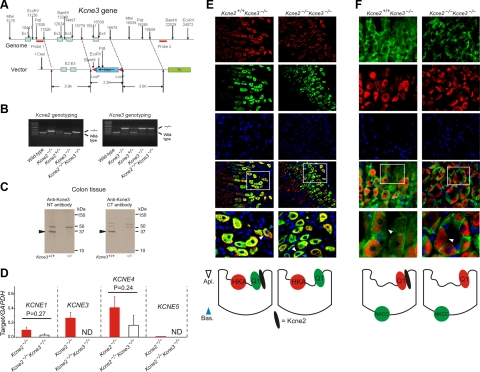
Kcne3 deletion restores apical trafficking of Kcnq1 in Kcne2−/− mice. A) Targeting strategy for genomic deletion of Kcne3 from mice. B) Agarose gels showing PCR genotyping of wild-type, heterozygous, and homozygous single-knockout Kcne2−/− and Kcne3−/− mice, and double-knockout Kcne2−/−Kcne3−/− mice. C) Western blots of Kcne3 protein in membrane fractions from colonic crypts of Kcne3+/+ and Kcne3−/− mice as indicated, using an in-house antibody raised against a Kcne3 N-terminal epitope (left panel), and a commercial antibody (Alomone) raised against a Kcne3 C-terminal epitope (right panel). Migration distance of molecular mass markers is indicated at right. Arrows indicate band at 37 kDa unique to wild-type tissue. D) qRT-PCR analysis of remodeling of the fundic Kcne expression profile by targeted deletion of both Kcne2 and Kcne3; expression level expressed as a ratio to that of reference gene GAPDH amplified in parallel each time; n = 10 mice/single gene deletion genotype; n = 5 mice/double gene deletion genotype. ND, not determined (for Kcne3, not measured in Kcne2−/−Kcne3−/− mice; for Kcne5, unable to detect signal conforming to quality controls as described in Materials and Methods). Error bars = sem. E, F) Top: exemplar IF colabeling of Kcne2+/+ Kcne3−/− and Kcne2−/−Kcne3−/− gastric glands as indicated. Bottom two IF panels are merged views of the 3 panels above; bottom merged panel shows expanded view of the boxed region in the top merged panel. Blue arrowheads, PC basolateral side; white arrowheads, PC apical side. Counterstained with DAPI (blue). Representative results from ≥2 mice, 3–5 sections/mouse/genotype. Bottom: cartoons summarizing IF data. E) Kcnq1 (green) and HKA β subunit (red). F) Kcnq1 (red) and NKCC1 (green). Width of view (except bottom merge): 100 μm (E); 50 μm (F).
To generate litters of Kcne2+/−Kcne3+/− mice, male Kcne2−/− mice were bred with female Kcne3−/− mice. All double-heterozygous mice appeared superficially normal and were interbred to yield the Kcne2−/−Kcne3−/− and Kcne2+/− Kcne3−/− mice used in experiments. Genotyping for Kcne2 was performed by PCR using the following oligonucleotide primers: 5′-CTGGAGGTAGCCAAATGGAGGAAG-3′, 5′-TCCTGCCAATC TTCCACGATGTAC-3′, and 5′-CGCTCCCGATTCGCAGCGCATC-3′, which generated a wild-type band of 382 bp and a knockout band of 680 bp. Genotyping for Kcne3 was performed by PCR using the following oligonucleotide primers: 5′-CTATTCTACACGCACTGTGGGATG-3′, 5′-CGTTGGAAGTCT CCATAGCAACAG-3′, and 5′-CGCTCCCGATTCGCAGCGCATC-3′, which generated a wild-type band of 280 bp and a knockout band of 1000 bp.
Quantitative RT-PCR (qRT-PCR)
Tissue extraction
Mice were euthanized by CO2 asphyxiation. Stomachs were excised and washed in PBS, and the fundus was removed. Tissue was flash-frozen in liquid nitrogen and stored at −80°C until use. In preparation for RNA extraction, frozen tissue sections were submerged overnight or for 8 h in RNAlater Ice (Ambion, Austin, TX, USA) at −20°C.
RNA extraction
RNA was extracted from 30 mg of tissue with RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. Tissue homogenization was achieved using a pestle grinder system (Fisher Scientific, Hampton, NH, USA). RNA yield and purity (A260/A280) were assessed by NanoDrop 2000 spectrophotometer (ThermoScientific, Waltham, MA, USA). RNA samples with A260/A280 absorbance ratios between 1.80 and 2.10 were considered acceptable for cDNA synthesis.
cDNA synthesis
cDNA was synthesized from 1 μg of RNA with Quantitect Reverse Transcriptase (Qiagen) according to the manufacturer's protocol. To remove genomic DNA, template RNA was mixed with gDNA Wipeout Buffer (Qiagen) and incubated at 42°C for 2 min. Quantitect Reverse Transciptase containing an RNase inhibitor and Quantiscript RT Buffer containing Mg2+ and dNTPs were then added to the genomic DNA elimination reaction and incubated at 42°C for 15 min. The reverse transcription reaction was inactivated with a 3-min incubation at 95°C. Synthesized cDNA was analyzed immediately thereafter by qPCR or stored at −20°C until use.
Targeting information
qRT-PCR was conducted adhering as closely as possible to MIQE guidelines (19). Primer pairs for target gene Kcne1 [National Center for Biotechnology Information (NCBI) GeneID 16509] produced an amplicon of 108 bp; match position of the expected sequence was number 1 out of 123 Basic Local Alignment Search Tool (BLAST; U.S. National Institutes of Health, Bethesda, MD, USA) matches. Primer pairs for target gene Kcne3 (NCBI GeneID 57442) produced an amplicon of 143 bp; match position of the expected sequence was number 1 out of 1001 BLAST matches. Primer pairs for target gene Kcne4 (NCBI GeneID 57814) produced an amplicon of 126 bp. Primer pairs for target gene Kcne5 (NCBI GeneID 66240) produced an amplicon of 113 bp; match position of the expected sequence was number 1 out of 428 BLAST matches. Primer pairs for reference gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH; NCBI Gene ID 14433) produced an amplicon of 123 bp; match position of the expected sequence is number 1 out of 251 BLAST matches.
Primer information
Primer sequences for qPCR analysis were acquired from the Harvard Medical School PrimerBank (Boston, MA, USA; ref. 20) and were as follows: Kcne1, forward 5′-ATGAGCCTGCCCAATTCCAC-3′ and reverse 5′-GAGCTGAGACTTACGAGCCA-3′; Kcne2, forward 5′-CACATTAGCCAATTTGACCCAGA-3′ and reverse 5′-GAACATGCCGATCATCACCAT-3′; Kcne3, forward 5′-CTTTGCTCGATGGAAGGGGAC-3′ and reverse 5′-GCTGTCGTTGAGAGGCGTC-3′; Kcne4, forward 5′-CTGAGGATGGAGCCTCTGAAC-3′ and reverse 5′-AGCAAATCGAAACGAGTCCTTC-3′; Kcne5, forward 5′-AGATCCGCTGTCCTCCTCATT-3′ and reverse 5′-GGGTTCTGACCTCTCATCATCTT-3′; and GAPDH, forward 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse 5′-TGTAGACCATGTAGTTGAGGTCA-3′. Primers (50-nm synthesis scale, desalted) were acquired from Invitrogen (Carlsbad, CA, USA).
Assay details
qPCR analysis was performed on the Roche Light Cycler 480 System using LightCycler 480 SYBR Green I Master Mix and LightCycler 480 96-well white plates (Roche Diagnostics, Indianapolis, IN, USA). Each reaction contained ∼75 ng of cDNA, 1 μl of PCR-grade water, 2 μl of 10 μM forward primer, 2 μl of 10 μM reverse primer, and 10 μl 2X Master Mix, which was comprised of dNTP mix, MgCl2, FastStart TaqDNA Polymerase, reaction buffer, and SYBR Green I dye.
Cycling conditions
Thermocycling parameters were as follows: for amplification, 1 cycle at 95°C (10 min); 45 cycles at 95°C (5 s), 68°C (5 s), and 72°C (25 s); for melting curve, 95°C (1 s), 65°C (1 s), 95°C (continuous); for cooling, 1 cycle 45°C (15 s).
Data analysis
Advanced relative quantification was used to obtain normalized changes in expression levels of target genes (Kcne1–5) relative to controls (GAPDH) using LightCycler 480 1.5 software. Primer pairs were previously validated by PrimerBank with amplification plots, dissociation curves, and 2% agarose gel analysis. Primer pair amplification efficiency was also established with calibration curves within the laboratory on LightCycler 480 equipment, and deemed satisfactory for experimentation. The calibration curve for GAPDH yielded a slope of −3.334 and efficiency of 1.995. The calibration curve for Kcne1 yielded a slope of −3.386 and efficiency of 1.974. The calibration curve for Kcne3 yielded a slope of −3.362 and efficiency of 1.983. The calibration curve for Kcne4 yielded a slope of −3.110 and efficiency of 2.097. The calibration curve for Kcne5 yielded a slope of −3.315 and efficiency of 2.003. Each sample was run in triplicate as a quality control measure, and triplicates varying from one another by >1 cycle were discarded. Melting curves were assessed for each reaction to verify the amplification of a single product. Final analysis of statistical significance was calculated using 1-way analysis of variance (ANOVA) test (Origin).
Semiquantitative RT-PCR
The observation of Kcne3 up-regulation in Kcne2−/− mouse fundus using qPCR was recapitulated using conventional semiquantitative RT-PCR on fundic cDNA, using different primer sequences to those used for qPCR, and an alternative reference gene: hypoxanthine-guanine phosphoribosyltransferase (HPRT). Briefly, RNA was extracted from 4 separate stomach fundi/genotype using an RNAeasy kit (Qiagen), then samples were diluted to give equal RNA concentrations, as assessed by spectrophotometry, before reverse-transcription to give cDNA as before (21). Primers used were as follows: HPRT, forward 5′-TGGAAAGAATGTCTTGATTGTTGA-3′ and reverse 5′-ACTTCGAGAGGTCCTTTTCACC-3′, which gives a 130-bp product; Kcne3, forward 5′-GGCTCTGAACACAACCCTTC-3′ and reverse 5′-TTTGTCCACTTTGCGTGAAC-3′, which gives a 205-bp product. Band densities of PCR products obtained with specific primers for HPRT transcript, run on a 1% agarose gel and stained with ethidium bromide, were measured using a Fluor-S MultiImager (Bio-Rad, Hercules, CA, USA) to confirm that the RNA-concentration-normalized samples each yielded similar amounts of this reference transcript. In parallel, cDNA samples from the same preps were amplified with Kcne3-specific primers, and optical density was measured. Results are expressed as mean optical density for each amplicon, with statistical analysis performed using 1-way ANOVA with statistical significance set at P < 0.05.
Immunostaining (IS) and immunofluorescence (IF)
KCNQ1 IS (Fig. 1C–E) was performed as we previously described (2). IF detection of HKA β, the Na+K+2Cl− cotransporter (NKCC1), KCNQ1, and KCNE3 was performed using a Discovery XT processor (Ventana Medical Systems, Tucson, AZ, USA). The primary antibody concentrations used were: 0.5 mg/ml anti-HKA β (mouse monoclonal; Affinity Bioreagents, Golden, CO, USA), 0.5 mg/ml anti-NKCC1 (goat polyclonal, Santa Cruz Biotechnology, Santa Cruz, CA, USA), and 1 mg/ml anti-KCNQ1 (rabbit or goat polyclonal; Chemicon, Temecula, CA, USA); in-house anti-KCNE3 serum was used at a 1:500 dilution after column-enriching IgG. Preceding the primary antibody incubation, the tissue sections were blocked for 30 min in 10% normal goat serum, 2% BSA in PBS, followed by 8 min avidin/biotin block. The primary antibody incubation (3 h) was followed by 32 min incubation with biotinylated anti-mouse IgG (Vectastain ABC kit; Vector Laboratories, Burlingame, CA, USA) for HKA β, 60 min incubation with biotinylated anti-goat IgG (Vectastain ABC kit) for NKCC1 (and for KCNQ1 for KCNE3 colocalization analysis), and biotinylated anti-rabbit or antibody at 1:200 dilution (Vectastain ABC kit) for KCNQ1 (for HKA and NKCC1 colocalization analysis). The secondary detection was performed with Streptavidin-HRP D (Ventana Medical Systems), followed by incubation with Tyramide-Alexa Fluor 488 (Invitrogen) or Tyramide Alexa Fluor 568 (Invitrogen). Stained slides were viewed with a Zeiss Axiovert 200 widefield microscope (Carl Zeiss, Oberkochen, Germany), and pictures were acquired using MetaMorph 7.1 software (Molecular Devices, Sunnyvale, CA, USA).
Western blotting and coimmunoprecipitation (co-IP)
For Western blotting, gastric membrane fractions were prepared as we previously described (2). Protein concentration of the supernatant was measured according to the Bradford method. Total protein (40 μg/lane) was loaded into a precast Tris-glycine 4–20% gel (Bio-Rad) and separated by electrophoresis. Proteins were then transferred onto a PVDF membrane (Bio-Rad) and blocked with 5% milk and 0.05% Tween-20 in PBS at 4°C on a rocker either for 1–2 h or overnight. Primary antibody incubations (4 h, room temperature, in 1% milk and 0.05% Tween-20 in PBS) were 1:1000 anti-KCNQ1 (Chemicon); 1:500 anti-KCNE3 (in-house anti-KCNE3 N terminus or anti-KCNE3 C terminus from Alomone Labs, Jerusalem, Israel); 1 mg/ml anti-NKCC1 (Santa Cruz Biotechnology). Membranes were washed 4 times, 20 min each, with antibody incubation buffer; incubated with the appropriate secondary antibodies (Bio-Rad), diluted 1:10,000 in buffer A, for 2 h at room temperature; then washed 4 times, 20 min each, with buffer A and once for 5 min with PBS. Membranes were incubated for 1 min with the SuperSignal ECL reagent (Pierce Biotechnology, Rockford, IL, USA), then exposed on BioMax Light Film (Kodak, Rochester, NY, USA) and developed using an RP X-OMAT processor (Kodak). For co-IPs, membrane fractions in buffer A—150 mM NaCl, 50 mM Tris-HCL (pH 7.4), 20 mM NaF, 10 mM NaVO4, 1 mM phenylmethylsulfonyl fluoride (Fisher Scientific), 1% Nonidet P-40 (Pierce), 1% CHAPS (Sigma, St. Louis, MO, USA), 1% Triton X-100 (Fisher Scientific), and 0.5% SDS (Sigma)—were precleared with Protein A Sepharose beads (Amersham Bioscience), incubated with antibodies raised against KCNQ1 or NKCC1, and precipitated with Protein A Sepharose beads; then beads were washed with buffer A, and bound proteins were eluted with SDS-PAGE loading buffer for Western blotting as above.
Histology
For histology and stomach mass quantification, mice were killed using CO2 asphyxiation (5–10/genotype). Stomachs and colons were removed postmortem, stomach mass was determined, and stomach and colon tissue was fixed in 10% neutral buffered formalin, processed by routine methods, and embedded in paraffin wax. Gastric mucosal and colonic epithelial sections were cut at 5-μm intervals, placed on positively charged Superfrost slides, stained with hematoxylin and eosin (H&E), and evaluated with an Olympus BX45 microscope (New York/New Jersey Scientific Inc., Middlebush, NJ, USA).
Whole-stomach pH measurements
Mice were killed by CO2 asphyxiation. Stomachs were ligated ex vivo at the esophageal and duodenal junctures and excised. Stomachs were then incubated for 1 h in oxygenated HEPES-buffered Ringer's solution with or without 300 μM histamine (Sigma). After 1 h incubation time, stomach contents were aspirated, and pH was measured using a microcombination pH probe (Microelectrodes Inc., Bedford, NH, USA).
RESULTS AND DISCUSSION
Kcne2 deletion reverses the polarity of Kcnq1 trafficking in PCs
Having previously determined that Kcnq1 expression in the gastric mucosa is increased after targeted deletion of Kcne2 (2), here we examined the effects of Kcne2 deletion on the intracellular localization of Kcnq1 in PCs. In 3-mo-old Kcne2−/− mouse gastric mucosa, Kcnq1 IS demonstrated an apparent sharply basolateral localization in PCs across the mucosa, in contrast to its diffuse staining in Kcne2+/+ PCs due to localization in the highly convoluted and invaginated apical membrane (Fig. 1C–E). Double IF staining confirmed that Kcnq1 was expressed in the apical side of PCs from 3-mo-old Kcne2+/+ mice, colocalizing with the H+/K+ATPase β subunit (HKA β), a PC apical membrane marker, but not with NKCC1, a marker for basolateral membrane in PCs (22) (Fig. 1F, G). In contrast, in age-matched Kcne2−/− mouse gastric sections, Kcnq1 was still expressed in PCs (which were identified by midgastric gland location and HKA β expression), but was colocalized with NKCC1 at the PC basolateral membrane (Fig. 1F, G). HKA β was still expressed in the apical membrane of PCs, as described previously (23), and did not colocalize with NKCC1 in either Kcne2+/+ or Kcne2−/− sections, demonstrating that Kcne2 deletion did not globally disrupt PC polarity (Fig. 1H).
Kcne2 deletion selectively up-regulates fundic Kcne3
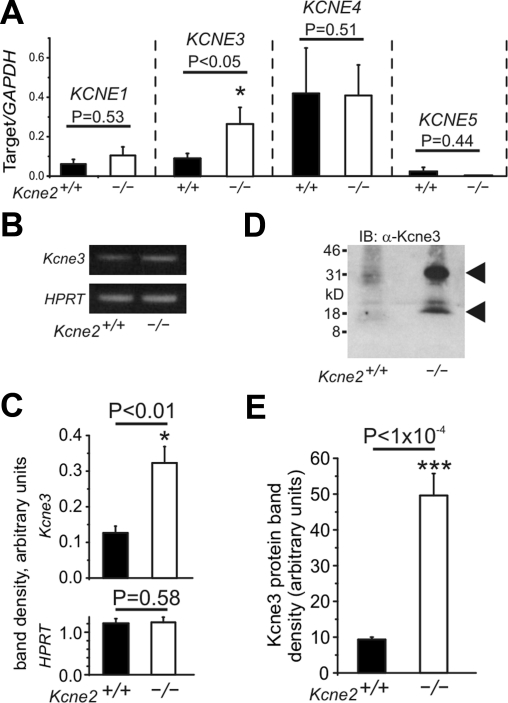
We considered two possible mechanisms underlying the observed switch in Kcnq1 location on Kcne2 deletion: passive, arising from homomeric Kcnq1 trafficking to the basolateral membrane in the absence of a required chaperone (postulated to be Kcne2) to target its expression to the apical side; or active, due to hijacking by another Kcne subunit. We therefore next investigated possible Kcne remodeling in Kcne2−/− gastric fundus tissue, using qRT-PCR analysis of transcripts for each of the four remaining Kcne genes, with GAPDH serving as a reference gene. Strikingly, we observed that fundic Kcne3 transcript expression was increased 3-fold at 3 mo of age by targeted deletion of Kcne2 (n=10 mice/genotype; P<0.05), whereas there were no significant changes in the fundic expression of transcripts for Kcne1, Kcne4, or Kcne5 (n=10 mice/genotype; P > 0.4) (Fig. 2A). The observation that Kcne3 mRNA was upregulated in Kcne2−/− fundus was recapitulated using conventional (semiquantitative) RT-PCR, with HPRT as a reference gene (n=4 mice/genotype; P<0.01; Fig. 2B, C). Notably, fundic Kcne3 protein expression was found to be increased 5-fold by Kcne2 deletion (n=7 independent preps, 21–35 mice/genotype, P<1×10−4; Fig. 2D, E).
Figure 2.
PC Kcne3 is selectively up-regulated in remodeling arising from targeted deletion of Kcne2. A) qRT-PCR analysis of remodeling of the fundic Kcne expression profile by targeted deletion of Kcne2; mRNA expression level expressed as a ratio to that of reference gene GAPDH; n = 10 mice/genotype/gene. B) Representative agarose gel of cDNA fragments for Kcne3 and the reference gene HPRT after semiquantitative RT-PCR from gastric mucosal lysates from Kcne2+/+ and Kcne2−/− mice as indicated. C) Mean band optical densities from samples as in panel B; n = 4 independent preps (and mice)/genotype. D) Representative Western blot of Kcne3 protein in membrane fractions from Kcne2+/+ and Kcne2−/− mouse fundus preparations as indicated, using anti-Kcne3 antibody. Migration distance of molecular mass markers is indicated at left. Arrows indicate expected sizes of nonglycosylated and fully glycosylated (mature) Kcne3. E) Band optical density from Kcne3 (mature form) Western blots of Kcne2+/+ and Kcne2−/− gastric mucosal membrane fractions, as in panel D; n = 7 independent preparations from 3–5 stomachs each; total of 21–35 stomachs/genotype. Error bars = sem.
Kcne3 forms PC basolateral complexes with Kcnq1 in the absence of Kcne2
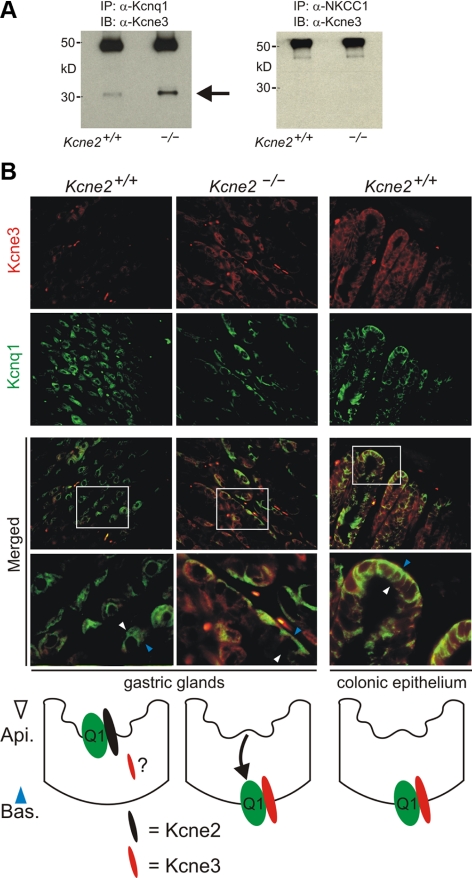
The fundic remodeling data (Fig. 2) suggested Kcne3 as the most likely Kcne candidate for diversion of Kcnq1 to the basolateral membrane in Kcne2−/− PCs. We adopted several biochemical and genetic approaches to test this hypothesis. First, we performed native co-IP studies using fundic tissue, and found greatly increased formation of Kcnq1–Kcne3 complexes in Kcne2−/− tissue compared to Kcne2+/+ tissue, with NKCC1 providing a negative control for Kcne3 co-IP (Fig. 3A). These data were supported by IF analyses, which indicated increased Kcne3 expression compared to Kcne2+/+ PCs, and basolateral colocalization of Kcne3 with Kcnq1, in Kcne2−/− PCs— similar to that observed in wild-type colonic epithelium (Fig. 3B).
Figure 3.
Remodeled Kcne3 forms basolateral complexes with Kcnq1 in Kcne2−/− PCs. A) Co-IPs showing complex formation of Kcne3 with Kcnq1 (left panel) but not with NKCC1 (right panel) from mouse fundic membrane fractions. IPs of fractions from Kcne2+/+ and Kcne2−/− mice were prepared using antibodies raised against Kcnq1 or NKCC1, and Western blots were performed with anti-Kcne3 antibody. Numbers indicate migration of molecular mass markers (kDa). Arrow indicates expected mature Kcne3 migration distance. Top bands are precipitated antibodies. Representative of n = 2 experiments/antibody, with each prep pooled from 3–5 mouse stomachs/genotype. B) Top: exemplar IF labeling of Kcne3 (red) and Kcnq1 (green) in Kcne2+/+ or Kcne2−/− gastric glands, or Kcne2+/+ colonic crypts, as indicated. Merged panels show merged views of the 2 panels above; bottom merged panel shows expanded view of the boxed region in the top merged panel. Yellow indicates colocalization. Width of view (except bottom merge): 100 μm. Representative of results from at least two mice, 3–5 sections/mouse/genotype. Bottom: cartoons summarizing IF data.
Kcne3 is necessary and sufficient for trafficking of Kcnq1 to the PC basolateral membrane
These observations were suggestive of an active role for Kcne3 in rerouting Kcnq1 to the PC basolateral membrane, but it was still possible that Kcnq1 could target to the PC basolateral side regardless of Kcne3, in the absence of Kcne2. To resolve this, we generated Kcne3−/− mice by targeted deletion of the Kcne3 gene (Fig. 4A), and then crossed them with Kcne2−/− mice to generate heterozygous, and ultimately double-knockout, Kcne2−/−Kcne3−/− mice. We confirmed their genotypes with PCR (Fig. 4B) and with Western blots from colon tissue, in which Kcne3 is known to be expressed (10) (Fig. 4C). Next, we determined whether there was remodeling of Kcne1, Kcne4, or Kcne5 due to concomitant Kcne2 and Kcne3 deletion, again using qRT-PCR. These experiments did not identify statistically significant changes in mRNA expression of these 3 genes; there was a general trend toward reduced expression, but considerable variability in expression within each genotype (Fig. 4D). Strikingly, IF studies showed that Kcnq1 colocalized in the apical compartment with HKA β, and not basolaterally with NKCC1, in Kcne2−/− Kcne3−/− mouse PCs (Fig. 4E, F). As we might have expected, Kcnq1 was also apically expressed in Kcne2+/+ Kcne3−/− mouse PCs (Fig. 4E, F). Thus, in PCs, Kcne2 is not required for apical localization of Kcnq1 in the absence of Kcne3. However, in the absence of Kcne2, Kcne3 is up-regulated and is necessary and sufficient to actively chaperone Kcnq1 to the PC basolateral membrane.
Kcne2 is required for Kcnq1 function at the PC apical membrane
Our findings suggested that in Kcne2−/−Kcne3+/+ mouse PCs, Kcnq1–Kcne3 channels could form an additional basolateral K+ efflux route, but that there would be no apical K+ recycling pathway. In contrast, in Kcne2−/−Kcne3−/− mouse PCs, homomeric Kcnq1 could potentially provide an apical K+ recycling pathway, but in the absence of Kcne2 its functionality might be limited, due to an inability to function efficiently at low pH and/or negative membrane potentials. There are several hypothetical consequences of these mistrafficking and subunit rearrangement events. Basolateral Kcnq1–Kcne3 channels in Kcne2−/−Kcne3+/+ mice could potentially restore some gastric acid secretion by alleviating PC K+ accumulation. In contrast, Kcne2−/− Kcne3−/− mice would exhibit restored gastric acidification if Kcnq1 was able to function alone at the apical membrane, but if homomeric Kcnq1 did not have this capability, Kcne2−/−Kcne3−/− mice would potentially have the most severe gastric pathology of all the genotypes. Kcne2+/+Kcne3−/− mice would be predicted to have normal gastric acidification.
We tested these hypotheses by measuring stomach mass to quantify gastric hyperplasia (one consequence of achlorhydria), and by quantifying gastric luminal pH. These studies yielded the striking finding that Kcne3 indeed affected gastric function and cell proliferation in Kcne2-depeleted mice, but not in mice with both Kcne2 alleles. Thus, 3-mo-old Kcne2−/−Kcne3−/− mice exhibited massive gastric hyperplasia, with twofold heavier stomachs than age-matched Kcne2−/−Kcne3+/+ mice, whereas Kcne2+/+Kcne3−/− mice had normal stomach mass (Fig. 5A, B). Notably, Kcne3 also prevented gastric hyperplasia in Kcne2+/−Kcne3+/+ mice, which had significantly smaller stomachs than those of Kcne2+/−Kcne3−/− mice (Fig. 5B). These data were supported by results from stomach lumen pH quantification, which indicated that Kcne2+/−Kcne3−/− mice had significantly less gastric acidification upon histamine stimulation than Kcne2+/−Kcne3+/+ mice, although Kcne3 did not affect the stomach lumen pH or response to histamine of Kcne2−/− mice. As expected, Kcne2+/+Kcne3−/− mice had normal stomach pH and response to histamine (Fig. 5C).
Figure 5.
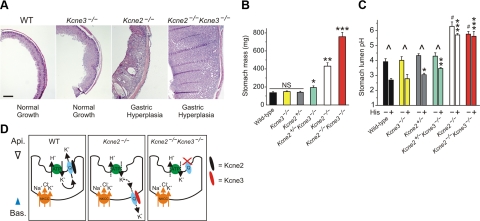
Kcne3 deletion exacerbates gastric hyperplasia and achlorhydria in Kcne2-deficient mice. A) Exemplar H&E-stained sections of gastric mucosa from 3-mo-old wild-type, Kcne3−/−, Kcne2−/− and double-knockout Kcne2−/−Kcne3−/− mice. Scale bar = 250 μm. Representative of ≥2 sections each from ≥2 mice/genotype. B) Mean stomach mass measured ex vivo from 3-mo-old mice, genotypes as indicated, n = 6–16. NS, nonsignificant. *P < 0.05; **P < 2 × 10−4; ***P < 4 × 10−5. C) Mean stomach lumen pH measured ex vivo from 3-mo-old mice, genotypes as indicated, with (+) or without (−) stimulation with 300 μM histamine (His); n = 3–5. *P < 0.05, **P < 0.03, ***P < 0.002 vs. all other His groups; #P < 0.01 vs. all other control groups; ^P < 0.05 vs. corresponding control group. D) Summary illustrating Kcne control of Kcnq1 trafficking in mouse PCs. Error bars = sem.
These findings are consistent with a novel model in which, in the absence of Kcne2, Kcne3 is upregulated and chaperones Kcnq1 to the PC basolateral side. When both Kcne3 and Kcne2 are deleted, homomeric Kcnq1 localizes to the apical membrane, but without restoration of gastric acidification. This indicates that Kcnq1 cannot function in the absence of both Kcne2 and Kcne3 in PCs, even if at the apical membrane, due either to inhibition by low extracellular pH, inability to constitutively activate, or both (Fig. 5D).
Implications of Kcne-directed polarized trafficking of Kcnq1
This study describes two main novel findings: discovery of the capacity of a KCNE subunit to act as a polar trafficking chaperone, and identification of KCNE subunit remodeling (and its functional consequences) in vivo. In native PCs, Kcnq1 probably localizes primarily in deeply invaginated sections of the apical membrane both at rest and when stimulated, although a fraction of it may be located in intracellular vesicles and move to the apical surface on secretagogue stimulation; in contrast, HKA is primarily located in intracellular vesicles until stimulation triggers its trafficking to the apical membrane (16, 24). Here, we show that Kcne2 deletion results in Kcnq1 residing basolaterally in PCs instead, and that Kcne3 is necessary and sufficient for this rerouting. In a previous study of MDCK cells, Kcnq1 was basolaterally located regardless of which Kcne subunit (subunits 1 through 5) it was heterologously coexpressed with (17), and we recently found that Kcnq1–Kcne2 channels are basolaterally located in thyrocytes (18). Furthermore, Kcne3 does not appear necessary for basolateral location of Kcnq1 in colonic epithelium, although potential remodeling of other Kcne subunits was not determined in that study (13). Clearly, the influence of Kcne subunits on Kcnq1 targeting in polarized cells is highly cell-type specific, perhaps due to differences in expression of proteins such as μ1B, an AP-1 clathrin adaptor complex that directs polarized trafficking (25–27).
The discovery of Kcne remodeling due to genetic disruption of another Kcne subunit has potentially profound implications for the etiology of Kcne-related disease states and for study of Kcne-knockout mice. In previous studies examining K+ channel α subunit gene deletion, a concern has been that functional redundancy exists given the similarity of some α subunits, e.g., Kv3.1 and Kv3.2 (28). Here, studying β subunits, we have unearthed a novel remodeling phenomenon, wherein the location of the α subunit in the absence of its regular β subunit partner is the polar opposite of that observed in wild-type mice—due to hijacking by a remodeled (upregulated), related β subunit. We suspect we have merely scratched the surface with respect to the prevalence of Kcne subunit remodeling in both model systems and in animal and human disease states in a variety of tissues, a hypothesis to be tested further in the future. The present findings highlight the importance not only of an apical localization per se for Kcnq1, but also the association with Kcne2 for full functionality at the apical side. Kcnq1–Kcne3 channels are acid-insensitive (29) and, like Kcnq1–Kcne2, are constitutively active, so one would assume they could provide an apical K+ recycling conduit if located there, but they could not rescue gastric acid secretion in Kcne2−/− mice because they were basolaterally located in parietal cells, although they partially restored function in Kcne2+/− mice. By the same token, even when apically located, Kcnq1 could not serve as a K+ recycling channel without Kcne2 in PCs—we speculate this is because homomeric Kcnq1 is voltage dependent and inhibited by acid. Our data suggest that Kcnq1 defaults to the apical side in the absence of Kcne2 or Kcne3, but further studies are required, perhaps adopting a proteomic approach together with the genetic models described here, to determine whether Kcnq1 requires an additional subunit to traffic apically in parietal cells, which is either upregulated on Kcne2 and Kcne3 double knockout, or simply permitted to associate with Kcnq1 only in the absence of these subunits. The mechanism underlying basolateral trafficking of Kcnq1– Kcne3 complexes in parietal cells is likely both Kcne3 and parietal cell dependent, and its elucidation will require use of chimeric Kcne subunits introduced either in vivo or into a suitable polarized parietal cell line; the former is more attractive because physiological consequences would be readily assayable. Our co-IP data suggest some (albeit relatively low-level) formation of Kcnq1–Kcne3 complexes even in wild-type gastric epithelium, which could correspond to complexes in gastric surface cells, or in chief cells, two gastric mucosal cell types suggested to express basolateral Kcnq1 and Kcne3; these cells are not located midgastric gland and do not express Kcne2 or HKA, and thus are easily distinguishable from PCs in IF studies (13, 29). Alternatively, Kcnq1–Kcne3 complexes could be occurring even in wild-type PCs, again albeit at relatively low levels. Further studies will identify what if any function these putative channels perform in PCs, and whether or not these are actually mixed Kcnq1–Kcne2–Kcne3 complexes, perhaps with functional characteristics we do not yet understand. This type of tripartite complex has been reported for Kcne1, Kcne3, and Kcnh3 in mouse brain (30).
Future work will determine which Kcne subunits or other factors control polarized trafficking of Kcnq1 in, e.g., the thyroid, and of other α subunits in polarized cells in general, together with a search for the molecular signals that cause, e.g., Kcne3 transcript up-regulation in Kcne2−/− mouse PCs. The extent to which perturbation of this polarized trafficking contributes directly to, or is reflective of compensatory remodeling in, the molecular etiology of human diseases of the epithelia and other related systems, will be explored—particularly in the light of our recent finding that Kcne2−/− mice develop gastritis cystica profunda and gastric neoplasia (31).
Acknowledgments
The authors are grateful for expert technical assistance from S. Backovic, L. Cohen-Gould (Director of the Electron Microscopy and Histology Core Facility at Weill Cornell Medical College), M. S. Jiao, the Molecular Cytology Core Facility of Memorial Sloan-Kettering Cancer Center, Sloan-Kettering Institute Mouse Genetics Core Facility, and Rockefeller University Transgenic Mouse Facility.
G.W.A. is thankful for support from the National Heart, Lung, and Blood Institute; the National Institutes of Health (NIH; R01 HL079275); the American Heart Association (grant-in-aid 0855756D); and an Irma T. Hirschl Career Scientist Award. K.P. is supported by an NIH predoctoral training grant (T32GM073546).
REFERENCES
- 1. Lee M. P., Ravenel J. D., Hu R. J., Lustig L. R., Tomaselli G., Berger R. D., Brandenburg S. A., Litzi T. J., Bunton T. E., Limb C., Francis H., Gorelikow M., Gu H., Washington K., Argani P., Goldenring J. R., Coffey R. J., Feinberg A. P. (2000) Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J. Clin. Invest. 106, 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roepke T. K., Anantharam A., Kirchhoff P., Busque S. M., Young J. B., Geibel J. P., Lerner D. J., Abbott G. W. (2006) The KCNE2 potassium channel ancillary subunit is essential for gastric acid secretion. J. Biol. Chem. 281, 23740–23747 [DOI] [PubMed] [Google Scholar]
- 3. Fujita A., Horio Y., Higashi K., Mouri T., Hata F., Takeguchi N., Kurachi Y. (2002) Specific localization of an inwardly rectifying K(+) channel, Kir4.1, at the apical membrane of rat gastric parietal cells; its possible involvement in K(+) recycling for the H(+)-K(+)-pump. J. Physiol. 540, 85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malinowska D. H., Sherry A. M., Tewari K. P., Cuppoletti J. (2004) Gastric parietal cell secretory membrane contains PKA- and acid-activated Kir2.1 K+ channels. Am. J. Physiol. Cell Physiol. 286, C495–C506 [DOI] [PubMed] [Google Scholar]
- 5. Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. (1996) K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature 384, 78–80 [DOI] [PubMed] [Google Scholar]
- 6. Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature 384, 80–83 [DOI] [PubMed] [Google Scholar]
- 7. Abbott G. W., Sesti F., Splawski I., Buck M. E., Lehmann M. H., Timothy K. W., Keating M. T., Goldstein S. A. (1999) MiRP1 forms IKr potassium channels with HERG and is associated with cardiac arrhythmia. Cell 97, 175–187 [DOI] [PubMed] [Google Scholar]
- 8. McCrossan Z. A., Abbott G. W. (2004) The MinK-related peptides. Neuropharmacology 47, 787–821 [DOI] [PubMed] [Google Scholar]
- 9. Panaghie G., Abbott G. W. (2007) The role of S4 charges in voltage-dependent and voltage-independent KCNQ1 potassium channel complexes. J. Gen. Physiol. 129, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schroeder B. C., Waldegger S., Fehr S., Bleich M., Warth R., Greger R., Jentsch T. J. (2000) A constitutively open potassium channel formed by KCNQ1 and KCNE3. Nature 403, 196–199 [DOI] [PubMed] [Google Scholar]
- 11. Tinel N., Diochot S., Borsotto M., Lazdunski M., Barhanin J. (2000) KCNE2 confers background current characteristics to the cardiac KCNQ1 potassium channel. EMBO J. 19, 6326–6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dedek K., Waldegger S. (2001) Colocalization of KCNQ1/KCNE channel subunits in the mouse gastrointestinal tract. Pflügers Arch. 442, 896–902 [DOI] [PubMed] [Google Scholar]
- 13. Preston P., Wartosch L., Gunzel D., Fromm M., Kongsuphol P., Ousingsawat J., Kunzelmann K., Barhanin J., Warth R., Jentsch T. J. (2010) Disruption of the K+ channel beta-subunit KCNE3 reveals an important role in intestinal and tracheal Cl- transport. J. Biol. Chem. 285, 7165–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vallon V., Grahammer F., Volkl H., Sandu C. D., Richter K., Rexhepaj R., Gerlach U., Rong Q., Pfeifer K., Lang F. (2005) KCNQ1-dependent transport in renal and gastrointestinal epithelia. Proc. Natl. Acad. Sci. U. S. A. 102, 17864–17869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Okada Y., Ueda S. (1984) Electrical membrane responses to secretagogues in parietal cells of the rat gastric mucosa in culture. J. Physiol. 354, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heitzmann D., Grahammer F., von Hahn T., Schmitt-Graff A., Romeo E., Nitschke R., Gerlach U., Lang H. J., Verrey F., Barhanin J., Warth R. (2004) Heteromeric KCNE2/KCNQ1 potassium channels in the luminal membrane of gastric parietal cells. J. Physiol. 561, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jespersen T., Rasmussen H. B., Grunnet M., Jensen H. S., Angelo K., Dupuis D. S., Vogel L. K., Jorgensen N. K., Klaerke D. A., Olesen S. P. (2004) Basolateral localisation of KCNQ1 potassium channels in MDCK cells: molecular identification of an N-terminal targeting motif. J. Cell Sci. 117, 4517–4526 [DOI] [PubMed] [Google Scholar]
- 18. Roepke T. K., King E. C., Reyna-Neyra A., Paroder M., Purtell K., Koba W., Fine E., Lerner D. J., Carrasco N., Abbott G. W. (2009) Kcne2 deletion uncovers its crucial role in thyroid hormone biosynthesis. Nat. Med. 15, 1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 [DOI] [PubMed] [Google Scholar]
- 20. Spandidos A., Wang X., Wang H., Dragnev S., Thurber T., Seed B. (2008) A comprehensive collection of experimentally validated primers for polymerase chain reaction quantitation of murine transcript abundance. BMC Genomics 9, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roepke T. K., Kontogeorgis A., Ovanez C., Xu X., Young J. B., Purtell K., Goldstein P. A., Christini D. J., Peters N. S., Akar F. G., Gutstein D. E., Lerner D. J., Abbott G. W. (2008) Targeted deletion of kcne2 impairs ventricular repolarization via disruption of I(K,slow1) and I(to,f). FASEB J. 22, 3648–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDaniel N., Pace A. J., Spiegel S., Engelhardt R., Koller B. H., Seidler U., Lytle C. (2005) Role of Na-K-2Cl cotransporter-1 in gastric secretion of nonacidic fluid and pepsinogen. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G550–G560 [DOI] [PubMed] [Google Scholar]
- 23. Courtois-Coutry N., Roush D., Rajendran V., McCarthy J. B., Geibel J., Kashgarian M., Caplan M. J. (1997) A tyrosine-based signal targets H/K-ATPase to a regulated compartment and is required for the cessation of gastric acid secretion. Cell 90, 501–510 [DOI] [PubMed] [Google Scholar]
- 24. Kaufhold M.-A., Krabbenhoft A., Song P., Engelhardt R., Riederer B., Fahrmann M., Klocker N., Biel W., Manns M., Hagen S. J., Seidler U. (2008) Localization, trafficking, and significance for acid secretion of parietal cell Kir4.1 and KCNQ1 K+ channels. Gastroenterology 134, 1058–1069 [DOI] [PubMed] [Google Scholar]
- 25. Matter K., Hunziker W., Mellman I. (1992) Basolateral sorting of LDL receptor in MDCK cells: the cytoplasmic domain contains two tyrosine-dependent targeting determinants. Cell 71, 741–753 [DOI] [PubMed] [Google Scholar]
- 26. Odorizzi G., Pearse A., Domingo D., Trowbridge I. S., Hopkins C. R. (1996) Apical and basolateral endosomes of MDCK cells are interconnected and contain a polarized sorting mechanism. J. Cell Biol. 135, 139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duffield A., Folsch H., Mellman I., Caplan M. J. (2004) Sorting of H,K-ATPase beta-subunit in MDCK and LLC-PK cells is independent of mu 1B adaptin expression. Traffic 5, 449–461 [DOI] [PubMed] [Google Scholar]
- 28. Porcello D. M., Ho C. S., Joho R. H., Huguenard J. R. (2002) Resilient RTN fast spiking in Kv3.1 null mice suggests redundancy in the action potential repolarization mechanism. J. Neurophysiol. 87, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 29. Heitzmann D., Warth R. (2008) Physiology and pathophysiology of potassium channels in gastrointestinal epithelia. Physiol. Rev. 88, 1119–1182 [DOI] [PubMed] [Google Scholar]
- 30. Clancy S. M., Chen B., Bertaso F., Mamet J., Jegla T. (2009) KCNE1 and KCNE3 beta-subunits regulate membrane surface expression of Kv12.2 K(+) channels in vitro and form a tripartite complex in vivo. PLoS ONE 4, e6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roepke T. K., Purtell K., King E. C., La Perle K. M., Lerner D. J., Abbott G. W. (2010) Targeted deletion of Kcne2 causes gastritis cystica profunda and gastric neoplasia. PLoS ONE 5, e11451. [DOI] [PMC free article] [PubMed] [Google Scholar]