Abstract
Target of rapamycin (TOR) is a conserved regulator of gene expression from yeast to humans. In budding yeast, TOR is associated with ribosomal DNA (rDNA) promoter, which is critical for ribosome biogenesis and transfer RNA (tRNA) synthesis. Whether mTOR behaves similarly in mammalian cells is unknown. Here, we report that mTOR is detected at several different promoters in human and murine cells, including that of rDNA and tRNA genes. The association of mTOR with these promoters is regulated by growth signals and sensitive to rapamycin. Together, our observations suggest that mTOR is closely involved in gene regulation at the promoters, which is a conserved mechanism to control RNA polymerase I- and III-dependent genes that are critical for protein synthesis and cell growth.
Keywords: mTOR, rDNA, Pol I, Pol III, tRNA
Introduction
The protein synthetic capacity of cells depends on the abundance of ribosomes and transfer RNAs (tRNAs). Transcription of ribosomal RNAs (rRNAs) and tRNAs by RNA polymerases I and III (Pol I and Pol III) accounts for as much as 80% nuclear transcriptional activity and is under tight control by growth factors and nutrients through TOR pathway.1 Overexpression of the products of Pol I- and Pol III-transcribed genes is commonly observed in transformed cells2,3 and can promote oncogenic transformation.4
TOR is a highly conserved serine/threonine protein kinase found in all eukaryotic organisms. It is a central component of a signaling pathway that regulates cell growth and metabolism in response to nutrient availability, growth factors and cellular stresses.5 TOR is known to regulate Pol I- and Pol III-transcribed genes in yeast and mammals. In yeast, TOR concomitantly binds to 5S and 35S rDNA promoter chromatins in a nutrient-dependent and rapamycin-sensitive manner, which is important for regulating Pol I- and Pol III-dependent genes, including 35S and 5S rRNAs and tRNAs.6–8 The mechanism by which mTOR regulates gene expression is less well understood. However, mTOR has been shown to be localized in the nucleus. A recent study found that mTOR is associated with the promoters of several mitochondrial biogenesis genes.9 These observations prompted us to investigate whether mTOR binds to the promoters of genes that are known to be regulated by mTOR pathway. We found that mTOR is associated with the promoter regions of Pol I- and Pol III-transcribed genes, which is regulated by growth factors and nutrients, and inhibited by rapamycin. These observations suggest an evolutionarily conserved mechanism for the cell to control ribosome biogenesis in response to environmental signals.
Results
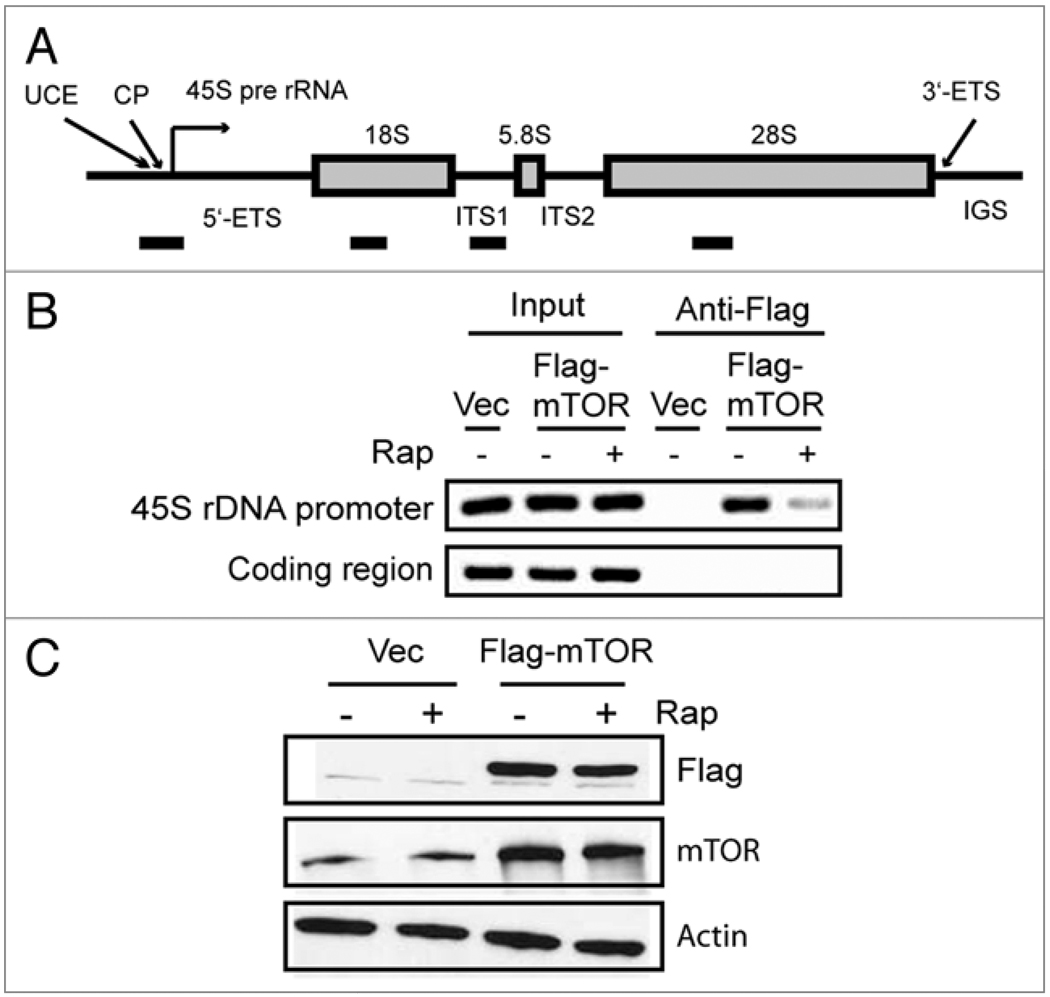
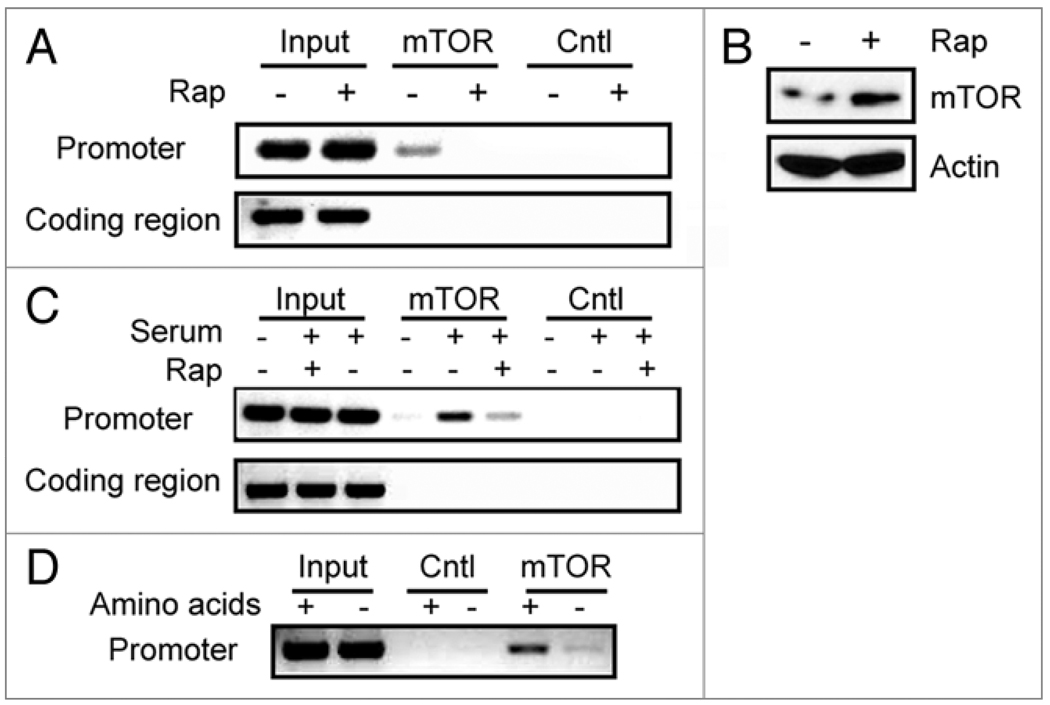
To facilitate the study of whether mTOR is associated with rDNA promoters in mammalian cells, we generated HEK293 cells stably expressed a Flag-tagged mTOR and preformed chromatin Immunoprecipitation (ChIP) assay using a Flag-specific antibody. We then analyzed the immunoprecipitated DNA with PCR primer pairs spanning the 45S rDNA promoter region (upstream control element and core promoter) and the coding region (28S) (Fig. 1A). In normal growth medium, Flag-mTOR was found to be associated with 45S rDNA promoter, but not the coding region (Fig. 1B). Rapamycin treatment substantially reduced the level of Flag-mTOR occupancy. Since rapamycin did not affect the protein levels of Flag-mTOR and endogenous mTOR (Fig. 1C), these results demonstrate that rapamycin causes the dissociation of mTOR from the rDNA promoter. To investigate whether the endogenous mTOR binds to rDNA promoter, we carried out ChIP assay in HEK293 cells using an mTOR-specific antibody.10 We detected the association of endogenous mTOR at 45S rDNA promoter, albeit at a lower level than Flag-mTOR, reflecting more abundant Flag-mTOR (Fig. 2A). With control IgG (Cntl), no signal was detected, demonstrating the specificity of the ChIP assay (Fig. 2A). Like Flag-mTOR, rapamycin blocked mTOR association with 45S rDNA promoter (Fig. 2A). Consistent with the earlier observation, rapamycin did not reduce endogenous mTOR protein level (Fig. 2B). Together, these results indicate that mTOR is specifically associated with 45S rDNA promoter in a rapamycin-sensitive manner.
Figure 1. Flag-tagged mTOR is associated with 45S rDNA promoter in HEK293 cells.
(A) Schematic representation of a mammalian rDNA gene. Primer pairs (solid bars) for the PCR-amplicons and their approximate positions are indicated. Abbreviations: UCE (upstream control element), CP (core promoter), 45S pre rRNA (45S precursor ribosomal RNA), ETS (external transcribed spacer), ITS (internal transcribed spacer), IGS (intergenic spacer). (B) Flag-mTOR binds to 45S rDNA promoter. HEK293 cells were stably transfected with empty vector (Vec) or vector carrying Flag-mTOR. Cells were treated with or without 20 nM rapamycin for 30 min. ChIP assay was conducted using a Flag antibody and PCR primer sets covering the 45S rDNA promoter and 28S coding regions. (C) Protein expression level of Flag-mTOR in the absence and presence of rapamycin. HEK293 cells were treated with rapamycin as described in (B) and subjected to western blot analysis. mTOR levels were determined by Flag and mTOR antibodies. Actin was used as a loading control.
Figure 2. Endogenous mTOR binds to 45S rDNA promoter.
(A) Endogenous mTOR is associated with 45S rDNA promoter in a rapamycin dependent manner. ChIP assay was performed in HEK293 as in Figure 1 except with an mTOR antibody. (B) mTOR protein level was not affected by rapamycin. (C) Growth factors promote mTOR association with 45S rDNA promoter in a rapamycin-sensitive manner. HEK293 cells were grown in serum-deprived medium for 24 hours, re-stimulated with serum in the absence or presence of 20 nM rapamycin. Binding of mTOR to 45S rDNA promoter and coding regions were determined by ChIP with an mTOR antibody. Rabbit IgG was used as a negative control (Cntl). (D) Amino acid starvation reduces the level of mTOR association with 45S rDNA promoter. HEK293 cells were starved with amino acids for 30 min and mTOR association with rDNA promoter was determined as (A).
Growth factors promote rDNA transcription through mTOR pathway.11 To determine whether growth factor regulates mTOR association with 45S rDNA promoter, HEK293 cells were starved from serum for 24 hours and then re-stimulated with serum in the absence or presence of rapamycin. As shown in Figure 2C, only little mTOR was detected at 45S rDNA promoter upon serum starvation. Addition of serum substantially increased mTOR binding to rDNA promoter (Fig. 2C). The presence of rapamycin blocked serum-induced mTOR association with 45S rDNA promoter (Fig. 2C). We further found that amino acid starvation inhibited the ability of mTOR to bind to 45S rDNA promoter (Fig. 2D). These results demonstrate that the recruitment of mTOR to 45S rDNA promoter is stimulated by growth factors and nutrients, which is inhibited by rapamycin.
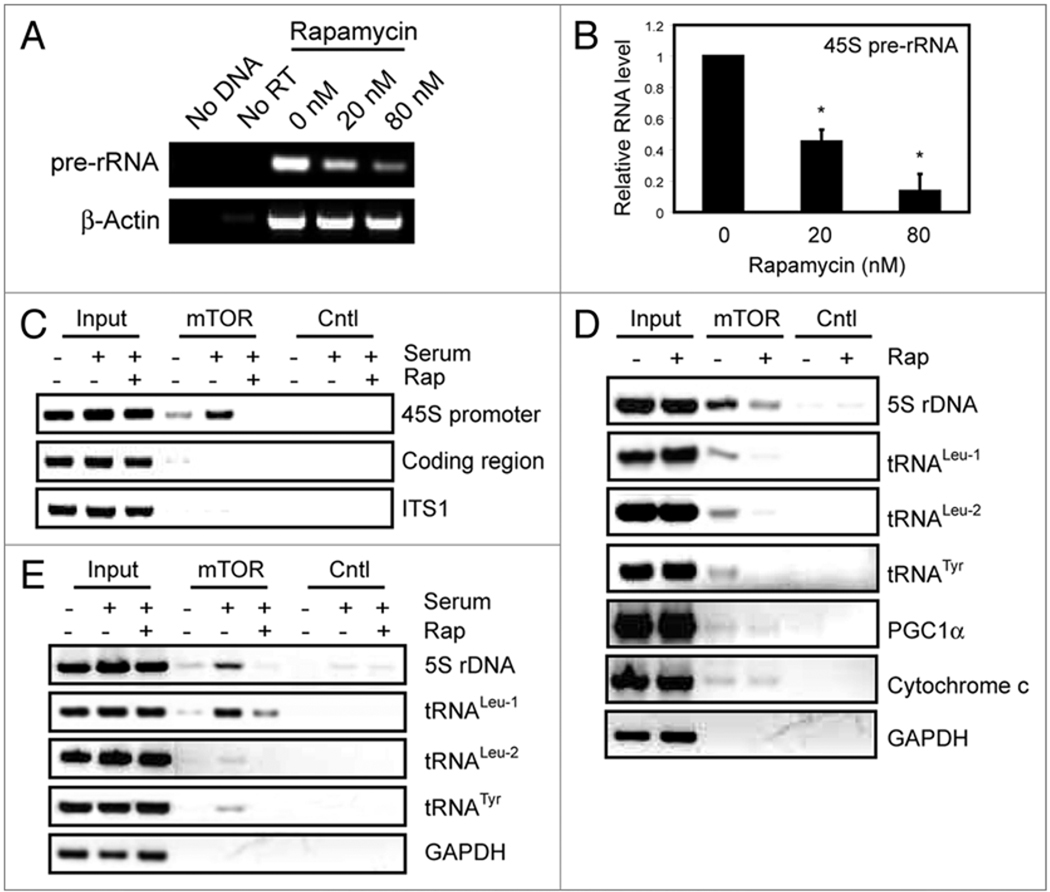
To confirm the above observation, we examined the DNA binding activity of mTOR in β-TC6 cell, a mouse pancreatic β cell line that exhibits sensitive nutrient-induced insulin secretion.12 In this cell line, the transcription of 45S rDNA is especially sensitive to rapamycin (Fig. 3A and B). In agreement with data from HEK293 cells, we found that serum promoted mTOR association with 45S rDNA promoter in a rapamycin-sensitive manner (Fig. 3C). mTOR was detected at the 45S rDNA promoter, but not the coding region and internal transcribed spacer (ITS1). In yeast TOR is associated with both Pol I- and Pol III-transcribed genes.6–8 Considering that mTOR regulates Pol III-transcribed genes,13 it is possible that mTOR also associates with these genes, including 5S rRNA and tRNA genes to coordinate ribosome biogenesis, protein synthesis and cell growth. To test this, we analyzed the presence of endogenous mTOR at different Pol III-transcribed genes. As shown in Figure 3D, mTOR was found to be associated with 5S rDNA, tRNALeu and tRNATyr genes. In contrast, mTOR was not detected at GAPDH. Similar to 45S rDNA, rapamycin treatment reduced the ability of mTOR to bind to Pol III-transcribed genes (Fig. 3D). In addition, serum promoted mTOR association with these Pol-III transcribed genes (Fig. 3E). A recent study showed that in skeletal muscle cells, mTOR binds to the promoters of PGC1α and cytochrome c.9 In line with this previous report, we detected the presence of mTOR at these promoters, which was insensitive to rapamycin treatment (Fig. 3D). Together, these results demonstrate that mTOR is associated with Pol I- and Pol-III transcribed genes in β-TC6 cells, which is regulated by growth signals in a rapamycin-sensitive manner.
Figure 3. mTOR binds to the promoters of Pol I- and Pol III-transcribed genes in β-TC6 cells.
(A) rDNA transcription is highly sensitive to rapamycin. β-TC6 cells were treated with rapamycin for 2 hours. The levels of 45S pre-rRNA and β-Actin mRNA were determined by RT-PCR. β-Actin was served as loading control. No reverse transcriptase treatment (No RT) and no DNA were used as negative controls. Primers that recognize the 5' ETS of pre-rRNA was used to measure the rate of rDNA transcription. (B) Quantification of the RT-PCR result. (C) mTOR is associated with Pol I-transcribed gene in a serum-dependent and rapamycin-sensitive manner. β-TC6 cells were grown in serum-deprived medium for 24 hours, re-stimulated with serum in the absence or presence of 20 nM rapamycin. Binding of mTOR to 45S rDNA promoter, coding regions and ITS1 were determined by ChIP with an mTOR antibody. Rabbit IgG was used as a negative control (Cntl). (D) Rapamycin inhibits mTOR association with Pol III-transcribed genes. β-TC6 cells were treated with or without 20 nM rapamycin and mTOR association with different genes was determined by ChIP assay. GAPDH was used as a negative control. (E) Growth factors promote mTOR association with Pol III-transcribed genes. β-TC6 cells were grown in serum-deprived medium for 24 hours, re-stimulated with serum in the absence or presence of 20 nM rapamycin. Binding of mTOR to the Pol III-transcribed genes were determined by ChIP. Rabbit IgG was used as a negative control (Cntl).
Discussion
In the present study, we show that mTOR is a chromatin-associated kinase that binds to the Pol I-transcribed 45S rDNA promoter and Pol III-transcribed genes including 5S rRNA and tRNAs under normal growth conditions in two mammalian cell lines, HEK293 and β-TC6. Inhibition of mTOR by rapamycin or starvation from growth factors and nutrients leads to its dissociation from these genes. In contrast, addition of serum to the starved cells promotes mTOR recruitment to these chromatin regions. We further demonstrated that the rapamycin-induced mTOR dissociation from the rDNA promoter is correlated with the reduction of rRNA synthesis, suggesting that the chromatin-binding activity of mTOR is critical for regulation of rDNA transcription. Such mTOR-chromatin binding, which is responsive to extra-cellular nutrient and growth factor signals, may provide a mechanism to rapidly respond to environmental changes. Our data presented here further indicate that the ability of mTOR binding to the Pol I- and Pol III-transcribed genes is evolutionarily conserved.
The molecular mechanism by which mTOR regulates Pol I- and Pol III-dependent transcription at the chromatin level is currently unknown. Recent studies in yeast have revealed that the chromatin-associated TOR may modulate the phosphorylation of factors responsible for regulation of Pol I and Pol III transcription machineries,8,14 and condensation of the rDNA repeats.15,16 Since the rDNA structural arrangement as well as the Pol I and Pol III machineries are highly conserved, it would be interesting to investigate whether mTOR also regulates these processes in mammalian cells. Transcription of 45S rDNA requires Pol I transcription initiation factors including TIF-IA, UBF and SL-1.17 Interestingly, it has been reported that mTOR regulates the phosphorylation and activation of TIF-IA and UBF.18,19 It will be interesting to determine whether chromatin-association is required for mTOR to regulate these transcription factors. With respect to Pol III-transcription, Maf1 has been shown to be a conserved Pol III repressor that integrates different growth signals to regulate Pol III activity.20–24 A recent study revealed that TOR interacts with Maf1 and phosphorylates Maf1 in yeast.8 In addition, TOR cooperates with Sch9, the yeast S6K1 homolog to provide optimal regulation of Maf1.25,26 It will also be of great interest to investigate whether mammalian Maf1 is a substrate for mTOR and/or S6K1.
Transcription by Pol I and Pol III is frequently hyper-activated in tumor cells, which can lead to uncontrolled growth. Thus understanding the regulatory mechanisms has important implications in cancer etiology and therapy. mTOR is currently viewed predominantly as a classic cytoplasmic kinase. The finding that mTOR is associated with chromatins provides a strong evidence that mTOR has important nuclear functions. Consistent with this view, a recent report has demonstrated that in skeletal muscle cells, mTOR binds to the promoters of the nuclear encoded mitochondrial genes, PGC-1α and cytochrome c, for the control of mitochondrial oxidative function through a YY1-PGC-1α transcription complex.9 Emerging evidence indicates that signaling kinases can have unorthodox roles in gene regulation. For example, certain kinases such as MAPK and PKA can serve as transcriptional activators and chromatin structure regulators that are responsible for recruitment of RNA polymerases to promoter sites.27–29 These observations, together with the present study, suggest that mTOR is closely involved in all three RNA polymerases through its association with the promoter regions of the target genes.
Materials and Methods
Cell culture, construct, transfection and antibodies
Mammalian cell lines used in this study were purchased from American Type Culture Collection (Manassas, VA) and maintained as recommended by the supplier. Flag-mTOR plasmid has been described previously.30 For stable transfections, HEK293 cells were transfected with Flag-mTOR plasmid or empty vector, and clones were selected with 300 µg/ml G418 (Invitrogen) and expanded as single colonies. The N-terminal mTOR antibody was described and characterized previously.10 Flag and Actin antibodies were purchased from Sigma-Aldrich and Cell Signaling Technologies, respectively.
ChIP assay
Chromatin immunoprecipitation assay was performed according to Upstate protocol. Sheared DNA fragments were immunoprecipitated with anti-Flag or anti-mTOR antibody. Sequences of primers used for PCR are as follows: human 45S rDNA promoter (upstream control element + core promoter), 5'-AGG AGC GCG GCC GGC TAG CC-3' and 5'-CCC TGC GTG TGG CAC GGG C-3'; human 28S coding region, 5'-GCG ACC TCA GAT CAG ACG TG-3' and 5'-CTT AAC GGT TTC ACG CCC TC-3'; mouse 45S rDNA promoter, 5'-GCG GTT TTC TTT CAT TGA CC-3' and 5'-AGG CTG GAC AAG CAA AAC AG-3'; mouse 18S coding region; 5'-TGC CAG GTA GCA TAT GCT TG-3' and 5'-CCG GGT TGG TTT TGA TCT GA-3'; mouse internal transcribed spacer, 5'-AAG GTT TCC GTA GGT GAA CC-3' and 5'-GAC GGG TCA AAA ACC CGT AA-3'; mouse 5S rDNA, 5'-GGC CAT ACC ACC CTG AAC GC-3' and 5'-CAG CAC CCG GTA TTC CCA GG-3'; mouse tRNALeu1 (chr. 7 Leu-TAG), 5'-CTG GAT TTA GGC TCC AGT CT-3' and 5'-CCT CCT ATC TTA GGG TCT GA-3'; tRNALeu-2 (ch. 14 Leu-TAG), 5'-TAA GGC GCT GGA TTT AGG CT-3' and 5'-GTG GTC ATG ACA CGT GTT TG-3'; tRNATyr (chr.3 Tyr-GTA), 5'-CCT TCG ATA GCT CAG CTG GT-3' and 5'-GTG GTA GTA CAC CCG TAC TC-3'; GAPDH, 5'-CAC CCA GAA GAC TGT GGA TG-3' and 5'-CTA TCC TTG TCC CAA GTC AC-3'. Primers for PGC1α and cytochrome c were used as described.9 PCR conditions for all primers were as follows: 94°C for 3 min, followed by 32–35 cycles at 94°C for 30 s, 55–58°C for 45 s and 72°C for 45 s.
Western blot analysis, RNA extraction and RT-PCR analysis
Western blot was done as previously described.10,31 Total RNA was extracted with RNA STAT-60 (TEL-TEST) according to manufacture’s instructions. RNA was reverse-transcribed to cDNA using a RETROscript Kit (Applied Biosystems) and then amplified by PCR as described in the manufacture’s instructions. The primer sequences and PCR conditions for pre-rRNA and β-Actin were used as described.32,33
Acknowledgements
This work was supported by NIH R01 CA123391 (X.S.Z.).
Abbreviations
- Pol I
RNA polymerase I
- mTOR
mammalian target of rapamycin
- rDNA
ribosomal DNA
- tRNA
transfer RNA
- ETS
external transcribed spacer
- ITS
internal transcribed spacer
- IGS
intergenic spacer
References
- 1.Warner J. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.White R. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol. 2005;6:69–78. doi: 10.1038/nrm1551. [DOI] [PubMed] [Google Scholar]
- 3.Johnson DL, Johnson SAS. CELL BIOLOGY: RNA Metabolism and Oncogenesis. Science. 2008;320:461–462. doi: 10.1126/science.1158680. [DOI] [PubMed] [Google Scholar]
- 4.Marshall L, Kenneth N, White R. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 5.Tsang C, Qi H, Liu L, Zheng X. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today. 2007;12:112–124. doi: 10.1016/j.drudis.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Tsang C, Watkins M, Bertram P, Zheng X. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature. 2006;442:1058–1061. doi: 10.1038/nature05020. [DOI] [PubMed] [Google Scholar]
- 7.Tsang C, Zheng X. TOR-in(g) the nucleus. Cell Cycle. 2007;6:25–29. doi: 10.4161/cc.6.1.3675. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y, Tsang C, Zheng XS. Mechanisms of regulation of RNA polymerase III-dependnt transcription by TORC1. EMBO J. 2009 doi: 10.1038/emboj.2009.179. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham J, Rodgers J, Arlow D, Vazquez F, Mootha V, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 10.Drenan RM, Liu X, Bertram PG, Zheng XFS. FKBP12-Rapamycin-associated Protein or Mammalian Target of Rapamycin (FRAP/mTOR) Localization in the Endoplasmic Reticulum and the Golgi Apparatus. J Biol Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- 11.James MJ, Zomerdijk JCBM. Phosphatidylinositol 3-Kinase and mTOR Signaling Pathways Regulate RNA Polymerase I Transcription in Response to IGF-1 and Nutrients. J Biol Chem. 2004;279:8911–8918. doi: 10.1074/jbc.M307735200. [DOI] [PubMed] [Google Scholar]
- 12.Poitout V, Stout LE, Armstrong MB, Walseth TF, Sorenson RL, Robertson RP. Morphological and functional characterization of beta TC-6 cells—an insulin-secreting cell line derived from transgenic mice. Diabetes. 1995;44:306–313. doi: 10.2337/diab.44.3.306. [DOI] [PubMed] [Google Scholar]
- 13.Woiwode A, Johnson SAS, Zhong S, Zhang C, Roeder RG, Teichmann M, et al. PTEN Represses RNA Polymerase III-Dependent Transcription by Targeting the TFIIIB Complex. Mol Cell Biol. 2008;28:4204–4214. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claypool JA, French SL, Johzuka K, Eliason K, Vu L, Dodd JA, et al. Tor Pathway Regulates Rrn3p-dependent Recruitment of Yeast RNA Polymerase I to the Promoter but Does Not Participate in Alteration of the Number of Active Genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang C, Bertram P, Ai W, Drenan R, Zheng X. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003;22:6045–6056. doi: 10.1093/emboj/cdg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsang C, Li H, Zheng X. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007;26:448–458. doi: 10.1038/sj.emboj.7601488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsang CK, Zheng XFS. Control of ribosome bio-synthesis by the target of rapamycin (TOR) Recent Research Developments in Molecular and Cellular Biology. 2004;5:135–147. [Google Scholar]
- 18.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, et al. mTOR-Dependent Regulation of Ribosomal Gene Transcription Requires S6K1 and Is Mediated by Phosphorylation of the Carboxy-Terminal Activation Domain of the Nucleolar Transcription Factor UBF{dagger} Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson S, Zhang C, Fromm J, Willis I, Johnson D. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26:367–379. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Goodfellow S, Graham E, Kantidakis T, Marshall L, Coppins B, Oficjalska-Pham D, et al. Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J Mol Biol. 2008;378:481–491. doi: 10.1016/j.jmb.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 22.Boguta M, Czerska K, Zoladek T. Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene. 1997;185:291–296. doi: 10.1016/s0378-1119(96)00669-5. [DOI] [PubMed] [Google Scholar]
- 23.Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, et al. Maf1p, a Negative Effector of RNA Polymerase III in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:5031–5040. doi: 10.1128/MCB.21.15.5031-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis I, Moir R. Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci. 2007;32:51–53. doi: 10.1016/j.tibs.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Wei Y, Zheng X. TORC1 association with rDNA chromatin as a mechanism to co-regulate Pol I and Pol III. Cell Cycle. 2009;8:3802–3803. doi: 10.4161/cc.8.23.10039. [DOI] [PubMed] [Google Scholar]
- 26.Wei Y, Zheng X. Sch9 partially mediates TORC1 signaling to control ribosomal RNA synthesis. Cell Cycle. 2009;8 doi: 10.4161/cc.8.24.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow C, Davis R. Proteins kinases: chromatin-associated enzymes? Cell. 2006;127:887–890. doi: 10.1016/j.cell.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Edmunds JW, Mahadevan LC. CELL SIGNALING: Protein Kinases Seek Close Encounters with Active Genes. Science. 2006;313:449–451. doi: 10.1126/science.1131158. [DOI] [PubMed] [Google Scholar]
- 29.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated Signal Transduction Kinases Frequently Occupy Target Genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 30.Choi J, Bertram P, Drenan R, Carvalho J, Zhou H, Zheng X. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 2002;3:988–994. doi: 10.1093/embo-reports/kvf197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Zheng XFS. Endoplasmic Reticulum and Golgi Localization Sequences for Mammalian Target of Rapamycin. Mol Biol Cell. 2007;18:1073–1082. doi: 10.1091/mbc.E06-05-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Li Z, Haruna K, Li Z, Li Z, Semba K, et al. Early pre-implantation lethality in mice carrying truncated mutation in the RNA polymerase 1–2 gene. Biochem Biophys Rese Commun. 2008;365:636–642. doi: 10.1016/j.bbrc.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 33.Heit JJ, Apelqvist AA, Gu X, Winslow MM, Neilson JR, Crabtree GR, et al. Calcineurin/NFAT signaling regulates pancreatic beta-cell growth and function. Nature. 2006;443:345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]