Abstract
Cell-surface protein CD10 is a prognostic marker for diffuse large B-cell lymphoma (DLBCL), where high expression of CD10 is found in the germinal center B-cell (GCB) subtype and CD10 expression is low or absent in the activated B-cell (ABC) subtype. As compared with the GCB subtype, patients with ABC DLBCL have a poorer prognosis after standard treatment, and ABC tumor cells have higher NF-κB activity. Herein, we show that increased expression of the NF-κB target micro-RNA miR-155 is correlated with reduced expression of transcription factor PU.1 and CD10 in several B-lymphoma cell lines. Moreover, electromobility shift assays and luciferase reporter assays indicate that PU.1 can directly activate expression from the CD10 promoter. Expression of a DLBCL-derived mutant of the adaptor CARD11 (a constitutive activator of NF-κB) in the GCB-like human BJAB cell line or v-Rel in the chicken DT40 B-lymphoma cell line causes reduced expression of PU.1. The CARD11 mutant also causes a decrease in CD10 levels in BJAB cells. Similarly, overexpression of miR-155, which is known to down-regulate PU.1, leads to reduced expression of CD10 in BJAB cells. Finally, we show that CD10 expression is reduced in BJAB cells after treatment with the NF-κB inducer lipopolysaccharide (LPS). Additionally, miR-155 is induced by LPS treatment or expression of the CARD11 mutant in BJAB cells. These results point to an NF-κB-dependent mechanism for down-regulation of CD10 in B-cell lymphoma: namely, that increased NF-κB activity leads to increased miR-155, which results in decreased PU.1, and consequently reduced CD10 mRNA and protein.
Keywords: Gene Expression, Micro-RNA, NF-κB Transcription Factor, NF-κB, Signal Transduction, B-cell Lymphoma, CD10, DLBCL, PU.1, miR-155
Introduction
The NF-κB signaling pathway is misregulated in a variety of human diseases including many chronic inflammatory diseases and cancers. As such, an understanding of the molecular details of NF-κB-dependent gene networks has implications for improved disease diagnoses and therapies.
CD10, also known as the common acute lymphocytic leukemia antigen (CALLA) or neutral endopeptidase, is a cell-surface zinc metalloendopeptidase (1, 2). The ability of CD10 to cleave signal peptides at the cell surface can affect cell proliferation, differentiation, and migration (2–4). Expression of CD10 can be used as a diagnostic marker for a variety of cancers (5–9). Relevant to this study, CD10 is highly expressed in the germinal center B-cell (GCB)6 molecular subtype of diffuse large B-cell lymphoma (DLBCL), whereas CD10 expression is low in the activated B-cell (ABC) subtype of DLBCL (8, 9). ABC DLBCLs also have a high NF-κB gene expression profile and a poorer clinical prognosis as compared with GCB DLBCLs (10). As such, reduced expression of CD10 and high NF-κB activity are both correlated with a less favorable DLBCL patient outcome (10–12).
The correlation between high NF-κB activity and reduced CD10 expression has been observed in several other settings as well. We previously showed that overexpression of an activated mutant of the NF-κB family transcription factor REL in the GCB-like B-lymphoma cell line BJAB leads to reduced expression of CD10 (13). Infection of cells with Epstein-Barr virus (EBV) or human cytomegalovirus, both inducers of NF-κB, causes reduced expression of CD10 (14, 15). Taken together, such results suggested to us that NF-κB or a target of NF-κB is involved in repressing CD10 gene/protein expression in certain B-cell lymphomas.
Little is known about the control of CD10 transcription. Sequence analysis of the CD10 promoter/enhancer region revealed the presence of three consensus binding sites for transcription factor PU.1 (16). PU.1 is a member of the Ets family of transcription factors and is required for proper B-cell development and differentiation (17). Additionally, increased PU.1 expression has been correlated with the GCB subtype of DLBCL (8). Therefore, we were interested in investigating whether PU.1 contributes to the regulation of CD10 expression in B-cell lymphoma.
In this report, we provide evidence for a functional link between activation of NF-κB and reduced expression of CD10. We show that activation of NF-κB in the GCB-like DLBCL cell line BJAB leads to reduced CD10 expression. Our data are consistent with a pathway in which NF-κB-induced up-regulation of micro-RNA miR-155 leads to down-regulation of PU.1 protein levels, and consequently, reduced levels of CD10. Moreover, we show that there is a correlation between high miR-155 expression and low PU.1/CD10 levels in a variety of B-lymphoma cell lines. These results are significant in that they reveal a molecular mechanism for down-regulation of CD10, a key marker for predicting the clinical response of patients with DLBCL.
EXPERIMENTAL PROCEDURES
Cell Culture, DNA Constructs, and Stable Cell Line Generation
Human A293, A293T, Bosc23, BJAB, SUDHL-4, BL41, Daudi, IB4, RC-K8, and chicken DT40 cells (gift of Céline Gélinas) (18), were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with either 10 or 20% heat-inactivated fetal bovine serum (FBS) (Biologos) as described (19). The six human B-lymphoma cell lines used in this study are as follows: BJAB, EBV-negative, GCB-like DLBCL (20, 21); SUDHL-4, a B-lymphoma that has been characterized as both a follicular lymophoma (22) and GCB-like DLBCL (23); BL-41, EBV-negative Burkitt lymphoma (14); Daudi, EBV-positive, LMP1-negative Burkitt lymphoma (24); IB4, EBV-transformed lymphoblastoid cell line (14); and RC-K8, ABC-like DLBCL (25). For treatment of cells with lipopolysaccharide (LPS) (Sigma), BJAB cells were first cultured in DMEM containing 5% FBS for 24 h and then treated with 5 μg/ml of LPS for the indicated times.
All primers used in the study were purchased from Invitrogen and are described under supplemental Table S1. pcDNA-PU.1 was a gift of Barbara Nikolajczyk (Boston University Medical School) and has been described previously (26). To create pCD10-luc, a 717-bp region of the CD10 promoter, containing the three consensus PU.1 binding sites, was amplified by PCR from BJAB cell genomic DNA. The PCR product was digested with KpnI and XmaI, and this fragment was subcloned into the pGL3 promoter vector (Promega). For mutagenesis of the PU-1 site in the CD10 promoter, two overlapping primers were used that contained the desired mutations, as shown under supplemental Table S1.
Transfections were performed using polyethylenimine (Polysciences, Inc.). On the day of transfection, cells were washed twice with TD buffer (25 mm Tris-HCl, pH 7.4, 137 mm NaCl, 5 mm KCl, 0.7 mm Na2HPO4) and were then incubated with a plasmid DNA:polyethylenimine ratio of 1:3 in 300 μl of DMEM for 15 min at room temperature. For transfection of Bosc23 cells with pSIREN and pVpack vectors, a plasmid DNA:polyethylenimine ratio of 2:3 in 300 μl of DMEM was used. The DNA mixture was added to 2 ml (35-mm plate) or 4.5 ml (60-mm plate) of DMEM containing 10% FBS and this mixture was then added to the cells. The next day, the medium was replaced with fresh DMEM containing 10% FBS. For reporter gene assays, cells were lysed 24 h later.
The retroviral vector pMSCV has been described previously (27). pMSCV-CARD11mut10 was created by subcloning a BamHI fragment containing the CARD11 mutant 10 cDNA (a gift of Georg Lenz, National Cancer Institute) into BglII-digested pMSCV. pMSCV-BIC was created by subcloning a BsrGI-EcoRI fragment from the BIC cDNA (a gift of Ricardo Aguiar, The University of Texas Health Science Center at San Antonio) into BsrGI-EcoRI-digested pMSCV. For overexpression of shRNAs, the retroviral vector pSIREN-RetroQ was used (a gift of Ulla Hansen, Boston University). The shRNA against HMGN1 (28) was excised from this vector and a BamHI-EcoRI fragment from the multiple cloning site of pSL1180 (Amersham Biosciences) was subcloned in to create pSIREN-MCS. All shRNAs (supplemental Table S1) were ligated into pSIREN-MCS using the BamHI-EcoRI sites. The shRNA against PU.1 (29) and the control shRNA (30) have been previously described (also see supplemental Table S1). The retroviral, doxycycline-inducible system has been previously described (31). pRetroX-tight-CARD11mut10HA-Hyg was created by subcloning a BamHI/NotI fragment containing CARD11mut10 from the pMSCV-CARD11mut10 vector described above.
Virus stocks were generated by transfecting A293T cells with pMSCV, pMSCV-CARD11mut10, pMSCV-BIC, pRetroX-tight-Hyg, or pRetroX-tight-CARD11mut10HA-Hyg plus helper plasmid pcL10a1, as described previously (13, 27). Virus stocks for pSIREN-RetroQ vectors were generated in Bosc23 cells using helper vectors pVpack-GP and pVpack-VSV (28). Two days later, virus was harvested. Two ml of virus (in the presence of 8 μg/ml of Polybrene) was used to infect 106 BJAB cells using the spin infection method (13). Two days later, cells were selected with 2.5 μg/ml of puromycin (Sigma) or 400 μg/ml of hygromycin (Sigma), with fluid changing every 5 days in media containing puromycin or hygromycin for 2–4 weeks.
Semi-quantitative Reverse Transcriptase PCR (RT-PCR) and Real Time PCR
Total RNA was isolated using TRIzol (Invitrogen). Semi-quantitative RT-PCR was performed as described previously (32) with the primer sets listed under supplemental Table S1. Semi-quantitative RT-PCR for miR-155 and 5S ribosomal RNAs was performed by first reverse transcribing the RNA using specific stem loop primers, which contain an annealing site for the universal reverse primer (supplemental Table S1) and the TaqMan miRNA Reverse Transcriptase kit (Applied Biosystems) as described previously (33). miR-155 and 5S sequences were then amplified using a forward-specific primer and a universal reverse primer (supplemental Table S1). Real time PCR was performed using the 7900HT Fast real time PCR system (Applied Biosystems) with the Power SYBR Green PCR Master Mix (Applied Biosystems). Quantification of miR-155 RNA by real time PCR was performed three times with triplicate samples and values were normalized to the 5S values. The final values represent relative expression (miR-155/5S) as compared with control samples.
Western Blotting
Western blotting was performed as described (25). Whole cell extracts were prepared in AT buffer (20 mm HEPES, pH 7.9, 1 mm EDTA, 1 mm EGTA, 20 mm Na4P2O7, 1 mm DTT, 1% (v/v) Triton X-100, 20% (w/v) glycerol, 1 mm Na3VO4, 1 μg/ml of PMSF, 1 μg/ml of leupeptin, 1 μg/ml of pepstatin). Samples were boiled in SDS sample buffer (62.5 mm Tris-HCl, pH 6.8, 3.2% (w/v) SDS, 10% (w/v) glycerol, 5% (v/v) β-mercaptoethanol, 0.1% (w/v) bromphenol blue). Samples containing equal amounts of protein were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Micron Separation Inc.). Antibodies against PU.1 (sc-352), CD10 (sc-58939), HA (sc-805), and β-tubulin (sc-9104) were purchased from Santa Cruz Biotechnology. The BCL2 antibody was obtained from BD Transduction Laboratories (number 610538). Anti-REL antiserum (number 265) was a kind gift of Nancy Rice, and antiserum against v-Rel was described previously (27). Nitrocellulose filters were incubated with primary antiserum for 2–18 h at room temperature or 4 °C. The appropriate horseradish peroxidase-labeled secondary antiserum was added and immunoreactive proteins were detected with the SuperSignal Dura West Extended Duration Substrate chemiluminescence detection system (Pierce).
Electromobility Shift Assays (EMSAs)
EMSAs were performed using 10 μg of nuclear extract prepared from A293 cells, A293 cells transfected with pcDNA-PU.1, and BJAB cells stably transduced with MSCV, MSCV-CARD11mut10, or MSCV-RELΔTAD1 as described previously (13, 25). Nuclear extracts were incubated with 2 μg of poly(dI-dC), and 32P-labeled PU.1 or NF-κB site probes (supplemental Table S1) in binding buffer (25 mm Tris-HCl, pH 7.4, 100 mm KCl, 0.5 mm EDTA, 6.25 mm MgCl2, 0.5 mm DTT, 10% (w/v) glycerol) in a final reaction volume of 50 μl, as previously described (25). DNA-binding reactions were carried out for 30 min at room temperature. For supershifts, 3 μl of anti-PU.1 antibody (sc-352X, Santa Cruz Biotechnology) was incubated with the protein-DNA complexes for an additional 1 h on ice. Samples were resolved on 5% nondenaturing polyacrylamide gels. Gels were dried and protein-DNA complexes were detected by autoradiography.
Reporter Gene Assays
A293 cells in 35-mm plates were transfected with 1 μg of a pGL3-based CD10 promoter luciferase reporter plasmid (pCD10-luc or pCD10mutPU-1-luc), 0.5 μg of normalization plasmid pRSV-βgal, and 0.5 μg of pcDNA or pcDNA-PU.1 using polyethylenimine, as described above. Luciferase activity was measured using the Luciferase Assay System according to the manufacturer's instructions (Promega). Luciferase values were normalized to β-galactosidase values in all assays, as described previously (25).
RESULTS
PU.1 and CD10 Protein Levels Are Inversely Correlated with miR-155 Levels in Several B-lymphoma Cell Lines
CD10 mRNA and protein levels are low in the less favorable ABC subtype of DLBCL (8, 9, 11, 12), which also has increased NF-κB activity and increased levels of the NF-κB target gene product miR-155 (23, 34–38), a micro-RNA that is processed from a non-coding RNA known as BIC (B-cell integration cluster gene). In contrast, GCB-like DLBCL primary tumors and cell lines express readily detectable levels of CD10 mRNA and protein, but have lower NF-κB activity and lower levels of miR-155 (13, 36, 37). One direct target of miR-155 repression is the mRNA encoding transcription factor PU.1 (39, 40).
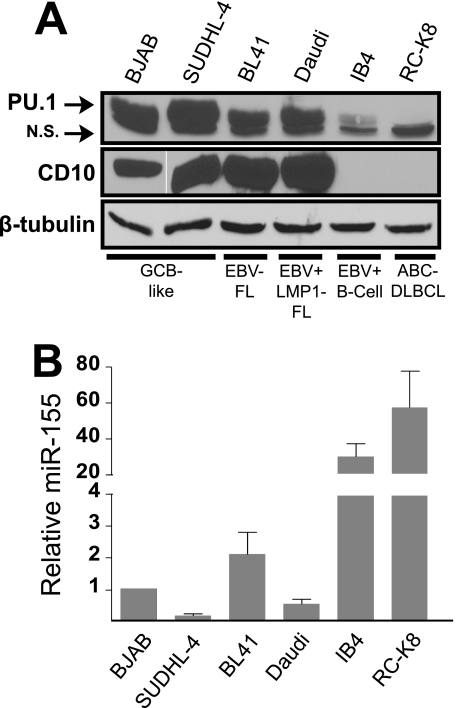
Based on the above findings, we analyzed the levels of PU.1, CD10, and miR-155 among a panel of six B-lymphoma-like cell lines with varying levels of NF-κB-dependent gene expression. Two cell lines, IB4 (14) and RC-K8 (25), have gene expression profiles consistent with high levels of NF-κB activity; four cell lines, BJAB (13, 21), SUDHL-4 (23), BL41 (41), and Daudi, are not known to have high NF-κB-driven gene expression. PU.1 and CD10 proteins were expressed at readily detectable levels in the four cell lines with low NF-κB activity, whereas PU.1 and CD10 protein levels were extremely low in the high NF-κB activity IB4 and RC-K8 cell lines (Fig. 1A). Moreover, as judged by qPCR, miR-155 was present at low levels in the four lymphoma cell lines with low NF-κB activity (Fig. 1B). In contrast, miR-155 was present at increased levels in the high NF-κB activity IB4 and RC-K8 cell lines, which had miR-155 levels ∼30–50 times higher than BJAB cells (Fig. 1B). These results indicate that PU.1 and CD10 levels are low in transformed B-lymphoid cell lines where miR-155 levels and NF-κB activity are high (e.g. IB4 and RC-K8).
FIGURE 1.
Elevated miR-155 expression correlates with reduced PU.1 and CD10 expression levels in several B-lymphoma cell lines. A, whole cell extracts were prepared from six transformed B-lymphoid cell lines (BJAB, SUDHL-4, BL41, Daudi, IB4, and RC-K8). Details of the lymphoma cell lines are indicated as follows: GCB-like, germinal center B-cell diffuse large B-cell lymphoma; EBV− FL, Epstein-Barr virus-negative follicular lymphoma; EBV+ LMP1− FL, Epstein-Barr virus-positive, latent membrane protein 1-negative follicular lymphoma; EBV+ B-cell, Epstein-Barr virus-transformed B-lymphoblastoid cell line; and ABC DLBCL, activated B-cell diffuse large B-cell lymphoma. Western blotting was performed for PU.1, CD10, and β-tubulin. B, miR-155 levels were quantified using real time qPCR. The levels of miR-155 in each cell line are relative to expression in BJAB cells (1.0). Error bars indicate S.E. N.S., nonspecific band.
PU.1 Binds to a Site in the CD10 Promoter to Increase Transcription
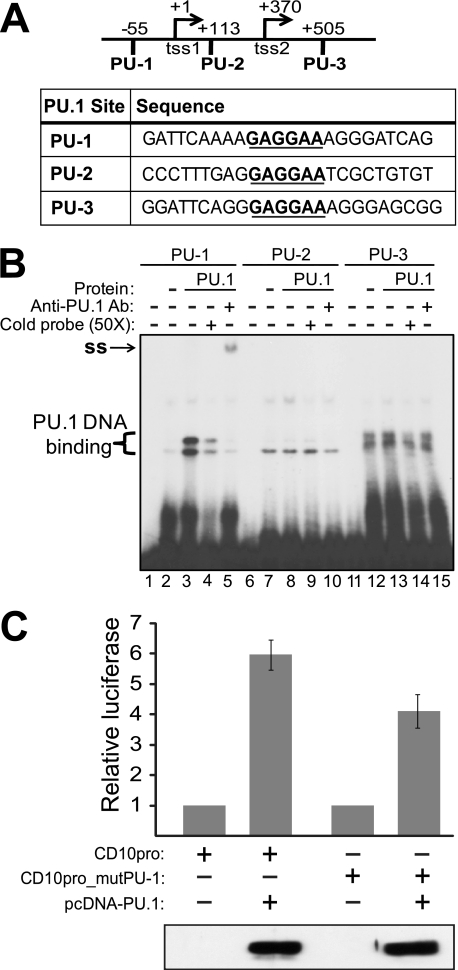
Based on this direct correlation between PU.1 and CD10 protein levels in six B-lymphoma-like cell lines and the presence of three consensus PU.1 binding sites (GAGGAA) in the CD10 upstream region (16) (Fig. 2A), we next sought to determine whether the CD10 gene might be a direct transcriptional target for PU.1. We first assessed the ability of PU.1 to bind to the CD10 promoter by performing EMSAs using DNA probes containing each of the three putative PU.1 binding sites (PU-1, PU-2, and PU-3). Nuclear extracts were prepared from control A293 cells (Fig. 2B, lanes 2, 7, and 12) or A293 cells overexpressing PU.1 (Fig. 2B, lanes 3, 8, and 13), and these extracts were incubated with each of the three PU.1 site probes (PU-1, PU-2, or PU-3). We found that extracts from cells overexpressing PU.1 could specifically bind to the first PU.1 site (PU-1), whereas these extracts showed no binding to the PU-2 or PU-3 probes above the background binding seen with control A293 extracts (Fig. 2B). Additionally, binding to the PU-1 site was competed by an excess of cold probe (Fig. 2B, lane 4), and the PU-1 site protein-DNA complex was supershifted by a PU.1-specific antibody (Fig. 2B, lane 5).
FIGURE 2.
PU.1 binds to a site in the CD10 promoter/enhancer region, which is important for PU.1-induced transactvation of the CD10 promoter. A, the CD10 promoter region has five transcription start sites; the two that are most 5′ are indicated in the figure (tss1 and tss2) (see also Ref. 16). Shown are the three putative PU.1 binding sites (PU-1, PU-2, and PU-3) and the sequences used for EMSA analysis. The PU.1 core sequence is indicated with bold underline. B, nuclear extracts (Protein) from non-transfected A293 cells (−) or A293 cells overexpressing PU.1 (PU.1) were incubated with 32P-labeled DNA probes (PU-1, PU-2, and PU-3). Lanes 1, 6, and 11 contain probe alone (no extract). Where indicated (lanes 4, 9, and 14), a 50-fold excess of cold probe was used to compete the radiolabeled probe. Where indicated (lanes 5, 10, and 15), a PU.1-specific antibody was included to supershift (ss) the protein-DNA complex. C, A293 cells were transfected with pCD10-luc or pCD10mut-luc (with the PU-1 site mutated) and either vector alone (pcDNA) or pcDNA-PU.1; a plasmid encoding β-galactosidase was also included for transfection normalization. Cells were harvested and both luciferase activity and β-galactosidase activity were measured. Relative luciferase activity is indicated ± S.E. Averages are from three assays performed with triplicate samples. Shown below the graph is a Western blot for PU.1, which was performed on representative samples from a single transfection.
To assess the ability of PU.1 to activate transcription from the CD10 promoter, we placed a portion of the CD10 promoter region containing the three consensus PU.1 binding sites upstream of a luciferase cassette (pCD10-luc). A293 cells were co-transfected with pCD10-luc and an expression plasmid for PU.1 or a vector control, and we then measured the relative luciferase activity. Co-transfection of the PU.1 expression plasmid resulted in an ∼6-fold increase in expression from pCD10-luc as compared with the vector control (Fig. 2C). Mutation of the first PU.1 site (PU-1) in the CD10 promoter reduced the ability of PU.1 to transactivate the CD10 reporter plasmid by ∼35% (Fig. 2C). Western blotting confirmed that PU.1 was overexpressed in cells transfected with the pcDNA-PU.1 expression plasmid (Fig. 2C).
Expression of a Constitutively Active Form of CARD11 in BJAB Cells Causes Down-regulation of CD10 and PU.1 Protein Expression and Up-regulation of miR-155
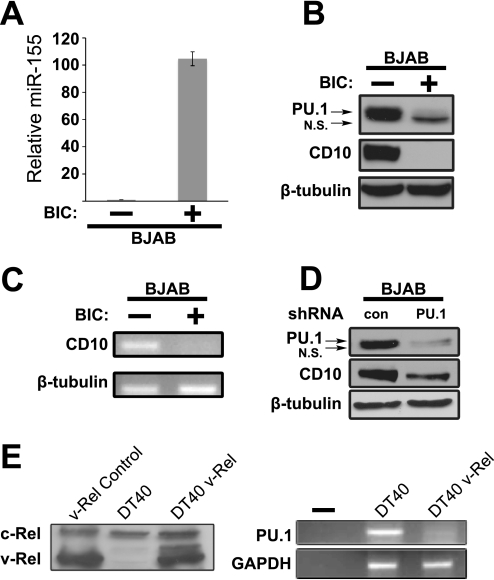
Mutations in the scaffolding protein CARD11, which cause it to be a constitutive activator of NF-κB, have been identified in a subset of patients with DLBCL, primarily of the ABC subtype (42). To determine whether expression of one such CARD11 mutant (mut10) (see Ref. 42) could affect CD10 expression, we used retroviral transduction to create a stable BJAB cell line expressing HA-tagged CARD11mut10. Western blotting confirmed that HA-CARD11mut10 was expressed in retrovirally transduced BJAB cells, but no such protein was detected in the parallel control cells (Fig. 3A). CD10 and PU.1 protein levels were both decreased in BJAB cells expressing HA-CARD11mut10 as compared with non-transduced BJAB or MSCV vector-transduced cells (Fig. 3A). Consistent with the ability of CARD11mut10 to activate NF-κB (42), BCL2, the product of an NF-κB target gene and an ABC-like marker, showed increased protein expression in BJAB-CARD11mut10 cells (Fig. 3A). Finally, miR-155 levels were increased by ∼6-fold in BJAB-CARD11mut10 cells as compared with BJAB or BJAB-MSCV control cells (Fig. 3A). To further demonstrate that CARD11mut10 protein expression could lead to reduced CD10 expression, we created an inducible expression system for CARD11mut10 protein (using a tet-ON retroviral transduction system (31)) in BJAB cells. Induction of CARD11mut10 expression for 7 days led to increased miR-155 and reduced CD10 protein (Fig. 3B).
FIGURE 3.
Expression of the B-lymphoma CARD11mut10 protein can decrease the expression of PU.1 and CD10 in BJAB cells. A, anti-HA, anti-CD10, anti-PU.1, anti-BCL2, and anti-β-tubulin Western blotting of extracts from non-transduced BJAB cells (−) or BJAB cells stably transduced with MSCV or MSCV-HA-CARD11mut10. miR-155 was quantified from the indicated cell lines using real time PCR as described in the legend to Fig. 1B. B, BJAB cells were transduced with a doxycycline-inducible retroviral vector for HA-CARD11mut10. In the absence (−) or presence (+) of doxycycline (Dox), the levels of HA-CARD11mut10 and CD10 were analyzed by Western blotting and the levels of miR-155 by qPCR. Non-transduced BJAB cells served as a negative control. C, nuclear extracts from BJAB-MSCV control cells or BJAB cells expressing HA-CARD11mut10 were incubated with 32P-labeled DNA probes (PU-1, top; NF-κB, bottom). Lane 1 in each case contains probe alone (no extract). As a size control for PU-1 binding, an extract from A293 cells overexpressing PU.1 was included (top panel, last lane). As an NF-κB DNA binding control, a nuclear extract from BJAB cells expressing RELΔTAD1 (13) was included (bottom panel, last lane).
We next compared nuclear DNA-binding activity for PU.1 and NF-κB in BJAB-MSCV versus BJAB-CARD11mut10 cells (Fig. 3C). Control BJAB-MSCV cells had considerable nuclear PU-1 site DNA-binding activity, which was clearly reduced in BJAB-CARD11mut10 cells. Conversely, nuclear NF-κB DNA-binding activity was increased in BJAB-CARD11mut10 cells as compared with control BJAB-MSCV cells, which is consistent with the reported ability of CARD11mut10 to activate NF-κB (42). As we previously showed (13), BJAB cells expressing an activated REL mutant (RELΔTAD1) also have increased levels of nuclear κB site DNA-binding activity (Fig. 3C).
CD10 Protein Levels Are Affected by Alterations in the Expression of miR-155 or PU.1
We next used retroviral transduction (with vector MSCV-BIC) to create BJAB cell lines that overexpressed miR-155. Overexpression of miR-155 (by ∼105-fold; Fig. 4A) in BJAB cells led to reduced levels of PU.1 and CD10 proteins and mRNA (Fig. 4, B and C). As shown in Fig. 4D, shRNA-mediated knockdown of PU.1 expression in BJAB cells also led to reduced expression of the CD10 protein.
FIGURE 4.
Alterations in the cellular levels of miR-155 or PU.1 affect the expression of CD10. A, micro-RNA-specific real time PCR was performed to assess the levels of miR-155 in BJAB cells transduced with a pMSCV-puro (−) or pMSCV-BIC (+) expression vector. Values were normalized to the 5 S rRNA levels and represent the mean values of three experiments performed with three samples ± S.E. B, the protein levels of PU.1, CD10, and β-tubulin were compared in the same cell types as described in the legend to Fig. 4A by Western blotting. C, the mRNA levels of CD10 and β-tubulin were compared in the same cell types as described in the legend to Fig. 4A by RT-PCR, agarose gel electrophoresis, and staining with ethidium bromide. D, Western blots for PU.1, CD10, and β-tubulin were compared in BJAB cells expressing an shRNA directed against PU.1 or a scrambled shRNA (con) that does not recognize any human sequence (negative control). E, whole cell extracts were made from DT40 and DT40-v-Rel cells (18), and anti-v-Rel Western blotting was performed (left panel); a v-Rel-transformed chicken spleen cell line served as the positive control. In the right panel, PU.1 and GAPDH transcript levels were analyzed by RT-PCR. N.S., nonspecific band.
The oncogenic NF-κB transcription factor v-Rel has recently been shown to up-regulate miR-155 (43). Similar to our results in BJAB cells, expression of v-Rel in the avian DT40 B-lymphoma cell line was also correlated with a reduction of PU.1 mRNA levels (Fig. 4E).
CD10 Expression Is Down-regulated in BJAB Cells following Treatment with Lipopolysaccharide
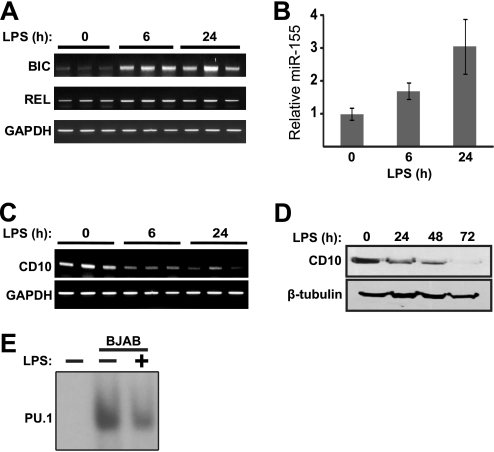
NF-κB activity and miR-155 expression are both induced by LPS in macrophages (34, 44). Therefore, to determine whether there is normal association between activation of NF-κB and reduced expression of CD10, we treated BJAB cells with LPS, and then measured expression of miR-155 and CD10 as well as PU-1 site DNA-binding activity. Treatment of BJAB cells with LPS for 6 h caused a significant increase in BIC mRNA levels, as assessed by semi-quantitative RT-PCR, and BIC mRNA remained elevated after 24 h treatment (Fig. 5A). REL and GAPDH mRNA levels remained unchanged following LPS treatment. By real time PCR, we found that miR-155 levels increased by ∼2-fold after 6 h of LPS treatment (Fig. 5B) and by ∼3-fold at 24 h, as compared with untreated cells (Fig. 5B).
FIGURE 5.
BIC/miR-155 is up-regulated and CD10 is down-regulated upon treatment of BJAB cells with LPS. BJAB cells were treated with 5 μg/ml of LPS for the indicated amounts of time (h). A, RT-PCR was performed for BIC, REL, and GAPDH transcripts on RNA samples obtained after treatment of BJAB cells with LPS. For each time point, RT-PCR from three independent samples are shown. B, micro-RNA-specific real time PCR was performed to assess the levels of miR-155 in BJAB cells. Values were normalized to the 5S rRNA levels and represent the mean ± S.E. of three experiments performed with three samples. C, RT-PCR was performed using primers for CD10 or GAPDH. For each time point, RT-PCR from three independent samples are shown. D, Western blotting was performed on whole cell extracts using antibodies against CD10 and β-tubulin. E, BJAB cells were treated with LPS for 72 h and nuclear extracts were analyzed by EMSA using the PU-1 site from the CD10 promoter (see Fig. 2B).
LPS treatment of BJAB cells also led to a decrease in CD10 transcript levels within 6 h and these remained decreased at 24 h as assessed by RT-PCR (Fig. 5C). Additionally, LPS treatment resulted in a time-dependent decrease in CD10 protein levels over a 72-h period (Fig. 5D). Finally, by EMSA, we found that BJAB cells treated with LPS for 72 h had decreased nuclear PU.1 DNA-binding activity as compared with untreated BJAB cells (Fig. 5E).
DISCUSSION
In this report, we describe for the first time a molecular link between increased NF-κB activity and reduced expression of CD10, both of which are poor prognostic markers for B-cell lymphoma. Our results suggest that increased NF-κB activity in B-lymphoma cells can lead to increased expression of micro-RNA miR-155, which then causes decreased PU.1 protein levels and consequently, reduced transcription from the CD10 promoter (leading to reduced CD10 protein levels). Or, more simply, ↑NF-κB → ↑miR-155 → ↓PU.1 → ↓CD10. The evidence for and implications of this model are discussed below.
Expression of CD10, a cell-surface zinc metalloendopeptidase, can be used to classify the two main molecular subtypes of DLBCL: CD10 is expressed at higher levels in patients with GCB DLBCL than in ones with ABC DLBCL (11, 12). On the other hand, high nuclear NF-κB activity is associated with the ABC subtype of DLBCL, which also carries a worse clinical outcome than GCB subtype DLBCL, which has lower NF-κB activity (8, 23). In this report, we demonstrate that increased NF-κB activity, as induced by expression of a lymphoma-derived mutant CARD11 protein or LPS, leads to reduced CD10 levels in the GCB-like B-lymphoma cell line BJAB. Moreover, two B-lymphoma-like cell lines with high NF-κB activity (IB4, due to expression of the LMP1 protein of EBV (41), and RC-K8, due to inactivating mutations in IκBα (25)) express little to no CD10 protein (Fig. 1A), whereas four B-lymphoma cell lines not known to have high NF-κB activity (BJAB (21), SUDHL-4 (23), BL-41 (14), Daudi (25, 45, 46)) have high CD10 levels.
By immunocytochemistry, PU.1 and CD10 protein levels have been shown to have a positive correlation in several B-cell lymphomas (47). By Western blotting, we also find a direct correlation between CD10 and PU.1 protein levels in six B-lymphoma cell lines (Fig. 1A). Moreover, CD10 protein expression and nuclear PU.1 DNA-binding activity in BJAB cells are considerably reduced by expression of CARD11mut10 (Fig. 3, A and C).
Our results show that PU.1 can activate a reporter plasmid containing sequences from the CD10 promoter (Fig. 2C); however, the sites required for this activation are not clear. In the promoter region of CD10 there are three putative PU.1 binding sites that contain the same core sequence GAGGAA (Fig. 2A); however, PU.1 was only able to bind one of these predicted sites (PU-1) in vitro (Fig. 2B). Although the core sequence GAGGAA is known to be required for efficient PU.1 binding, bases outside of this core motif can also affect proper PU.1 binding (48, 49). Mutation of the PU-1 binding site only partially reduced PU.1-induced activation of the CD10 promoter (Fig. 2C), and mutation of all three predicted PU.1 consensus sites did not further reduce PU.1-induced transactivation (data not shown). The residual PU.1-induced activation of the CD10 promoter in the presence of a mutated PU-1 site may be mediated through non-consensus PU.1 binding sites in the CD10 promoter. Indeed, there are six AAGGAA sites within the CD10 promoter fragment we have used, and PU.1 has been shown to bind to some AAGGAA sequences (50, 51). One of these AAGGAA sites (PU-4) is found within the first region of the CD10 promoter (nucleotides 1–470) and five of these sites are clustered in the second region of the promoter (nucleotides 449–717) (supplemental Fig. S1A). Reporter plasmids containing each region can be weakly induced by PU.1 (supplemental Fig. S1B), and the sum of the PU.1-induced activation of each region of the CD10 promoter is approximately equal to the induction of the full 717-bp CD10 promoter region (supplemental Fig. S1B). These results suggest that non-consensus (AAGGAA) PU.1 sites play a role in PU.1-induced activation of CD10 transcription.
By several criteria, we show that high miR-155 levels correlate with activation of NF-κB and reduced levels of PU.1 and CD10 in B-lymphoma cell lines. First, transformed B-lymphoid cell lines with high NF-κB activity have high miR-155 levels and low PU.1 and CD10 levels, whereas B-lymphoma cell lines with low NF-κB/miR-155 levels have increased levels of PU.1 and CD10 (Fig. 1). Second, overexpression of miR-155 leads to reduced levels of PU.1 and CD10 in BJAB cells (Fig. 4, A–C). Third, miR-155 levels are up-regulated in BJAB cells by two inducers of NF-κB: expression of a lymphoma-derived, constitutively active CARD11 mutant (Fig. 3A) or by treatment with LPS (Fig. 5B). Fourth, overexpression of v-Rel in the chicken B-lymphoma cell line DT40 leads to decreased PU.1 mRNA levels (Fig. 4E), and v-Rel has been shown to increase miR-155 expression (43). Fifth, shRNA-mediated knockdown of PU.1 leads to reduced CD10 protein levels in BJAB cells (Fig. 4D). Our results are consistent with previous reports showing that miR-155 is up-regulated in macrophages in response to various cytokines or inducers of Toll-like receptors, including LPS (52), and that the EBV LMP1 protein can activate miR-155 expression through an NF-κB binding site in the BIC promoter (53).
Micro-RNAs base pair to the 3′ UTR of target mRNAs, which cause translational repression, sometimes through mRNA degradation (54). PU.1 mRNA has been shown to be directly targeted by miR-155 in B cells (40, 55). Reduced PU.1 expression caused by miR-155 is important for the proper production of IgG1 antibodies (40), and miR-155 knock-out mice have a defect in the B-cell humoral response to antigen due to impaired germinal center formation (56, 57). Moreover, PU.1 knock-out mice have defects in the development of certain B-cell lineages (17). In general, there is little known about the downstream transcriptional network of PU.1 or how the expression of PU.1 target genes is important in immune function and regulation. We now show that PU.1 can directly increase transcription from the CD10 promoter. CD10 acts to cleave peptides in the extracellular matrix and thus has been thought to regulate B cells by activating and/or repressing factors that promote differentiation (2). Further investigation of the role of CD10 in the germinal center and its function in B-cell differentiation may reveal the normal function of miR-155-mediated regulation of PU.1.
Transgenic mice overexpressing miR-155 in B cells develop lymphoblastic leukemias and lymphomas (59). Of note, these miR-155 transgenic mice also have an expanded population of B cells with low CD10 expression (58). Moreover, elevated levels of miR-155 are present in various types of B-cell lymphoma (34), including ABC-like DLBCL and, as we show here, the IB4 and RC-K8 cell lines (Fig. 1B). Inhibition of NF-κB, which would be expected to reduce miR-155 expression, has been shown to specifically inhibit the proliferation of ABC-like DLBCL cell lines, including RC-K8 (23, 25).
The results from the present study shed light on a B-lymphoma gene network controlled indirectly by NF-κB. This pathway is of particular interest because less clinically favorable DLBCLs (e.g. the ABC subtype) can have several types of gene mutations that endow them with constitutive activation of NF-κB and reduced expression of CD10 (11, 42, 59, 60). Future studies will aim to determine whether CD10 affects B-lymphoma pathogenesis and may also focus on determining whether the NF-κB → CD10 pathway is involved in other cancer types, given that CD10 is a prognostic marker for some types of solid tumors (6, 7).
Supplementary Material
Acknowledgments
We thank Siloe Alvarado for technical assistance, and Dan Starczynowski and Mike Garbati for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant CA047763 and American Recovery and Reinvestment Act of 2009 supplement Grant CA047763021S3 (to T. D. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- GCB
- germinal center B-cell
- DLBCL
- diffuse large B-cell lymphoma
- ABC
- activated B-cell
- miR
- micro-RNA
- BIC
- B-cell integration cluster
- qPCR
- quantitative PCR
- LMP1
- latent membrane protein-1.
REFERENCES
- 1. Shipp M. A., Vijayaraghavan J., Schmidt E. V., Masteller E. L., D'Adamio L., Hersh L. B., Reinherz E. L. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 297–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salles G., Chen C. Y., Reinherz E. L., Shipp M. A. (1992) Blood 80, 2021–2029 [PubMed] [Google Scholar]
- 3. Sunday M. E., Hua J., Torday J. S., Reyes B., Shipp M. A. (1992) J. Clin. Invest. 90, 2517–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sumitomo M., Shen R., Nanus D. M. (2005) Biochim. Biophys. Acta 1751, 52–59 [DOI] [PubMed] [Google Scholar]
- 5. Ishimaru F., Potter N. S., Shipp M. A. (1996) Exp. Hematol. 24, 43–48 [PubMed] [Google Scholar]
- 6. Langner C., Ratschek M., Rehak P., Schips L., Zigeuner R. (2004) Histopathology 45, 460–467 [DOI] [PubMed] [Google Scholar]
- 7. Deschamps L., Handra-Luca A., O'Toole D., Sauvanet A., Ruszniewski P., Belghiti J., Bedossa P., Couvelard A. (2006) Hum. Pathol. 37, 802–808 [DOI] [PubMed] [Google Scholar]
- 8. Alizadeh A. A., Eisen M. B., Davis R. E., Ma C., Lossos I. S., Rosenwald A., Boldrick J. C., Sabet H., Tran T., Yu X., Powell J. I., Yang L., Marti G. E., Moore T., Hudson J., Jr., Lu L., Lewis D. B., Tibshirani R., Sherlock G., Chan W. C., Greiner T. C., Weisenburger D. D., Armitage J. O., Warnke R., Levy R., Wilson W., Grever M. R., Byrd J. C., Botstein D., Brown P. O., Staudt L. M. (2000) Nature 403, 503–511 [DOI] [PubMed] [Google Scholar]
- 9. Wright G., Tan B., Rosenwald A., Hurt E. H., Wiestner A., Staudt L. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9991–9996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Staudt L. M., Dave S. (2005) Adv. Immunol. 87, 163–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hans C. P., Weisenburger D. D., Greiner T. C., Gascoyne R. D., Delabie J., Ott G., Müller-Hermelink H. K., Campo E., Braziel R. M., Jaffe E. S., Pan Z., Farinha P., Smith L. M., Falini B., Banham A. H., Rosenwald A., Staudt L. M., Connors J. M., Armitage J. O., Chan W. C. (2004) Blood 103, 275–282 [DOI] [PubMed] [Google Scholar]
- 12. Iqbal J., Joshi S., Patel K. N., Javed S. I., Kucuk C., Aabida A., d'Amore F., Fu K. (2007) Indian J. Cancer 44, 72–86 [DOI] [PubMed] [Google Scholar]
- 13. Chin M., Herscovitch M., Zhang N., Waxman D. J., Gilmore T. D. (2009) Oncogene 28, 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carter K. L., Cahir-McFarland E., Kieff E. (2002) J. Virol. 76, 10427–10436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Phillips A. J., Tomasec P., Wang E. C., Wilkinson G. W., Borysiewicz L. K. (1998) Virology 250, 350–358 [DOI] [PubMed] [Google Scholar]
- 16. Ishimaru F., Shipp M. A. (1995) Blood 85, 3199–3207 [PubMed] [Google Scholar]
- 17. Medina K. L., Pongubala J. M., Reddy K. L., Lancki D. W., Dekoter R., Kieslinger M., Grosschedl R., Singh H. (2004) Dev. Cell 7, 607–617 [DOI] [PubMed] [Google Scholar]
- 18. Gupta N., Delrow J., Drawid A., Sengupta A. M., Fan G., Gélinas C. (2008) Cancer Res. 68, 808–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang M. C., Bardhan S., Pace E. A., Rosman D., Beutler J. A., Porco J. A., Jr., Gilmore T. D. (2006) Biochem. Pharmacol. 71, 634–645 [DOI] [PubMed] [Google Scholar]
- 20. Menezes J., Leibold W., Klein G., Clements G. (1975) Biomedicine 22, 276–284 [PubMed] [Google Scholar]
- 21. Ngo V. N., Davis R. E., Lamy L., Yu X., Zhao H., Lenz G., Lam L. T., Dave S., Yang L., Powell J., Staudt L. M. (2006) Nature 441, 106–110 [DOI] [PubMed] [Google Scholar]
- 22. Vaughn C. P., Crockett D. K., Lin Z., Lim M. S., Elenitoba-Johnson K. S. (2006) Proteomics 6, 3223–3230 [DOI] [PubMed] [Google Scholar]
- 23. Davis R. E., Brown K. D., Siebenlist U., Staudt L. M. (2001) J. Exp. Med. 194, 1861–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Contreras-Salazar B., Ehlin-Henriksson B., Klein G., Masucci M. G. (1990) J. Virol. 64, 5441–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalaitzidis D., Davis R. E., Rosenwald A., Staudt L. M., Gilmore T. D. (2002) Oncogene 21, 8759–8768 [DOI] [PubMed] [Google Scholar]
- 26. Pongubala J. M., Van Beveren C., Nagulapalli S., Klemsz M. J., McKercher S. R., Maki R. A., Atchison M. L. (1993) Science 259, 1622–1625 [DOI] [PubMed] [Google Scholar]
- 27. Gilmore T. D., Jean-Jacques J., Richards R., Cormier C., Kim J., Kalaitzidis D. (2003) Virology 316, 9–16 [DOI] [PubMed] [Google Scholar]
- 28. Zhu N., Hansen U., (2007) Mol. Cell. Biol. 27, 8859–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagel S., Scherr M., Kel A., Hornischer K., Crawford G. E., Kaufmann M., Meyer C., Drexler H. G., MacLeod R. A. (2007) Cancer Res. 67, 1461–1471 [DOI] [PubMed] [Google Scholar]
- 30. Scherr M., Battmer K., Ganser A., Eder M. (2003) Cell Cycle 2, 251–257 [PubMed] [Google Scholar]
- 31. Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., Bujard H. (1995) Science 268, 1766–1769 [DOI] [PubMed] [Google Scholar]
- 32. Leeman J. R., Weniger M. A., Barth T. F., Gilmore T. D. (2008) Oncogene 27, 6770–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Nucleic Acids Res. 33, e179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eis P. S., Tam W., Sun L., Chadburn A., Li Z., Gomez M. F., Lund E., Dahlberg J. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3627–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lawrie C. H., Soneji S., Marafioti T., Cooper C. D., Palazzo S., Paterson J. C., Cattan H., Enver T., Mager R., Boultwood J., Wainscoat J. S., Hatton C. S. (2007) Int. J. Cancer 121, 1156–1161 [DOI] [PubMed] [Google Scholar]
- 36. Rai D., Karanti S., Jung I., Dahia P. L., Aguiar R. C. (2008) Cancer Genet. Cytogenet. 181, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rahadiani N., Takakuwa T., Tresnasari K., Morii E., Aozasa K. (2008) Biochem. Biophys. Res. Commun. 377, 579–583 [DOI] [PubMed] [Google Scholar]
- 38. Kluiver J., Poppema S., de Jong D., Blokzijl T., Harms G., Jacobs S., Kroesen B. J., van den Berg A. (2005) J. Pathol. 207, 243–249 [DOI] [PubMed] [Google Scholar]
- 39. Faraoni I., Antonetti F. R., Cardone J., Bonmassar E. (2009) Biochim. Biophys. Acta 1792, 497–505 [DOI] [PubMed] [Google Scholar]
- 40. Vigorito E., Perks K. L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P. P., Miska E. A., Rodriguez A., Bradley A., Smith K. G., Rada C., Enright A. J., Toellner K. M., Maclennan I. C., Turner M. (2007) Immunity 27, 847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cahir-McFarland E. D., Carter K., Rosenwald A., Giltnane J. M., Henrickson S. E., Staudt L. M., Kieff E. (2004) J. Virol. 78, 4108–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lenz G., Davis R. E., Ngo V. N., Lam L., George T. C., Wright G. W., Dave S. S., Zhao H., Xu W., Rosenwald A., Ott G., Muller-Hermelink H. K., Gascoyne R. D., Connors J. M., Rimsza L. M., Campo E., Jaffe E. S., Delabie J., Smeland E. B., Fisher R. I., Chan W. C., Staudt L. M. (2008) Science 319, 1676–1679 [DOI] [PubMed] [Google Scholar]
- 43. Bolisetty M. T., Dy G., Tam W., Beemon K. L. (2009) J. Virol. 83, 12009–12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Derudder E., Laferté A., Ferreira V., Mishal Z., Baud V., Tarantino N., Körner M. (2003) Biochem. Biophys. Res. Commun. 308, 744–749 [DOI] [PubMed] [Google Scholar]
- 46. Bernal-Mizrachi L., Lovly C. M., Ratner L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9220–9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Torlakovic E., Malecka A., Myklebust J. H., Tierens A., Aasheim H. C., Nesland J. M., Smeland E., Kvaløy S., Delabie J. (2005) J. Pathol. 206, 312–319 [DOI] [PubMed] [Google Scholar]
- 48. Li S. L., Schlegel W., Valente A. J., Clark R. A. (1999) J. Biol. Chem. 274, 32453–32460 [DOI] [PubMed] [Google Scholar]
- 49. Smith M. F., Jr., Carl V. S., Lodie T., Fenton M. J. (1998) J. Biol. Chem. 273, 24272–24279 [DOI] [PubMed] [Google Scholar]
- 50. Kwon U. K., Yen P. H., Collins T., Wells R. A. (2006) Biochem. Biophys. Res. Commun. 344, 146–154 [DOI] [PubMed] [Google Scholar]
- 51. Eisenbeis C. F., Singh H., Storb U. (1993) Mol. Cell. Biol. 13, 6452–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baltimore D., Boldin M. P., O'Connell R. M., Rao D. S., Taganov K. D. (2008) Nat. Immunol. 9, 839–845 [DOI] [PubMed] [Google Scholar]
- 53. Gatto G., Rossi A., Rossi D., Kroening S., Bonatti S., Mallardo M. (2008) Nucleic Acids Res. 36, 6608–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chekulaeva M., Filipowicz W. (2009) Curr. Opin. Cell Biol. 21, 452–460 [DOI] [PubMed] [Google Scholar]
- 55. Martinez-Nunez R. T., Louafi F., Friedmann P. S., Sanchez-Elsner T. (2009) J. Biol. Chem. 284, 16334–16342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Science 316, 604–608 [DOI] [PubMed] [Google Scholar]
- 57. Rodriguez A., Vigorito E., Clare S., Warrren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Science 316, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Compagno M., Lim W. K., Grunn A., Nandula S. V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A., Bhagat G., Chadburn A., Dalla-Favera R., Pasqualucci L. (2009) Nature 459, 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kato M., Sanada M., Kato I., Sato Y., Takita J., Takeuchi K., Niwa A., Chen Y., Nakazaki K., Nomoto J., Asakura Y., Muto S., Tamura A., Iio M., Akatsuka Y., Hayashi Y., Mori H., Igarashi T., Kurokawa M., Chiba S., Mori S., Ishikawa Y., Okamoto K., Tobinai K., Nakagama H., Nakahata T., Yoshino T., Kobayashi Y., Ogawa S. (2009) Nature 459, 712–716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.