Abstract
Heat shock factor 1 (HSF1) is the master switch for heat shock protein (HSP) expression in eukaryotes. A synthetic chemical library was screened to identify inhibitors of HSF1 using a luciferase reporter under the control of a heat shock element. A compound named KRIBB11 (N2-(1H-indazole-5-yl)-N6-methyl-3-nitropyridine-2,6-diamine) was identified for its activity in abolishing the heat shock-induced luciferase activity with an IC50 of 1.2 μmol/liter. When the cells were exposed to heat shock in the presence of KRIBB11, the induction of HSF1 downstream target proteins such as HSP27 and HSP70 was blocked. In addition, treatment of HCT-116 cells with KRIBB11 induced growth arrest and apoptosis. Markers of apoptosis, such as cleaved poly(ADP-ribose) polymerase, were detected after KRIBB11 treatment. Biotinyl-KRIBB11 was synthesized as an affinity probe for the identification of KRIBB11 target proteins. Using affinity chromatography and competition assays, KRIBB11 was shown to associate with HSF1 in vitro. Chromatin immunoprecipitation analysis showed that KRIBB11 inhibited HSF1-dependent recruitment of p-TEFb (positive transcription elongation factor b) to the hsp70 promoter. Finally, intraperitoneal treatment of nude mice with KRIBB11 at 50 mg/kg resulted in a 47.4% (p < 0.05) inhibition of tumor growth without body weight loss. Immunoblotting assays showed that the expression of HSP70 was lower in KRIBB11-treated tumor tissue than in control tissues. Because HSPs are expressed at high levels in a wide range of tumors, these results strengthen the rationale for targeting HSF1 in cancer therapy.
Keywords: Apoptosis, Cancer Therapy, Gene Expression, Oncogene, RNA Polymerase II, Signal Transduction, Transcription Elongation Factors, Transcription Factors, Heat Shock Factor 1, p-TEFb
Introduction
The heat shock response (HSR)4 was first reported in 1962 by Ritossa (1). Since then, many investigators have reported that the HSR is an evolutionarily conserved protective mechanism against a wide range of stresses, including heat shock, heavy metal, oxidative stress, fever, or protein misfolding (reviewed in Refs. 2, 3). The HSR is mediated by the heat shock factor family, which in mammalian cells is composed of three heat shock factors (HSF1, HSF2, and HSF4) that control the transcription of heat shock proteins (4, 5). Although HSF2 and HSF4 play a role in the HSR, HSF1 is the master regulator of the heat shock response in eukaryotes.
RNA polymerase II (pol II) transcribes all mRNAs and has an extended carboxyl-terminal domain (CTD) in its largest subunit. This CTD consists of multiple repeats of the hepta-peptide 1YSPTSPS7. Before heat shock induction, pol II associates with the heat shock promoters, transcribes 20–50 bases downstream of the transcription site, and stays there in an arrested inactive state (6, 7). Releasing pol II requires the recruitment and activation of HSF1. However, HSF1 alone is not sufficient to release arrested RNA pol II (8).
HSF1 is required to recruit a second factor, p-TEFb, a heterodimer of CDK9 and cyclin T (8). The artificial recruitment of p-TEFb to the hsp70 promoter is sufficient for the induction of the hsp70 gene in the absence of heat shock (8). Phosphorylation of pol II Ser-2 of CTD by p-TEFb is a critical rate-limiting step in releasing paused pol II into productive elongation of several inducible genes, including hsp70. Recruited p-TEFb phosphorylates Ser-2 of the pol II CTD as well as negative elongation factor and DRB sensitivity-inducing factor. Phosphorylation of these inhibitory factors releases them from pol II, thus stimulating pol II transcription elongation (9, 10).
HSF1 is localized in the cytoplasm as an inactive monomer. Both the DNA-binding domain and the transcription activation domain of this factor are repressed through intramolecular interactions and constitutive serine phosphorylation (11). Upon exposure to stress, HSF1 translocates into the nucleus, where it becomes a trimer, is inducibly phosphorylated, and binds heat shock response elements (HSEs) in the promoter of heat shock genes such as hsp90, hsp70, hsp47, and hsp27 (12, 13).
The transcriptional activity of HSF1 is positively or negatively regulated by phosphorylation at different sites (14). HSF1 is positively regulated by polo-like kinase I (15, 16) and calcium/calmodulin-dependent protein kinase II (17, 18). HSF1 is negatively regulated by protein kinase C (19), extracellular signal-regulated kinase (20, 21), and glycogen synthase kinase 3β (22). HSEs typically consist of multiple contiguous repeats of the pentameric sequence nGAAn (23). HSEs are also present in the promoters of multidrug resistance genes (24) and of superoxide dismutase (25).
Although HSPs are only induced transiently upon stress, HSPs are often constitutively overexpressed in tumors. The expression of hsp70 is induced by several oncogenes such as H-rasVal-12 (26), c-myc (27), c-myb, SV40 large T antigen, and adenovirus E1a (28). The tumorigenic factor heregulin-β1 leads to increased HSP expression through induced stabilization of HSF1 (29). In human tumors, mutations in p53 lead to enhanced transcription of the hsp70 gene through the loss of repression of its promoter (30). HSP27 is induced by HSF1, as well as the POU domain-containing protein Brn3a (31). However, the precise mechanisms responsible for the overexpression of HSPs in cancer cells are not known.
Dai et al. (32) reported that HSF1 knockdown has a minimal effect on normal primary human cells but significantly impairs proliferation of several human malignant cell lines. In addition, they showed that knockdown of HSF1 suppresses chemically induced skin cancer formation in mice, suggesting an essential role for HSF1 during transformation. Down-regulation of HSP70 was found to inhibit cell proliferation and induce apoptosis (33). Similar results were reported when HSP27 was down-regulated (34). In contrast, cells overexpressing HSP70 or HSP27 showed an increase in tumorigenicity when inoculated into mice (35, 36). Overexpression of HSP70 in the immortalized Rat-1 cell line confers transformation phenotypes to these cells, such as loss of contact inhibition and growth on soft agar (37). In addition, the development of T-cell lymphoma was induced by the overexpression of the human hsp70 gene in transgenic mice (38).
Geldanamycin (GA) belongs to the family of benzoquinone ansamycin antibiotics, and it selectively binds to the ATP-binding pocket of HSP90, disrupting HSP90-substrate interactions. GA-mediated inhibition of HSP90 leads to degradation of its client proteins. By disrupting the HSP90-Raf kinase interaction, GA treatment was shown to inhibit the activation of the ERK signaling pathway (39). HSP90 binds to and blocks the activation of HSF1 (40). However, the treatment of cancer cells with GA results in the disruption of the HSP90-HSF1 interaction, releasing HSF1 and promoting its nuclear localization and transcriptional activation of the hsp70 gene. This induction of HSP70 by GA confers cell resistance to GA-induced apoptosis (40).
Mutations cause an increased demand for molecular chaperone activity within cancer cells expressing abnormal protein variants with suboptimal folding characteristics. In this study, a screen for inhibitors of HSF1 capable of down-regulating chaperone activity was conducted. KRIBB11 was identified for its activity in abolishing the heat shock-dependent induction of the hsp70 gene through inhibition of HSF1. Affinity chromatography with biotinyl-KRIBB11 demonstrated a physical association between KRIBB11 and HSF1. Evidence that KRIBB11 exerts its inhibitory effect on HSF1 function by blocking HSF1-dependent p-TEFb recruitment to the hsp70 promoter is also presented. Finally, the treatment of nude mice with KRIBB11 resulted in a significant inhibition of tumor growth, confirming HSF1 as a potential therapeutic target.
EXPERIMENTAL PROCEDURES
Reagents
All chemicals used in the study, including 17-(allylamino)-17-demethoxygeldanamycin (17-AAG), DMSO, and monoclonal anti-β-tubulin antibody, were purchased from Sigma. Antibodies against HSF1, HSF2, HSP70, and human-specific monoclonal HSP70 antibody were purchased from StressGen. Antibodies against HSP90, HSP27, and PARP were purchased from Cell Signaling Technology. Human-specific tubulin antibody was purchased from Abcam (Cambridge, UK). Phospho-Ser-2 RNA polymerase II antibody was purchased from Bethyl Laboratories Inc. Antibodies against RNA polymerase II (N-20), cyclin T1 (T-18), CDK9 (H-169), and phospho-Ser-230 HSF1 were obtained from Santa Cruz Biotechnology. KRIBB11 (N2-(1H-indazole-5-yl)-N6-methyl-3-nitropyridine-2,6-diamine), KRIBB14 (N2-(4-(diethylamino)-3-fluorophenyl)-N6-methyl-3-nitropyridine-2,6-diamine), and DW11120 (N6-(4-aminobutyl)-N2-(1H-indazol-5-yl)-3-nitropyridine-2,6-diamine) were synthesized using previously reported methods (Patent KR 2001048570). To synthesize biotinyl-KRIBB11, N,N′-dicyclohexylcarbodiimide (200 mg, 1 mmol), dimethylaminopyridine (120 mg, 1 mmol), and hydroxybenzotriazole (135 mg, 1 mmol) were added to a solution of biotin (240 mg, 1 mmol) in 10 ml of N,N-dimethylformamide. This mixture was added to a solution of compound DW11120 (340 mg, 1 mmol) in N,N-dimethylformamide and stirred at room temperature for 5 h. After the reaction, crude products were extracted with ethyl acetate and washed with brine. Biotinyl-KRIBB11 products were purified by HPLC, and its structure was confirmed using NMR analysis.
Cell Culture
All cancer cell lines were originally obtained from ATCC. HCT-116 (human colon cancer) was maintained in McCoy's 5A (Invitrogen). A549 (small cell lung cancer cell) and Mia PaCa-2 (human pancreatic cancer cell) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen). HCT-15 (human colon cancer), SW-620 (human colon cancer), HT-29 (human colon cancer), MDA-MB-231 (human breast cancer), and PANC-1 (human pancreatic cancer cell) were cultured in RPMI 1640 medium (Invitrogen). All culture media were supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen). Cell cultures were maintained at 37 °C under 5% CO2 in an incubator.
Luciferase Reporter Construct and Dual-LuciferaseTM Reporter Assay
p(HSE)4-TA-Luc plasmid was constructed by inserting four copies of the palindromic HSE (5′-GAT CTA GAA CGT TCT AGA ACG TTC TAG AAC GTT CTA-3′) in front of the pTA-Luc promoter vector (Clontech). The activity of the reporter was measured using a Dual-LuciferaseTM reporter system (Promega, Madison, WI). HCT-116 cells were seeded at a density of 2 × 106 cells in 100-mm2 culture dishes. Cells were co-transfected with 18 μg of p(HSE)4-TA-Luc vector and 6 μg of pRL-TK containing the Renilla luciferase gene as an internal control vector. The transfection was carried out using TransFectin (Bio-Rad) according to the manufacturer's protocol. Five hours after transfection, cells were trypsinized and seeded onto sterilized, black-bottom, 96-well plates at a density of 1.5 × 104 cells per well. After incubation for 24 h, cells were pretreated with chemicals for 30 min, exposed to heat shock at 44 °C for 15 min, and then incubated further at 37 °C for 5 h. Firefly and Renilla luciferase activities were measured using a dual-light reporter gene assay kit (Promega) with a GloMaxTM 96 microplate luminometer (Promega).
Cell Proliferation Assay
Cells were seeded onto 96-well plates at a density of 6 × 103 cells per well in McCoy's 5A medium with 10% FBS. After 24 h, the medium was replenished with fresh complete medium containing chemicals or 0.1% DMSO. After incubation for 48 h, the cell proliferation reagent WST-1 (Dojindo, Japan) was added to each well. The amount of WST-1 formazan produced was measured at 450 nm using an ELISA reader (Bio-Rad).
Detection of KRIBB11-binding Proteins
HCT-116 cells were washed with PBS and then homogenized with a 27-gauge syringe in binding buffer (10 mm Tris-HCl (pH 7.4), 50 mm KCl, 5 mm MgCl2, 1 mm EDTA, and 0.1 mm Na3VO4). The cell lysate was centrifuged at 13,000 rpm for 30 min at 4 °C, and the supernatant was collected. The HCT-116 cell lysate supernatant was precleared by incubating with Dynabeads M-280 streptavidin (Invitrogen) for 30 min at 4 °C and captured by magnet separation. The cleared supernatants were incubated with biotinyl-KRIBB11 compound. After overnight incubation at 4 °C, proteins associated with the biotinyl-KRIBB11 compound were precipitated with Dynabeads M-280 streptavidin. Precipitated samples were separated by a magnet. Samples were washed with 1 ml of washing buffer containing 50 mm HEPES (pH 7.5), 50 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.1% Tween 20, 10% (v/v) glycerol, 1 mm NaF, 0.1 mm Na3VO4, and protease inhibitor mixture tablets (1 tablet/10 ml; Roche Applied Science). Samples were boiled in SDS-PAGE sample buffer, separated by 10% polyacrylamide gel, and immunoblotted with antibodies against HSF1, HSF2, HSP90, or CDK9.
Chromatin Immunoprecipitation (ChIP) Assay
HCT-116 cells were treated with either no heat or heated at 37 °C for 1 h in the presence of KRIBB11. After that, cells were fixed by adding formaldehyde (Sigma) to the medium to a final concentration of 1.5% for 15 min and glycine to a final concentration of 125 mm. Cells were scraped and centrifuged for 5 min at 240 × g at room temperature. Next, cells were washed with ice-cold phosphate-buffered saline (PBS) containing protease inhibitors (1 mm phenylmethylsulfonyl fluoride (PMSF), 1 μg/ml aprotinin, and 1 μg/ml pepstatin A). After centrifugation, cells were resuspended in SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl (pH 8.1), and protease inhibitors) and incubated for 10 min on ice. After incubation, chromatin was sheared by sonication. After removal of nuclear debris by centrifugation at 13,000 × g for 10 min at 4 °C, lysates were diluted 10-fold with ChIP Dilution Buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.1), 167 mm NaCl) and then precleared for 30 min using 20 μl of protein A/G plus agarose beads (Santa Cruz Biotechnology). Immunoprecipitation was carried out at 4 °C overnight, and immune complexes were collected with Protein A/G plus agarose beads. The antibodies used were anti-HSF1 (StressGen, SPA-901), anti-CDK9 (Santa Cruz Biotechnology, sc-8338 (H-169)) or preimmune rabbit serum to detect nonspecific interactions. After washing three times with Low Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), 150 mm NaCl), High Salt Immune Complex Wash Buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl (pH 8.1), 500 mm NaCl), LiCl Immune Complex Wash Buffer (0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris-HCl (pH 8.1)), and twice with TE Buffer (10 mm Tris-HCl (pH 8.1), 1 mm EDTA (pH 8.0)), immunocomplexes were eluted with Elution Buffer (1% SDS, 0.1 m NaHCO3). Protein-DNA cross-links were reverted by incubating at 65 °C for 4 h. After proteinase K digestion, DNA was extracted with a PCR purification kit (Bioneer). Real time RT-PCR analysis was performed with iQTM5 (Bio-Rad), using FastStart SYBR Green Master (Roche Applied Science) to prepare the reaction mixes. The primers used for real time PCR of human hsp70 genes were the following: HSF1 ChIP assay, HSP70A forward primer, 5′-CACTCCCCCTTCCTCTCAG-3′, and HSP70A reverse primer, 5′-TTCCCTTCTGAGCCAATCAC-3′; p-TEFb ChIP assay, HSP70A forward primer, 5′-GAAAAGGCGGGTCTCCGTGACG-3′, and HSP70A reverse primer, 5′-GGTTCGCTCTGGGAAGCCTTGG-3′. Relative quantities of hsp70 were normalized to the input DNA. Data were expressed as the mean ± S.D. of triplicate samples.
Knockdown of HSF1 Proteins Using siRNA
Human HSF1 small interfering RNAs were synthesized as duplexes (Samcheonri Oligo, Korea) as follows: siHSF1-1, 5′-AAGUACUUCAAGCACAACAACTT-3′ and siHSF1-3, 5′-GAACGACAGUGGCUCAGCAUU-3′. The negative control siRNA, 5′-CCUACGCCACCAAUUUCGU-3′, was purchased from Bioneer, Inc. (Daejeon, Korea). Cells were seeded at a density of 3 × 105 cells per well on 6-well plates and incubated for 18 h. Cells were then transfected with 100 nm of each siRNA and the control siRNA duplexes, described above, after incubation for 20 min with Oligofectamine RNAi Max (Invitrogen) in serum-free Opti-MEM (Invitrogen). Five hours after transfection, the medium was replaced with fresh medium containing 10% FBS and further incubated for 72 h.
Quantitation of mRNA Using Quantitative Real Time Reverse Transcription PCR
RNA was isolated using the RNeasy kit (Qiagen). Two micrograms of isolated RNA for each sample were reverse-transcribed with RevertAid first strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions. Real time PCR was performed using IQTM SYBR Green supermix (Bio-Rad) according to the manufacturer's instructions using an iQ5 real time PCR detection system. The following primers were used for RT-PCR: HSP70 forward primer, 5′-ACCAAGCAGACGCAGATCTTC-3′, and HSP70 reverse primer, 5′-CGCCCTCGTACACCTGGAT-3′; HSP47 forward primer, 5′-CGCCATGTTCTTCAAGCCA-3′, and HSP47 reverse primer, 5′-CATGAAGCCACGGTTGTCC-3′; HSP27 forward primer, 5′-GGCATTTCTGGATGTGAGCC-3′, and HSP27 reverse primer, 5′-AGCAGGCAGGACATAGGTGC-3′; GAPDH forward primer, 5′-GGGAGCCAAAAGGGTCATCATCTC-3′, and GAPDH reverse primer, 5′-CCATGCCAGTGAGCTTCCCGTTC-3′. Relative quantities of hsp70 mRNA were normalized against GAPDH mRNA.
Western Blotting
Lysates were prepared using RIPA buffer as described previously (41). A volume of lysate corresponding to 30 μg was subjected to SDS-PAGE and transferred to a PVDF membrane (Millipore). Proteins were detected with the indicated primary antibodies. The secondary antibodies used were horseradish peroxidase-conjugated goat anti-rabbit, anti-mouse, anti-rat, and goat anti-mouse IgG (Jackson ImmunoResearch). The blots were developed with an enhanced chemiluminescence detection reagent (Roche Applied Science).
FACS Analysis
HCT-116 cells were treated with KRIBB11 at various concentrations for 48 h. Cells were then harvested by trypsinization, fixed with 70% chilled ethanol, and preserved at −20 °C before FACS analysis. Fixed cells were washed twice with phosphate-buffered saline (PBS) solution before being suspended in 500 μl of PBS and treated with 100 mg/ml RNase A at 37 °C for 30 min. Propidium iodide was then added to a final concentration of 50 mg/ml for DNA staining, and 20,000 fixed cells were analyzed on a FACSCalibur system (BD Biosciences). Cell cycle distribution was analyzed using the ModFit program (BD Biosciences).
Nude Mouse Xenograft Assay
All animal work was carried out in accordance with the guidelines and under the approval of the Institutional Review Committee for Animal Care and Use, Korea Research Institute of Bioscience and Biotechnology. Seven-week-old female inbred specific pathogen-free Balb/c nude mice were obtained from Charles River Laboratories (Japan), housed under sterile conditions with 12-h light/dark cycles, and fed food and water ad libitum. For the evaluation of the in vivo anti-tumor activity of KRIBB11, HCT-116 cells (0.3 ml of 4 × 107 cells/ml) were implanted subcutaneously into the right flank of the mice on day 0. KRIBB11 was dissolved in 10% dimethylacetamide, 50% PEG300, and 40% distilled water. When the size of tumors reached 72.2 mm3, the compound was administered intraperitoneally at a dose of 50 mg/kg/day for 18 days. Tumor volumes were estimated by using the formula length (mm) × width (mm) × height (mm)/2. To determine the toxicity of the compound, the body weight of tumor-bearing animals was recorded. On day 18, the mice were sacrificed, and the tumors were weighed.
RESULTS
HSF1-targeted Screening Identifies KRIBB11 as Inhibitor of HSF1 Activity
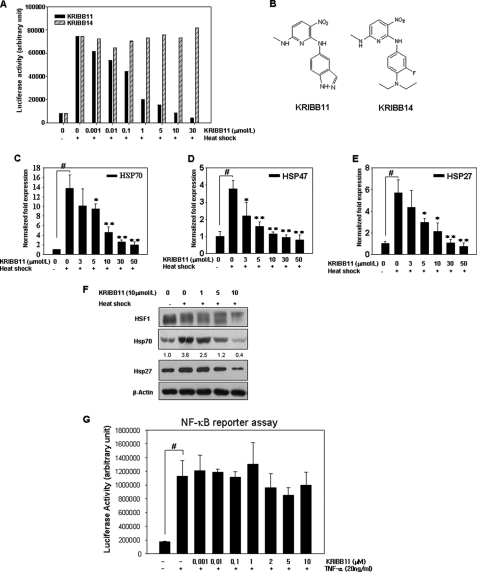
A heat shock-dependent luciferase reporter plasmid, p(HSE)4-TA-Luc, was constructed by introducing four copies of palindromic HSE into the pTA-Luc vector. To confirm that p(HSE)4-TA-Luc could be used to identify inhibitors of HSF1, the effect of heat shock on the reporter was tested by co-transfecting HCT-116 cells with a p(HSE)4-TA-Luc reporter and an internal control vector constitutively expressing pRL-TK to normalize for transfection efficiency and exposing cells to heat shock at 44 °C for 15 min. After 5 h of recovery at 37 °C, luciferase activity was measured. In response to heat shock, approximately a 10-fold increase in luciferase activity was observed in HCT-116 cells (Fig. 1A). Following the validation of the p(HSE)4-TA-Luc reporter in the cell-based assay, ∼6,230 compounds obtained from the Korea Chemical Bank, a collection of chemicals with known chemical structures, were screened. KRIBB11 was identified as a hit compound (Fig. 1B) and was therefore characterized further.
FIGURE 1.
KRIBB11 inhibits HSF1 activity in a concentration-dependent manner. A, HCT-116 cells were transfected with the p(HSE)4-TA-Luc reporter plasmid and treated with or without heat in the presence of different concentrations (0.001–30 μmol/liter) of KRIBB11 or KRIBB14. Reporter assay was carried out as described under “Experimental Procedures.” B, structure of KRIBB11 and KRIBB14. C–E, KRIBB11 inhibited heat shock-induced transcription of hsp70, hsp47, and hsp27 in a concentration-dependent manner. HCT-116 cells were treated with KRIBB11 (3–50 μmol/liter) for 30 min at 37 °C, exposed to heat shock at 43 °C for 1 h, and then incubated further at 37 °C for 1 h; quantitative analysis of mRNA levels of hsp70, hsp47, and hsp27 was carried out using real time PCR. The expression of each mRNA was normalized against the GAPDH gene. Each experiment was repeated three times, and each value is a mean ± S.D. Statistical significance (p value) was determined with an unpaired t test. #, p < 0.01 versus no heat control. *, p < 0.05; **, p < 0.01 versus heat control. F, KRIBB11 inhibited heat-induced HSP70 and HSP27 expression in HCT-116 cells. HCT-116 cells were treated with the indicated concentrations of KRIBB11 for 30 min, exposed to heat shock at 43 °C for 1 h, and then incubated further at 37 °C for 5 h. Whole cell lysates were analyzed by Western blotting as described under “Experimental Procedures.” G, HeLa cells expressing the pNF-κB-Luc reporter were pretreated for 30 min with the indicated concentrations of KRIBB11 and then stimulated for 16 h with TNF-α (20 ng/ml). Luciferase activity was determined with a GloMaxTM 96 microplate luminometer (Promega). Each experiment was repeated two times, and each value is the mean ± S.D. Statistical significance (p value) was determined with an unpaired t test. #, p < 0.01 versus control.
The effect of KRIBB11 on heat shock-induced luciferase activity in HCT-116 cells transfected with p(HSE)4-TA-Luc was investigated. As shown in Fig. 1A, KRIBB11 inhibited heat shock-induced luciferase activity in a concentration-dependent manner, with 50% inhibition at 1.2 μmol/liter. However, even though KRIBB14 (Fig. 1B) is a structural analog of KRIBB11, it did not inhibit heat shock-induced luciferase activity. This result suggests that the specific structure of KRIBB11 is important for its HSF1 inhibitory activity.
KRIBB11 Inhibits Heat Shock-dependent Induction of HSP mRNAs and Proteins
Because KRIBB11 inhibited HSF1-dependent reporter activity, the inhibitory effect of KRIBB11 on the endogenous hsp70, hsp47, and hsp27 promoter activities was investigated. For this purpose, HCT-116 cells were subjected to heat shock at 43 °C for 1 h in the presence or absence of KRIBB11 and incubated further at 37 °C for 1 h to allow recovery. After isolation of total RNA, hsp70 mRNA expression was evaluated by quantitative real time reverse transcription-PCR. As shown in Fig. 1C, heat shock treatment caused a 13-fold increase in hsp70 mRNA expression relative to the non-heat shock condition. Pretreatment of HCT-116 cells with KRIBB11 blocked heat shock-induced hsp70 mRNA expression in a concentration-dependent manner, with 70% inhibition at 10 μmol/liter. Similarly, KRIBB11 inhibited hsp47 and hsp27 mRNA expression in a concentration-dependent manner (Fig. 1, D and E). In accordance with its effect on mRNA expression, KRIBB11 also significantly down-regulated HSF1 downstream target proteins such as HSP70 and HSP27 in a concentration-dependent manner (Fig. 1F). Heat shock induces hyperphosphorylation of HSF1, resulting in a shift in its mobility on SDS gels. To exclude the possible nonspecific transcriptional inhibitory activity of KRIBB11, its effect on NF-κB activity was tested. NF-κB regulates the transcription of various inflammatory cytokines as well as anti-apoptotic genes. A pNF-κB-Luc plasmid for an NF-κB luciferase reporter assay was obtained from Stratagene (La Jolla, CA) and used as described previously (42). As shown in Fig. 1G, NF-κB reporter activity was stimulated by treating cells with 20 ng/ml TNF-α. However, pretreatment with high concentrations of KRIBB11 weakly inhibited TNF-α-dependent NF-κB reporter activity. This result suggests that KRIBB11 is not a general transcription inhibitor.
KRIBB11 Inhibits Cancer Cell Proliferation
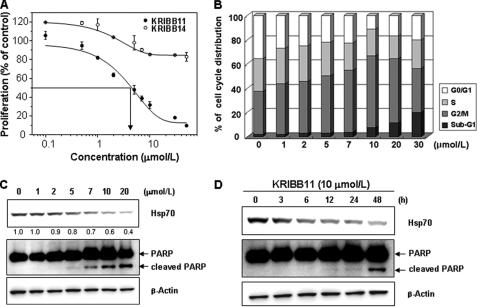
The inhibition of HSF1 activity by KRIBB11 and the consequent down-regulation of HSP70 and HSP27 led to the speculation that KRIBB11 could inhibit the proliferation of cancer cells. To evaluate the effect of KRIBB11 on the growth of cancer cells, HCT-116 cells were treated with different concentrations of KRIBB11 (0–50 μmol/liter) for 48 h (Fig. 2A). KRIBB11 exhibited a dose-dependent inhibition of HCT-116 cell growth over a broad range of concentrations, with an IC50 of 5 μmol/liter, where IC50 is the inhibitor concentration at which a 50% inhibition of cell growth is observed. The effect of KRIBB11 on the proliferation of various other tumor cell lines was also analyzed; these cell lines and the IC50 value for each are as follows: HCT-15 (5 μmol/liter), Mia-PaCa-2 (3 μmol/liter), SW-620 (4 μmol/liter), HT-29 (3 μmol/liter), A549 (5 μmol/liter), and MDA-MB-231 (8 μmol/liter).
FIGURE 2.
KRIBB11 inhibits proliferation by arresting the cell cycle at the G2/M phase. A, HCT-116 cells were treated with 0.1% DMSO or different concentrations of KRIBB11 or KRIBB14. After incubation for 48 h, a cell proliferation assay was carried out as described under “Experimental Procedures.” Proliferation is expressed as the percentage of KRIBB11-treated cells compared with the 0.1% DMSO-treated cells. Each value is the mean ± S.D. B, HCT-116 cells were treated with the indicated concentrations of KRIBB11 or vehicle solvent (0.1% DMSO) for 48 h. After incubation, cells were subjected to FACSCalibur analysis. Relative percentages of cells in the sub-G1 (<2 N), G2, M, and G0/G1 phases were determined by using the ModiFit program (BD Biosciences). C, Western blot analysis of whole cell extracts of HCT-116 cells treated with the indicated concentrations of KRIBB11 or vehicle solvent for 48 h (0.1% DMSO). D, HCT-116 cells were treated with 0.1% DMSO or KRIBB11 (10 μmol/liter) for 3–48 h, and whole cell lysates were analyzed by Western blotting as described under “Experimental Procedures.”
KRIBB11 Arrests the Cell Cycle at G2/M Phase and Induces Apoptosis
Based on the KRIBB11-mediated inhibition of cancer cell proliferation, the phase of the cycle affected by KRIBB11 was evaluated. HCT-116 cells were treated with KRIBB11 at different concentrations for 48 h and subjected to FACS analysis. KRIBB11 caused an increase in the proportion of G2/M phase cells in a concentration-dependent manner, with a maximum increase detected at 10 μmol/liter (Fig. 2B). At KRIBB11 concentrations higher than 10 μmol/liter, an increase in the proportion of sub-G1 phase cells was observed. PARP is involved in DNA repair following environmental stress, and cleavage of PARP is a marker of apoptosis. The increase in the proportion of the sub-G1 population in KRIBB11-treated cells suggested that KRIBB11 may induce PARP cleavage. In accordance with the FACS analysis, PARP cleavage was detected in HCT-116 cells treated with KRIBB11 (Fig. 2C). To analyze the time dependence of the induction of apoptosis by KRIBB11, HCT-116 cells were treated with 10 μmol/liter of KRIBB11 for different times. As shown in Fig. 2D, treating HCT-116 cells with KRIBB11 for 48 h increased PARP cleavage.
KRIBB11 Binds to HSF1
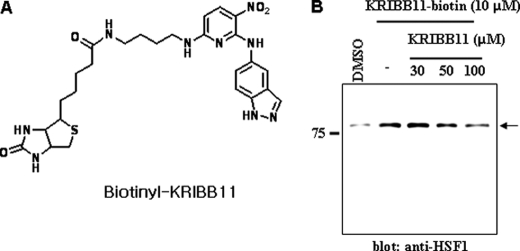
Target identification is a major challenge for phenotype-based chemical genetic studies. In this study, a biochemical purification approach using immobilized KRIBB11 was used. For this, biotinyl-KRIBB11 was synthesized using the method described under “Experimental Procedures.” The synthesized biotinyl-KRIBB11 had weak activity compared with KRIBB11 (IC50 of 20 versus 5 μm, respectively). Cell extracts of HCT-116 cells were incubated with biotinyl-KRIBB11. The bound proteins were precipitated with Dynabeads M-280 streptavidin and resolved using SDS-PAGE. Because of the low sensitivity of Coomassie Brilliant Blue staining, Western blot analysis with a specific HSF1 antibody revealed that HSF1 protein was present in the biotinyl-KRIBB11 (Fig. 3), indicating that HSF1 is a binding target of KRIBB11. When the interaction between HSF1 and biotinyl-KRIBB11 was challenged by excess KRIBB11, the interaction between HSF1 and biotinyl-KRIBB11 decreased (Fig. 3). Biotinyl-KRIBB11 may indirectly associate with HSF1 through HSF2 or HSP90, which are known to interact with HSF1. Therefore, binding of biotinyl-KRIBB11 to HSF2, HSP90, or CDK9 was tested using specific antibodies. HSF2, HSP90, and CDK9 were not detected in the biotinyl-KRIBB11 (data not shown). These results suggested that HSF1 might be a target protein for KRIBB11.
FIGURE 3.
KRIBB11 binds to HSF1. A, structure of biotinyl-KRIBB11. B, 300 μg of total protein was incubated with 10 μm biotinyl-KRIBB11 and competed with KRIBB11 at an indicated concentration. Proteins were captured with 10 μl of Dynabeads M-280 streptavidin and eluted by boiling in SDS-PAGE sample buffer. The eluted proteins were resolved by 10% SDS-PAGE and blotted with an anti-HSF1-specific antibody.
KRIBB11 Does Not Inhibit Heat Shock-induced Recruitment of HSF1 to the hsp70 Promoter
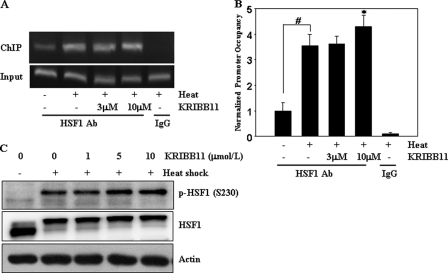
HSF1 is recruited to the promoters of heat shock genes upon heat shock. Because KRIBB11 binds to HSF1, the effect of KRIBB11 on the recruitment of HSF1 to the hsp70 promoter was assessed by ChIP analysis of HCT-116 cell extracts prepared from heat-shocked cells treated with KRIBB11. An upstream primer set, centered at −120, detects the binding of HSF1 to heat shock elements. As shown in Fig. 4, A and B, HSF1 association with the hsp70 promoter was significantly increased by heat shock. However, treating cells with KRIBB11 did not inhibit heat shock-induced HSF1 binding to the hsp70 promoter.
FIGURE 4.
KRIBB11 did not inhibit heat shock-induced HSF1 binding to the hsp70 promoter or phosphorylation of HSF1 Ser-230. A and B, ChIP analysis of HSF1 binding to the hsp70 promoter. HCT-116 cells were incubated with the indicated concentrations of KRIBB11, heat-shocked at 43 °C for 1 h, and analyzed for recruitment of HSF1 to the hsp70 promoter by the ChIP assay as described under “Experimental Procedures.” ChIP-enriched DNAs using preimmune IgG or anti-HSF1 antibody, as well as input DNAs, were prepared. Quantification of the DNA fragment of the hsp70 gene (−216 to −24) was done using PCR (A) or real time PCR (B). The ChIP assay was done three times, and similar results were obtained. Relative promoter occupancy is expressed as fold induction compared with the control prepared from samples that were not heat treated. Error bars indicate ± S.D. Statistical significance (p value) was determined with an unpaired t test. #, p < 0.01 versus no heat control. *, p < 0.05 versus heat control. C, phosphorylation of HSF1 on serine 230. HCT-116 cells were incubated with the indicated concentrations of KRIBB11, heat-shocked at 43 °C for 1 h, and then incubated at 37 °C for 5 h. HCT-116 cell lysates were analyzed by immunoblotting using an anti-phospho-HSF1 Ser-230 antibody as described under “Experimental Procedures.”
KRIBB11 Does Not Inhibit Heat Shock-induced Phosphorylation of HSF1 Ser-230
HSF1 is regulated by multisite phosphorylation. Twelve Ser residues in HSF1 were found to be phosphorylated upon heat shock (43). Ser-230 was phosphorylated by the calcium/calmodulin-dependent protein kinase II, which is important for the transcriptional activity of HSF1 (18). The possible inhibition of HSF1 Ser-230 phosphorylation by KRIBB11 was therefore analyzed. As shown in Fig. 4C, HSF1 Ser-230 was significantly phosphorylated after heat shock. In addition, heat shock-induced phosphorylation of HSF1 Ser-230 was maintained even after KRIBB11 treatment (Fig. 4C). These results implied that treating cells with KRIBB11 did not inhibit positive phosphorylation of HSF1 Ser-230.
KRIBB11 Inhibits Heat Shock-induced Recruitment of pTEFb to the hsp70 Promoter
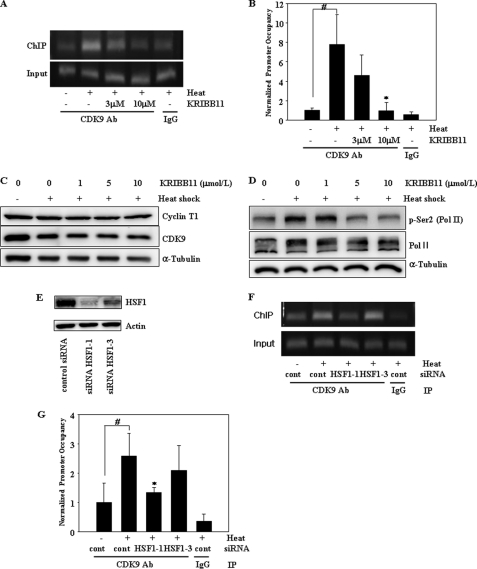
p-TEFb is a heterodimer of CDK9 and cyclin T. Like HSF1, p-TEFb is rapidly recruited to the hsp70 promoter upon heat shock, and this recruitment was blocked in an HSF1 mutant, suggesting that HSF1 plays an essential role in the recruitment of p-TEFb (8). The artificial recruitment of the p-TEFb hybrid protein to the hsp70 promoter is sufficient to activate transcription in the absence of heat shock stress. These results suggest that p-TEFb is essential for the induction of heat shock genes and HSF1 functions by recruiting p-TEFb to the promoters of these genes. In this study, ChIP assay was used to assess whether KRIBB11 binding to HSF1 could block the HSF1-dependent recruitment of p-TEFb to the hsp70 promoter. A primer set, centered at +45, contains the region of the paused pol II and was used to detect p-TEFb association. As shown in Fig. 5, A and B, p-TEFb association with the hsp70 promoter was significantly increased by heat shock. As expected, treating cells with KRIBB11 inhibited p-TEFb recruitment to the hsp70 promoter. Heat shock-induced association of p-TEFb with the hsp70 promoter was almost abolished by the treatment of cells with 10 μmol/liter of KRIBB11. The lack of an association between p-TEFb and the hsp70 promoter under these conditions could be due to the down-regulation of CDK9 or cyclin T expression by KRIBB11. To exclude this possibility, the effect of KRIBB11 on the expression of CDK9 and cyclin T1 was analyzed. As shown in Fig. 5C, treating cells with KRIBB11 did not decrease CDK9 or cyclin T1 expression.
FIGURE 5.
KRIBB11 inhibits heat shock-induced recruitment of p-TEFb to the hsp70 promoter and phosphorylation of RNA pol II CTD Ser-2. A and B, ChIP analysis of CDK9 of the p-TEFb subunit. HCT-116 cells were incubated with the indicated concentrations of KRIBB11, heat-shocked at 43 °C for 1 h, and then analyzed for the recruitment of p-TEFb to the hsp70 promoter by ChIP assay as described under “Experimental Procedures.” ChIP-enriched DNAs using preimmune IgG or anti-CDK9 antibody, as well as input DNAs, were prepared. Quantification of the DNA fragment of the hsp70 gene (−26 to +115) was done using PCR (A) or real time PCR (B). The ChIP assay was repeated three times. Relative promoter occupancy is expressed as fold induction compared with the control prepared without heat shock. Error bars indicate ± S.D. Statistical significance (p value) was determined with an unpaired t test. #, p < 0.01 versus no heat control. *, p < 0.05 versus heat control. C, expression of cyclin T1 and CDK9. HCT-116 cells were incubated with the indicated concentrations of KRIBB11, heat-shocked at 43 °C for 1 h, and then incubated at 37 °C for 5 h. Equal amounts of lysates were analyzed by Western blotting using anti-cyclin T1 or anti-CDK9 antibodies as described under “Experimental Procedures.” D, phosphorylation of RNA pol II CTD Ser-2. HCT-116 cells were incubated with the indicated concentrations of KRIBB11 and heat-shocked at 43 °C for 1 h, and then incubated at 37 °C for 5 h. HCT-116 lysates were analyzed by Western blotting using anti-phospho-Ser-2 or RNA pol II antibody as described under “Experimental Procedures.” E, knockdown of HSF1 by siRNA. HCT-116 cells were transfected with the indicated siRNAs for 72 h. siRNA-transfected cells were lysed, and equal amounts of protein were subjected to immunoblotting with the indicated antibodies. F and G, ChIP analysis of CDK9 of the p-TEFb subunit. HCT-116 cells were transfected with the indicated siRNAs for 72 h, heat-shocked at 43 °C for 1 h, and then analyzed for recruitment of p-TEFb to the hsp70 promoter by ChIP assay as described under “Experimental Procedures.” ChIP-enriched DNAs using preimmune IgG or anti-CDK9 antibody, as well as input DNAs, were prepared. Quantification of the DNA fragment of the hsp70 gene (−26 to +115) was done using PCR (F) or real time PCR (G). Statistical significance (p value) was determined with an unpaired t test. #, p < 0.05 versus no heat control. *, p < 0.05 versus heat control.
KRIBB11 Inhibits p-TEFb-dependent Phosphorylation of pol II CTD Ser-2
Because the CDK9 subunit of p-TEFb phosphorylates pol II CTD Ser-2 and KRIBB11 inhibits p-TEFb recruitment to the hsp70 promoter, it was important to test whether KRIBB11 could inhibit the phosphorylation of pol II CTD Ser-2. As shown in Fig. 5D, phosphorylation of CTD Ser-2 was significantly increased by heat shock. In addition, as expected, heat shock-induced phosphorylation of CTD Ser-2 was significantly inhibited by treating cells with KRIBB11. This result is consistent with the inhibitory effect of KRIBB11 on the recruitment of p-TEFb to the hsp70 promoter. RNA pol II appeared as two bands because of the use of the polyclonal pol II (N-20) antibody, which binds to hypo-phosphorylated RNA pol II (lower band) and hyperphosphorylated RNA pol II (upper band).
Knockdown of HSF1 Inhibits Heat Shock-induced Recruitment of pTEFb to the hsp70 Promoter
To prove that HSF1 is essential for p-TEFb recruitment to the hsp70 promoter, ChIP analysis was performed on HCT-116 cell extracts prepared from heat-shocked cells treated with HSF1 siRNA. Two different siRNAs, siRNA HSF1–1 and siRNA HSF1–3, were used. Three days after transfection, expression of HSF1 was more efficiently knocked down by siRNA HSF1–1 than by siRNA HSF1–3 (Fig. 5E). As shown in Fig. 5, F and G, heat shock-induced recruitment of p-TEFb was inhibited by knockdown of HSF1. This result confirms that HSF1 is important for p-TEFb recruitment to the hsp70 promoter and that its inhibition by either KRIBB11 or HSF1 siRNA impairs HSF1-dependent p-TEFb recruitment to the promoter of heat shock genes.
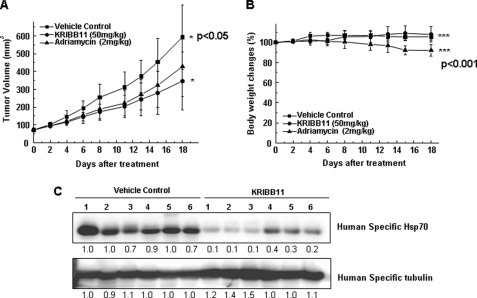
KRIBB11 Inhibits the Growth of HCT-116 Cells in BALB/c Nude Mice
HCT-116 tumor xenografts in nude mice were used to investigate the inhibitory activity of KRIBB11 on tumor growth in vivo. HCT-116 cells were implanted subcutaneously into the right flank of nude mice. When the tumor volume reached 72.2 mm3 (13 days after implantation), the indicated compounds were administered intraperitoneally at a dose of 50 mg/kg of KRIBB11 or 2 mg/kg of adriamycin per day. Because there is no available anticancer drug that specifically targets HSF1, adriamycin was used as a positive control compound. Adriamycin interacts with DNA by intercalation and is commonly used in the treatment of a wide range of cancers. To determine the toxicity of the compound, the body weight of tumor-bearing mice was measured. On day 18, the mice were sacrificed, and the tumors were removed and weighed. Mice treated with KRIBB11 showed a 47.4% (p < 0.05) decrease in tumor volume compared with control mice (Fig. 6A). Similarly, when adriamycin was used as a positive control compound, tumor volume was decreased by 31.7%. There was no change in body weight when KRIBB11 was used at 50 mg/kg (Fig. 6B). However, when adriamycin was used at 2 mg/kg, a loss of 13.2% (p < 0.001) of body weight was observed. To confirm that KRIBB11 suppressed the growth of HCT-116 tumors through the inhibition of HSF1 activity in vivo, HSP70 protein levels were measured in tumor tissues from both KRIBB11- and control-treated mice. As shown in Fig. 6C, HSP70 protein levels were significantly decreased in tumors from mice treated with KRIBB11, as compared with mice treated with vehicle.
FIGURE 6.
KRIBB11 inhibits the growth of HCT-116 cells in a nude mouse xenograft regression model. A, for the evaluation of the in vivo anti-tumor activity of KRIBB11, HCT-116 cells were implanted subcutaneously into the right flank of nude mice on day 0. Compounds were dissolved in 10% dimethylacetamide, 50% PEG300, and 40% distilled water and were administered intraperitoneally for 18 days at a concentration of 50 mg/kg for KRIBB11 or 2 mg/kg for adriamycin per day. The results shown were obtained from one assay using 18 mice (six mice for each compound). Tumor volumes were estimated by the formula length (mm) × width (mm) × height (mm)/2. B, body weight was measured on each indicated day. C, lysates were prepared from tumor tissues of six mice for each compound, and whole cell lysates were analyzed by Western blotting as described under “Experimental Procedures.” Statistical significance (p value) was determined with an unpaired t test. *, p < 0.05 versus control; ***, p < 0.001 versus control.
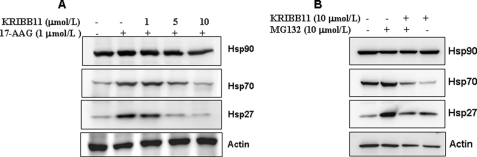
KRIBB11 Inhibits the HSP90- or Proteasome Inhibitor-mediated Induction of HSP70 and HSP27
The chaperone HSP90 is a promising cancer drug target, and several inhibitors of HSP90 are currently under development (44). HSP90 binds HSF1 and blocks its activation. However, the treatment of cancer cells with HSP90 inhibitors disrupts the HSF1-HSP90 interaction, causing the release of HSF1 and the induction of the expression of other HSPs, especially HSP70 and HSP27. The induction of HSP70 and HSP27 by the HSP90 inhibitor GA confers resistance to apoptosis and chemotherapy. Inhibition of HSF1 by KRIBB11 could therefore result in the blocking of GA-induced HSP70 and HSP27 expression. To examine this possibility, HCT-116 cells were treated with 17-AAG in the absence or presence of KRIBB11, and the expression of HSP70 and HSP27 was evaluated by Western blotting (Fig. 7A). Treatment of cells with 17-AAG induced HSP70 and HSP27 expression. In contrast, pretreatment with KRIBB11 abolished 17-AAG-induced HSP70 and HSP27 expression. The proteasome has recently been identified as a target for cancer treatment, and the proteasome inhibitor bortezomib was approved for the treatment of multiple myeloma (45). Similar to 17-AAG, proteasome inhibitor treatment results in the induction of the expression of HSPs, therefore reducing the antitumor activity of the inhibitor. The ability of KRIBB11 to block the proteasome inhibitor-dependent induction of HSPs was tested. As expected, co-treatment of cells with the proteasome inhibitor MG132 and KRIBB11 abolished the induction of HSP70 and HSP27 (Fig. 7B). This result implies that KRIBB11 can be used not only as a single agent but also in combination with GA or bortezomib.
FIGURE 7.
KRIBB11 inhibits the induction of HSPs by HSP90 or a proteasome inhibitor. HCT-116 cells were pretreated with the indicated concentrations of KRIBB11 or vehicle solvent (0.1% DMSO) for 30 min and then exposed to 1 μmol/liter 17-AAG or 10 μmol/liter MG132 for 5 h at 37 °C, and whole cell lysates were analyzed by Western blotting as described under “Experimental Procedures.”
DISCUSSION
Cancer cells are characterized by genetic mutations or epigenetic changes in oncogenes and require increased activity of these oncogenes for tumor initiation and progression. This dependence of cancer cells has been called “oncogene addiction” (46). Many drug screening programs have focused on these oncogenes. However, cancer cells are still heavily dependent on specific signaling pathways that are independent of oncogenic pathways (47). One of these is the “molecular chaperone” pathway (48). Mutations present in cancer cells can result in suboptimal conformation for cellular proteins. In addition, cancer cells are exposed to high levels of reactive oxygen species (49), which makes them highly dependent on molecular chaperone activity; this is one example of “non-oncogene addiction.”
Dai et al. (32) reported that knockdown of HSF1 could inhibit the viability of malignant cancer cell lines. They used HSF1 shRNA constructs packaged as lentiviral particles and enriched only for cells that stably expressed the shRNA. After puromycin selection, the viability of several cancer cell lines, but not that of a normal cell line, was significantly reduced. In the case of HeLa cervix cancer cells, viability was inhibited more than 90% by HSF1 shRNA (32).
In this study, the use of a luciferase reporter screening assay, which measured molecular chaperone activity, resulted in the identification of KRIBB11 as a novel inhibitor of HSF1 (Fig. 1). KRIBB11 was found to inhibit the transcriptional activity of HSF1 and the proliferation of HCT-116 cells in a dose-dependent manner. This result is consistent with the report by Dai et al. (32).
Lee et al. (16) reported that HSF1 is a mitotic regulator and that the knockdown of HSF1 causes defective mitotic progression. As expected, treating HCT-116 cells with KRIBB11 increased the proportion of G2/M phase cells in a concentration-dependent manner (Fig. 2B). These results confirmed that KRIBB11 is an inhibitor of HSF1 function during the mitotic progression in cancer cells.
The major technical challenge during cell-based screening is the identification of the target protein of the bioactive molecule (50). In this study, proteins purified by KRIBB11 affinity chromatography were resolved by SDS-PAGE and visualized by Coomassie Blue staining. However, the levels of signaling proteins were relatively low compared with the levels of structural proteins and were hardly detectable by Coomassie Blue staining. The staining of bound protein(s) with specific and sensitive antibodies for HSF1 signaling proteins revealed that biotinyl-KRIBB11 associated with HSF1 (Fig. 3B). The specificity of the interaction was confirmed by competition assays with free KRIBB11. An excess amount of free KRIBB11 inhibited the binding of HSF1 to immobilized biotinyl-KRIBB11. HSF1 can associate with several proteins such as HSF2 and HSP90. It is therefore possible that biotinyl-KRIBB11 indirectly associates with HSF1 through HSF2 or HSP90. To exclude this possibility, the binding of biotinyl-KRIBB11 to HSF2, HSP90, or CDK9 was tested. However, HSF2, HSP90, and CDK9 were not detected in the biotinyl-KRIBB11. Although the possibility of indirect association cannot be completely excluded, these results support that HSF1 may be a target protein for KRIBB11.
As mentioned above, the recruitment of p-TEFb to the hsp70 gene is essential for heat shock gene expression, and HSF1 is necessary for p-TEFb recruitment (8). The activity of KRIBB11 in blocking the recruitment of p-TEFb to the hsp70 promoter was therefore tested. As expected, exposing HCT-116 cells to heat shock significantly increased the association between p-TEFb and the hsp70 gene, and this association was abolished by treating cells with KRIBB11 (Fig. 5, A and B). This result implies that KRIBB11 blocked HSF1-dependent p-TEFb recruitment. In addition, the inhibitory effect of KRIBB11 on HSF1-dependent p-TEFb recruitment was confirmed by analyzing p-TEFb-dependent phosphorylation of Ser-2 of the pol II CTD (Fig. 5D). Currently, it is not known whether HSF1 interacts with CDK9 directly. Park et al. (51) reported that upon heat shock HSF1 directly interacts with Mediator, which is responsible for the recruitment of p-TEFb.
Several HSF1 inhibitors (for review see Ref. 52) have been investigated, including quercetin (53), QC12 (54), KNK437 (55), Stresgenin (56), triptolide (57), Emunin (58), and NZ28 (58). Among these inhibitors, triptolide shows the most potent activity against HSF1, and it was used to show the antitumor effect of HSF1 inhibition in vivo. However, triptolide inhibits not only HSF1 but also NF-κB and AP-1 (59). Therefore, it is not clear whether the anticancer activity of triptolide is a result of HSF1 inhibition. Quercetin is a naturally occurring flavonoid that inhibits multiple target proteins, including HSF1, NF-κB, several kinases, and CYP3A4. In addition, most of these inhibitors have not been well characterized. This study showed the physical association between KRIBB11 and HSF1 and the functional inhibition of HSF1-dependent p-TEFb recruitment by KRIBB11. To our knowledge, KRIBB11 is the first direct inhibitor of HSF1 to be reported.
Using a mouse xenograft model, KRIBB11 treatment decreased tumor volume by 47% compared with untreated control mice (Fig. 6). In addition, HSP70 protein levels were significantly decreased in tumors from mice treated with KRIBB11, supporting the notion that KRIBB11 exerts its in vivo antitumor activity through HSF1 inhibition. Furthermore, there was no change in body weight when KRIBB11 was used at 50 mg/kg. However, when adriamycin was used at 2 mg/kg, a 13% loss of body weight was observed, indicating cytotoxicity. Adriamycin is a drug commonly used in a wide range of cancers, including many types of carcinoma and hematological malignancies. Combination therapies are important in the treatment of cancer to reduce the development of resistance. The results of this study clearly showed that the inhibition of HSF1 by KRIBB11 resulted in a reduction in the GA- and MG-132-induced expression of HSPs (Fig. 7), supporting the potential value of HSF1 for combination therapy. This study also reinforces the viewpoint that it should be possible to develop clinical applications for molecules such as KRIBB11 that circumvent the oncogene addiction of cancer cells by targeting molecular chaperones (48).
This work was supported by the KRIBB Research Initiative Program, the National Chemical Genomics Research Program, and the Center for Biological Modulators of the 21st Century Frontier Research Program. Y. J. Yoon, J. S. Lee, B.-M. Kwon, and D. C. Han have applied for a patent using a part of this work.
- HSR
- heat shock response
- 17-AAG
- 17-N-allylamino-17-demethoxygeldanamycin
- GA
- geldanamycin
- HSE
- heat shock element
- HSF1
- heat shock factor 1
- HSP
- heat shock protein
- PARP
- poly(ADP-ribose) polymerase
- p-TEFb
- positive transcription elongation factor b
- pol
- polymerase
- CTD
- carboxyl-terminal domain.
REFERENCES
- 1. Ritossa F. (1962) Experientia 18, 571–573 [Google Scholar]
- 2. Morimoto R. I. (2008) Genes Dev. 22, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shamovsky I., Nudler E. (2008) Cell. Mol. Life Sci. 65, 855–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rabindran S. K., Giorgi G., Clos J., Wu C. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 6906–6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakai A., Tanabe M., Kawazoe Y., Inazawa J., Morimoto R. I., Nagata K. (1997) Mol. Cell. Biol. 17, 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rougvie A. E., Lis J. T. (1988) Cell 54, 795–804 [DOI] [PubMed] [Google Scholar]
- 7. Brown S. A., Imbalzano A. N., Kingston R. E. (1996) Genes Dev. 10, 1479–1490 [DOI] [PubMed] [Google Scholar]
- 8. Lis J. T., Mason P., Peng J., Price D. H., Werner J. (2000) Genes Dev. 14, 792–803 [PMC free article] [PubMed] [Google Scholar]
- 9. Saunders A., Core L. J., Lis J. T. (2006) Nat. Rev. Mol. Cell Biol. 7, 557–567 [DOI] [PubMed] [Google Scholar]
- 10. Renner D. B., Yamaguchi Y., Wada T., Handa H., Price D. H. (2001) J. Biol. Chem. 276, 42601–42609 [DOI] [PubMed] [Google Scholar]
- 11. Kline M. P., Morimoto R. I. (1997) Mol. Cell. Biol. 17, 2107–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sarge K. D., Murphy S. P., Morimoto R. I. (1993) Mol. Cell. Biol. 13, 1392–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westwood J. T., Wu C. (1993) Mol. Cell. Biol. 13, 3481–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holmberg C. I., Tran S. E., Eriksson J. E., Sistonen L. (2002) Trends Biochem. Sci. 27, 619–627 [DOI] [PubMed] [Google Scholar]
- 15. Kim S. A., Yoon J. H., Lee S. H., Ahn S. G. (2005) J. Biol. Chem. 280, 12653–12657 [DOI] [PubMed] [Google Scholar]
- 16. Lee Y. J., Kim E. H., Lee J. S., Jeoung D., Bae S., Kwon S. H., Lee Y. S. (2008) Cancer Res. 68, 7550–7560 [DOI] [PubMed] [Google Scholar]
- 17. Taylor D. M., De Koninck P., Minotti S., Durham H. D. (2007) Mol. Cell. Neurosci. 34, 20–33 [DOI] [PubMed] [Google Scholar]
- 18. Holmberg C. I., Hietakangas V., Mikhailov A., Rantanen J. O., Kallio M., Meinander A., Hellman J., Morrice N., MacKintosh C., Morimoto R. I., Eriksson J. E., Sistonen L. (2001) EMBO J. 20, 3800–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holmberg C. I., Roos P. M., Lord J. M., Eriksson J. E., Sistonen L. (1998) J. Cell Sci. 111, 3357–3365 [DOI] [PubMed] [Google Scholar]
- 20. Chu B., Zhong R., Soncin F., Stevenson M. A., Calderwood S. K. (1998) J. Biol. Chem. 273, 18640–18646 [DOI] [PubMed] [Google Scholar]
- 21. Knauf U., Newton E. M., Kyriakis J., Kingston R. E. (1996) Genes Dev. 10, 2782–2793 [DOI] [PubMed] [Google Scholar]
- 22. Xavier I. J., Mercier P. A., McLoughlin C. M., Ali A., Woodgett J. R., Ovsenek N. (2000) J. Biol. Chem. 275, 29147–29152 [DOI] [PubMed] [Google Scholar]
- 23. Xiao H., Perisic O., Lis J. T. (1991) Cell 64, 585–593 [DOI] [PubMed] [Google Scholar]
- 24. Vilaboa N. E., Galán A., Troyano A., de Blas E., Aller P. (2000) J. Biol. Chem. 275, 24970–24976 [DOI] [PubMed] [Google Scholar]
- 25. Yoo H. Y., Chang M. S., Rho H. M. (1999) J. Biol. Chem. 274, 23887–23892 [DOI] [PubMed] [Google Scholar]
- 26. Stanhill A., Levin V., Hendel A., Shachar I., Kazanov D., Arber N., Kaminski N., Engelberg D. (2006) Oncogene 25, 1485–1495 [DOI] [PubMed] [Google Scholar]
- 27. Kingston R. E., Baldwin A. S., Jr., Sharp P. A. (1984) Nature 312, 280–282 [DOI] [PubMed] [Google Scholar]
- 28. Kingston R. E., Cowie A., Morimoto R. I., Gwinn K. A. (1986) Mol. Cell. Biol. 6, 3180–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoang A. T., Huang J., Rudra-Ganguly N., Zheng J., Powell W. C., Rabindran S. K., Wu C., Roy-Burman P. (2000) Am. J. Pathol. 156, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Agoff S. N., Hou J., Linzer D. I., Wu B. (1993) Science 259, 84–87 [DOI] [PubMed] [Google Scholar]
- 31. Lee S. A., Ndisang D., Patel C., Dennis J. H., Faulkes D. J., D'Arrigo C., Samady L., Farooqui-Kabir S., Heads R. J., Latchman D. S., Budhram-Mahadeo V. S. (2005) Cancer Res. 65, 3072–3080 [DOI] [PubMed] [Google Scholar]
- 32. Dai C., Whitesell L., Rogers A. B., Lindquist S. (2007) Cell 130, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo F., Rocha K., Bali P., Pranpat M., Fiskus W., Boyapalle S., Kumaraswamy S., Balasis M., Greedy B., Armitage E. S., Lawrence N., Bhalla K. (2005) Cancer Res. 65, 10536–10544 [DOI] [PubMed] [Google Scholar]
- 34. Rocchi P., Beraldi E., Ettinger S., Fazli L., Vessella R. L., Nelson C., Gleave M. (2005) Cancer Res. 65, 11083–11093 [DOI] [PubMed] [Google Scholar]
- 35. Jäättelä M. (1995) Int. J. Cancer 60, 689–693 [DOI] [PubMed] [Google Scholar]
- 36. Garrido C., Fromentin A., Bonnotte B., Favre N., Moutet M., Arrigo A. P., Mehlen P., Solary E. (1998) Cancer Res. 58, 5495–5499 [PubMed] [Google Scholar]
- 37. Volloch V. Z., Sherman M. Y. (1999) Oncogene 18, 3648–3651 [DOI] [PubMed] [Google Scholar]
- 38. Seo J. S., Park Y. M., Kim J. I., Shim E. H., Kim C. W., Jang J. J., Kim S. H., Lee W. H. (1996) Biochem. Biophys. Res. Commun. 218, 582–587 [DOI] [PubMed] [Google Scholar]
- 39. Schulte T. W., Blagosklonny M. V., Romanova L., Mushinski J. F., Monia B. P., Johnston J. F., Nguyen P., Trepel J., Neckers L. M. (1996) Mol. Cell. Biol. 16, 5839–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zou J., Guo Y., Guettouche T., Smith D. F., Voellmy R. (1998) Cell 94, 471–480 [DOI] [PubMed] [Google Scholar]
- 41. Shin K. D., Lee M. Y., Shin D. S., Lee S., Son K. H., Koh S., Paik Y. K., Kwon B. M., Han D. C. (2005) J. Biol. Chem. 280, 41439–41448 [DOI] [PubMed] [Google Scholar]
- 42. Lee J. H., Koo T. H., Hwang B. Y., Lee J. J. (2002) J. Biol. Chem. 277, 18411–18420 [DOI] [PubMed] [Google Scholar]
- 43. Guettouche T., Boellmann F., Lane W. S., Voellmy R. (2005) BMC Biochem. 6, 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Solit D. B., Chiosis G. (2008) Drug Discov. Today 13, 38–43 [DOI] [PubMed] [Google Scholar]
- 45. Sánchez-Serrano I. (2006) Nat. Rev. Drug Discov. 5, 107–114 [DOI] [PubMed] [Google Scholar]
- 46. Weinstein I. B. (2002) Science 297, 63–64 [DOI] [PubMed] [Google Scholar]
- 47. Luo J., Solimini N. L., Elledge S. J. (2009) Cell 136, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Solimini N. L., Luo J., Elledge S. J. (2007) Cell 130, 986–988 [DOI] [PubMed] [Google Scholar]
- 49. Szatrowski T. P., Nathan C. F. (1991) Cancer Res. 51, 794–798 [PubMed] [Google Scholar]
- 50. Burdine L., Kodadek T. (2004) Chem. Biol. 11, 593–597 [DOI] [PubMed] [Google Scholar]
- 51. Park J. M., Werner J., Kim J. M., Lis J. T., Kim Y. J. (2001) Mol. Cell 8, 9–19 [DOI] [PubMed] [Google Scholar]
- 52. Whitesell L., Lindquist S. (2009) Expert Opin. Ther. Targets 13, 469–478 [DOI] [PubMed] [Google Scholar]
- 53. Elia G., Amici C., Rossi A., Santoro M. G. (1996) Cancer Res. 56, 210–217 [PubMed] [Google Scholar]
- 54. Mulholland P. J., Ferry D. R., Anderson D., Hussain S. A., Young A. M., Cook J. E., Hodgkin E., Seymour L. W., Kerr D. J. (2001) Ann. Oncol. 12, 245–248 [DOI] [PubMed] [Google Scholar]
- 55. Yokota S., Kitahara M., Nagata K. (2000) Cancer Res. 60, 2942–2948 [PubMed] [Google Scholar]
- 56. Akagawa H., Takano Y., Ishii A., Mizuno S., Izui R., Sameshima T., Kawamura N., Dobashi K., Yoshioka T. (1999) J. Antibiot. 52, 960–970 [DOI] [PubMed] [Google Scholar]
- 57. Westerheide S. D., Kawahara T. L., Orton K., Morimoto R. I. (2006) J. Biol. Chem. 281, 9616–9622 [DOI] [PubMed] [Google Scholar]
- 58. Zaarur N., Gabai V. L., Porco J. A., Jr., Calderwood S., Sherman M. Y. (2006) Cancer Res. 66, 1783–1791 [DOI] [PubMed] [Google Scholar]
- 59. Leuenroth S. J., Crews C. M. (2008) Cancer Res. 68, 5257–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]