Abstract
A subset of poly ADP-ribose polymerases (PARP) that also contain macro domains regulate transcription. One such macro PARP, PARP-14 alters interleukin 4 (IL-4) and Stat6-dependent transcription. Stat6, activated by IL-4 plays an important role in T helper cell immunity and B cell responses. Here we define the mechanism by which PARP-14 regulates Stat6-activated transcription. Under non-stimulating conditions, PARP-14 recruits HDAC 2 and 3 to IL-4 responsive promoters. In the presence of IL-4, PARP-14 promotes efficient binding of Stat6 to its target genes. Moreover, HDAC 2 and 3 are released from the promoter with an IL-4 signal, this is aided by the ADP-ribosylation of the HDACs by PARP-14. The HDACs and PARP-14 get replaced by coactivators containing HAT activity. Based on these observations we put forth a mechanism in which PARP-14 functions as a transcriptional switch for Stat6-dependent gene induction. Thus, in the absence of a signal PARP-14 acts as a transcriptional repressor by recruiting HDACs. In contrast, in the presence of IL-4 the catalytic activity of PARP-14 facilitates Stat6 binding to the promoter, and release of HDACs so as to activate transcription.
Keywords: Interleukin, STAT Transcription Factor, Transcription Coactivators, Transcription Regulation, Transcription Repressor, Interleukin-4, PARP-14, Stat6, Transcriptional Switch
Introduction
The poly ADP-ribose polymerase (PARP)2 superfamily of proteins is characterized by the presence of the PARP catalytic domain and consists of 17 members (1). These proteins are implicated in a number of cellular processes including, DNA damage repair, transcription regulation, telomere cohesion, and energy metabolism (1, 2). The PARP domain catalyzes the transfer of ADP-ribose moieties from NAD to protein acceptors (1). Recently, a new nomenclature for this family of proteins was proposed based on the structure of the catalytic domain, termed the ADP-ribosyltransferases diphtheria toxin-like (ARTD) family of proteins (3). Within the PARP/ARTD family, there are three members that contain macro domains. These domains were originally found in the non-classical histone macroH2A (mH2A) (4, 5). One of these proteins, PARP-14/ARDT8/CoaSt6/BAL2 was shown to regulate interleukin-4 dependent transcription (6). To avoid confusion and as the official gene symbol for this protein is Parp14, here we are using the old PARP family nomenclature.
Interleukin 4 (IL-4) is a pleiotropic cytokine that plays an important role in both T and B cell function. IL-4 promotes naïve T cells to differentiate into the Th2 phenotype, which plays an important role in immunity against extracellular parasites, humoral immunity, and allergy. IL-4 also acts as growth factor for B cells and promotes immunoglobulin class switching (7). IL-4 uses the Jak-Stat pathway to mediate these functions, specifically it activates the transcription factor Stat6 (8). Stat6 is found in the cell in its latent form, upon cytokine stimulation it is phosphorylated by the Jak kinases, this leads to dimerization and translocation of Stat6 to the nucleus where it binds to canonical DNA elements to induce transcription (9). In B cells, the targets of Stat6 include the germline epsilon (Iϵ, precursor for IgE) promoter and the gene for low affinity IgE Fc receptor (Fcer2a) (10). Stat6 recruits a number of coactivators at the promoter to activate transcription efficiently. These include HATs (histone acetyl transferase) such as, CBP/p300 and the nuclear coactivators (NCoA) (11–15). The protein p100 (TSN) has been shown to enhance Stat6-dependent transcription by bridging Stat6 to RNA polymerase II (16). We have demonstrated that PARP-14 potently and specifically enhances Stat6 dependent transcription (6). The PARP catalytic domain found in PARP-14 is enzymatically active, and it uses NAD as a substrate to transfer ADP-ribose onto itself and p100 (17). We have previously shown that this enzymatic activity of PARP-14 is required for its enhancement of Stat6-mediated transcription (17). However, the exact molecular mechanism by which the ADP-ribosyl transferase reaction impacts Stat6 dependent transcription is not known. Here we provide evidence that PARP-14 is important for the binding of Stat6 to the promoter it activates.
In addition to containing the PARP catalytic domain, PARP-14 contains three copies of the macro domains that were first identified in the non-classical histone macroH2A (mH2A) (5). Like core histone H2A, mH2A is also associated with nucleosomes and replaces H2A in three percent of vertebrate nucleosomes (4). mH2A participates in the inactivation of the X chromosome (Xi) and depletion of mH2A in female cells results in the reactivation of genes on Xi (18, 19). There is some evidence that macro domains associate with histone deacetylases (HDACs). Thus, mH2A may participate in transcriptional repression by recruiting HDACs (20). All of these observations indicate that macro domains are transcription repressors rather than activators. It is possible that the macro domains found in PARP-14 may also function to repress transcription. However, we have shown that PARP-14 enhances Stat6-dependent transcription instead of repressing it (6). To reconcile this paradox here we present evidence that PARP-14 does function as a repressor first by recruiting HDAC 2 and 3. However, in the presence of IL-4 the ADP-ribosyl transferase activity of PARP-14 is activated, which results in relieving its repressive function.
EXPERIMENTAL PROCEDURES
Cell Lines and Mice
293T and M12 B cells were cultured in DMEM and RPMI, respectively, supplemented by 10% FBS as previously described (6). Parp14−/− mice on a C57BL/6 background were a kind gift from Dr. Mark Boothby. 6–8-week-old mice were used for the entire study. Mice were maintained in a pathogen-free condition, and all studies were approved by Indiana University School of Medicine and the Institutional Animal Care and Use Committee. B cells were isolated from spleens using the MACS system (Miltenyi Biotec), according to the manufacturer's protocol and cultured in IMDM-10% FBS. The cells were treated with 5–10 ng/ml of IL-4 (PeproTech), 10 or 50 μm PJ34 (Alexis Biochemicals) and 5 nm TSA (Sigma-Aldrich) as indicated in the figures. For the ChIP and expression analysis of the Iϵ gene, the cells were treated with 2 μg/ml anti-CD40 (BD Biosciences).
Chromatin Immunoprecipitation
The cells were fixed with 1% formaldehyde for 10 min and quenched by adding 0.125 m glycine. The chromatin was sonicated to average length of 200–1000 bp. Samples were precleared with protein A beads and immunoprecipitated with anti-Stat6 (Santa Cruz Biotech), anti-PARP-14 (6), anti-HDAC2 (Santa Cruz Biotech), anti-HDAC3 (Santa Cruz Biotech), anti-acetyl H3 (Upstate Biotech), anti-acetyl H4 (Upstate Biotech), anti-p300 (Santa Cruz Biotech), anti-NCoA1 (Santa Cruz Biotech), anti-NCoA3 (Santa Cruz Biotech), or isotype control (Upstate Biotech). The immune complexes were collected on protein A beads, and the beads were washed three times in immunoprecipitation buffer. Samples were treated with proteinase K and cross-linking was reversed by heating at 65 °C for 4 h. DNA was extracted using phenol-chloroform and ethanol and analyzed for Fcer2a and ~Iϵ gene fragments by SYBR green qPCR. The specific primers used for PCR analysis are listed in supplemental Table S1. The immunoprecipitated fragments were expressed as a percentage of the total chromatin used in the sample.
DNA Affinity Precipitation Assay (DAPA)
Cell extracts were prepared from M12 cells or 293T cells transfected with PARP-14 cDNA. 500 μg of protein extract were incubated with double-stranded biotinylated oligonucleotides corresponding to the Fcer2a and Iϵ promoter regions (supplemental Table S1) immobilized on streptavidin-agarose beads (Millipore) and 100 μg of salmon sperm DNA. The reaction was carried out for 2 h in pull-down buffer containing 25 mm HEPES, 15 mm NaCL, 0.5 mm DTT, 0.1 mm EDTA, 10% glycerol, and 0.5% Nonidet P-40 at 4 °C. The beads were then washed with pull-down buffer to remove nonspecific binding. The proteins bound to the oligonucleotides were then identified by Western blotting.
Gene Expression Analysis
Total RNA was isolated using the TRIzol reagent (Invitrogen). cDNA was prepared using the SuperScript First-Strand cDNA synthesis system (Invitrogen). Quantitative RT-PCR (qRT-PCR) for Fcer2a and Iϵ transcripts was performed by the comparative threshold cycle method and normalized to Gapdh or Hprt. The primers used for these analyses are listed in supplemental Table S1.
Transfections, Immunoprecipitation, GST Pull-down Assays, Immunoblotting, and Reporter Assays
293T cells were transfected using the calcium phosphate precipitation method and whole cell extracts were prepared. 5 μg of the indicated antibodies were used for immunoprecipitation along with protein G beads. The immunoprecipitates were equally divided, resolved on SDS-PAGE, blotted onto PVDF membranes and probed with the indicated antibodies. GST-tagged proteins were expressed in the BL-21 bacterial strain and immobilized onto glutathione beads. These were then incubated with extracts from cells that were transfected with the indicated HDAC constructs. After 1 h of binding the beads were washed and the proteins bound to the beads were eluted in SDS-PAGE-loading dye and analyzed by Western blotting or Coomassie staining as indicated. M12 cells were transfected with the indicated plasmids by electroporation as described previously (6). After 24 h, the cells were divided and were untreated or treated with IL-4 for 24 h, and both luciferase and β-galactosidase assays (Promega) were performed according to the manufacturer's protocol.
PARP Assay
PARP assays were performed essentially as described earlier (17). Briefly, the reaction was performed with immunoprecipitated PARP-14 or its variants on protein G beads and in PARP reaction buffer (20 μl) containing 10 μCi of 32P-labeled NAD (800 Ci/mmol) (Perkin Elmer), 50 μm cold NAD, 50 mm Tris-Cl, pH 8.0, 4 mm MgCl2, and 0.2 mm dithiothreitol for 30 min. The reaction was stopped by washing the beads with PBS and adding loading dye. To evaluate ADP-ribosylation, the proteins were then resolved on SDS-PAGE and autoradiographed.
Flow Cytometry
Cells were fixed with 1.5% formaldehyde for 10 min and permeabilized with 100% cold methanol for 10 min. The cells were then stained with PE-conjugated anti-pStat6 and FITC-conjugated anti-B220 (BD Biosciences) and analyzed by flow cytometry.
RESULTS
PARP-14 and Its Enzymatic Activity Promote the Binding of Stat6 to Target Genes
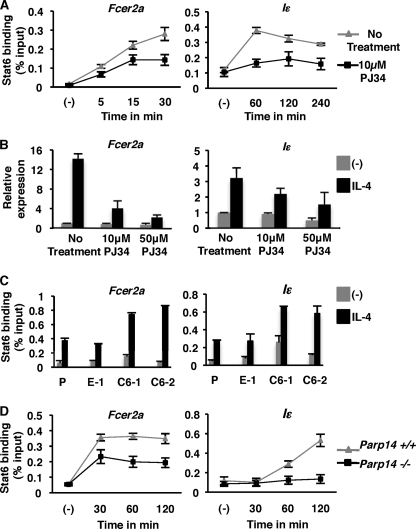
It has been previously demonstrated that Stat6-dependent transcription is enhanced by PARP-14 and the enzymatic activity associated with it (6, 17). To further investigate the exact molecular mechanism by which ADP-ribosylation promotes Stat6 transcription, we tested the binding of Stat6 to IL-4-responsive promoters in the presence of a PARP inhibitor. The M12 B cell line was pretreated with PJ34, a PARP inhibitor for 30 min before incubation with IL-4. The binding of Stat6 to two IL-4 responsive promoters (Fcer2a and Iϵ) was then determined by chromatin immunoprecipitation (ChIP) at different intervals after IL-4 treatment. Stat6 binding to the Fcer2a promoter region was observed as early as 5-min post-IL-4 treatment. The maximum response was observed 30 min after the addition of IL-4. Importantly, pretreatment of cells with PJ34 resulted in reduced Stat6 binding to the Fcer2a promoter (Fig. 1A). A similar trend was observed for the Iϵ promoter (Fig. 1A). It is possible that the reduced binding of Stat6 to its promoter elements in the presence PJ34 was due to decreased Stat6 phosphorylation. To test the effect of inhibition of PARP activity on Stat6 phosphorylation we treated M12 cells similar to the ChIP experiments outlined above, and performed flow cytometric analysis with an antibody specific for phospho-Stat6. We did not observe any changes in the phosphorylation of Stat6 when the cells were treated with PJ34 (supplemental Fig. S1a). Taken together, these data indicate that PARP activity promotes the binding of Stat6 to targets genes independent of the proximal signaling events that activate Stat6. To evaluate if reduced Stat6 binding to the Fcer2a and Iϵ promoters in the presence of PJ34 resulted in a corresponding decrease in the expression of these genes, we performed quantitative RT-PCR for the Fcer2a and Iϵ transcripts. When cells were treated with IL-4, with increasing amounts of PJ34, we observed a dose-dependent inhibition of Fcer2a and Iϵ induction (Fig. 1B). These data indicate that the maximal binding of Stat6 to the promoters it activates requires PARP activity, and this correlates to the level of transcription induced by Stat6.
FIGURE 1.
PARP-14 and the enzyme activity associated with it dictate the DNA binding ability of Stat6. A, M12 cells were either untreated or pretreated with PJ34 (10 μm) for 30 min, after which IL-4 was added for the indicated times. Nuclear extracts from these cells were then used for ChIP with anti-Stat6 and evaluated for the presence of either the Fcer2a or germline epsilon (Iϵ) promoter by qPCR. B, total RNA was extracted from M12 cells that were treated as indicated. Transcripts for either Fcer2a or Iϵ were quantified using qRT-PCR. The data plotted is the relative expression of Fcer2a normalized to Hprt and compared with untreated samples, relative expression of Iϵ is normalized to Gapdh and compared with untreated controls. C, parental M12 cells (P), two M12 cell lines overexpressing PARP-14 (C6-1 and C6-2) and an empty vector control (E-1) that were generated and published earlier (6) were treated with IL-4 and Stat6 ChIPs were performed similar to A. D, B cells were isolated from either Parp14−/− or wild-type littermate controls and were treated with IL-4 for the indicated time points. ChIP experiments similar to A were performed on nuclear extracts obtained from these cells. The data plotted for each panel is the mean (±S.E.) of three independent experiments.
We have previously shown that ectopic expression of PARP-14 in M12 B cells results in the increased expression of CD23 (6). To test whether the increase in CD23 expression in these M12 cell lines was due to increased Stat6 binding to its promoter element, we performed Stat6 ChIP experiments in the absence and presence of IL-4. Consistent with the increase in CD23 expression (6) we observed that there was increased binding of Stat6 to both Fcer2a and Iϵ promoters in cell lines overexpressing PARP-14 (C6-1 and C6-2) as compared with the controls (Fig. 1C and Ref. 6). Taken together, these data indicate that the expression level of PARP-14 and the enzyme activity associated with it play an important role in the binding efficiency of Stat6 to its cognate promoter elements.
PJ34 is a broad spectrum inhibitor of PARP catalytic activity and, thus, from the above experiments we are unable to determine the role PARP-14 specifically plays in the binding of Stat6 to the promoters it activates. Therefore, to determine if PARP-14 affects Stat6 binding to its cognate promoters, we performed ChIP experiments on B cells isolated from PARP-14 deficient animals (21) and littermate controls with intact PARP-14 expression. As expected the binding of Stat6 to both the Fcer2a and Iϵ promoter region was observed after IL-4 treatment in B cells isolated from wild type mice (Fig. 1D). Similar to our observation with PJ34 treatment, lack of PARP-14 expression resulted in a significant decrease in the binding of Stat6 to both the Fcer2a and Iϵ promoters (Fig. 1D). Particularly, the binding of Stat6 to the Iϵ promoter was completely lost in the absence of PARP-14 expression (Fig. 1D, right panel). The IL-4-dependent phosphorylation of Stat6 in B cells with and without PARP-14 expression was compared by flow cytometric analysis and no differences were observed (supplemental Fig. S1b). These data are consistent with our previous results with PJ34 indicating that, neither PARP activity nor PARP-14 influences Stat6 phosphorylation. We further tested the expression of Fcer2a and Iϵ transcripts in WT and Parp14−/− mice that were left untreated or treated with the PARP inhibitor. As expected the expression of both transcripts were decreased in PARP-14-deficient animals as compared with WT (supplemental Fig. S2). Consistent with our Stat6 binding data, the Iϵ gene expression was more dependent on PARP-14. Fcer2a expression was more affected by the presence of PJ34 than the absence of PARP-14 suggesting that other PARP enzymes may also play a role in regulating Fcer2a. However, it seems unlikely that other PARPs are involved in regulating Iϵ, as PJ34 was unable to significantly reduce the expression of Iϵ in Parp14−/− B cells (supplemental Fig. S2). Taken together, these data indicate that the efficient binding of Stat6 to its promoter elements and therefore, the transcription competence is dependent on both the expression of PARP-14 and the enzyme activity associated with it.
PARP-14 Is Associated with Stat6-responsive Promoters in the Absence of IL-4 and Disassociates upon IL-4 Treatment
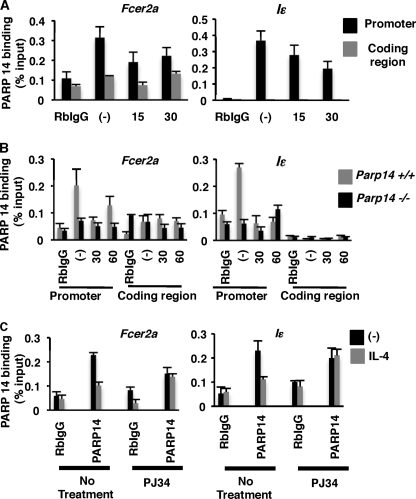
We have shown thus far that PARP-14 and the catalytic activity associated with it dictates the efficiency by which Stat6 binds to the promoter element it activates. However, it is not known whether PARP-14 regulates Stat6 mediated transcription indirectly or directly by binding to the same promoter DNA sequence as Stat6. To test the DNA binding capacity of PARP-14 we first performed DNA affinity precipitation assays (DAPA) with 50 bp biotinylated double-stranded oligonucleotides corresponding to the Fcer2a and Iϵ promoter region flanking the Stat6 binding site. Cell extracts from M12s treated in the presence or absence of IL-4 were incubated with the oligonucleotides immobilized on beads. After extensive washing of the beads, proteins bound to the oligonucleotides were probed with anti-Stat6. Treatment with IL-4 increased binding of Stat6 to both oligonucleotides (supplemental Fig. S3a). We performed similar experiments with 293T cell extracts transfected with PARP-14 or a vector control. A robust association of PARP-14 to both the promoter fragments was observed (supplemental Fig. S3b). These data indicate that PARP-14 associates with DNA that contains a Stat6 binding element. To investigate whether PARP-14 associates with native Stat6 responsive promoters we performed ChIP experiments with an antibody directed against PARP-14. M12 cells were treated with IL-4 for varying lengths of time and ChIPs were performed using anti-PARP-14 antibody, followed by qPCR to quantify DNA sequences corresponding to either, the Fcer2a and Iϵ promoters, or the coding sequences of these genes. We observed that under non-stimulating conditions PARP-14 associated with both of the Stat6 responsive promoters tested. Moreover, this association was lost when the cells were treated with IL-4 (Fig. 2A, black bars). PARP-14 was bound only to the promoter region of Fcer2a and Iϵ and not the coding region of these two genes (Fig. 2A, gray bars). These results indicate that PARP-14 regulates Stat6-mediated transcription by using a mechanism in which it is bound to the promoter in the absence of IL-4 and is no more required at the promoter in the presence of IL-4. To validate the specificity of the anti-PARP-14 antibody for our ChIP experiments, we isolated B cells from PARP-14-deficient animals and their wild-type littermates and performed similar ChIP experiments. Our data are consistent with our M12 studies where we observed that PARP-14 was associated with DNA in the absence of IL-4 and this association was lost upon IL-4 treatment (Fig. 2B, gray bars). Importantly, we observed no positive ChIP signal in PARP-14-deficient samples (Fig. 2B, black bars).
FIGURE 2.
PARP-14 is bound to Stat6-responsive promoters under non-stimulating conditions and has less affinity for DNA in the presence of IL-4. A, M12 cells were treated with IL-4 for the indicated time points, and nuclear extracts were prepared. These were then used for ChIP with an antibody raised against PARP-14. Primers specific for the either the Fcer2a or Iϵ promoters (black bars), or their respective coding regions (gray bars) were used to determine their presence by qPCR. B, similar experiments using B cells isolated for Parp14+/+ or Parp14−/− mice were performed. C, M12 cells were treated with IL-4 and PJ34 as indicated and PARP-14 ChIPs similar to A were performed. Means (±S.E.) from three independent experiments are plotted for each panel.
Previously, we have shown that PARP-14 specifically enhances Stat6 but not Stat1-dependent gene induction (6). Therefore, we next tested whether the association of PARP-14 under non-stimulating conditions was specific for Stat6-responsive promoters. Similar PARP-14 ChIP experiments were performed and the immunoprecipitated DNA was analyzed for the presence of DNA elements corresponding to the Stat1-responsive Irf1 promoter and Stat4-responsive Il18r1 promoter (22, 23). Consistent with our previous finding the association of PARP-14 was specific for Stat6-responsive genes and no association was observed for either, Stat1 or Stat4 responsive promoters (supplemental Fig. S3). Taken together, these results suggest that PARP-14 specifically participates in regulating Stat6 but not Stat1- or Stat4-responsive promoters. PARP-14 is bound to the promoter in the absence of a signal, and then in the presence of an activating signal, PARP-14 exits the promoter before transcription initiates.
In the previous section we have shown that the PARP enzyme activity plays an important role in Stat6-dependent gene expression and, now we present evidence that PARP-14 disassociates from Stat6-responsive promoters upon IL-4 stimulation. Therefore, next we wanted to determine if the disassociation of PARP-14 from DNA was dependent on the PARP catalytic activity. To test this, cells were treated with a combination of IL-4 and PJ34 as indicated (Fig. 2C) and PARP-14 ChIPs were performed as before. PARP-14 was unable to disassociate from both the Fcer2a and Iϵ promoters upon IL-4 treatment in the presence of PJ34 (Fig. 2C). These data indicate that to regulate Stat6-dependent transcription PARP-14 uses a mechanism in which it leaves the promoter with a cytokine signal, and this function requires PARP enzymatic activity.
Histone Deacetylases Are Recruited by PARP-14 and Regulate Stat6-dependent Transcription
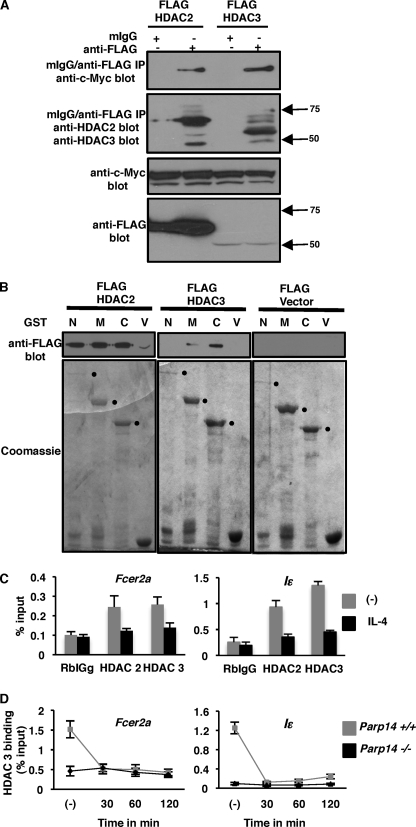
Our finding that PARP-14 is associated with Stat6-responsive promoters in the absence of IL-4, and is disassociated following stimulation, suggests that PARP-14 may initially act as a repressor or recruit repressors to prevent transcription under non-stimulating conditions. However, in the presence of an appropriate signal the repressive function of PARP-14 may no longer be required and thus, it may need to disassociate from the promoter for transcription to be activated. As outlined in the introduction, PARP-14 contains three iterations of the macro domains, and similar macro domains found in macroH2A are known to associate with HDACs (20). Thus, we evaluated if the class I HDACs, that are predominantly nuclear, associate with PARP-14. We co-expressed Flag-tagged HDAC and Myc-tagged PARP-14 in 293T cells and performed co-immunoprecipitation experiments. When the cell extracts were subjected to immunoprecipitation with anti-Flag or an isotype control antibody we observed positive signals for HDAC 2 and 3 only with anti-Flag. Importantly, when the same immunoprecipitates were probed with anti-Myc we observed positive signals corresponding to the molecular weight of PARP-14 only in the anti-FLAG IPs (Fig. 3A). These results indicate that PARP-14 associates with HDAC 2 and 3. To test which regions of PARP-14 associated with HDAC 2 and 3, we performed pull-down experiments with the N (aa 1–813), middle (aa 800–1395), or C (aa 1389–1817) portion of PARP-14 (17) tagged with GST and HDAC 2 and 3. Bacterially expressed regions of PARP-14 tagged with GST or GST alone were immobilized on glutathione-agarose beads and incubated with cell extracts transfected with Flag-tagged HDAC 2 or 3. After extensive washing of the beads the bound proteins were subjected to Western blotting and probed with antibodies specific for the Flag epitope. GST by itself was unable to pull-down either HDAC 2 or 3. However, HDAC2 showed association with all three regions of PARP-14, and HDAC3 was brought down by the middle and C-terminal of PARP-14 (Fig. 3B). Taken together, these data indicate that PARP-14 associates with HDAC 2 and 3 and the N-terminal and middle portion of PARP-14 participate in its ability to bind the two HDACs.
FIGURE 3.
HDAC 2 and 3 partner with PARP-14 and associate with promoters activated by Stat6. A, 293T cells were co-transfected with the indicated FLAG-tagged HDAC constructs and c-Myc-tagged PARP-14. Cell extracts obtained from these cells were subjected to immunoprecipitation with either anti-FLAG or an isotype control. The immunoprecipitates were then probed with the indicated antibodies. To confirm expression from transfected plasmids straight Westerns were performed as indicated. B, indicated regions of PARP-14 tagged with GST were immobilized onto glutathione agarose beads and incubated with extracts from cells transfected with the indicated HDAC constructs. The proteins pulled down by the beads were then evaluated by Western blotting and Coomassie staining as indicated. C, nuclear extracts of M12 cells treated with or without IL-4 were subjected to ChIP using the indicated antibodies. The presence of promoter fragments corresponding to either Fcer2a or Iϵ in the immunoprecipitates were evaluated as in Fig. 1A. D, B cells isolated from Parp14+/+ and Parp14−/− mice were treated with IL-4 for the indicated times and ChIPs using HDAC 3 antibody were performed as in C. The plotted graphs are the mean (± S.E.) from three independent experiments.
Thus far, our data are consistent with a hypothesis that HDAC 2 and 3 through PARP-14 may repress Stat6-dependent transcription under non-stimulating conditions. If this was truly the case then we should observe the regulation of IL-4- and Stat6-dependent transcription by HDAC 2 and 3. To test this, M12 cells were co-transfected with a Stat6-responsive reporter construct and either, an empty vector, or one encoding HDAC 2 or 3. The transfected cells were treated with IL-4 or nothing and the reporter activity was determined. In the presence of both HDAC 2 and 3, the IL-4-mediated induction of the reporter was decreased by at least 50% (supplemental Fig. S4a). To further evaluate if HDACs play a role in IL-4-dependent gene induction we used an alternate approach. M12 cells were treated with a HDAC inhibitor, TSA (trichostatin A), the expression of Fcer2a and Iϵ transcripts was evaluated by qRT-PCR after IL-4 treatment. Both the Fcer2a and Iϵ transcripts were significantly increased in the presence of TSA (supplemental Fig. S4b). Our reporter assays along with the experiments with TSA strongly indicate that HDAC 2 and 3 regulate IL-4- and Stat6-dependent transcription.
If HDAC 2 and 3 repress basal transcription from an IL-4 responsive promoter then these molecules should be present at the promoter under non-stimulating conditions. To test this we performed ChIP experiments with the HDAC 2 and 3 antibodies and examined the presence of Fcer2a and Iϵ promoter elements in the immunoprecipitates. As expected we observed that both HDAC 2 and 3 were bound to the Fcer2a and the Iϵ promoters in the absence of an IL-4 signal (Fig. 3C, gray bars). This association was lost when the cells were incubated with IL-4 (Fig. 3C, black bars). Thus far, our biochemical and functional data with HDAC 2 and 3 are highly suggestive that these molecules participate in regulation of transcription mediated by Stat6 and this may occur via the association of the HDACs with PARP-14. To confirm that the HDACs were binding to the Stat6-responsive promoters via PARP-14, we compared HDAC3 association with the promoters in B cells isolated from either wild type or PARP-14-deficient mice. Our HDAC3 ChIP analysis showed that it was bound to DNA under non-stimulating conditions in B cells isolated from wild-type mice, and this association was lost upon IL-4 stimulation (Fig. 3D). Importantly, HDAC3 did not show any association with Stat6-responsive promoters in B cells isolated from PARP-14 knock-out mice. These data strongly indicate that HDAC3 binds to Stat6-responsive genes through its interaction with PARP 14.
PARP-14 ADP-ribosylates Itself and HDAC 2 and 3
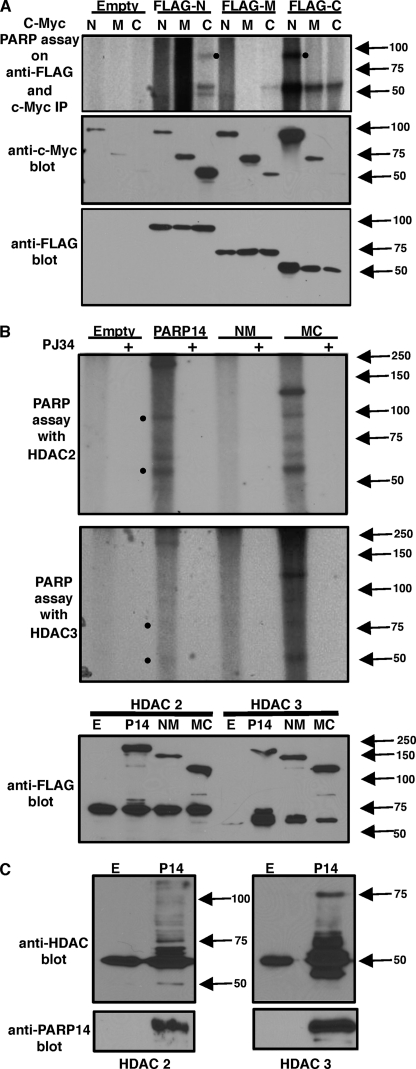
As we have shown that the ADP-ribosylation enzymatic activity is quite important for regulating Stat6-dependent transcription. Therefore, next we wanted to investigate the exact mechanism by which the enzyme activity associated with PARP-14 impacts transcription. Previously we have demonstrated that PARP-14 ADP-ribosylates p100 (17), a coactivator that is recruited by Stat6 (16). Indeed, it has been demonstrated that besides auto-modifying itself PARP-1 ADP-ribosylates other transcription factors to regulate transcription (24). Thus, first we tested if other portions of PARP-14 besides its C-terminal act as acceptors of ADP-ribose moieties. We have previously shown that the regions of PARP-14 that contain the C-terminal get ADP-ribosylated (17). We used those same plasmid constructs containing either FLAG-tagged N-terminal (N), middle region (M), or C-terminal (C), and cotransfected them with c-Myc-tagged N, M, or C of PARP-14. The resultant cell extracts were subjected to immunoprecipitation with anti-FLAG and anti-c-Myc. Portions of the immunoprecipitate were subjected to immunoblot analysis to verify the precipitation of the various proteins. Another portion of the immunoprecipitate was used for PARP assay with radiolabeled (32P) NAD. All of the samples that contained the C-terminal region of PARP-14 showed a positive signal on the autoradiograph (Fig. 4A). Importantly, a positive signal corresponding to the size of the N terminus of PARP-14 was observed in the samples that contained both the N and C termini of PARP-14 (Fig. 4A). These results indicate that in addition to the C terminus, the N terminus of PARP-14 also acts as an acceptor for ADP-ribose residues.
FIGURE 4.
PARP-14 ADP-ribosylates itself on the N-terminal, as well as, HDAC 2 and 3 get modified by PARP-14. A, indicated c-Myc-tagged PARP-14 variants were coexpressed with FLAG-tagged N-terminal, middle region, and C-terminal of PARP-14 in 293T cells. The resultant extracts were subjected to immunoprecipitation with anti-c-Myc and anti-FLAG and the immunoprecipitate was divided into three portions. One set of samples was incubated with 32P NAD (PARP assay) and autoradiographed after resolving the proteins on SDS-PAGE. The other fractions of the immunoprecipitate were probed with the indicated antibodies. B, indicated FLAG-tagged PARP-14 constructs were cotransfected with either FLAG-tagged HDAC 2 or 3. After immunoprecipitating both the proteins with anti-FLAG, PARP assays were performed similar to A and probed with anti-FLAG as indicated. C, similar to B PARP assays were performed on immunoprecipitates containing PARP-14 and HDACs with cold NAD. Instead of autoradiography the samples were probed with antibodies specific for HDAC 2 and 3 as indicated.
We have shown earlier that HDAC 2 and 3 are recruited by PARP-14. Thus, next we tested if HDAC 2 and 3 act as substrates for PARP-14. FLAG-tagged PARP-14, NM and MC were coexpressed along with FLAG-tagged HDAC 2 or 3. The extracts obtained from these transfections were then immunoprecipitated with anti-FLAG and PARP assays and Western blots were carried out. We also performed PARP assays in the presence of PJ34. Besides observing a 32P-positive signal corresponding to full-length PARP-14 and its MC version, we also observed a radiolabeled band corresponding to the size of HDAC 2 and 3 in these lanes (Fig. 4B). Other higher molecular weight bands were also observed in these lanes. Importantly, these signals were absent in the PARP-14 NM sample, which lacks the PARP catalytic domain and, the samples that were treated with the PARP inhibitor (Fig. 4B). To verify that indeed the higher molecular weight bands were modified HDAC 2 and 3, we performed similar experiments with PARP-14 and HDAC 2 and 3. However, instead of performing radioactive PARP assays we performed PARP assays with cold NAD and probed the resultant blots with HDAC 2 and 3 specific antibodies (Fig. 4C). Consistent with our assay with 32P NAD, we observed higher molecular weight species of HDAC 2 and 3 (Fig. 4C). Taken together these data strongly indicate that PARP-14 is able to ADP-ribosylate both HDAC 2 and 3.
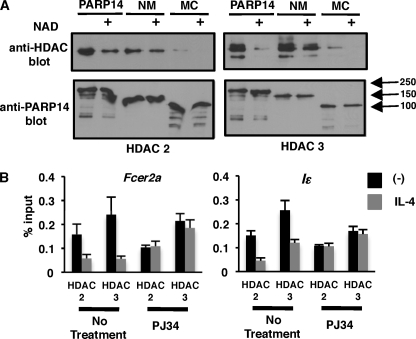
Association between HDACs and PARP-14 Is Lost in the Presence of NAD
We showed earlier that PARP-14 associates with HDAC 2 and 3. Further, we have demonstrated that PARP-14 is able to ADP-ribosylate itself at the N terminus and, both HDAC 2 and 3. It is possible that once both PARP-14 and the HDACs are ADP-ribosylated they are unable to interact due to the presence of negatively charged ADP-ribose residues on both these molecules. To test this hypothesis we coexpressed FLAG-tagged HDACs with c-Myc-tagged PARP-14, the NM or MC version in 293T cells. The resultant extracts were used for immunoprecipitation with anti-c-Myc and the immunoprecipitate was divided equally. One portion was treated with cold NAD for the PARP reaction to proceed and the other was left untreated. After the PARP reaction was completed, the immunoprecipitates were washed thoroughly to remove all unbound proteins. The immunoprecipitated proteins were then probed with anti-HDAC 2 and 3, respectively, to determine the amount of HDACs that coimmunoprecipitated with PARP-14 (Fig. 5A). Consistent with our previous data we observed strong association between PARP-14 and the HDACs in the absence of NAD (Fig. 5A). Notably, this association was significantly reduced when the immunoprecipitates were treated with NAD. When the NM version of PARP-14 lacking the PARP catalytic domain was used for a similar assay, there was very little difference in the extent of association between PARP-14 and HDACs in the absence or presence of NAD (Fig. 5A). However, the MC version of PARP-14 that contains the PARP catalytic domain showed a similar profile like full-length PARP-14. The MC version of PARP-14 was unable to coimmu-noprecipitate the HDACs to the same extent as full-length PARP-14 or NM suggesting that the N-terminal of PARP-14 is more important to mediate the association between the two proteins (Fig. 5A). With this data and the data presented in the previous sections we conclude that PARP-14 recruits HDAC 2 and 3 to Stat6-responsive promoters. Once the enzyme activity of PARP-14 is induced by IL-4-activated Stat6, PARP-14 ADP-ribosylates itself as well as the HDACs. The ADP-ribosylated HDACs are no longer able to associate with PARP-14 and this allows for the HDACs to exit the promoter region.
FIGURE 5.
A, HDAC 2 and 3 do not associate with PARP-14 in the presence of NAD. The indicated c-Myc-tagged PARP-14 variants were coexpressed with FLAG-tagged HDAC 2 or 3 in 283T cells. The resultant extracts were immunoprecipitated with anti-c-Myc and divided equally. One portion was treated with NAD and the other was left untreated as indicated. After the completion of the PARP reaction the immunoprecipitate was washed thoroughly and probed with the indicated antibodies. B, disassociation of HDAC 2 and 3 from Stat6-responsive promoters upon IL-4 stimulation requires PARP catalytic activity. M12 cells were treated with PJ34 and IL-4 as indicated, and HDAC 2 and 3 ChIPs were performed as indicated in Fig. 3C.
If indeed this were the case then inhibiting PARP activity during IL-4 stimulation would preclude HDACs to disassociate from the promoter. To address this scenario we carried out ChIP experiments with anti-HDAC 2 and 3-like before and included samples in which the cells were also treated with PJ34 to inhibit PARP catalytic activity (Fig. 5B). Inhibiting PARP activity resulted in HDAC 2 and 3 to remain on the promoter in the presence of IL-4 (Fig. 5B). These data are consistent with the molecular mechanism that we have proposed above.
Sequence of Molecular Events at Stat6-responsive Promoters upon IL-4 Stimulation
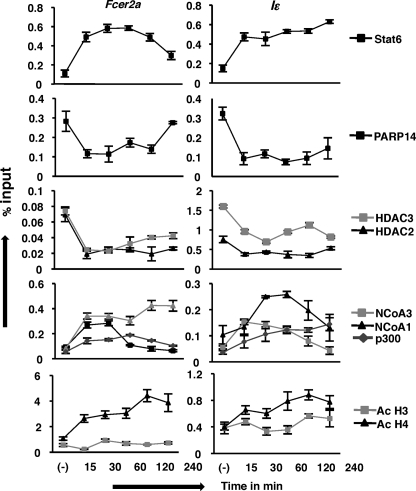
Previously, it has been demonstrated that histone acetyl transferases such as CBP/p300, NCoA-1, and NCoA-3 promote Stat6-dependent transcription (11, 14, 15). However, the exact sequence of events by which IL-4- and Stat6-responsive genes are activated is not known. Therefore, to delineate the exact molecular mechanism that occurs at the Stat6-responsive promoter, we performed a series of ChIPs at various time points after IL-4 stimulation with antibodies raised against the indicated molecules for two IL-4-responsive genes (Fig. 6). As expected, under non-stimulating conditions Stat6 was not bound to the promoters. Consistent with our previous result, PARP-14, HDAC 2 and 3 were bound to DNA in the absence of IL-4. Fifteen minutes after IL-4 treatment we observed the association of Stat6 to both the promoters tested and the loss of association of PARP-14 and the HDACs. These events coincided with the association of the p300, NCoA-1 and 3 to the promoters and the acetylation of histone H3 and H4. For the most part, both promoters showed a similar pattern for protein-DNA interaction, however, some differences between Fcer2a and Iϵ were observed. For example, for the Fcer2a promoter-less of Stat6 was bound at 4-h post IL-4 treatment, and this overlapped with the re-association of PARP-14. However, for the Iϵ promoter Stat6 did not disassociate from the promoter nor was there significant re-association of PARP-14 at the 4 h time point. For both Fcer2a and Iϵ, once the HDACs disassociated from the DNA they did not re-associate even after 4 h of IL-4 treatment. These data indicate that there is a reciprocal association of Stat6 and PARP-14 at these promoters, and confirms previous data that HDACs are present at the promoter only when PARP-14 is present. The association of the HATs with the Fcer2a and Iϵ promoters was also distinct for each of the genes. NCoA-1 associated transiently with the Fcer2a promoter for 30 min after IL-4 treatment and was no more bound at the 1 h time point. In contrast, for the Iϵ promoter, NCoA-1 showed a more sustained binding which was lost after 4 h of IL-4 treatment. NCoA-3 on the other hand, bound quickly to the Fcer2a promoter after IL-4 treatment and the binding persisted for 4 h. The Iϵ promoter showed a different pattern, as NCoA-3 bound to DNA only after 30 min of IL-4 induction and started to fall off the promoter starting at 2 h. The maximum binding of p300 to the DNA was observed at 1 h IL-4 treatment after which it waned for the Fcer2a gene. On the other hand, for the Iϵ gene there was a gradual increase in the association of p300 that increased until the last time point tested. The acetylation status of histone H3 and H4 mirrored the association of the HATs with the promoter, though H4 demonstrated a greater increase in acetylation. Taken together, these data define the dynamic molecular events that occur at a Stat6-responsive promoter upon IL-4 treatment.
FIGURE 6.
Molecular events involved in the transcription from the Fcer2a and Iϵ promoters. M12 cells were treated with IL-4 for the indicated time intervals. ChIP studies were performed using the indicated antibodies. The pulled-down DNA was quantified for the presence of Stat6-responsive elements found in the Fcer2a and Iϵ genes by qPCR. The results are plotted as a mean (±S.E.) from three independent experiments.
DISCUSSION
PARP-14 Functions as a Molecular Switch
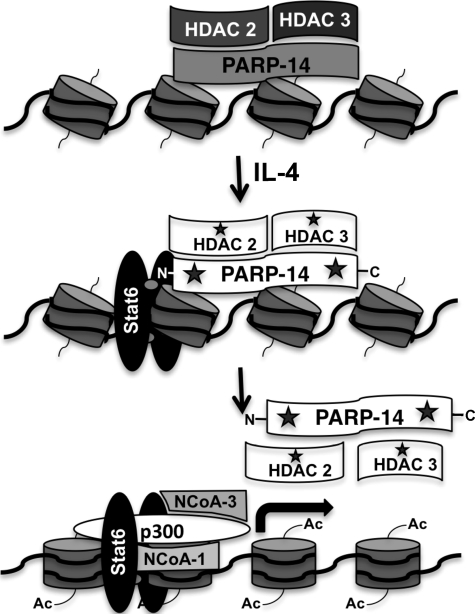
Based on the observations presented here we propose a mechanism by which PARP-14 regulates Stat6- and IL-4-mediated transcription. Our data are consistent with a model wherein PARP-14 functions as a transcriptional switch, first functioning to keep transcription off under non-stimulating conditions by binding to the promoter and recruiting HDAC 2 and 3 to the promoter (Fig. 7). In the presence of an IL-4 signal and activated Stat6, it is likely that the enzymatic activity of PARP-14 is activated, and this converts it from a repressor to an activator (Fig. 7). Our previous data indicating higher PARP-14 enzymatic activity in the presence of IL-4 activated Stat6 supports this conclusion (17). We have also previously demonstrated that the enzymatic activity associated with PARP-14 is required for its enhancement function (17). Moreover, here we provide evidence that this enzymatic activity is required for efficient binding of Stat6 to its cognate DNA element (Fig. 1). All of these observations indicate that the ADP ribosyl transferase function of PARP-14 is extremely important for its transcriptional enhancement function. It is intriguing to speculate that once the enzymatic activity of PARP-14 is activated this is a signal for PARP-14 and HDACs to exit the promoter making the promoter accessible to the histone acetyl transferases. Indeed, we provide data here that PARP-14 ADP-ribosylates its N terminus and the HDACs (Fig. 4), these modifications most likely cause PARP-14 and HDACs to disassociate from each other and the promoter (Figs. 2 and 5). Based on our model PARP-14 acts as a repressor first, therefore, the prediction would be that loss of PARP-14 expression should result in hyperexpression of Stat6-responsive genes under non-stimulating conditions. But our data do not show this result, we observe no differences in the expression of Fcer2a and Iϵ under non-stimulating conditions in the presence or absence of PARP-14 expression (supplemental Fig. S2). We speculate that PARP-9 which has the same domain architecture as PARP-14 but does not contain PARP catalytic activity (26), compensates for the repressive but not the activation potential of PARP-14 in PARP-14-deficient cells. Consistent with the mechanism we are proposing, Ouatathni et al. (25) have shown that for the inducible Hsp70.1 and Hsp70.2 promoters, PARP-1 that is bound to these promoters is inactive under basal conditions. In the presence of a heat shock signal PARP-1 gets auto-modified and this leads to the release of both PARP-1 and mH2A from the promoter (25). This group also demonstrated that mH2A associated with HDAC1. As PARP-14 contains both the macro domain and the PARP catalytic domain it may utilize a similar mechanism as observed with PARP-1 and mH2A for the Hsp70 promoters. Nevertheless, here we provide evidence for a unique mechanism in which a single molecule, PARP-14 functions first to switch off transcription under basal conditions, however, in the presence of a signal the catalytic activity associated with PARP-14 allows for the transcription to be activated.
FIGURE 7.
PARP-14 functions as a transcriptional switch for Stat6 dependent transcription. Conceptual model of how PARP-14 may regulate Stat6-mediated transcription. Under non-stimulating conditions PARP-14 is bound to Stat6-responsive promoters and recruits HDAC 2 and 3 and keeps the gene silent. Upon IL-4 stimulation, Stat6 is activated and binds to its promoter element and induces the PARP-14 enzymatic activity indicated by the star symbol. PARP-14 then modifies itself on the N terminus and HDAC 2 and 3 in the complex, again represented as stars. This results in the disassociation of PARP-14 and the HDACs from the promoter, which allows for p300, NCoA-1, and NCoA-3 to be recruited to the promoter and acetylation of the histones.
DNA Binding Ability of PARP-14
Our ChIP analysis with anti-PARP-14 indicates that it has the ability to associate with DNA. We have shown that PARP-14 associates with promoter elements rather than the coding sequence of the gene (Fig. 2). Moreover, PARP-14 is bound to IL-4- and Stat6-responsive promoters but not Stat1- or Stat4-responsive genes (supplemental Fig. S3). These data indicate that PARP-14 exhibits specificity to the DNA it associates with. The specific ability of PARP-14 to associate with DNA may be dictated by the direct binding of PARP-14 to a particular DNA sequence. Or, other factors associated with a specific DNA sequences may recruit PARP-14 to a particular region of a gene. A search for conserved domains in PARP-14 did not reveal a known DNA binding motif. The only conserved domains that were found in PARP-14 were the macro domains, the WWE domain and the PARP catalytic domain (6). Neither of these domains has been reported to have any DNA binding ability. The N-terminal of PARP-14 is rather large and it is possible that this region of PARP-14 contains a unique DNA binding domain. This will need to be further investigated experimentally or by structural analysis. The other alternative that other DNA binding factors recruit PARP-14 to Stat6-responsive genes is an intriguing thought. It is known that both C/EBP and NF-kB collaborate with Stat6 to induce transcription (10). It is possible that these factors are already present at the promoter and they are the ones that associate with PARP-14 to bring it to the promoter under non-stimulating conditions. Further investigations need to be conducted to better understand the mechanism by which PARP-14 associates with DNA. However, we provide strong evidence that there is a dynamic association and disassociation of PARP-14 with Stat6 promoters and, that this interaction is required for PARP-14 to regulate IL-4-dependent transcription.
Modulation of Stat6 Activity in the Presence of PARP Inhibitors
Here we have demonstrated that in the presence of PARP inhibitors the efficiency at which Stat6 binds to the promoter it activates is compromised (Fig. 1A). Correspondingly, the expression of these genes is also reduced in the presence of PARP inhibitors (Fig. 1B). These data suggest that PARP inhibitors may be employed in disease states in which the pathogenesis is caused by Stat6 and IL-4. It is well established that IL-4 and Stat6 play a major role in the progression of asthma. IL-4 and Stat6 are responsible for the differentiation of naïve T helper (Th) cells to a Th2 phenotype that produce IL-4, IL-5, and IL-13 and promote an asthmatic response (7). Moreover, both IL-4 and Stat6 play a major role in immunoglobulin class switching in B cells. The IgE isotype is one of the major antibodies produced with an IL-4 and Stat6 signal. IgE is a central player in allergic hypersensitivity (7). As we have demonstrated that PARP-14 enhances IL-4 and Stat6 function (6), it is intriguing to speculate that PARP-14 may also be involved in Th2 differentiation and the pathogenesis of asthma. Indeed we have preliminary evidence that indicates that inhibiting PARP activity during differentiation of T helper cells reduces the ability of these cells to produce Th2 cytokines.3 Moreover, here we provide evidence that indicates that the germline transcription for the precursor of IgE (Iϵ) is reduced in the presence of PARP inhibitors (Fig. 1B), as well as, in the absence of PARP-14 expression (supplemental Fig. S2). Consistent with our Iϵ expression data, our experiments with Parp14-deficient cells indicates that the binding of Stat6 to the Iϵ promoter is significantly compromised when compared with cells with intact PARP-14 expression (Fig. 1C). Taken together these data indicate that targeting PARP activity may be a potential therapy for asthma and allergy. Nevertheless, here we provide insight into the molecular mechanism by which a unique macroPARP enzyme regulates transcription.
Supplementary Material
Acknowledgments
We thank Mark Kaplan for critical review of our manuscript, Mark Boothby for providing us with the Parp14−/− mice, Ed Seto for providing us with the HDAC 2 and 3 expression plasmids.
This work was funded, in whole or in part, by National Institutes of Health Grant HL093105, the Showalter Foundation Grant, and the Biomedical Research Grant awarded by the Indiana University School of Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Table S1.
S. Goenka, unpublished observations.
- PARP
- poly ADP-ribose polymerase
- DAPA
- DNA affinity precipitation assay
- HDAC
- histone deacetylase
- ARTD
- ADP-ribosyltransferases diphtheria toxin-like
- NCoA
- nuclear coactivators.
REFERENCES
- 1. Schreiber V., Dantzer F., Ame J. C., de Murcia G. (2006) Nat. Rev. Mol. Cell Biol. 7, 517–528 [DOI] [PubMed] [Google Scholar]
- 2. Kim M. Y., Zhang T., Kraus W. L. (2005) Genes Dev. 19, 1951–1967 [DOI] [PubMed] [Google Scholar]
- 3. Hottiger M. O., Hassa P. O., Luscher B., Schuler H., Koch-Nolte F. (2010) Trends Biochem. Sci. 35, 208–219 [DOI] [PubMed] [Google Scholar]
- 4. Ladurner A. G. (2003) Mol. Cell 12, 1–3 [DOI] [PubMed] [Google Scholar]
- 5. Pehrson J. R., Fried V. A. (1992) Science 257, 1398–1400 [DOI] [PubMed] [Google Scholar]
- 6. Goenka S., Boothby M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4210–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelms K., Keegan A. D., Zamorano J., Ryan J. J., Paul W. E. (1999) Annu. Rev. Immunol. 17, 701–738 [DOI] [PubMed] [Google Scholar]
- 8. Schindler C. (1999) Exp. Cell Res. 253, 7–14 [DOI] [PubMed] [Google Scholar]
- 9. Wurster A. L., Tanaka T., Grusby M. J. (2000) Oncogene. 19, 2577–2584 [DOI] [PubMed] [Google Scholar]
- 10. Tinnell S. B., Jacobs-Helber S. M., Sterneck E., Sawyer S. T., Conrad D. H. (1998) Int. Immunol. 10, 1529–1538 [DOI] [PubMed] [Google Scholar]
- 11. Gingras S., Simard J., Groner B., Pfitzner E. (1999) Nucleic. Acids. Res. 27, 2722–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McDonald C., Reich N. C. (1999) J Interferon Cytokine Res. 19, 711–722 [DOI] [PubMed] [Google Scholar]
- 13. Goenka S., Marlar C., Schindler U., Boothby M. (2003) J. Biol. Chem. 278, 50362–50370 [DOI] [PubMed] [Google Scholar]
- 14. Litterst C. M., Pfitzner E. (2001) J. Biol. Chem. 276, 45713–45721 [DOI] [PubMed] [Google Scholar]
- 15. Arimura A., vn Peer M., Schröder A. J., Rothman P. B. (2004) J. Biol. Chem. 279, 31105–31112 [DOI] [PubMed] [Google Scholar]
- 16. Yang J., Aittomäki S., Pesu M., Carter K., Saarinen J., Kalkkinen N., Kieff E., Silvennoinen O. (2002) EMBO J. 21, 4950–4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goenka S., Cho S. H., Boothby M. (2007) J. Biol. Chem. 282, 18732–18739 [DOI] [PubMed] [Google Scholar]
- 18. Costanzi C., Pehrson J. R. (1998) Nature 393, 599–601 [DOI] [PubMed] [Google Scholar]
- 19. Chadwick B. P., Valley C. M., Willard H. F. (2001) Nucleic. Acids Res. 29, 2699–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chakravarthy S., Gundimella S. K., Caron C., Perche P. Y., Pehrson J. R., Khochbin S., Luger K. (2005) Mol. Cell. Biol. 25, 7616–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cho S. H., Goenka S., Henttinen T., Gudapati P., Reinikainen A., Eischen C. M., Lahesmaa R., Boothby M. (2009) Blood 113, 2416–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Q., Thieu V. T., Kaplan M. H. (2007) EMBO J. 26, 2052–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thieu V. T., Yu Q., Chang H. C., Yeh N., Nguyen E. T., Sehra S., Kaplan M. H. (2008) Immunity 29, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Butler A. J., Ordahl C. P. (1999) Mol. Cell. Biol. 19, 296–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ouararhni K., Hadj-Slimane R., Ait-Si-Ali S., Robin P., Mietton F., Harel-Bellan A., Dimitrov S., Hamiche A. (2006) Genes Dev. 20, 3324–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aguiar R. C., Takeyama K., He C., Kreinbrink K., Shipp M. A. (2005) J. Biol. Chem. 280, 33756–33765 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.