Abstract
Macrophages play a central role in the balance and efficiency of the immune response and are at the interface between innate and adaptive immunity. Their phenotype is a delicate equilibrium between the M1 (classical, pro-Th1) and M2 (alternative, pro-Th2) profiles. This balance is regulated by cytokines such as interleukin 13 (IL-13), a typical pro-M2-Th2 cytokine that has been related to allergic disease and asthma. IL-13 binds to IL-13 receptor α1 (IL13Rα1), a component of the Type II IL-4 receptor, and exerts its effects by activating the transcription factor signal transducer and activator of transcription 6 (STAT6) through phosphorylation. MicroRNAs are short (∼22 nucleotide) inhibitory non-coding RNAs that block the translation or promote the degradation of their specific mRNA targets. By bioinformatics analysis, we found that microRNA-155 (miR-155) is predicted to target IL13Rα1. This suggested that miR-155 might be involved in the regulation of the M1/M2 balance in macrophages by modulating IL-13 effects. miR-155 has been implicated in the development of a healthy immune system and function as well as in the inflammatory pro-Th1/M1 immune profile. Here we have shown that in human macrophages, miR-155 directly targets IL13Rα1 and reduces the levels of IL13Rα1 protein, leading to diminished activation of STAT6. Finally we also demonstrate that miR-155 affects the IL-13-dependent regulation of several genes (SOCS1, DC-SIGN, CCL18, CD23, and SERPINE) involved in the establishment of an M2/pro-Th2 phenotype in macrophages. Our work shows a central role for miR-155 in determining the M2 phenotype in human macrophages.
Keywords: Gene Expression, Gene Regulation, Immunology, Inflammation, Interleukin, Macrophage, MicroRNA, STAT Transcription Factor
Introduction
Macrophages are key players at the interface between innate and adaptive immunity. They arise from circulating monocytes that are recruited to tissues by different stimuli. They work as phagocytes and antigen-presenting cells, promoting inflammation and its resolution. Macrophages present a wide range of phenotypic profiles and defense mechanisms depending on the tissue context and the stimuli present (pathogens, cytokines, apoptotic cells, and so forth). They are generally classified into two main types: M1 (classically activated) and M2 (alternatively activated) macrophages. Classically activated (M1) macrophages are a result of an exposure to pro-Th1 cytokines, whereas alternatively activated (M2) macrophages are generated in a pro-Th2 environment (1). Classically activated macrophages are specialized in defense against intracellular pathogens, and upon stimulation with pro-inflammatory stimuli (interferon-γ or LPS), they promote inflammation, causing tissue damage. By contrast, alternative activation of macrophages is induced by a broader range of stimuli including interleukin 4 (IL-4), interleukin 13 (IL-13), interleukin 10 (IL-10), or glucocorticoids, and alternative macrophages are specialized in defense against extracellular pathogens, promoting tissue repair and the resolution of the inflammatory process (2). Regardless of this classification, one of the most remarkable characteristics of macrophages is their plasticity and heterogeneity, depending on the specific task carried out. This is reflected by their ability to reverse their phenotype and reprogram their M1/pro-Th1 and M2/pro-Th2 gene expression profiles, presenting in between phenotypic profiles and a constituting a heterogeneous population (1). The present study has focused on alternatively activated macrophages generated by IL-4 and IL-13 (2).
IL-13 is a typical Th2 type cytokine that, together with IL-4, drives and modulates the immune response. First described as a Th1 down-regulator (3), its role as an active immune mediator has been described and distinguished from those of IL-4 by several studies (4–7). Interleukin 13 is a key cytokine in the defense against gastrointestinal nematodes (8) and plays a central role in some chronic inflammatory diseases such as asthma and ulcerative colitis (9, 10). Interestingly, and underscoring the role of IL-13 in asthma, mice lacking the IL-13 receptor α1 chain (IL13Rα1)2 showed a complete absence of allergen-induced airway hyper-reactivity and mucus hypersecretion (6). IL13Rα1 is an essential component of the Type II IL-4 receptor, which consists of heterodimers of IL4Rα and IL13Rα1 chains. Both IL-4 and IL-13 bind to the Type II receptor, but only IL-4 can bind to the Type I receptor. Therefore, the binding of IL-13 depends solely on the presence of IL13Rα1 (11, 12). Engagement of these receptors leads to phosphorylation and activation of Janus tyrosine kinases (JAK) proteins, believed to be bound to these cytokine receptors in unstimulated cells. The active phospho-JAK proteins phosphorylate the IL4Rα chain, providing docking sites for STAT6. Once bound to the receptor, STAT6 is also phosphorylated by JAKs, which causes its activation, dimerization, and translocation to the nucleus, where it exerts its transcriptional roles (13).
Since their relatively recent discovery (14), miRNAs have been shown to play important biological roles in different contexts: during development, cell differentiation, and immune regulation and also in pathologies such as cancer (15–17). They are small non-coding RNAs of ∼22 nucleotides that regulate gene expression upon binding to the 3′-UTRs (untranslated regions) of their target mRNAs (18). MicroRNAs are firstly transcribed as immature primary miRNAs that are processed in the nucleus into ∼70-nucleotide hairpin pre-miRNAs by Drosha proteins. Pre-miRNAs are then exported to the cytoplasm, where Dicer proteins process them into mature miRNA*-miRNA complexes. The leading strand, miRNA (as opposed to miRNA*, the discarded strand) is loaded into the RNA-induced silencing complex, where it guides Argonaute proteins toward their target mRNAs (19). The selectivity of miRNA action is given by the nucleotides 2–7 at their 5′ end (the “seed region”) that pairs to its complementary site in the targeted 3′-UTR by Watson-Crick interactions directing the RNA-induced silencing complex action (18). The inhibitory activity of microRNAs ensues by blocking the target mRNA translation into protein and/or the degradation of the mRNA (20).
MicroRNAs have proven to be key in modulating the immune system (21).The microRNA focus of this study, microRNA-155 (miR-155), has been extensively studied in immunology and inflammation (21–23). Two different knock-out models have been generated showing that mice lacking miR-155 present an abnormal immune function with aberrant B and T cell repertoires and defective antigen-presenting cells (16, 24). Several reports present miR-155 as a key player in B cell responses (17) and in dendritic cell function (25, 26). Moreover, miR-155 expression levels increase during inflammation in classically (pro-Th1) activated macrophages (21, 27) and have been clearly linked to a pro-Th1 bias because knock-out mice for miR-155 have a pro-Th2 unbalanced T cell repertoire (16). Therefore, it is well established that miR-155 plays a central role not only in the development of a healthy immune system but also as a pro-Th1 microRNA.
Using in silico analysis (miRanda (28), RNAHybrid (29), and the PITA algorithm (30), we identified IL13Rα1 as a putative target of miR-155. We have shown for the first time that indeed miR-155 down-regulates the effects of the Th2 cytokines IL-4 and IL-13, providing a straightforward link between miR-155 and the Th1/Th2 balance. Our work shows that miR-155 directly targets IL13Rα1 3′-UTR, reducing the levels of IL13Rα1 protein. By doing so, miR-155 affects the IL-4- and IL-13-dependent phosphorylation of STAT6. We finally show that miR-155 levels modulate the response of human macrophages to IL-13, leading to a change in their genetic profile. Therefore, miR-155 contributes to the Th1/Th2 equilibrium, favoring a pro-Th1/classical activation of macrophages by reducing the expression of several pro-Th2/IL-13-dependent genes.
EXPERIMENTAL PROCEDURES
Cell Culture
THP1-155 Cells
This cell line was generated as described before (26). Briefly, THP-1 cells were doubly transduced with a doxycycline-inducible (Tet-on) lentiviral system described elsewhere (31), in which miR-155 transgene is under the control of a tetracycline response element. Lentiviral vectors were kindly provided by Prof. Didier Trono, and cells were maintained in RPMI 10% FBS (Invitrogen). To induce miR-155 expression, 2.5 μg/ml doxycycline (Sigma) was added or not to the medium and renewed daily over 96 h of culture. Cytokine treatments were performed as follows. Firstly, cells were incubated or not with doxycycline for 96 h, and then they were starved for an extra 12-h period and then stimulated or not with IL-4 (Immunotools) or IL-13 (R&D Systems). Cell extracts were collected when indicated, after the treatment.
HeLa Cells
HeLa cells were maintained in DMEM 10% FBS.
Macrophages
Monocytes were obtained from buffy coats from healthy donors over a density gradient (Ficoll-PaqueTM PLUS, GE Healthcare) following the manufacturer's instructions. Briefly, peripheral blood mononuclear cells were isolated by centrifugation over Ficoll-Paque, and monocytes were isolated from the peripheral blood mononuclear cell fraction using CD14 magnetic microbeads (Miltenyi). Cells were maintained in RPMI 10% FBS supplemented with 500 units/ml GM-CSF (Immunotools) to allow differentiation.
Vector Constructs
For pcDNA BIC, the genomic region encompassing miR-155 was cloned into pcDNA 3.1 expression vector as described previously (26). IL13Rα1 3′-UTR was amplified using plasmid DNA (HmiT009700-MT01, gene Copoeia) as template and the following primers: IL13RA1_3′-UTR forward, 5′-GGC TGT TAG GGG CAG TGG AG-3′ and IL13RA1_3′-UTR reverse, 5′-CAG AGC CTT GGC TGG CTG G-3′. The product was cloned in pCR2.1 TOPO TA (Invitrogen), from where it was removed using BamHI/NotI. After blunt-ending with Klenow DNA polymerase, the product was cloned into the NotI site in pRLTK (Promega); this construct was named henceforth pRLTK__WT_3′-UTR_IL13RA1.
Mutagenesis was performed on pRLTK__WT_3′-UTR_IL13RA1 using QuikChange site-directed mutagenesis (Stratagene) following the manufacturer's instructions. For pRLTK_MUT1_3′-UTR_IL13RA1, we used the primers IL13RA1 3′-UTR MUT1 forward, 5′-CTG CTA CTC AAG TCG GTA CCA CTG TGT CTT TGG TTT GTG CTA GGC CCC-3′ and IL13RA1 3′-UTR MUT1 reverse, 5′-GGG GCC TAG CAC AAA CCA AAG ACA CAG TGG TAC CGA CTT GAG TAG CAG-3′. In the case of pRLTK__MUT2_3′-UTR_IL13RA1, the primers used were IL13RA1 3′-UTR MUT2 forward, 5′-CCA TGT GAG GGT TTT CAG GGC CGA TAT TTG TGC ATT TTC TAA ACA G-3′ and IL13RA1 3′-UTR MUT2 reverse, 5′-CTG TTT AGA AAA TGC ACA AAT ATC GGC CCT GAA AAC CCT CAC ATG G-3′. Normalization was performed using pGL3 (Promega).
RT-qPCR
Total RNA was extracted using TRI Reagent (Ambion). Reverse transcription was performed using the high capacity cDNA reverse transcription kit (Applied Biosystems). MicroRNA detection was performed using TaqMan microRNA assays (Applied Biosystems). For real-time PCR (qPCR), we employed TaqMan® universal PCR master mix, No AmpErase® UNG in a 7900HT fast real-time PCR system machine (both from Applied Biosystems). IL13RA1 qPCR detection was performed using PerfectProbe from PrimerDesign (Southampton, UK) and normalized against GAPDH from the same manufacturer. Genes assayed in the IL-13/STAT6 pathway were TaqMan® gene expression assays (Applied Biosystems).
Western Blotting
Total protein lysates were subjected to SDS-PAGE under reducing conditions and transferred onto an Immobilon polyvinylidene difluoride membrane (Millipore Corp., Bedford, MA). Antibodies used were: anti-IL13Rα1 (antibody sc-27861, Santa Cruz Biotechnology); anti-STAT6 (antibody 9362, Cell Signaling Technology); anti-phospho-STAT6 (antibody 9361, Cell Signaling Technology); and anti-β-actin antibody loading control (ab8227, Abcam).
Transfections
To determine direct targeting of IL13Rα1 by miR-155, constructs pRLTK_WT_3′-UTR_IL13RA1, pRLTK_MUT1_3′-UTR_IL13RA1, or pRLTK_MUT2_3′-UTR_IL13RA1 were transfected into HeLa cells employing TransIT-LT1 reagent (Mirus) following the manufacturer's instructions. pcDNA BIC or control pcDNA 3.1 empty vector were co-transfected to check miR-155 activity on the 3′-UTR of IL13Rα1. pGL3 (Promega) was used as normalizing vector. Luminometry was performed using a Dual-Glo kit (Promega). Experiments were performed three times in triplicates. Statistical differences were determined using Student's t test and GraphPad Prism software.
To reduce the levels of miR-155 in primary macrophages, 100 nm anti-miR-155 inhibitor or Anti-miR inhibitors–negative control #1 (Applied Biosystems) were transfected in human monocytes at day 0 as described previously (26). Briefly, cells were plated onto 96-well flat bottom plates at a cell density of 5 × 105 cells/ml with GM-CSF, and oligonucleotides were added at a final concentration of 100 nm and then kept in culture. On day 3, IL-13 was added or not with a renewal dose of GM-CSF, and 24 h later, RNA was collected and subjected to analysis.
RESULTS
miR-155 Directly Targets IL13Rα1
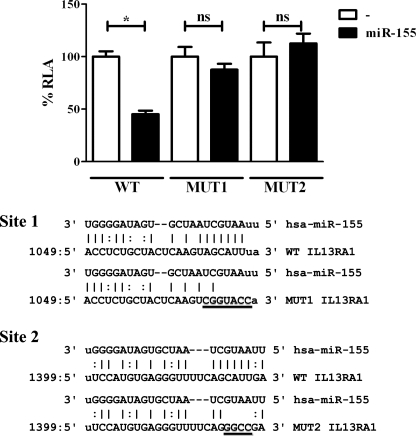
By performing bioinformatic analysis (miRanda, RNAHybrid, and PITA algorithm), we found that IL13Rα1 has two putative binding sites for miR-155 in the 3′-UTR, and hence, it was predicted to be a direct target for miR-155 (Fig. 1). Both miR-155-IL13RA1-3′-UTR pairs are shown in supplemental Fig. S1, and their free energy is within the range of validated miRNA target pairs (32). Site 1 is located between nucleotides 1049 and 1071 of the 3′-UTR (seeding region is a 7-mer), and site 2 is located between nucleotides 1399 and 1424 (seeding region is an 8-mer that includes a G:U wobble pair). The direct targeting by miR-155 of IL13Rα1, a component of the Type II IL-4 receptor, could provide a crucial link between miR-155 and its role as pro-Th1 microRNA; by reducing signaling via the Type II IL-4 receptor, miR-155 would reduce the ability of a cell to respond to the classical pro-Th2 cytokines IL-4 and IL-13. To test IL13Rα1 as a direct target for miR-155, we employed a Dual Luciferase assay in HeLa cells. For this purpose, the 3′-UTR sequence of IL13Rα1 harboring the predicted binding sites for miR-155 was cloned into the Renilla luciferase reporter vector pRLTK (named pRLTK_WT_3′-UTR_IL13RA1). To test the contribution of the predicted binding sites for miR-155 action, each site was mutated by site-directed mutagenesis. Mutations (shown in Fig. 1) were designed according to the predicted abrogation of miR-155 binding using bioinformatics (RNAHybrid). The constructs were named pRLTK_MUT1_3′-UTR_IL13RA1 (mutant in site 1) and pRLTK_MUT2_3′-UTR_IL13RA1 (mutant in site 2). Co-transfection of each Renilla luciferase construct with an expression vector for miR-155 (pcDNA BIC) allowed determination of the effects of this microRNA on the 3′-UTR of IL13Rα1 as well as mapping its binding. When miR-155 was co-transfected with the reporter construct that harbored the wild type 3′-UTR of IL13Rα1 (Fig. 1, WT), the expression of the Renilla luciferase construct was reduced to less than 50% of its activity. When the putative sites for miR-155 binding were mutated individually, both mutants failed to be regulated by miR-155 (Fig. 1, MUT1 and MUT2). Thus, it was concluded that miR-155 directly targets IL13Rα1 and that it binds to the 3′-UTR of IL13Rα1 in positions 1049–1071 and 1399–1424 and that both sites are necessary for miRNA action.
FIGURE 1.
miR-155 directly targets the 3′-UTR of IL13Rα1. HeLa cells were co-transfected with a Renilla luciferase construct harboring an IL13Rα1 3′-UTR fragment containing the predicted binding sites for miR-155 (wild type, WT) and either an empty expression vector (−) or an miR-155-overexpressing vector (miR-155). MUT1 and MUT2 correspond to mutants in each one of the predicted sites, site 1 and site 2, respectively. One of three independent experiments is shown. ns = not significant, *, p ≤ 0.05. Error bars indicate S.D. RLA, relative luciferase activity.
miR-155 Regulates the Expression of IL13Rα 1 in Human Monocytes
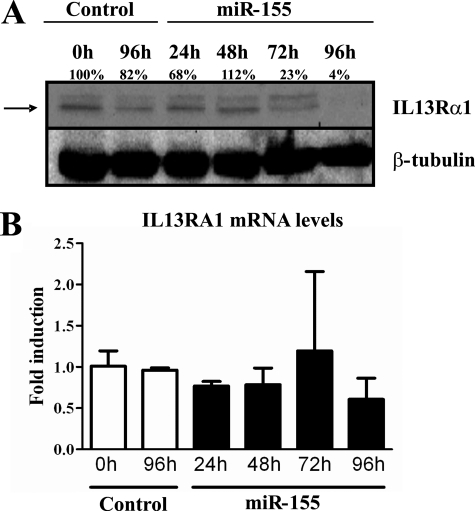
Having established that miR-155 directly targets the IL13Rα1 3′-UTR, we aimed to determine the effects of miR-155 on the expression of this receptor chain in human monocytes. It is known that miR-155 is up-regulated by several pro-Th1 factors during the inflammatory response and that it plays a role in the Th1 response in different cell types such as dendritic cells (25, 26) and macrophages (21, 27). We wondered whether miR-155 could modulate the M2/alternative activation of macrophages by decreasing IL13Rα1 expression. This would also suggest a role for miR-155 in the differentiation of a classically activated pro-Th1 macrophage. For this purpose, we chose a previously established and validated monocytic cell line, THP1-155 (26). The THP1-155 cell line is a doubly transduced cell line in which an miR-155 transgene is under the control of a Tet-On response element, a regulatory system described elsewhere (31). Thus, miR-155 expression can be induced upon the addition of tetracycline or a derivative, doxycycline. This system allows miR-155 up-regulation to be isolated from other events that are also triggered by inflammatory stimuli (e.g. LPS) that lead to miR-155 overexpression (21). In this cellular system, we could determine whether increased levels of miR-155 have an effect on the expression of IL13Rα1 both at the protein and at the mRNA levels. Cells were treated with doxycycline for 96 h, allowing miR-155 overexpression to occur. Cells were collected at 24-h intervals during the course of the treatment, for RNA and protein analysis. miR-155 expression was checked by qPCR and showed a 3.5-fold increase (supplemental Fig. S4). miR-155 effects on IL13Rα1 expression were examined at the protein level by Western blotting protein extracts (Fig. 2A). The protein levels of IL13Rα1 (lower band) were down-regulated in parallel with the up-regulation observed for miR-155. Densitometric analysis determined that IL13Rα1 protein levels were reduced to 4% when cell extracts were compared at time 96 and 0 h of treatment (Fig. 2A). We also determined mRNA levels for IL13Rα1 by RT-qPCR and found that the levels did not vary significantly during the course of the treatment (Fig. 2B).
FIGURE 2.
Overexpression of miR-155 reduces the levels of IL13Rα1 protein. THP1-155 cells were treated with doxycycline (miR-155) or not (Control) during the course of 96 h to allow miR-155 overexpression. Cells were collected in intervals of 24 h and subjected to protein and RNA extraction. A, cell lysates were subjected to Western blotting for IL13Rα1 protein detection (upper panel, lower band indicated by arrow) and normalized against β-tubulin (lower panel). B, total RNA was extracted, and mRNA levels of IL13Rα1 were determined by RT-qPCR. Shown is one experiment out of three independent ones. Statistical analysis of Western blots is shown in supplemental Fig. S5. Error bars indicate S.D.
Altogether, these findings show that miR-155 regulates the expression of IL13Rα1 in human monocytes. Up-regulation of miR-155 led to a down-regulation of IL13Rα1 at the protein level, whereas its mRNA levels remained stable. These findings suggest that miR-155 is acting by blocking the translation of IL13Rα1 mRNA rather than promoting its degradation in THP1-155 cells. These results, together with the molecular link previously shown between the 3′-UTR of IL13Rα1 and miR-155, prove that IL13Rα1 is a direct target for miR-155 in human monocytes.
miR-155 Reduces the IL-13- and IL-4-dependent Phosphorylation of STAT6
Having shown that miR-155 directly targets IL13Rα1 in monocytes (Figs. 1 and 2), we expected that the miR-155-dependent reduction of IL13Rα1 would impair the ability of macrophages to respond to IL-13 and IL-4. Both cytokines bind to the Type II IL-4 receptor, composed of heterodimers of IL13Rα1 and IL4Rα, but only IL-4 can bind to Type I receptors (12). Upon binding to the Type II receptor, IL-4 and IL-13 initiate a signaling cascade that causes phosphorylation and activation of STAT6, which is then able to perform its transcriptional roles (13). An impaired response to IL-13 and IL-4, cytokines that promote M2/alternative activation in macrophages, would bias the macrophages toward a more M1/classical activation. Hence, we predicted that by down-regulating the levels of IL13Rα1, miR-155 would modulate the downstream signaling cascade and reduce STAT6 phosphorylation.
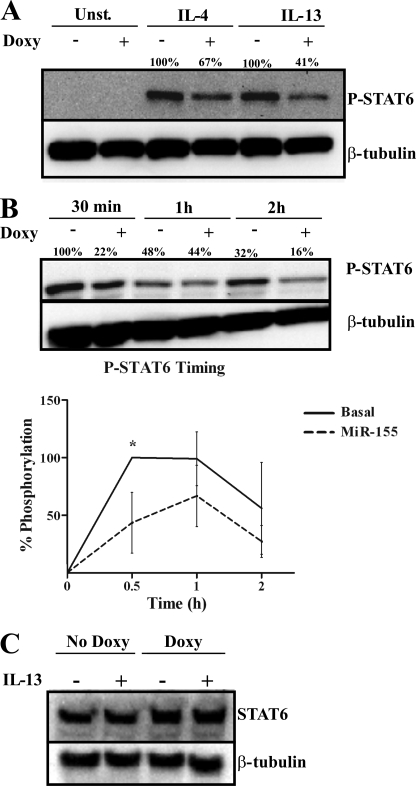
To test the effects of miR-155 levels on STAT6 phosphorylation, we used THP1-155 cells, in which miR-155 overexpression can be induced upon the addition of doxycycline. Moreover, STAT6 phosphorylation can be seen as an indicator of the status of the Th2 pathway (13). THP-1–155 cells were treated (+Doxy) or not (−Doxy) with doxycycline for 5 days to allow miR-155 overexpression. Cells were then starved overnight before stimulation with IL-13 (10 ng/ml) or IL-4 (250 units/ml). When miR-155 was up-regulated (Fig. 3A, +Doxy), IL-4- and IL-13-induced STAT6 phosphorylation was reduced below basal levels (−Doxy). In the case of IL-13 stimulation, the reduction in phospho-STAT6 levels was more pronounced (41% remaining) than in the case of IL-4 (67% phospho-STAT6 remaining). These data point to a more pronounced inhibitory effect by miR-155 on the IL-13 pathway than on the IL-4 one. This could be due to signaling from the Type I IL-4 receptor in the case of IL-4 as IL-13-dependent STAT6 phosphorylation depends solely on its binding to IL13Rα1 (12). We therefore aimed to dissect the role of miR-155 on IL-13 signaling through STAT6 phosphorylation as it relies on the presence of the direct target of miR-155 IL13Rα1.
FIGURE 3.
Overexpression of miR-155 reduces STAT6 phosphorylation. THP1-155 cells were treated with doxycycline (+Doxy) or not (−Doxy) during 96 h to overexpress miR-155. Cells were then starved overnight and stimulated with either IL-4 or IL-13 or not stimulated (Control) and lysed at the indicated times. A, analysis of STAT6 phosphorylation (P-STAT6) after 30 min of treatment was performed by Western blotting and normalized against β-tubulin. B, THP1-155 cells were stimulated with IL-13 or not, collected after 30 min, 1 h, and 2 h, and subjected to Western blotting. The lower panel shows the percentage of STAT6 phosphorylation (P-STAT6) in this panel plotted against time of treatment as analyzed by densitometry (three independent experiments shown, *, p ≤ 0.05). Error bars indicate S.D. C, THP1-155 cells treated or not with doxycycline for 96 h (overexpressing or not miR-155, respectively) were subjected to analysis of total STAT6 content by Western blotting and normalized against β-tubulin expression. Shown is one experiment out of three independent ones. Statistical analysis of Western blots is shown in supplemental Fig. S6.
We then assayed the duration and timing of the effects of miR-155 on IL-13-dependent STAT6 signaling. For this purpose, THP1-155 cells were treated or not with doxycycline over 96 h, respectively allowing (or not) miR-155 overexpression to occur. After this, cells were starved overnight before stimulation with IL-13. Cells were collected and lysed 30 min, 1 h, and 2 h after IL-13 stimulation, and protein extracts were subjected to Western blotting to detect the course of STAT6 phosphorylation.
In cells with basal levels of miR-155 (−Doxy), IL-13-induced STAT6 phosphorylation reached a peak after 30 min and decreased over the course of 2 h, (Fig. 3B, upper panel). In cells overexpressing miR-155 (+Doxy), STAT6 phosphorylation was reduced when compared with non-overexpressing cells (−Doxy), confirming the results shown in Fig. 3A. Quantification of STAT6 phosphorylation (P-STAT6)/β-tubulin (Fig. 3B, lower panel) showed that the peak of STAT6 phosphorylation was abrogated when miR-155 was overexpressed (+Doxy) To determine whether this effect could reflect a direct targeting of STAT6 by miR-155, we analyzed the levels of total STAT6 protein in Western blots of the same cell extracts. STAT6 protein levels were not affected by overexpression of miR-155 (Fig. 3C); therefore, miR-155 is acting at an upstream step of the IL-13 signaling pathway, most likely by targeting IL13Rα1, as shown in Figs. 1 and 2. Our results suggest that miR-155 is reducing the STAT6-dependent alternative activation of macrophages.
miR-155 Regulates IL13Rα1 and STAT6 Phosphorylation in Human Macrophages
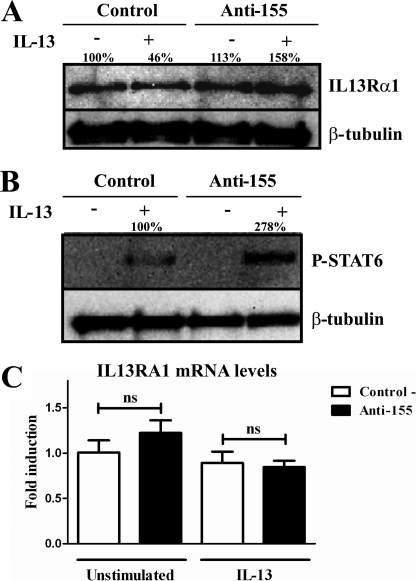
Having shown that miR-155 directly targets IL13Rα1 and that up-regulation of miR-155 in THP1-155 cells reduces the expression of IL13Rα1 with diminished downstream phosphorylation of STAT6, we next investigated in a “reverse model” whether down-regulation of miR-155 would increase IL13Rα1 levels and STAT6 activation in human primary macrophages. Human macrophages were transfected with either specific anti-miR-155 oligonucleotides or a negative control. Three days after transfection, cells were treated (or not) with IL-13 and collected after 2 h. Prior to this treatment, a group of cells was collected to determine expression of miR-155. Supplemental Fig. S9 shows that miR-155 was effectively knocked down. Cell lysates collected after 2 h of treatment were subjected to Western blot detection to determine the levels of IL13Rα1 protein. Down-regulation of miR-155 resulted in increased expression of IL13Rα1 protein, both in IL-13-treated cells and in unstimulated cells (Fig. 4A), thus confirming that miR-155 regulates IL13Rα1 in primary macrophages. The presence of phospho-STAT6 was also assessed in Western blots of cell lysates. As expected, the IL-13-dependent phosphorylation of STAT6 was affected by miR-155 levels (Fig. 4B); when miR-155 was blocked (Anti-155), the phosphorylated and active form of STAT6 showed a 2.7-fold increase when compared with cells transfected with anti-miR control. Interestingly, the levels of mRNA of IL13Rα1 remained constant (Fig. 4C), suggesting that miR-155 targeting effects are due to the blocking of translation of IL13Rα1 mRNA into protein. These data show that miR-155 down-regulates the levels of IL13Rα1 in human macrophages, thereby reducing the phosphorylation of STAT6.
FIGURE 4.
miR-155 down-regulation increases IL13Rα1 protein expression and STAT6 phosphorylation. In a reverse model in human macrophages, cells were transfected with blocking oligonucleotides against miR-155 (Anti-155) or a negative control (Control). On day 3 of culture macrophages were stimulated with IL-13 or not and collected after 30 min. A, cell lysates were subjected to Western blotting to detect IL13Rα1 normalized against β-tubulin. B, cell lysates were used to determine STAT6 phosphorylation (P-STAT6) levels normalizing against β-tubulin. C, RNA was extracted from the same collected cells, and mRNA of IL13Rα1 was determined by RT-qPCR. One of three independent experiments is shown. ns = not significant. Shown is one experiment out of three independent ones. Statistical analysis of Western blots is shown in supplemental Figs. S7 and S8. Error bars indicate S.D.
miR-155 Regulates the IL-13 Cascade in Human Macrophages
Importantly, IL13Rα1 is not only a key component in the IL-13 cascade but is also a marker for alternative activation of macrophages (2), suggesting a possible role for miR-155 in the “M type” or “Th” profile of these cells. miR-155 has been shown to play an important role in the inflammatory profile of macrophages (21), and it is known that STAT6 is the main mediator in the Th2 signaling cascade triggered by IL-13 (13). Our results show that miR-155 directly targets IL13Rα1 (Fig. 1) and that the levels of miR-155 regulate the expression of IL13Rα1 in macrophages (Figs. 2 and 4) with consequent effects on the phosphorylation of STAT6 (Figs. 3 and 4). We next aimed to test the influence of miR-155 levels on the transcriptional profile of macrophages stimulated with IL-13, i.e. M2 or alternatively activated macrophages (2).
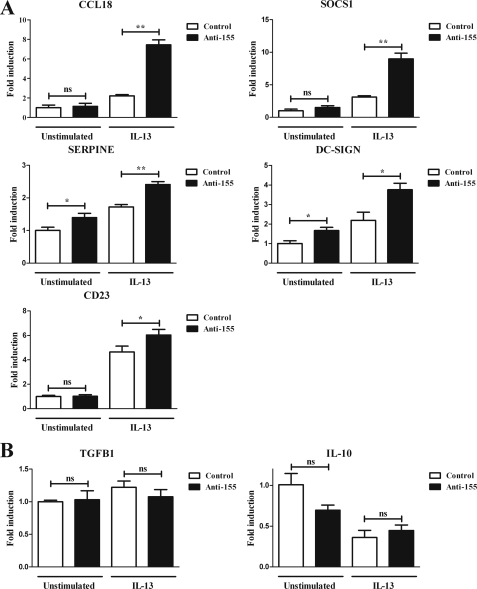
Human primary monocytes were transfected with specific anti-miR-155 inhibitors or control anti-miR oligonucleotides. Three days after transfection, cells were stimulated or not with IL-13 for 24 h and lysed, and RNA was extracted and analyzed by RT-qPCR. Based on our previous results, we hypothesized that reducing the levels of miR-155 would lead to an increase in the IL-13-dependent expression of several target genes in the IL-13 cascade. To test this, we determined the expression of genes known to be involved in the STAT6 cascade and/or alternative activation of macrophages (2, 13). Stimulation with IL-13 increased the levels of expression of SOCS1, DC-SIGN, CCL18, CD23, and SERPINE (Fig. 5A). This occurred in cells transfected with both control anti-miR and active anti-miR-155, although as expected, the up-regulation was significantly greater in the cells in which miR-155 was knocked down (Fig. 5). miR-155 levels did not seem to affect significantly the basal expression of these genes with the exception of DC-SIGN and SERPINE. We also assayed other genes that showed no statistical difference, including TGFβ1 and IL-10. (Fig. 5B). Thus, we can conclude that microRNA-155 directly targets IL13Rα1 in human macrophages, that it has an effect on the activation of STAT6, and that miR-155 levels play a key role in the IL-13 pathway, contributing to the expression profile and activation of macrophages.
FIGURE 5.
Down-regulation of miR-155 increases the transcription of several STAT6/IL-13-dependent genes. Human macrophages were transfected with anti-miR-155 oligonucleotides (Anti-155) or a negative control (Control). On day 3 of culture, cells were stimulated with or without IL-13 and collected 24 h after stimulation to analyze mRNA expression by RT-qPCR analysis. The genes assayed were grouped as genes dependent on the IL-13/STAT6 signaling, CCL18, SOCS1, CD23, SERPINE, and DC-SIGN (A) and as genes not affected by IL-13 treatment, TGFB1 and IL-10 (B). One of three independent experiments is shown. ns = not significant, *, p ≤ 0.05, **, p ≤ 0.01. Error bars indicate S.D.
DISCUSSION
In this work, we have shown that microRNA-155 modulates the response of human macrophages to IL-13, a crucial cytokine in the programming of Th2 responses (8), and we have shown that miR-155 regulates the IL-13-dependent expression profile of these cells. We also provide evidence regarding the mechanism underlying this role, showing that miR-155 directly targets IL13Rα1, a key component of the Type II IL-4 receptor (11). To study the role of miR-155 in the immune profile of monocytes/macrophages, we used a previously generated cell line, THP1-155 cells (26), and primary human macrophages in which we blocked miR-155 expression. Thus, our study provides a molecular mechanism by which miR-155 regulates the response of human macrophages to IL-13.
From our bioinformatic analysis (miRanda, RNAHybrid, and PITA algorithm) IL13Rα1 was predicted as a putative target of miR-155, with two binding sites mapped at positions 1049 (site 1) and 1399 (site 2) in the 3′-UTR of IL13Rα1 mRNA. To determine the role of these potential binding sites, we used a Renilla luciferase reporter assay to show that miR-155 directly targets the 3′-UTR of IL13Rα1 (Fig. 1). Interestingly, both binding sites contributed to the inhibitory role of miR-155 because when either site was mutated, the effect of miR-155 was almost completely abrogated. Both miRNA target pairs present free energy values (supplemental Fig. S1) just below the −20 kcal/mol cut-off suggested by Watanabe et al. (32) and above other strong target sites for miR-155 studied by us (PU.1 and SMAD2, ∼−26 kcal/mol, (26, 44)). Additionally, both sites are conserved across several species (supplemental Figs. S2 and S3), although the conservation is not as wide as in the previously mentioned examples PU.1 and SMAD2. Thus, these relatively weaker binding regions could explain why miR-155 needs both site 1 and site 2 intact to block expression of IL13Rα1 and point toward a possible cooperation between both sites, which has also been suggested to lead to translational repression of the target gene rather than mRNA degradation (33, 34).
We next performed a series of experiments in a cell model, THP1-155, that allows experimental augmentation of miR-155 levels without the use of inflammatory stimuli (such as LPS) that could affect other pathways (21). It was shown that miR-155 up-regulation led to a decrease of IL13Rα1 at the protein level, whereas its mRNA levels remained stable (Fig. 2, A and B). This suggested that miR-155 could be blocking the translation of IL13Rα1 mRNA in these cells.
We next aimed to establish the role of miR-155 in the IL-13 signaling pathway by determining the activation of STAT6. STAT6 is the main mediator in the IL-13/IL-4 signaling pathway, becoming phosphorylated and active upon stimulation with these cytokines (12). The overexpression of miR-155 in THP1-155 led to diminished STAT6 phosphorylation without affecting total STAT6 protein levels (Fig. 3, A–C). Both IL-4 and IL-13 signaling cascades seemed to be affected, which can be explained by the fact that both cytokines share and can signal through the Type II IL-4 receptor, which consists of dimers of IL4Rα and IL13Rα1. Importantly, only IL-4 can also signal through the Type I IL-4 receptor, whereas IL-13 requires the presence of IL13Rα1 to exert its effects. The specific dependence on IL13Rα1 of the IL-13 pathway could explain the observation that IL-13 signaling appeared to be more affected than that induced by IL-4 (Fig. 3A). After this experimental observation, we decided to focus on the effect of miR-155 on IL-13 signaling.
Having investigated the effects of miR-155 in the first series of experiments by augmenting its expression, we went on to confirm our results with a reverse experimental approach in which miR-155 was knocked down by transfection of anti-miR-155 oligonucleotides in human primary macrophages. As expected, the reduction of miR-155 led to an increase in IL13Rα1 protein (Fig. 4A) and in the IL-13-dependent phosphorylation of STAT6 (Fig. 4B). Because IL-13 triggers an M2 or a pro-Th2 phenotype (8), we next determined the impact of miR-155 on several genes that are markers of alternative activation/pro-Th2 profiling and/or phospho-STAT6-dependent genes (2, 13). After reducing the levels of miR-155, IL-13 signaling was increased (Fig. 4), so we hypothesized an increase in the IL-13-dependent expression of several target genes in the IL-13 cascade. Among the genes assayed when miR-155 was knocked down, SOCS1, DC-SIGN, CCL18, CD23, and SERPINE showed a significant increase in their IL-13-dependent gene expression (Fig. 5). DC-SIGN and SERPINE also showed a decrease in their basal expression levels, probably due to a parallel effect of miR-155 on factors that control their expression, such as PU.1 and SMAD2, known to regulate DC-SIGN (35), and SERPINE (36), respectively, and that are directly targeted by miR-155 (26, 44). DC-SIGN is a typical marker for alternatively activated macrophages (2), and it is involved in the binding and recognition of pathogens by the immune system. We have previously described the inhibitory effects of miR-155 on DC-SIGN expression in dendritic cells (26). Thus, the increased expression of DC-SIGN when miR-155 is down-regulated (Fig. 5) suggests that miR-155 is probably affecting pathogen binding ability also in macrophages.
Our results showed that miR-155 blockade leads to increased STAT6 signaling. Because STAT6 is a key transcription factor involved in the generation of Th2 cells, it could be supposed that the pro-Th2-biased phenotype of miR-155-deficient mice might involve STAT6 regulation. Moreover, signaling through STAT6 also involves SHIP-1 (12), a repressor of M2 macrophage differentiation, and SHIP-1 has been demonstrated to be a direct target of miR-155 (37). In addition to this, miR-155 was recently reported to be inhibited by IL-10 (22), an anti-inflammatory cytokine that also promotes alternative activation of macrophages (2). Together, all these data suggest a central role for miR-155 in the acquisition and modulation of the M1/M2 and Th1/Th2 profiles. By targeting IL13Rα1 and modulating the STAT6 cascade, miR-155 would shift the immune profile toward a more pro-Th1 phenotype; thus, the regulation of miR-155 levels is key to exert and develop an appropriate and balanced immune response.
This central role seems to be confirmed by the fact that miR-155 affects several genes that are important to the human immune balance. CCL18 is a cytokine associated with Th2 profiling that has also been shown to be secreted by tumor-associated macrophages (38), macrophages that display an M2-like profile (39); CD23, which binds to IgE complexes, promoting an inflammatory response, has been shown to play an important role in allergy and antigen presentation, also being a typical marker of M2 macrophages; SOCS1 a pro-Th2 protein, inhibits the Janus kinase/signal transducer, blocking the signaling of pro-Th1 stimuli such as LPS (40, 41). SOCS1 is reportedly affected by miR-155 by direct targeting (15, 23), and our data suggest that miR-155 also affects SOCS1 indirectly by reducing the ability of IL-13 to stimulate SOCS1 gene expression.
Importantly, several of the genes affected by miR-155, CCL18, CD23, and SOCS1, together with IL-13, have been shown to play a key role in the pathogenesis of allergy and asthma (9, 42, 43), characterized by an exacerbated Th2 profile and lung remodeling, features also shown by miR-155-deficient mice (16). Additionally, miR-155-mediated down-regulation of the IL-13- and TGFβ- (44) dependent regulation of SERPINE underscores the role of miR-155 in pro-fibrotic processes that could also explain the previously mentioned lung remodeling, an important feature in asthma and other lung diseases.
Together, all these data position miR-155 at the heart of immune regulation and with an important role in the pathogenesis of diseases such as asthma, with a pro-fibrotic remodeling component. This is in line with our previous observation that miR-155 can decrease the response of macrophages to TGFβ (44), which indicated that this microRNA affects not only the immune functions of these cells but also their role in repair and remodeling. The broad implications of these biological functions position miR-155 as a crucial molecule for the fine-tuning of a healthy balance in the immune system and justify future research on the clinical implications that might link miR-155 with several pathologies.
Supplementary Material
Acknowledgment
We thank Professor Peter Friedmann for critical review of the manuscript and for help in the discussion of the results.
This work was supported by Medical Research Council (MRC) Grant G0801984 (to T. S.-E. and to F. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
- IL13Rα1
- interleukin 13 receptor α1
- DC-SIGN
- dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin
- miR
- microRNA
- qPCR
- quantitative PCR.
REFERENCES
- 1. Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. (2004) Trends Immunol. 25, 677–686 [DOI] [PubMed] [Google Scholar]
- 2. Martinez F. O., Helming L., Gordon S. (2009) Annu. Rev. Immunol. 27, 451–483 [DOI] [PubMed] [Google Scholar]
- 3. Minty A., Chalon P., Derocq J. M., Dumont X., Guillemot J. C., Kaghad M., Labit C., Leplatois P., Liauzun P., Miloux B., et al. (1993) Nature 362, 248–250 [DOI] [PubMed] [Google Scholar]
- 4. Hogan S. P., Mould A., Kikutani H., Ramsay A. J., Foster P. S. (1997) J. Clin. Invest. 99, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Punnonen J., Aversa G., Cocks B. G., McKenzie A. N., Menon S., Zurawski G., de Waal Malefyt R., de Vries J. E. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramalingam T. R., Pesce J. T., Sheikh F., Cheever A. W., Mentink-Kane M. M., Wilson M. S., Stevens S., Valenzuela D. M., Murphy A. J., Yancopoulos G. D., Urban J. F., Jr., Donnelly R. P., Wynn T. A. (2008) Nat. Immunol. 9, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kolodsick J. E., Toews G. B., Jakubzick C., Hogaboam C., Moore T. A., McKenzie A., Wilke C. A., Chrisman C. J., Moore B. B. (2004) J. Immunol. 172, 4068–4076 [DOI] [PubMed] [Google Scholar]
- 8. Wynn T. A. (2003) Annu. Rev. Immunol. 21, 425–456 [DOI] [PubMed] [Google Scholar]
- 9. Wills-Karp M. (2004) Immunol. Rev. 202, 175–190 [DOI] [PubMed] [Google Scholar]
- 10. Fuss I. J., Strober W. (2008) Mucosal Immunol. 1, Suppl. 1, S31–S33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hershey G. K. K. (2003) J. Allergy Clin. Immunol. 111, 677–690 [DOI] [PubMed] [Google Scholar]
- 12. Jiang H., Harris M. B., Rothman P. (2000) J. Allergy Clin. Immunol. 105, 1063–1070 [DOI] [PubMed] [Google Scholar]
- 13. Hebenstreit D., Wirnsberger G., Horejs-Hoeck J., Duschl A. (2006) Cytokine Growth Factor Rev. 17, 173–188 [DOI] [PubMed] [Google Scholar]
- 14. Lee R. C., Feinbaum R. L., Ambros V. (1993) Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 15. Jiang S., Zhang H. W., Lu M. H., He X. H., Li Y., Gu H., Liu M. F., Wang E. D. (2010) Cancer Res. 70, 3119–3127 [DOI] [PubMed] [Google Scholar]
- 16. Rodriguez A., Vigorito E., Clare S., Warren M. V., Couttet P., Soond D. R., van Dongen S., Grocock R. J., Das P. P., Miska E. A., Vetrie D., Okkenhaug K., Enright A. J., Dougan G., Turner M., Bradley A. (2007) Science 316, 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vigorito E., Perks K. L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P. P., Miska E. A., Rodriguez A., Bradley A., Smith K. G., Rada C., Enright A. J., Toellner K. M., Maclennan I. C., Turner M. (2007) Immunity 27, 847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 19. Eulalio A., Huntzinger E., Izaurralde E. (2008) Cell 132, 9–14 [DOI] [PubMed] [Google Scholar]
- 20. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 21. O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCoy C. E., Sheedy F. J., Qualls J. E., Doyle S. L., Quinn S. R., Murray P. J., O'Neill L. A. (2010) J. Biol. Chem. 285, 20492–20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu L. F., Thai T. H., Calado D. P., Chaudhry A., Kubo M., Tanaka K., Loeb G. B., Lee H., Yoshimura A., Rajewsky K., Rudensky A. Y. (2009) Immunity 30, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thai T. H., Calado D. P., Casola S., Ansel K. M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J. L., Schmidt-Supprian M., Rajewsky N., Yancopoulos G., Rao A., Rajewsky K. (2007) Science 316, 604–608 [DOI] [PubMed] [Google Scholar]
- 25. Ceppi M., Pereira P. M., Dunand-Sauthier I., Barras E., Reith W., Santos M. A., Pierre P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2735–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martinez-Nunez R. T., Louafi F., Friedmann P. S., Sanchez-Elsner T. (2009) J. Biol. Chem. 284, 16334–16342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Worm J., Stenvang J., Petri A., Frederiksen K. S., Obad S., Elmén J., Hedtjärn M., Straarup E. M., Hansen J. B., Kauppinen S. (2009) Nucleic Acids Res. 37, 5784–5792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008) Nucleic Acids Res. 36, D149–D153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. (2004) RNA 10, 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. (2007) Nat. Genet. 39, 1278–1284 [DOI] [PubMed] [Google Scholar]
- 31. Wiznerowicz M., Trono D. (2003) J. Virol. 77, 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Watanabe Y., Yachie N., Numata K., Saito R., Kanai A., Tomita M. (2006) Gene 365, 2–10 [DOI] [PubMed] [Google Scholar]
- 33. Doench J. G., Sharp P. A. (2004) Genes Dev. 18, 504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) PLoS Biol. 2, e363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Domínguez-Soto A., Puig-Kröger A., Vega M. A., Corbí A. L. (2005) J. Biol. Chem. 280, 33123–33131 [DOI] [PubMed] [Google Scholar]
- 36. Gaspar N. J., Li L., Kapoun A. M., Medicherla S., Reddy M., Li G., O'Young G., Quon D., Henson M., Damm D. L., Muiru G. T., Murphy A., Higgins L. S., Chakravarty S., Wong D. H. (2007) Mol. Pharmacol. 72, 152–161 [DOI] [PubMed] [Google Scholar]
- 37. O'Connell R. M., Chaudhuri A. A., Rao D. S., Baltimore D. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 7113–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schutyser E., Struyf S., Proost P., Opdenakker G., Laureys G., Verhasselt B., Peperstraete L., Van de Putte I., Saccani A., Allavena P., Mantovani A., Van Damme J. (2002) J. Biol. Chem. 277, 24584–24593 [DOI] [PubMed] [Google Scholar]
- 39. Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. (2002) Trends Immunol. 23, 549–555 [DOI] [PubMed] [Google Scholar]
- 40. Kinjyo I., Hanada T., Inagaki-Ohara K., Mori H., Aki D., Ohishi M., Yoshida H., Kubo M., Yoshimura A. (2002) Immunity 17, 583–591 [DOI] [PubMed] [Google Scholar]
- 41. Nakagawa R., Naka T., Tsutsui H., Fujimoto M., Kimura A., Abe T., Seki E., Sato S., Takeuchi O., Takeda K., Akira S., Yamanishi K., Kawase I., Nakanishi K., Kishimoto T. (2002) Immunity 17, 677–687 [DOI] [PubMed] [Google Scholar]
- 42. de Nadaï P., Charbonnier A. S., Chenivesse C., Sénéchal S., Fournier C., Gilet J., Vorng H., Chang Y., Gosset P., Wallaert B., Tonnel A. B., Lassalle P., Tsicopoulos A. (2006) J. Immunol. 176, 6286–6293 [DOI] [PubMed] [Google Scholar]
- 43. Lee C., Kolesnik T. B., Caminschi I., Chakravorty A., Carter W., Alexander W. S., Jones J., Anderson G. P., Nicholson S. E. (2009) Clin. Exp. Allergy 39, 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Louafi F., Martinez-Nunez R. T., Sanchez-Elsner T. (2010) J. Biol. Chem. 285, 41328–41336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.