Abstract
Alpha actinins (ACTNs) are known for their ability to modulate cytoskeletal organization and cell motility by cross-linking actin filaments. We show here that ACTN4 harbors a functional LXXLL receptor interaction motif, interacts with nuclear receptors in vitro and in mammalian cells, and potently activates transcription mediated by nuclear receptors. Whereas overexpression of ACTN4 potentiates estrogen receptor α (ERα)-mediated transcription in transient transfection reporter assays, knockdown of ACTN4 decreases it. In contrast, histone deacetylase 7 (HDAC7) inhibits estrogen receptor α (ERα)-mediated transcription. Moreover, the ACTN4 mutant lacking the CaM (calmodulin)-like domain that is required for its interaction with HDAC7 fails to activate transcription by ERα. Chromatin immunoprecipitation (ChIP) assays demonstrate that maximal associations of ACTN4 and HDAC7 with the pS2 promoter are mutually exclusive. Knockdown of ACTN4 significantly decreases the expression of ERα target genes including pS2 and PR and also affects cell proliferation of MCF-7 breast cancer cells with or without hormone, whereas knockdown of HDAC7 exhibits opposite effects. Interestingly, overexpression of wild-type ACTN4, but not the mutants defective in interacting with ERα or HDAC7, results in an increase in pS2 and PR mRNA accumulation in a hormone-dependent manner. In summary, we have identified ACTN4 as a novel, atypical coactivator that regulates transcription networks to control cell growth.
Keywords: Coactivator Transcription, Gene Transcription, Hormones, Nuclear Receptors, Transcription Target Genes
Introduction
The alpha actinins (ACTNs)2 belong to a family of cytoskeletal proteins that bind actin filaments to maintain cytoskeletal structure and cell morphology (1). Among the four members of the family, ACTN2 and ACTN3 are expressed primarily in muscle, while ACTN1 and ACTN4 are ubiquitously expressed (2). All four members of the actinin family share high sequence homology with conserved functional domains including an N-terminal actin-binding domain containing two highly conserved calponin homology (CH1 and CH2) domains, a central domain consisting of four spectrin repeats (SR), two EF hand calcium-binding domains and a C-terminal calmodulin (CaM)-like domain (3). Although predominantly localized in the cytoskeleton, ACTN4 is also found in the nucleus of certain cell types and is capable of translocating to the nucleus in response to extracellular stimuli (4). In addition to its role in the cytosol, we have recently identified a novel function for ACTN4 in transcriptional regulation by myocyte enhancer factor 2 (MEF2) (5). Another study has also demonstrated an association between ACTN4 and nuclear factor κB (NF-κB) (6). These findings suggest that ACTN4 may play an unexpected role in transcriptional regulation.
Nuclear hormone receptors, including vitamin D receptor (VDR) and steroid hormone receptors such as estrogen receptors (ER) are ligand-activated transcription factors that control aspects of homeostasis, cell differentiation, proliferation, and development (7–9). Transcriptional regulation by nuclear receptors is thought to occur through the exchange of associated corepressors and coactivators. Ligand binding induces an allosteric change in the nuclear receptor, leading to dissociation of corepressor complexes and recruitment of coactivator proteins followed by transcriptional activation of target genes.
The hormone-induced interaction between nuclear receptors and coactivators is mediated through one or more copies of highly conserved signature sequence designated as a nuclear receptor interaction (NR) box. The NR box is comprised of a short α-helical LXXLL motif (where L is leucine and X can be any amino acid) (10). NR boxes and surrounding residues have been categorized into four classes depending on the amino acid residue present at the −1 and −2 positions upstream of the LXXLL motif. Among these classes, the first three were identified using the phage display approach and the fourth class was identified following analysis of naturally occurring motifs among coactivators (11). This motif is present in many nuclear receptor coactivators including p160 family of coactivators (NCoA 1, 2, and 3), histone acetyltransferases (CBP/p300) and p300/CBP-associated factor (PCAF) (12–15). The integrity of this motif is essential for the ability of the coactivators to potentiate transcriptional activation by NRs.
We have previously shown that the full-length ACTN4 and its isoform potentiate transcriptional activation by MEF2 through antagonizing HDAC7 (5). In this study, we focus on the full-length ACTN4 and provide evidence that ACTN4 has a broad role in transcriptional regulation including its role as a transcriptional coactivator for nuclear receptor-mediated transcription. Additionally, we demonstrate that ACTN4 potentiates transcriptional activity of ERα, in part, through its association with HDAC7.
EXPERIMENTAL PROCEDURES
Plasmid Construction
CMX-HA-ACTN4, CMX-HA-ACTN4 (LXXAA) and CMX-HA-ACTN4 (Δ831–869) expression plasmids were generated by site directed PCR mutagenesis according to the manufacturer's protocol (Stratagene). For the glutathione S-transferase (GST) constructs, full-length ACTN4 cDNA was PCR-amplified and subcloned into pGEX4T vector using standard techniques. Expression plasmids for nuclear receptors and reporters were generous gifts from Ron Evans (The Salk Institute, La Jolla, CA).
Antibodies and Chemicals
α-ACTN4 and α-HDAC7 antibodies have been previously described (5, 16). Anti-HA-conjugated α-horseradish peroxidase was purchased from Roche Applied Science. α-VDR (C-20), α-ERα (d-12), α-Lamin B (sc 6216), α-GAPDH (sc 25778), and α-HDAC1 (sc 7872) antibodies were purchased from Santa Cruz Biotechnology. Anti-α-tubulin (T-5168) antibody was purchased from Sigma-Aldrich. 1-(24R)-24, 25-dihydroxy-vitamin D3 (705861) and β-estradiol (E2, E8875) were purchased from Sigma-Aldrich.
Cell Culture
HEK293, CV-1, and MCF-7 cells were grown in Dulbecco's Eagle's modified medium (DMEM) supplemented with 10% fetal bovine serum, 50 units/ml penicillin G and 50 μg/ml streptomycin sulfate at 37 °C in 5% CO2.
In Vitro Protein-Protein Interaction Assays
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli DH5α strain, affinity purified and immobilized on glutathione-Sepharose 4B beads. In vitro pull-down assays were carried out using immobilized GST-ACTN4 (WT) or GST-ACTN4 (LXXAA) mutant with whole cell extracts expressing nuclear receptors in the absence or presence of vitamin D3 (100 nm) or E2 (10 nm) for 2 h at 4 °C. For GRIP1 and ACTN4 interaction, GST-ACTN4 (WT) was incubated with whole cell lysates expressing GRIP1. After extensive washes with NETN buffer (100 mm NaCl, 1 mm EDTA, 10 mm Tris-Cl, pH 8.0, 0.1% Nonidet P-40, 10% glycerol, and 1 mm dithiothreitol), SDS-PAGE sample buffer was added to the beads, boiled, and separated by 10% SDS-PAGE gel and immunoblotted with the indicated antibodies.
Coimmunoprecipitation
HEK293 cells were grown on 10-cm plates and transfected with indicated plasmids (10 μg of total DNA) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. After 48 h, the cells were washed with 1× PBS and resuspended in NETN buffer along with protease inhibitors. Lysed cells were centrifuged at 4 °C at 14,000 rpm for 10 min and the supernatant was collected and kept at −80 °C. Immunoprecipitations were performed using α-VDR antibodies with or without vitamin D3 for 12 h at 4 °C. The immunoprecipitated fractions were analyzed by immunoblotting using α-ACTN4 antibodies. For interaction between endogenous ACTN4 and overexpressed ERα in HEK293 cells, immunoprecipitations were carried out using α-ACTN4 antibodies in the presence or absence of 10 nm E2 for 4 h at 4 °C followed by immunoblotting with α-ERα antibodies.
Transient Transfection Reporter Assays
For reporter assays, CV-1 or MCF-7 cells were co-transfected with equal amounts of either 100 ng of VDRE-TK-Luc or ERE-TK-Luc with or without pCMX-ACTN1 or pCMX-ACTN4 along with 100 ng pCMX-β-gal in 200 μl Opti-MEM I using Lipofectamine 2000 (Invitrogen). The amount of DNA was kept constant (1 μg) by the addition of pCMX vector. After 5 h, the medium was replaced with DMEM supplemented with 10% charcoal stripped fetal bovine serum, 50 units/ml penicillin G, and 50 μg/ml streptomycin sulfate. After 24 h, medium was replaced with or without hormones as indicated. For all cell types, cells were harvested 48 h after transfection, and luciferase and β-galactosidase (β-gal) activities were measured according to the manufacturer's protocol using a luciferase assay system (Promega). Luciferase activity was normalized to the level of β-gal activity. Each reaction was performed in triplicate. The data are representative of at least three separate trials. For siRNA transfection, MCF-7 cells were transfected a control siRNA (siCtrl, 1027415), siACTN4-1 (SI02779973), or siACTN4-2 (SI02779980) at a final concentration of 10 nm using GeneSolution siRNA (Qiagen) according to the manufacturer's protocol. After 48 h, cells were split into 48-well plates and transfected with ERE-TK-Luc along with 100 ng of pCMX-β-gal plasmids as mentioned above for the luciferase assays.
Transient Transfections and RNA Analysis
MCF-7 cells were transfected with HA-ACTN4 (WT), HA-ACTN4 (LXXAA) mutant and HA-ACTN4 (Δ831–869) mutant along with GFP expression plasmids using Lipofectamine 2000 (Invitrogen). An aliquot of cells were used for mRNA analyses by qRT-PCR. An aliquot of cells were subjected to protein analyses. For protein analyses, whole cell extracts were prepared using NETN buffer (100 mm NaCl, 1 mm EDTA, 10 mm Tris-Cl (pH 8.0), 0.1% Nonidet P-40, 10% glycerol, and 1 mm dithiothreitol). SDS-PAGE sample buffer was added to the lysates, boiled, and separated by 10% SDS-PAGE gel and immunoblotted with the indicated antibodies.
Transient Transfections and Immunofluorescence
MCF-7 cells were transfected either with appropriate ACTN4 (WT) and/or ACTN4 mutant plasmids using Lipofectamine 2000 (Invitrogen) or with siACTN4 or siCtrl in 12-well culture plates. For siRNA transfection, cells were treated with or without 10 nm of E2 as indicated after 48 h, followed by immunostaining. For plasmid transfection, cells were subjected to immunostaining 24-h post-transfection. Transfected cells were fixed in 3.7% paraformaldehyde (PFA) in PBS for 30 min at room temperature and permeabilized in PBS with the addition of 0.1% Triton X-100 and 10% goat serum for 10 min. The cells were washed three times with PBS and incubated in a PBS-goat serum (10%) + 0.1% Tween-20 solution (ABB) for 60 min. Incubation with primary antibodies was carried out for 120 min in ABB. The cells were washed three times in PBS, and the secondary antibodies were added for 60 min in the dark, at room temperature in ABB. Cover slips were mounted to slides using Vectashield mounting medium with DAPI (H-1200, Vector Laboratories, Inc.) The primary antibodies used were: purified α-ACTN4 polyclonal, and α-HA and α-ERα mouse monoclonal antibodies (Santa Cruz Biotechnology). The secondary antibodies used are from Molecular Probes (α-mouse or α-rabbit Alexa Fluor 594 or α-mouse Alexa Fluor 488).
Subcellular Fractionation
Subcellular fractionation of MCF-7 cells was carried out according to a published protocol (17). MCF-7 cells were treated with 10 nm E2 for indicated periods of time. Nuclear and cytoplasm fractions were resolved on SDS-PAGE gel and immunoblotting was done with the indicated antibodies.
RNA Extraction and Quantitative Real-time PCR
Transient transfection of siRNA of ACTN4 was performed as described above. For siRNA transfection of HDAC7, MCF-7 cells were transfected with a non-targeting siRNA siCtrl (D-001810-01-50), siHDAC7-1 (J-009330-07), or siHDAC7-2 (J-009330-08) at a final concentration of 100 nm according to the manufacturer's protocol (Dharmacon). Forty-eight hours post transfection, cells were treated with 10 nm E2 or ethanol as a vehicle control for 24 h prior to harvesting RNA. Total RNA was extracted from MCF-7 cells according to the manufacturer's protocol (USB). cDNA was synthesized from 1 μg of total RNA according to the manufacturer's instructions (Invitrogen). Gene expression levels were determined by qRT-PCR using Real-time PCR System (Bio-Rad) with the following primers: GAPDH: 5′-GAAGGTGAAGGTCGGAGT-3′, 5′-GAAGATGGTGATGGGATTTC-3′; PR (progesterone receptor): 5′-CCATGTGGCAGATCCCACAGGAGTT-3′, 5′-TGGAAATTCAACACTCAGTGCC-3′; pS2: 5′-GAGAACAAGGTGATCTGCGCCC-3′, 5′-CCCACGAACGGTGTCGAAACA-3′. Relative changes in gene expression were calculated using the ΔΔCt method. Each value is representative of three replicates, and all the experiments were repeated twice.
Chromatin Immunoprecipitation Assays
MCF-7 cells were treated with 100 nm E2, and ChIP assays were performed according to our published protocol (18) except that α-HDAC7, α-ACTN4, and α-ERα antibodies were used.
Cell Proliferation Assay
MCF-7 cells were transfected with siRNA targeted against non-targeting siRNA (siCtrl), ACTN4 (siACTN4), or HDAC7 (siHDAC7). Forty-eight hours after transfection, cells were counted and 1,500 cells were plated into each well in a 96-cell plate. Cells were treated with either 10 nm E2 or ethanol as a vehicle control. The day 0 time point was taken 5 h after plating. Cell proliferation was measured at days indicated using CyQUANT NF Cell Proliferation Assay kit (Invitogen).
Statistical Analysis
Statistical analysis was performed using two-tailed Student's t test.
RESULTS
ACTN4 Is Localized in Both Nucleus and Cytoplasm of MCF-7 Cells
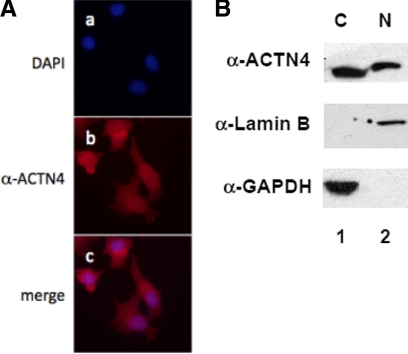
ACTN4 is thought to play a role in coordinating cytoskeletal architecture but several reports have suggested additional activities (5–6). To explore whether ACTN4 also plays a role in transcriptional regulation, we examined the subcellular distribution of endogenous ACTN4 in MCF-7 breast cancer cells by immunofluorescence microscopy (Fig. 1A) and Western blotting following fractionation of cell lysates (Fig. 1B). In both assays, we found that ACTN4 was predominantly present in the cytoplasm. However, a significant fraction of the ACTN4 was localized in the nucleus.
FIGURE 1.
Subcellular distribution of ACTN4 in MCF-7 cells. A, MCF-7 cells were grown and immunostained with α-ACTN4 antibodies, and the images were taken by immunofluorescence microscopy. DNA was visualized by DAPI staining. B, subcellular fractionation of MCF-7 cells was carried out followed by Western blotting with indicated antibodies.
ACTN4 Potentiates Nuclear Receptor-mediated Transcription
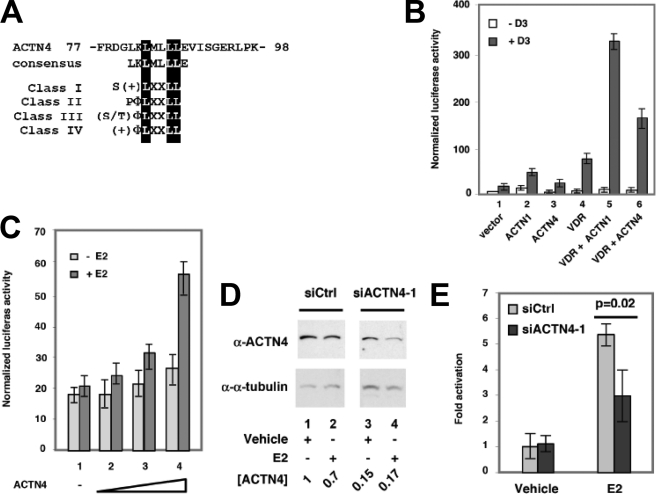
Sequence analysis indicated that ACTN4 harbors a putative nuclear receptor interacting motif, LXXLL, found in all four classes of nuclear receptor coactivators (Fig. 2A) (11). This observation raised the possibility that ACTN4 functions as a transcriptional co-activator for nuclear receptors. To test transcriptional response with nuclear receptors, we carried out transient transfection reporter assays using reporter constructs harboring vitamin D3 response element (VDRE) in CV-1 cells. We found that ectopic overexpression of ACTN1, ACTN4, or VDR alone modestly activated VDR-mediated transcription (Fig. 2B, lanes 2–4). However, co-expression of ACTN1 or ACTN4 with VDR stimulated hormone (vitamin D3) driven VDR-mediated transcription activity (lanes 5–6). We further investigated whether ACTN4 enhances the transcriptional activity of ERα in transient transcription assays in MCF-7 cells, which express ERα. As shown in Fig. 2C, ACTN4 increased the ERE (estrogen response element)-luciferase reporter activity in a hormone (E2) and dosage-dependent manner (lanes 2–4). No significant changes were observed in the reporter activity in the absence of hormone.
FIGURE 2.
ACTN4 potentiates transcriptional activation by nuclear hormone receptors. A, an alignment of the LXXLL nuclear receptor interacting motif of ACTN4. Consensus nuclear receptor interacting motifs of all four classes of coactivators are shown (11). B and C, ACTN1 and ACTN4 potentiate reporter activity of VDR in CV-1 cells (B) and ERα in MCF-7 cells (C). Transient transfection assays were conducted as described under “Experimental Procedures.” Each data point represents the mean and S.D. of the results from triplicate wells. D and E, MCF-7 cells were transiently transfected with control (siCtrl) or with ACTN4-1 siRNA (siACTN4-1). Forty-eight hours after transfection, cells were split for whole cell extract preparation (D), transient transfection reporter assays (E), or proliferation assays. Cells were harvested for whole cell extract preparation 24 h after split. The ACTN4 protein levels were examined by Western blot analyses, quantified, and normalized to α-tubulin protein levels. The protein level of siCtrl treated with vehicle is set to 1. E, knockdown of ACTN4 disrupts E2-driven ERE-mediated transcription activity. MCF-7 cells transfected with siCtrl or siACTN4-1 were subjected to transient transfection reporter assay as described in C. The reporter activity of siCtrl treated with vehicle is set to 1.
To determine the effects of endogenous ACTN4 in E2-mediated transcriptional activation, we performed knockdown experiments using two separate ACTN4 siRNAs. Knockdown of ACTN4 with siACTN4-1 or siACTN4-2 potently decreased its protein accumulation (Fig. 2D and supplemental Fig. S1A). However, the mRNA level of ACTN1 was only marginally affected (data not shown). For both siRNAs, we found that knockdown of ACTN4 significantly decreased E2-induced ERE-luciferase reporter activity (Fig. 2E and supplemental Fig. S1B). Furthermore, to investigate whether ACTN4 has an effect on the subcellular distribution of ERα we carried out immunostaining following ACTN4 knockdown using siACTN4. As shown in supplemental Fig. S2, we did not observe any significant change in the subcellular localization of ERα when ACTN4 was knocked down, indicating that ACTN4 does not affect the subcellular distribution of ERα. Taken together, our results indicate that ACTN1 and ACTN4 can function as transcriptional co-activators, though the degree of activation varies for different nuclear receptors.
ACTN4 Binding to Nuclear Hormone Receptors Requires the LXXLL Receptor Interaction Motif
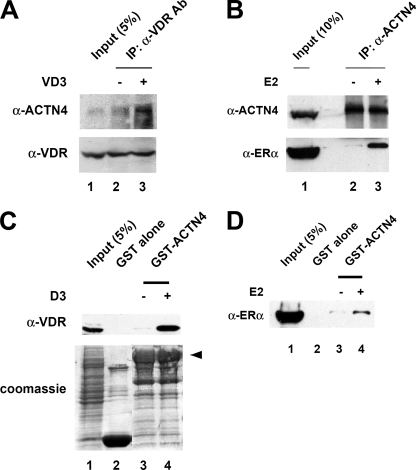
The observation that ACTN4 harbors a LXXLL motif suggested that ACTN4 potentiates transcriptional activity mediated by nuclear receptors through a physical interaction with nuclear receptors. To test this hypothesis, we first carried out immunoprecipitation experiments. HEK293 cells were transfected with a VDR expression plasmid and immunoprecipitations were carried out on whole cell lysates using α-VDR antibodies in the presence or absence of vitamin D3 followed by immunoblotting with α-ACTN4 antibodies. As shown in Fig. 3A, ACTN4 interacted with VDR in a hormone-dependent manner (lanes 2–3). To analyze the interaction between ACTN4 and ERα, whole cell extracts prepared from HEK293 cells overexpressing ERα were immunoprecipitated with α-ACTN4 antibodies and immunoblotted with α-ACTN4 and α-ERα antibodies. We found that E2 enhanced the association between ACTN4 and ERα (Fig. 3B, lanes 2–3, bottom panel). Furthermore, we verified the interaction between nuclear receptors and ACTN4 in vitro by GST pulldown assays. Bacterially purified immobilized GST-ACTN4 (WT) was incubated with whole cell lysates expressing VDR or ERα in the presence or absence of their respective ligands. As a control, little or no binding to GST alone was observed with either of the nuclear receptors while GST-ACTN4 interacted with these receptors in a ligand-dependent manner (Fig. 3, C and D, lanes 2–4). From these data we concluded that hormone enhances the association of ACTN4 with VDR and ERα in mammalian cells and in vitro.
FIGURE 3.
Hormone-dependent association of ACTN4 and nuclear receptors. A, interaction between ACTN4 and overexpressed VDR. HEK293 cells were transfected with a VDR expression plasmid. The cells were harvested 48 h post-transfection. Extracts were immunoprecipitated with α-VDR antibodies in the presence or absence of 100 nm of vitamin D3 followed by Western blotting with α-ACTN4 and α-VDR antibodies. Five percent input is shown. B, association between endogenous ACTN4 and overexpressed ERα in HEK293 cells. HEK293 cells were transfected with an ERα expression plasmid, whole cell lysates prepared and immunoprecipitated with α-ACTN4 antibodies in the presence or absence of 100 nm E2. The immunoprecipitated pellets were analyzed by immunoblotting with α-ERα and α-ACTN4 antibodies. Ten percent input is shown. C, in vitro interaction between ACTN4 and VDR. Lysates from HEK293 cells overexpressing VDR were incubated with immobilized GST or GST-ACTN4 in the presence or absence of 100 nm of vitamin D3. Pulldown fractions were subjected to Western blotting with α-VDR antibodies (top panel). Commassie Blue staining is shown to demonstrate equal loading for GST beads in each lane (bottom panel). The arrow shows the full-length GST-ACTN4 fusion protein. Lane 1 shows 5% of the input for pulldown assays. D, in vitro interaction of ACTN4 and ERα. Whole cell extracts overexpressing ERα were used in this assay. GST pulldown assays were carried out in the presence or absence of 100 nm of E2. Pulldown fractions were subjected to Western blotting with α-ERα antibodies. Five percent input is shown in lane 1.
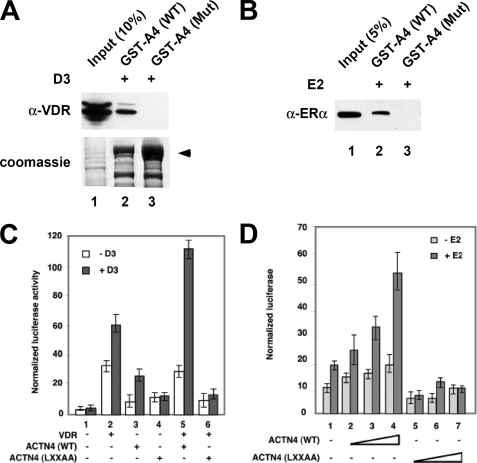
To further investigate whether the LXXLL motif of ACTN4 is required for its hormone-dependent interaction with nuclear receptors, we generated an ACTN4 (LXXAA) mutant where Leu-87 and Leu-88 were substituted by Ala and carried out GST pulldown assays. Fig. 4, A and B (lanes 2–3) show that the ACTN4 (LXXAA) mutant displayed reduced interactions with nuclear receptors, even in the presence of hormone. We next investigated whether disruption of the LXXLL motif affects ACTN4-mediated transcriptional activation in reporter assays using wild-type ACTN4 or the ACTN4 (LXXAA) mutant in CV-1 or MCF-7 cells. As demonstrated in Fig. 4, C and D, wild-type ACTN4 potentiated VDR- and ERα-mediated transcriptional activity in a hormone-dependent manner (lanes 5 and 2–4), whereas the ACTN4 (LXXAA) mutant showed a significant decrease in the ability to potentiate hormone-dependent transcriptional activity mediated by VDR or ERα (lanes 6 and 5–7). We also addressed whether the reduced activity of the ACTN4 (LXXAA) mutant was due to its mis-localization by carrying out immunofluorescence microscopy in MCF-7 cells. As shown in supplemental Fig. S3, the ACTN4 (LXXAA) mutant exhibited a similar subcellular distribution pattern to that of the wild-type protein. Taken together, we have identified that LXXAA motif in ACTN4 is required for its interaction with and transcriptional activity by VDR and ERα.
FIGURE 4.
LXXLL motif is essential for the ACTN4 to potentiate nuclear receptor-mediated transcriptional activity. A, ACTN4 (LXXAA) mutant loses its ability to interact with VDR. HEK293 cells were transfected with a VDR expression plasmid. 48-h post-transfection, the lysates were prepared and incubated with bacterially expressed GST-ACTN4 (WT) or with GST-ACTN4 (LXXAA) fusion proteins in the presence of 100 nm vitamin D3. Pulldown fractions were subjected to Western blotting with α-VDR -antibodies. The arrow indicates the full-length GST-ACTN4 fusion protein. Lane 1 shows 10% of the input for pulldown experiments. B, ACTN4 (LXXAA) mutant loses its ability to interact with ERα. GST pulldown assays were carried out as described in A, except ERα expression plasmids were used for transfection, and pulldowns were carried out in the presence of 100 nm of E2. Lane 1 shows 5% input. C, LXXLL motif is essential for VDR-mediated transcriptional activation. Expression plasmids for ACTN4 (WT) or the ACTN4 (LXXAA) mutant were co-transfected with or without expression plasmids for VDR along with a reporter construct harboring a VDRE in CV-1 cells. Reporter assays were carried out as described in Fig. 2B. D, ACTN4 (LXXAA) loses its ability to potentiate ERα activity. The experiment was performed as described in Fig. 2C. Each data point represents the mean and S.D. of results from triplicates.
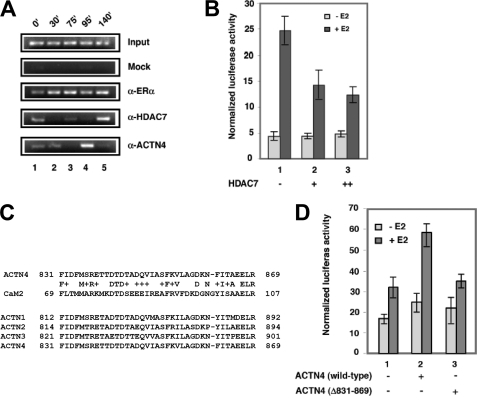
To further dissect the role of ACTN4 in ERα-mediated transcriptional activation, we examined whether ACTN4 associates with the ERα target gene promoter by ChIP assays in MCF-7 cells. We have previously shown that ACTN4 interacts with HDAC7 (5). Therefore we also examined the recruitment of HDAC7 to the promoter of the pS2 gene, a well-studied ERα target gene. As shown in the Fig. 5A, ERα was recruited to the pS2 promoter at all time points (lanes 2–5), while ACTN4 and HDAC7 were recruited in distinct patterns (lanes 2–5). A faint signal for the former was detected at 30′ and a strong signal at 95′. For HDAC7, a weak signal was visible at 75′ and a strong signal at 140′. Moreover, when ACTN4 was most abundant at the pS2 promoter, HDAC7 was absent and vice versa (lane 4 versus lane 5). This observation raised the possibility that HDAC7 may also regulate ERα-mediated transcriptional activation. To test this possibility, we performed transient transfection reporter assays to determine the effects of overexpressing HDAC7 on ERE-mediated reporter activity. Fig. 5B shows that HDAC7 is capable of repressing E2-induced ERE reporter activity in a dose- and hormone-dependent manner (lanes 1–3).
FIGURE 5.
ACTN4 associates with the pS2 promoter. A, association of ERα, ACTN4 and HDAC7 with the pS2 promoter. Chromatin prepared from MCF-7 cells treated with 10 nm E2 for the indicated time periods was subjected to chromatin immunoprecipitation assays as detailed under “Experimental Procedures,” with antibodies against ERα, HDAC7, and ACTN4. B, effects of overexpressing HDAC7 on ERE reporter activity. MCF-7 cells were transfected with reporter constructs harboring an ERE and β-gal along with increasing amounts of an HDAC7 expression plasmid. Luciferase reporter assays were performed as described in Fig. 2C. C, an alignment between the ACTN4 CaM-like domain and parts of calmodulin (CaM2). The CaM-like domains of all four ACTNs are similar to each other. D, ACTN4 (Δ831–869) loses its ability to potentiate ERE reporter activity. The reporter assays were (panels B and D) were performed as described in Fig. 2C.
We have previously mapped an HDAC7-interacting domain in ACTN4 to 38 residues between amino acids 831 and 869 (5). This region also corresponds to the CaM-like domain of ACTN4, which is highly conserved in the other family members (Fig. 5C). Unexpectedly, deletion of amino acids 831–869 (Δ831–869) resulted in a greater nuclear distribution (supplemental Fig. S4) and loss of the ability to potentiate ERE reporter activity (Fig. 5D). In summary, these data indicate that ACTN4 associates with an endogenous ERα target, the pS2 promoter and is capable of stimulating transcription of an ERE reporter construct in the presence of hormone in MCF-7 cells.
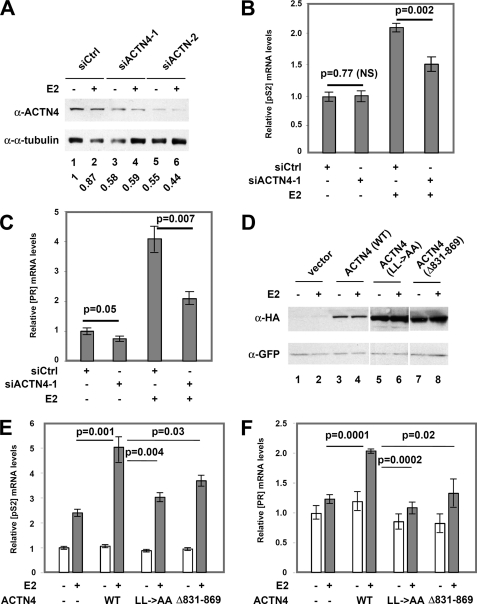
Knockdown of ACTN4 Decreased the Expression of Several Endogenous ERα Target Genes
To determine the biological function of ACTN4 in the transcriptional regulation of estrogen responsive genes, we knocked down ACTN4 by two separate siRNAs in MCF-7 cells and determined the effects on the expression of selected estrogen responsive genes. Transfection of siACTN4 into MCF-7 cells reduced ACTN4 by ∼40–60% at both the mRNA (data not shown) and protein levels (Fig. 6A), respectively. Knockdown of ACTN4 by siACTN4-1 did not affect pS2 mRNA accumulation in the absence of E2, while significantly inhibited E2-induced increases in pS2 transcript (Fig. 6B). Knockdown of ACTN4 by siACTN4-2 led to decreases in pS2 mRNA in the presence or absence of E2 (supplemental Fig. S5A). Similarly, transient transfection of siACTN4-1 or siACTN4-2 decreased PR mRNA in the absence and presence of E2 (Fig. 6C and supplemental Fig. S5B). We also determined whether overexpression of ACTN4 had an effect on pS2 and PR mRNA accumulation. We overexpressed ACTN4 wild-type, LXXAA, and Δ831–869 in MCF-7 cells. A GFP expression plasmid was co-transfected for normalization purposes. The expression levels of wild-type and mutant ACTN4s were monitored by Western blotting (Fig. 6D). Real-time PCR was carried out to determine the expression levels of pS2 and PR mRNA. We found that wild-type increased pS2 mRNA levels 2-fold in a hormone-dependent manner, whereas overexpression of mutants LXXAA and Δ831–869 had little effect (Fig. 6E). Overexpression of wild-type or mutant ACTN4s resulted in a similar effect on PR mRNA expression (Fig. 6F). Taken together, these data demonstrate that ACTN4 positively regulates expression of ERα target genes, pS2 and PR.
FIGURE 6.
Knockdown of ACTN4 reduces expression of endogenous ERα regulated genes. A, knockdown of ACTN4 in MCF-7 cells. Western blot analysis of ACTN4 protein levels in MCF-7 cells following transfection of control siRNA (siCtrl) or siRNAs targeting ACTN4 (siACTN4-1) after treatment with or without 10 nm E2 for 24 h. Quantification of ACTN4 level after normalization to α-tubulin is shown below the blot with siCtrl level treated with vehicle being set to 1. B and C, mRNA expression levels of pS2 (B) and PR (C) in MCF-7 cells following ACTN4 knockdown by siACTN4-1. RNA was isolated as described under “Experimental Procedures,” and relative expression levels of pS2 and PR were analyzed by qRT-PCR. The mRNA levels of pS2 and PR were normalized to GAPDH mRNA expression levels. NS represents no significant change compared with control siRNA. D, MCF-7 cells were transfected with an expression plasmid for HA-ACTN4 (WT), HA-ACTN4 (LXXAA), or HA-ACTN4 (Δ831–869) along with a GFP expression plasmid. The GFP expression plasmid was included for normalization purposes. An aliquot of the samples was used to prepare total cell lysates for Western blotting (D) or total RNA for real-time PCR (E and F). E, mRNA expression level of pS2 in ACTN4-overexpressing cells. F, mRNA expression of PR in ACTN4-overexpressing cells. The mRNA level of pS2 and PR in MCF-7 cells overexpressing vector alone treated with vehicle is set to 1.
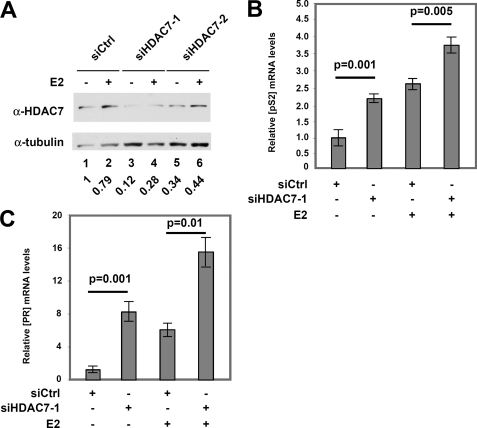
siRNA Targeting of HDAC7 Enhances the Expression of Selected Endogenous ERα Target Genes
Our previous data suggested that HDAC7 antagonizes ACTN4 function (Fig. 5). To investigate the role of HDAC7 in ERα-mediated transcription, we tested whether knockdown of HDAC7 has effects on the expression of ERα target genes. HDAC7 was knocked down by siHDAC7–1 and siHDAC7–2 by 80 and 40%, respectively (Fig. 7A). Knockdown of HDAC7 by either siRNA led to a significant increase in pS2 mRNA expression in the absence or presence of E2 (Fig. 7B and supplemental Fig. S6A). Similarly, PR mRNA levels were up-regulated when HDAC7 was knocked down with or without E2 (Fig. 7C and supplemental Fig. S6B). These observations demonstrate that in contrast to ACTN4, HDAC7 negatively regulates the expression of pS2 and PR, and this regulation is independent of the presence of E2.
FIGURE 7.
HDAC7 knockdown enhances the expression of endogenous ERα-regulated genes. A, knockdown of HDAC7 in MCF-7 cells. Western blot analysis of HDAC7 protein levels in MCF-7 cells following transfection of control siRNA (siCtrl) or siRNA targeting HDAC7 (siHDAC7) after treatment with or without 10 nm E2 for 24 h. HDAC7 protein levels are normalized to α-tubulin and are shown below the figure with the siCtrl treated with vehicle being set to 1. B and C, mRNA expression level of estrogen responsive genes pS2 and PR in MCF-7 cells following HDAC7–1 knockdown. RNA was isolated as described under “Experimental Procedures,” and relative expression levels of pS2 and PR mRNA levels were determined by qRT-PCR. The mRNA levels of ps2 and PR were normalized to GAPDH mRNA expression levels.
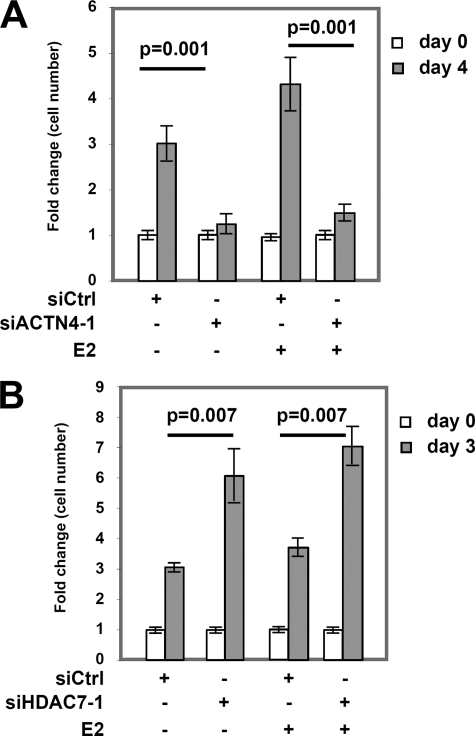
The fact that ACTN4 and HDAC7 regulate transcriptional activation by ERα suggests that ACTN4 and HDAC7 may play a role in E2-mediated regulation of cell proliferation. We found that transient knockdown of ACTN4 significantly decreased MCF-7 cell proliferation in an E2-independent manner (Fig. 8A and supplemental Fig. S7A). By contrast, knockdown of HDAC7 increased MCF-7 cell proliferation regardless of the presence of E2 (Fig. 8B and supplemental Fig. S7B). In conclusion, these data indicate that ACTN4 and HDAC7 play important roles in multiple cellular activities including gene expression and cell proliferation.
FIGURE 8.
The effects of ACTN4 or HDAC7 knockdown on MCF-7 cell proliferation. A, knockdown of ACTN4 decreases MCF-7 cell proliferation in E2-independent manner. MCF-7 cells transfected with siRNA as described in Fig. 2, D and E were counted, split evenly into 96-well plates and treated with or without 10 nm E2 for the indicated times. Cell proliferation was measured at the days indicated. Error bars represent ± S.D. The knockdown efficiency of ACTN4 is shown in Fig. 2D. B, cell proliferation assays were conducted as described in A except that an HDAC7 siRNA-1 was used. The knockdown efficiency of HDAC7 is shown in Fig. 7A.
DISCUSSION
In this study, we demonstrate that ACTN4 potentiates transcriptional activity mediated by VDR and ERα, while knockdown of ACTN4 decreased E2-induced reporter activity and the expression of endogenous ERα target genes, pS2 and PR. ChIP assays established a physical association between ACTN4 and the pS2 promoter and that E2 induced this interaction. Finally, we showed that an ACTN4 mutant defective in interacting with HDAC7 loses its ability to potentiate the activity of an ERE-driven reporter. These observations support a model in which ACTN4 plays an important role in transcriptional regulation, in addition to its well-established function in maintaining cytoskeletal architecture.
By immunofluorescence microscopy, we show that ACTN4 is localized in both the nucleus and cytoplasm in MCF-7 cells. This result is consistent with a recent observation that ACTN4 localizes to the nucleus during G1 and G2 phases in HeLa cells (19), implying a nuclear role of ACTN4. The ability of ACTN4 to shuttle between the nucleus and cytoplasm depends on a functional nuclear export sequence (NES), CRM1, and its spectrin repeats (SRs). We also tested the possibility that E2 potentiates nuclear import of ACTN4 in MCF-7 cells by immunofluorescence microscopy and Western blotting analyses and found that E2 had little effect on subcellular distribution of endogenous ACTN4 (supplemental Fig. S8). Interestingly, the ACTN4 mutant, Δ831–869, exhibited increased nuclear staining compared with the wild-type protein (supplemental Fig. S4). It should be noted that this region of ACTN4 contains a CaM-like domain. These data suggest that the CaM-like domain negatively regulates nuclear import or positively regulates nuclear export of ACTN4. Further investigation will be needed to elucidate how extracellular signaling may regulate nucleocytoplasmic shuttling of ACTN4.
Actin-binding proteins other than ACTN4 have been shown to play roles in other cellular processes including transcriptional regulation, in addition to their ability to maintain cytoskeletal architect or cell morphology (20–22). The simplest explanation for what ACTN4 is doing in the nucleus is binding nuclear actin. This may globally affect nuclear structure, architecture or volume. Our data indicate that E2 does not alter ACTN4 shuttling, thus the E2-dependent effects are not due to global nucleocytoskeletal changes. We also show that ACTN4 physically associates with VDR and ERα in a hormone- and NR box-dependent manner (Figs. 3 and 4). Furthermore, we demonstrate that ACTN4 is recruited to the pS2 promoter (Fig. 5A). These observations argue for a more direct role of ACTN4 in E2-mediated transcriptional regulation. Indeed, several studies have indicated that actin and actin-related proteins can associate with chromatin remodeling complexes, suggesting a role for these proteins in transcriptional regulation (22–24).
ACTN4 harbors a conserved CaM-like domain (Fig. 5C) that is essential for its interaction with HDAC7 (5). Notably, removal of this domain significantly abolishes the ability of ACTN4 to potentiate E2-mediated transcription (Figs. 5D and 6, E and F). As described earlier, the lack of activation activity is not due to exclusion of this mutant from the nucleus (supplemental Fig. S4). Intriguingly, our ChIP data suggest that maximal associations of ACTN4 and HDAC7 with the pS2 promoter are mutually exclusive (Fig. 5A), suggesting that the interaction between ACTN4 and HDAC7 on the pS2 promoter is likely to be transient and regulated. Based on these data, we propose a model in which ACTN4 activates E2-induced transcription, in part, through its association with HDAC7. The interaction leads to the displacement of one of the partners from the promoter.
Several lines of evidence suggest that ACTN4 has a broader role as a transcriptional regulator than originally anticipated. We have previously shown that ACTN4 potentiates MEF2-mediated transcriptional activation. In this study, we further demonstrate that knockdown of ACTN4 decreased E2-induced activity of an ERE-driven reporter, while overexpression of ACTN4 increased it, implying a positive role of ACTN4 in E2-dependent transcriptional activation. However, knockdown of ACTN4 also resulted in decreases in mRNA levels of pS2 and PR, regardless of the presence of E2, implying that ACTN4 is also required for ERα-independent transcriptional activation of pS2 and PR. One plausible explanation for these observations is that ACTN4 is required for induction of pS2 and PR mRNA by other transcription factors. Lastly, it has been previously shown that ACTN4 physically interacts with NF-κB (6), though the functional significance of this association is not clear. In light of our findings, we speculate that ACTN4 may regulate NF-κB transcriptional activity.
Our data showing that knockdown of ACTN4 impaired transcriptional activation by ERα both in transient transfection studies as well as endogenous expression of E2-induced target genes suggest that ACTN4 is required for optimal ERα-mediated transcriptional activation. ACTN4 does not possess any known histone-modifying or chromatin-remodeling activity. One possible mechanism by which ACTN4 could potentiate transcription is through an association with histone acetyltransferases, histone methyltransferases, or chromatin-remodeling complexes. A previous report suggested that GRIP1 interacts with ACTN2, a member of the ACTN protein family (25). However, we were unable to detect a similar association between ACTN4 and GRIP1 under conditions in which ACTN4 interacts with VDR by GST pulldown assays (supplemental Fig. S9). Of note, we found that ACTN4 associates with the arginine methyltransferase CARM1 and that an ACTN4 splice variant interacts with several transcriptional coactivators.3 Moreover, it has been recently shown that ACTN4 associates with the INO80 chromatin-remodeling complex (19). Together, these data suggest that ACTN4 might regulate transcription through multiple mechanisms including an association with histone-modifying enzymes and chromatin-remodeling complexes.
Gene amplification and overexpression of the transcription coactivator SRC family proteins are associated with oncogenesis (26). In this report, we present evidence that ACTN4 enhances MCF-7 cell proliferation because knockdown of ACTN4 decreases MCF-7 cell proliferation (Fig. 8). This growth suppressive effect is independent of the presence of ERα ligands and suggests that ACTN4 may promote MCF-7 cell cycle progression independently of its role in potentiating expression of ERα target genes. We speculate that ACTN4 mediates transcriptional activation of other transcription factors such as NF-κB to control cell proliferation. Alternatively, but not exclusively, ACTN4 cytoskeletal functions may underlie its effect on proliferation. Further investigation will be necessary to distinguish these two possibilities.
Supplementary Material
Acknowledgments
We thank Dr. Samols and Daniel Factor for their comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK078965, HL093269, and the Pardee Foundation (to H.-Y. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S9.
S. Khurana, S. Chakraborty, X. Zhao, and H.-Y. Kao, manuscript in preparation.
- ACTN
- alpha actinin
- CaM
- calmodulin
- HDAC
- histone deacetylase
- CH
- calponin homology
- VDR
- vitamin D receptor
- NR
- nuclear receptor.
REFERENCES
- 1. Djinovic-Carugo K., Gautel M., Ylänne J., Young P. (2002) FEBS Lett. 513, 119–123 [DOI] [PubMed] [Google Scholar]
- 2. Otey C. A., Carpen O. (2004) Cell Motil. Cytoskeleton 58, 104–111 [DOI] [PubMed] [Google Scholar]
- 3. Djinović-Carugo K., Young P., Gautel M., Saraste M. (1999) Cell 98, 537–546 [DOI] [PubMed] [Google Scholar]
- 4. Honda K., Yamada T., Endo R., Ino Y., Gotoh M., Tsuda H., Yamada Y., Chiba H., Hirohashi S. (1998) J. Cell Biol. 140, 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chakraborty S., Reineke E. L., Lam M., Li X., Liu Y., Gao C., Khurana S., Kao H. Y. (2006) J. Biol. Chem. 281, 35070–35080 [DOI] [PubMed] [Google Scholar]
- 6. Babakov V. N., Petukhova O. A., Turoverova L. V., Kropacheva I. V., Tentler D. G., Bolshakova A. V., Podolskaya E. P., Magnusson K. E., Pinaev G. P. (2008) Exp. Cell Res. 314, 1030–1038 [DOI] [PubMed] [Google Scholar]
- 7. Mangelsdorf D. J., Evans R. M. (1995) Cell 83, 841–850 [DOI] [PubMed] [Google Scholar]
- 8. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Francis G. A., Fayard E., Picard F., Auwerx J. (2003) Annu Rev. Physiol. 65, 261–311 [DOI] [PubMed] [Google Scholar]
- 10. Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997) Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- 11. Chang C., Norris J. D., Grøn H., Paige L. A., Hamilton P. T., Kenan D. J., Fowlkes D., McDonnell D. P. (1999) Mol. Cell. Biol. 19, 8226–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G. (1998) Genes Dev. 12, 3357–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., Yamamoto K. R. (1998) Genes Dev. 12, 3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. (1998) Nature 395, 137–143 [DOI] [PubMed] [Google Scholar]
- 15. Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 16. Gao C., Li X., Lam M., Liu Y., Chakraborty S., Kao H. Y. (2006) FEBS Lett. 580, 5096–5104 [DOI] [PubMed] [Google Scholar]
- 17. Rana B., Mischoulon D., Xie Y., Bucher N. L., Farmer S. R. (1994) Mol. Cell. Biol. 14, 5858–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng X., Kao H. Y. (2009) J. Biol. Chem. 284, 36395–36404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumeta M., Yoshimura S. H., Harata M., Takeyasu K. (2010) J. Cell Sci. 123, 1020–1030 [DOI] [PubMed] [Google Scholar]
- 20. Olave I. A., Reck-Peterson S. L., Crabtree G. R. (2002) Annu. Rev. Biochem. 71, 755–781 [DOI] [PubMed] [Google Scholar]
- 21. Blessing C. A., Ugrinova G. T., Goodson H. V. (2004) Trends Cell Biol. 14, 435–442 [DOI] [PubMed] [Google Scholar]
- 22. Chen M., Shen X. (2007) Curr. Opin. Cell Biol. 19, 326–330 [DOI] [PubMed] [Google Scholar]
- 23. Miralles F., Visa N. (2006) Curr. Opin. Cell Biol. 18, 261–266 [DOI] [PubMed] [Google Scholar]
- 24. Zheng B., Han M., Bernier M., Wen J. K. (2009) FEBS J. 276, 2669–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang S. M., Huang C. J., Wang W. M., Kang J. C., Hsu W. C. (2004) J. Mol. Endocrinol. 32, 481–496 [DOI] [PubMed] [Google Scholar]
- 26. Xu J., Wu R. C., O'Malley B. W. (2009) Nat. Rev. Cancer Nat. Rev. Cancer 9, 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.