Abstract
Integrin-dependent cell invasion of some pathogenic bacteria is mediated by surface proteins targeting the extracellular matrix protein fibronectin (FN). Although the structural basis for bacterial FN recognition is well understood, it has been unclear why proteins such as streptococcal SfbI contain several FN-binding sites. We used microcalorimetry to reveal cooperative binding of FN fragments to arrays of binding sites in SfbI. In combination with thermodynamic analyses, functional cell-based assays show that SfbI induces conformational changes in the N-terminal 100-kDa region of FN (FN100kDa), most likely by competition with intramolecular interactions defining an inactive state of FN100kDa. This study provides insights into how long range conformational changes resulting in FN activation may be triggered by bacterial pathogens.
Keywords: Adhesion, Bacteria, Biophysics, Calorimetry, Cell Migration, Extracellular Matrix Proteins, Fibronectin, Protein-Protein Interactions, Natively Unfolded Proteins, Streptococcus pyogenes
Introduction
Invasion of nonprofessional phagocytes is a common virulence mechanism among bacterial pathogens. It involves the subversion of cellular uptake mechanisms, cell signaling and cytoskeletal dynamics (1, 2). For Staphylococcus aureus and Streptococcus pyogenes, adhesion to, and internalization by, epithelial and endothelial cells is mediated by cell wall-attached fibronectin-binding proteins (FnBPs)4 FnBPA and SfbI, respectively. FnBPs recruit fibronectin (FN) to the bacterial surface where it forms a molecular bridge between bacteria and host cell integrins (3, 4).
FN occurs ubiquitously in vertebrate tissues either in its soluble form or as an insoluble component of fibrillar matrix networks. FN is a disulfide-linked homodimer consisting almost entirely of three types of domains or modules, FNI, FNII, and FNIII (Fig. 1A). FN interacts with other extracellular components and different integrin subfamilies, most notably α5β1, αvβ3, and α4β1 (5). The major integrin-binding site in FN, the Arg-Gly-Asp (RGD) motif, is located in the 10th FNIII module (10FNIII). RGD may be cryptic in plasma FN and only exposed upon fibrillogenesis (6–8). Understanding of FN activation is hampered by the lack of information on its in-solution conformation. Important progress has been made with the discovery of long range interactions between FNI modules and module 3FNIII in an N-terminal 100-kDa FN fragment (FN100kDa) (Fig. 1A) (9). Although FN100kDa lacks the RGD motif, it is a soluble construct containing cryptic sites that are also observed in full-length FN. Neither FN nor FN100kDa stimulate fibroblast migration into collagen gels (9, 10), which depends on motogenic Ile-Gly-Asp (IGD) motifs (located on 7FNI and 9FNI) and αvβ3 integrins (11). In contrast, a truncated oncofetal FN isoform known as migration-stimulating factor and the very similar proteolytic N-terminal 70-kDa FN fragment (FN70kDa) (Fig. 1A) are efficient triggers of cell migration (10, 12). The IGD sites appear to be partially exposed in an FN100kDa mutant (FN100kDa-R222A). FN100kDa-R222A lacks a salt bridge between 4FNI and 3FNIII that stabilizes a closed (inactive) FN100kDa conformation and is proposed to be broken upon FN activation (9).
FIGURE 1.
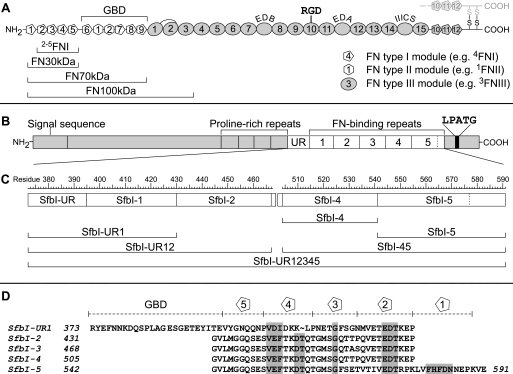
FN fragments and SfbI constructs. A, modular structure of FN (one subunit). Types I–III modules are shown as pentagons, hexagons, and ovals, respectively. The locations of the GBD, alternatively spliced sites (curved labels), and the major integrin-binding site (RGD) are shown on top. FN fragments used in this study are indicated below the diagram. B, molecular organization of SfbI. UR, upstream FN-binding region; LPATG, cell wall anchor. C, map of SfbI constructs used in this study with residue numbers corresponding to UniProt entry Q01924. D, alignment of SfbI FN-binding repeats. Residues that are highly conserved in repeats from different species are shaded gray. Boundaries of FNI-specific motifs and the GBD-binding region (SfbI-UR) are shown on top.
Like other FnBPs, SfbI from S. pyogenes is a mosaic protein (13). Its FN binding activity is confined to a series of repeats (SfbI-1 to SfbI-5) and a unique upstream FN-binding region (SfbI-UR) (Fig. 1B) (14). The FN-binding repeats are natively unfolded in their unbound state (15), target the 30-kDa N-terminal domain of FN (FN30kDa), and adopt a well defined structure upon binding. The in-solution structure of an FnBP peptide in complex with the N-terminal module pair 1FNI-2FNI of FN led to the tandem β-zipper model of bacterial FN binding (16), which is well supported by further molecular structures (17) and in-solution binding experiments (18, 19). Each repeat forms a linear array of nFNI-specific β-strand motifs (Fig. 1D) complementing the triple-stranded β-sheets of either four or five consecutive FNI modules (2–5FNI or 1–5FNI) in FN30kDa. The combination of β-zipper segments in a repeat results in specific and tight binding to FN30kDa.
In contrast to the FN-binding repeats, SfbI-UR (Fig. 1B) targets the gelatin-binding domain of FN (GBD). It significantly enhances the efficiency of SfbI-dependent uptake of S. pyogenes by epithelial cells (20). SfbI-UR is likely to bind mainly to modules 8FNI and 9FNI, indicating a nonlinear arrangement of FN modules forming the binding site for streptococci (21).
Staphylococcal FnBPs contain up to 11 FN-binding repeats, whereas their number in SfbI varies between strains from 2 to 7 (repeat definition according to the tandem β-zipper model) (22, 23). Recruitment of several copies of FN to FnBPs may be required for clustering of host cell integrins. Conceptually, this could be achieved either by multiple binding of FN to each of a few copies of an FnBP or by recruitment of FN to a large number of copies of a surface protein containing a single binding site (24). It is unclear why FnBPs with multiple sites have evolved. We combined a systematic quantitative analysis of FN-SfbI interactions by isothermal titration calorimetry (ITC) with a sensitive cell migration assay to investigate the purpose and consequences of multiple FN bindings by bacteria. FN fragments interact tightly and cooperatively to the array of binding sites in SfbI. ITC and cell migration assays indicate furthermore that multiple binding induces a conformational change in FN100kDa. Structural data provide a rationale for this finding.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of SfbI Constructs
Cloning, expression, and purification of SfbI-4 and SfbI-5 have been described previously (16). SfbI-45, SfbI-UR1, and SfbI-UR12 were produced in a similar fashion. The respective DNA segments were amplified by PCR from S. pyogenes strain MGAS6180 genomic DNA using the following primers: SfbI-45 forward, 5′-CGGGATCCCCCGGAGTGCTGATGGGAGGCCAAAGTGAGTCTTTGAATTTACTAAAGACACTCAAACAGGCATGAGTGGTCAAACAACTCCTCA-3′, and SfbI-45 reverse, 5′-GGAATTCTCATTCCACTTTGGGCTCATTATTGTCAAAATGGAACACTAACTTCGGACGGGTATCTTCAACAATGGTCACTGTTTCACTGAA-3′, and SfbI-45 linker, 5′-GCGACAACTCCTCAGGTTGAGACAGA-3′; SfbI-UR1 and SfbI-UR12 forward, 5′-GCGGGATCCCCCGTTATGAGTTTAAC-3′, and SfbI-UR1 reverse, 5′-GCGGAATTCTCACATATTTCCTGAAAAACCTGT-3′, and SfbI-UR12 reverse, 5′-GCGGAATTCTCATCCTGGGTGTTTCCTTTGTCTT-CTGTCTCAACCTGAGGAGT-3′.
Restriction enzyme cleavage sites were engineered at the 5′ ends of the appropriate primer pairs to facilitate directional cloning. After PCR amplification, the products were analyzed by agarose gel electrophoresis, purified using QIAquick PCR purification kit (Qiagen), digested with BamHI/EcoRI (New England Biolabs) and ligated into digested pGEX5X-1 plasmids (GE Healthcare). Plasmids were introduced into Escherichia coli strains JM101 or XL1 Blue (SfbI-45) by heat shock. Transformed cells were grown overnight on Lysogeny Broth (LB) agar plates containing 100 μg/ml ampicillin. Colonies were screened for the presence of inserts by restriction enzyme digestion and agarose gel electrophoresis. Plasmid inserts were confirmed by sequencing. Protein expression and purification were achieved as described previously for SfbI-4 and SfbI-5 (16).
Cloning, Expression, and Purification of SfbI-UR12345
The coding region of the full fibronectin-binding domain (SfbI-UR12345) of SfbI (S. pyogenes DSM2071) was cloned using the Ek/LIC cloning kit (Novagen). The coding region was amplified by PCR using the following primers: SfbI-UR12345 forward, 5′-GACGACGACAAGATGCGTTATGAGTTTAACAATAAAGACC-3′, and SfbI-UR12345 reverse, 5′-GAGGAGAAGCCCGGTTATTCCACTTTGGGCTC-3′. The amplified product was treated with LIC-qualified T4 DNA polymerase and ligated into the vector pET-41 Ek/LIC. The plasmid was transformed into BL21D pLysS E. coli, and positive clones were confirmed by DNA sequencing. Transformed BL21D pLysS cells were grown in LB containing 100 mg/liter kanamycin at 37 °C. Protein expression was induced at an A600 of 0.4–0.6 with 1 mm isopropyl 1-thio-β-d-galactopyranoside, and cultures were incubated for 4 h at 37 °C. Cells were harvested by centrifugation and resuspended in phosphate-buffered saline (PBS). Phenylmethanesulfonyl fluoride was added (10 μm), and cells were sonicated. Cell debris was removed by centrifugation. The supernatant was applied to glutathione-Sepharose beads. The column was washed with 500 ml of cleavage buffer (50 mm NaCl, 20 mm Tris, 2 mm CaCl2, pH 7.4), and the GST-fused SfbI-UR12345 peptide was eluted with 150 ml of 10 mm glutathione in the same buffer, pH 8.5. Protein-containing fractions were concentrated to 1 ml by ultrafiltration. The expression tag was cleaved from SfbI-UR12345 over 36 h using recombinant enterokinase (Novagen) (1 unit per 500 μg of protein). SfbI-UR12345 was purified using reversed-phase high pressure liquid chromatography using a C4 matrix (Phenomenex). Electrospray ionization and MALDI-TOF-TOF mass spectrometry were used to confirm the identity of the purified polypeptide.
FN Fragments
FN fragments FN30kDa and FN70kDa were purchased from Sigma. Cloning, expression, and purification of 2–5FNI, FN100kDa and FN100kDa-R222A have been described previously (9, 25).
Nuclear Magnetic Resonance Spectroscopy
NMR experiments were performed at 25 °C on a 500-MHz Bruker DRX500 spectrometer. The instrument was controlled by and spectra were processed with Topspin (Bruker). A 20 μm sample of SfbI-UR12345 was prepared in 10 mm phosphate buffer, pH 6.5, containing 10% D2O. A one-dimensional 1H spectrum was acquired using a standard proton pulse sequence with Watergate water suppression. 512 scans were accumulated with a spectral resolution of 0.4 Hz/point. The sample was freeze-dried and dissolved in 0.6 ml of D2O. This cycle was repeated two more times and a one-dimensional spectrum of the final sample was recorded without water suppression.
Circular Dichroism Spectroscopy
Far UV (240 to 190 nm) circular dichroism spectra of a solution of 5 μm SfbI-UR12345 in 10 mm phosphate buffer, pH 6.5, were recorded on a Jasco J-810 spectropolarimeter in a 0.05-cm path length quartz cell. Spectra were averaged from six scans and corrected using a buffer blank. Secondary structure calculations were carried out using the DichroWeb server (26).
ITC and Data Analysis
ITC experiments were carried out using a VP-ITC instrument (MicroCal) in PBS, pH 7.4, at 37 °C. Protein concentrations were determined using absorbance at 280 nm and predicted extinction coefficients (ProtParam, ExPASy). Concentrations of SfbI constructs lacking absorbance at 280 nm (SfbI-4, SfbI-5, and SfbI-45) were based on dry weight of the polypeptides. All FN fragment solutions were dialyzed overnight. SfbI constructs were dissolved in PBS, and the pH was adjusted with small amounts of 1 m NaOH to match the buffer. Cell and syringe solutions were degassed at 25 °C for 15 min. Titrations were performed as follows: one injection of 2 μl followed by injections of 4–6 μl at an injection speed of 0.5 μl/min. The stirring speed was 307 rpm; the delay between the injections was 4–6 min. To take into account heats of dilution, blank titrations were performed by injecting ligand solution into PBS, and the averaged heat of dilution was subtracted from the main experiment. Blank titrations were omitted in cases of saturated binding where the residual signal of the last injections was used to determine the heat of dilution. Raw data were processed using MicroCal Origin software. The base line was adjusted and integrations were carried out manually. Integrated data were analyzed using SEDPHAT (27). Concentrations were corrected using SEDPHAT (fraction of incompetent binding partner). Data were fitted to AB (one site) and ABB (two sites) models. To obtain apparent thermodynamic parameters for SfbI-UR12345 binding, the two-site model of Origin was used. Origin was also used to simulate ITC data.
Cell Migration Studies
Human foreskin fibroblast lines were established in our laboratory from healthy male donors and cultured as described previously (28). Type I collagen was extracted from rat tail tendons and used to make 2-ml collagen gels in 35-mm plastic tissue culture dishes as described previously (28). Collagen gels were overlaid with 1 ml of either serum-free minimum Eagle's medium or serum-free minimum Eagle's medium containing four times the final concentration of SfbI constructs and 0.4 ng/ml FN100kDa or FN100kDa-R222A (∼4 pm). For control samples, SfbI peptides and/or FN fragments were omitted. Confluent stock cultures of fibroblasts were trypsinized, pelleted by centrifugation, and resuspended in growth medium containing 4% (v/v) donor calf serum at 2 × 105 cells/ml, and 1-ml aliquots were added to the overlaid gels. Considering the 2-ml volume of gel, 1 ml of medium overlay and 1 ml of cell inocula, this procedure gave a final concentration of 1% (v/v) serum. The assay cultures were incubated for 4 days, and the percentage of fibroblasts found within the gel matrix was then ascertained by microscopic observation of 10 randomly selected fields in each of two duplicate cultures, as described previously (28).
RESULTS
SfbI Constructs and FN Fragments
Six different SfbI constructs (Fig. 1, B and C) were expressed as cleavable GST fusion proteins. In addition to the two C-terminal repeats (SfbI-4 and SfbI-5), a third single-site construct SfbI-UR1, comprising SfbI-UR and the 1st repeat, was made. SfbI-45 and SfbI-UR12 contained two consecutive binding sites each. Finally, the full-length FN-binding domain (SfbI-UR12345) of an SfbI variant containing the upstream FN-binding region and five repeats was produced.
Four FN fragments were used in this study (Fig. 1A). Although FN30kDa represents the complete binding site for FN-binding repeats, it was necessary to use larger FN fragments containing the GBD to allow binding studies of SfbI constructs, including the upstream region (SfbI-UR1, SfbI-UR12, and SfbI-UR12345). FN70kDa consists of FN30kDa and GBD. FN100kDa was produced by transient mammalian cell expression (25). This fragment resembles an alternatively spliced mRNA variant of FN (29, 30) and comprises FN30kDa, GBD, and 1–3FNIII (Fig. 1A). The FNIII modules do not take part in the interaction with FnBPs but have been shown to be involved in long range intramolecular interactions with FN30kDa defining a closed conformation of FN100kDa (9). To elucidate the role of the first type I FN module (1FNI) in multiple binding of FN to FnBPs, a truncated FN30kDa construct, 2–5FNI, was also used (25).
FN-binding Sites of SfbI Are Not Equivalent
All cell and syringe solution concentrations used in the ITC experiments are listed in the supplemental Table 1. Thermodynamic parameters derived from all experiments are presented in Table 1. SfbI-4, the construct chosen to represent the central repeats of SfbI (100% identical to SfbI-2 and 95% identical to SfbI-3), bound to FN30kDa, FN70kDa, and FN100kDa with 1:1 stoichiometry (supplemental Fig. 1). Binding, as was the case for all other interactions investigated in this study, was associated with large exothermic changes in enthalpy (ΔH) and large unfavorable changes in entropy (ΔS). Binding of FN30kDa, FN70kDa, and FN100kDa to SfbI-5 yielded very similar thermodynamic parameters indicating that the binding site for SfbI-5, i.e. 1–5FNI, is fully available in each case (supplemental Fig. 1 and Table 1). The dissociation constant (Kd) for FN30kDa was higher (166 nm) than the values measured for FN70kDa and FN100kDa (37 and 32 nm, respectively). This may be due to different structural contexts in which the FNI modules are found in FN30kDa and in the larger fragments. SfbI-5 binding was characterized by more exothermic ΔH, more unfavorable ΔS, and significantly higher affinities (Kd ∼2 nm) compared with SfbI-4 (Table 1). This reflects the additional interactions between 1FNI and its unique binding site in SfbI-5 (Fig. 1D).
TABLE 1.
Thermodynamic parameters obtained by fitting experimental data to AB or ABB hetero-association models (SEDPHAT)
Experiments 19–21 were analyzed using a two-site model (Microcal Origin) (N1 = 1, N2 = 4). The parameters shown for these experiments are apparent values.
| No. | SfbI construct | Binding sitea | FN fragment | Modeb | Kd | ΔH | −TΔS | ΔG |
|---|---|---|---|---|---|---|---|---|
| nm | kcal mol−1 | kcal mol−1 | kcal mol−1 | |||||
| 1 | SfbI-4 | FN30kDa | 166.09 | −30.98 | 21.36 | −9.62 | ||
| 2 | SfbI-5 | FN30kDa | 1.42 | −45.30 | 32.74 | −12.56 | ||
| 3 | SfbI-UR1 | FN70kDa | 3.30 | −70.02 | 57.98 | −12.04 | ||
| 4 | SfbI-4 | FN70kDa | 37.44 | −32.41 | 21.87 | −10.54 | ||
| 5 | SfbI-5 | FN70kDa | 2.53 | −40.33 | 28.13 | −12.20 | ||
| 6 | SfbI-UR1 | FN100kDa | 1.85 | −66.73 | 54.34 | −12.39 | ||
| 7 | SfbI-4 | FN100kDa | 32.00 | −36.79 | 26.15 | −10.64 | ||
| 8 | SfbI-5 | FN100kDa | 1.77 | −42.69 | 30.26 | −12.42 | ||
| 9 | SfbI-4 | 2–5FNI | 304.69 | −27.51 | 18.26 | −9.25 | ||
| 10 | SfbI-5 | 2–5FNI | 12.96 | −29.91 | 18.72 | −11.19 | ||
| 11 | SfbI-UR12 | SfbI-UR1 | FN70kDa | S | 0.07 | −78.39 | 64.01 | −14.38 |
| 11 | SfbI-2 | FN70kDa | S | 3.82 | −38.90 | 26.95 | −11.95 | |
| 12 | SfbI-UR1 | FN70kDa | C | 0.09 | −80.36 | 66.13 | −14.23 | |
| 12 | SfbI-2 | FN70kDa | C | 3.41 | −32.62 | 20.60 | −12.02 | |
| 13 | SfbI-UR1 | FN100kDa | S | 0.06 | −63.97 | 49.51 | −14.46 | |
| 13 | SfbI-2 | FN100kDa | S | 3.24 | −27.55 | 15.50 | −12.05 | |
| 14 | SfbI-UR1 | FN100kDa | C | 0.36 | −67.83 | 54.43 | −13.40 | |
| 14 | SfbI-2 | FN100kDa | C | 8.57 | −25.32 | 13.87 | −11.45 | |
| 15 | SfbI-45 | SfbI-4 | FN30kDa | S | 4.48 | −32.41 | 20.56 | −11.85 |
| 15 | SfbI-5 | FN30kDa | S | 0.32 | −50.49 | 37.02 | −13.47 | |
| 16 | SfbI-4 | FN30kDa | C | 12.25 | −37.42 | 26.19 | −11.23 | |
| 16 | SfbI-5 | FN30kDa | C | 0.05 | −45.40 | 30.81 | −14.58 | |
| 17 | SfbI-4/5 | FN70kDa | C | 2.66 | −41.51 | 29.34 | −12.17 | |
| 18 | SfbI-4/5 | FN100kDa | C | 1.55 | −34.98 | 22.48 | −12.50 | |
| 19 | SfbI-UR12345 | SfbI-4 | FN30kDa | S | (4) | (−33.04) | (21.12) | (−11.92) |
| 19 | SfbI-5 | FN30kDa | S | (0.05) | (−47.25) | (32.56) | (−14.69) | |
| 20 | SfbI-4/5 | 2–5FNI | S | (188) | (−33.56) | (24.01) | (−9.55) | |
| 21 | SfbI-UR1 | FN100kDa | S | (0.16) | (−105) | (92) | (−13) | |
| 21 | SfbI-2 | FN100kDa | S | (3.7) | (−30.5) | (18.5) | (−12) |
a Binding sites within multiple site constructs are assigned according to the expected order of binding based on single site experiments except for experiment 21 where it is based on Kd values obtained in experiments 13 and 18.
b C indicates ligand in cell; S indicates ligand in syringe.
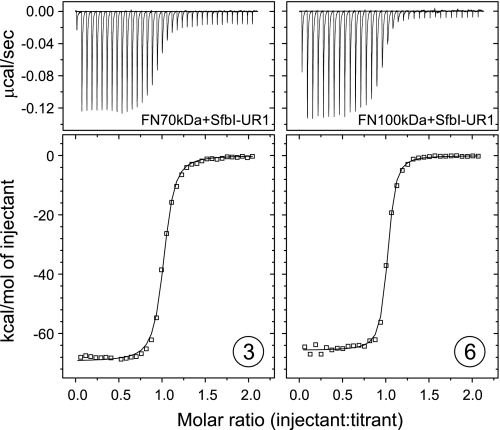
SfbI-1 was previously found to have low affinity for FN30kDa (18). This repeat, however, together with the upstream FN-binding region forms a larger FN-binding site SfbI-UR1, i.e. the complete binding region for FN70kDa (Fig. 1D). Binding of FN70kDa to SfbI-UR1 resulted in an isotherm representing a 1:1 interaction with a Kd of ∼3 nm (Fig. 2, 3). As can be expected for the transition of an entirely unstructured protein of 58 residues to its bound state, ΔS is highly unfavorable (−192 cal mol−1 K−1). The remarkable ΔH of −70 kcal/mol is probably a consequence of the enthalpy/entropy compensation commonly observed in biomolecular interactions (31). It reflects the formation of a large binding interface and extensive conformational rearrangements that the natively unfolded FN-binding sites of SfbI will undergo by forming an extended interface along arrays of FNI modules. In comparison with FN30kDa binding to SfbI-1 (18) and FN70kDa binding to other repeats (Table 1), the thermodynamic parameters obtained for SfbI-UR1 indicate that both domains of FN70kDa (FN30kDa and GBD) take part in the interaction. FN100kDa binding to SfbI-UR1 resembled FN70kDa binding (Fig. 2, 6), implying that the complete binding site for SfbI is available in both FN70kDa and FN100kDa.
FIGURE 2.
ITC data obtained for the injection of SfbI-UR1 into solutions of FN70kDa (left) and FN100kDa (right). The top panels show raw data representing the response to injections, and the bottom panels show integrated heats of injections (□) and the best fit (—) to the AB hetero-association model (SEDPHAT). Circled numbers refer to the list of experiments in Table 1.
The results obtained for isolated FN-binding sites are of little biological relevance. Therefore, FN binding to the complete FN-binding region of SfbI (SfbI-UR12345) was investigated. As a full quantitative thermodynamic analysis of cooperative five-site binding is not feasible due to the lack of a suitable binding model, two-site binding was explored using two constructs representing the N- and C-terminal pairs of binding sites in SfbI-UR12345.
Cooperative Binding of FN Fragments to SfbI-45
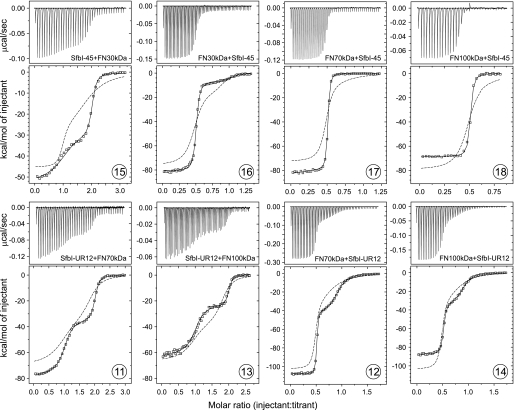
SfbI-45 includes the repeats SfbI-4 and SfbI-5. ITC experiments involving multiple binding sites can be conducted in two modes giving rise to nonidentical titration curves. The binding partner containing multiple sites can either be present in the cell (referred to as the “ligand in syringe” experiment) or in the syringe (“ligand in cell”). Injection of FN30kDa into a SfbI-45 solution (ligand in syringe) yielded a two-step transition curve (Fig. 3, 15). The relative affinities of the binding sites appear to be preserved in SfbI-45, i.e. SfbI-5 binds more strongly and with a larger ΔH than SfbI-4. The ΔH for the stronger site in SfbI-45 is more exothermic by about −5 kcal/mol when compared with FN30kDa binding to SfbI-5. The affinity is slightly enhanced, as reflected by a subnanomolar Kd (∼0.3 versus 2 nm). On the other hand, the ΔH for the second transition, reflecting the weaker site, is very similar to the value observed for isolated SfbI-4. The Kd for this site, however, is lower by a factor of nearly 40 (4.5 versus 170 nm).
FIGURE 3.
ITC data for the interaction of two-site SfbI constructs with FN fragments. The top panels show raw data representing the response to injections, and the bottom panels show integrated heats of injections (□) and the best fit (—) to the ABB hetero-association model (SEDPHAT). The dashed traces are simulated isotherms based on thermodynamic parameters obtained in single-site experiments. Circled numbers refer to the list of experiments in Table 1.
The experiment was also carried out in the ligand in cell mode (Fig. 3, 16). The experiment also yielded a two-step titration curve. Although there were slight differences in the thermodynamic parameters derived from the two experimental modes, the sum of the ΔH values for both binding events was the same in both experiments (−80 kcal/mol), and both experiments clearly show mutually enhanced (cooperative) binding of FN30kDa to the two sites in SfbI-45 when compared with single sites. This enhancement in binding affinity and enthalpy is evident when experimental data are compared with simulated ITC curves that are based on data obtained with isolated sites and the assumption of independent binding (dashed traces in Fig. 3).
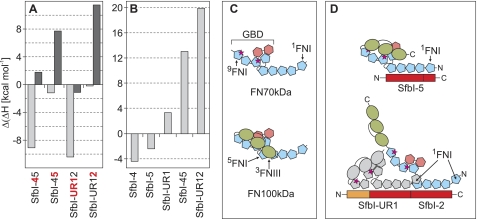
FN70kDa bound to SfbI-45 with 2:1 stoichiometry resulting in an ITC curve with only one transition (Fig. 3, 17) reflecting indistinguishable binding sites within SfbI-45. The sum of the ΔH contributions of both binding sites (−83 kcal/mol) is considerably larger than the sum of ΔH values measured in single site experiments (−63 kcal/mol, Fig. 4A). Based on the observed Kd value (3 nm), binding of FN70kDa to SfbI-4 within SfbI-45 is enhanced in comparison with isolated SfbI-4 (Kd = 37 nm). The equivalent experiment with FN100kDa revealed an unexpected difference to FN70kDa (Fig. 3, 18). Although the cooperativity of binding of FN100kDa to the SfbI-4 site is apparent and very similar to the effect observed for FN70kDa, the interaction of FN100kDa with SfbI-45 is enthalpically diminished in comparison with both single-site binding (Fig. 4A) and FN70kDa binding to SfbI-45 (Fig. 4B).
FIGURE 4.
Differences in SfbI binding to FN70kDa and FN100kDa. A, differences in ΔH observed for single site and double site binding of FN70kDa (light gray) and FN100kDa (dark gray) (ligand in cell mode). Positive values indicate less exothermic binding to sub-sites (red) in two-site constructs. B, differences in ΔH observed for FN70kDa and FN100kDa binding to SfbI constructs (ligand in cell mode). Positive values indicate enthalpically diminished (less exothermic) binding of FN100kDa. C, models for the arrangement of modules in FN70kDa and FN100kDa. The models are in agreement with structural data (44) and long range interactions in FN100kDa (9). Motogenic IGD sites (magenta stars) on 7FNI and 9FNI are exposed in FN70kDa but cryptic in FN100kDa due to the proposed closed conformation. D, binding of FN100kDa to SfbI results in a conformational change and exposure of motogenic sites. Although single site binding (shown for SfbI-5, top) does result in some FN100kDa activation, multiple binding (shown for SfbI-UR12, bottom) may facilitate this process through interactions involving binding of 1FNI to the adjacent ligand. Furthermore, SfbI-UR may support activation of FN100kDa through binding to the GBD.
Cooperative Binding to SfbI-UR12
Multiple binding of FN fragments involving the upstream FN-binding region was explored using the construct SfbI-UR12. ITC experiments with both FN70kDa and FN100kDa for both ligand in syringe (Fig. 3, 11 and 13) and ligand in cell (Fig. 3, 12 and 14) modes showed two clearly distinct transitions. Although the ΔH values obtained for FN70kDa were generally higher or comparable with single site enthalpies, the experiments with FN100kDa reflected enthalpically neutral or diminished binding in comparison with single sites (Fig. 4A). The enthalpy changes for FN100kDa were less exothermic by ∼10 kcal/mol per site than the ones observed for FN70kDa (Fig. 4B). Irrespective of the experimental mode, binding of the FN fragments to both sites within SfbI-UR12 was mutually cooperative.
Full-length FN-binding Domain of SfbI Is Natively Unfolded
SfbI-UR12345 is a 23.9-kDa polypeptide that, based on secondary structure predictions and data obtained for other FN-binding repeats (15), was expected to be natively unfolded. One-dimensional 1H nuclear magnetic resonance (NMR) spectra in water and in deuterated water (D2O) were acquired (supplemental Fig. 2A). The poor signal dispersion of these spectra indicates the lack of tertiary structure in SfbI-UR12345. A circular dichroism spectrum displayed the characteristics of an unfolded polypeptide (supplemental Fig. 2B). Deconvolution using two different algorithms resulted in secondary structure compositions with mean values for α-helix (1.5%), β-strand (5%), β-turns (4%), and unordered structure (89%) that are in good agreement with a protein largely lacking secondary structure.
FN Fragment Binding to SfbI-UR12345
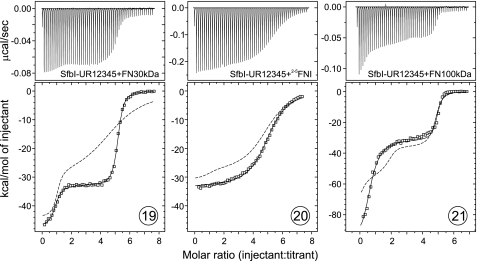
Injection of FN30kDa into an SfbI-UR12345 solution resulted in a two-step titration curve (Fig. 5, 19) with transitions occurring at molar ratios of ∼1:1 and 5:1 (FN30kDa:SfbI-UR12345). There are no models available for the analysis of ITC data for five nonidentical sites. However, given the two-step nature of the binding, data were analyzed with a two-site model (Microcal Origin). The thermodynamic parameters derived were very similar to values obtained for FN30kDa binding to SfbI-45. The experiment demonstrated the availability of five binding sites in SfbI-UR12345, four of which were indistinguishable. Based on the ΔH values, it can be assumed that the first transition reflects binding to the unique high affinity site SfbI-5, whereas the second step in the titration curve represents subsequent binding to the other four repeats. Binding of FN30kDa to all repeats in SfbI-UR12345 was cooperative in comparison with single repeats (Table 1, simulated data in Fig. 5).
FIGURE 5.
ITC data obtained for the injection of FN fragments into SfbI-UR12345 solutions. The top panels show raw data representing the response to injections, and the bottom panels show integrated heats of injections (□) and the best fit (—) to the two-site model (Origin). The dashed traces are simulated isotherms based on thermodynamic parameters obtained in single-site experiments.
A two-step titration curve was also obtained for FN100kDa and SfbI-UR12345 (Fig. 5, 21). Saturation of all binding sites in SfbI-UR12345 was achieved at a molecular ratio of ∼5:1 (FN100kDa:SfbI-UR12345). Fitting the data to a two-site model yielded thermodynamic parameters that are unlikely to be very accurate due to the unsuitability of the model and the uncertainty of ΔH estimation for the first transition. However, the estimated Kd values and the ΔH for the second transition of the titration are in good agreement with the SfbI-UR12 experiments and signify cooperative binding. The highly exothermic ΔH observed for the first transition cannot be explained. Interpretation of the curve is further complicated by the presence of two sites within SfbI-UR12345, SfbI-UR1 and SfbI-5, which have very similar high affinities for FN100kDa. The order of binding to these sites cannot be predicted. The apparent stoichiometry (∼0.6) for the first transition could be a consequence of the simultaneous titration of these sites, which give rise to different enthalpy changes.
Role of 1FNI in Cooperativity
FN-binding repeats are contiguous arrays of FNI-binding motifs. Structural analyses revealed the presence of very few spacer residues between FNI-binding motifs and between repeats (17). Binding of FN (fragments) to the linear array of five sites in SfbI-UR12345 therefore will inevitably result in close spatial proximity of the bound ligands, even if the resulting molecular assembly may not be linear in shape (supplemental Fig. 3). In this context, the lack of a motif for binding to the most N-terminal FNI module in FN (1FNI) in all but the last repeat of SfbI is interesting. When neighboring sites are occupied, 1FNI will likely be oriented such that the adjacent (downstream) 5FNI-binding motif is available. Such a conformation could be easily adopted given the highly flexible link between the first two FNI modules (32). Close spacing of FN (fragments) bound to SfbI may quite possibly result in ligand-ligand interactions, as indicated in Fig. 4D. Such an interaction, even if weak, could contribute the additional free energy in multiple binding observed in ITC experiments.
To test this hypothesis, interactions of SfbI with 2–5FNI (FN30kDa lacking 1FNI) were studied. 2–5FNI bound to both SfbI-4 and SfbI-5 with ΔH values comparable with the ones obtained for SfbI-4 interactions with other FN fragments (Table 1, supplemental Fig. 1, 9 and 10), confirming the presence of a complete binding site for SfbI-4 in this four-module construct. The interaction was slightly attenuated in comparison with SfbI-4 binding to FN30kDa (Kd 305 versus 166 nm). 2–5FNI binding to SfbI-5 was comparably strong but clearly weaker than FN30kDa binding (13 versus 2 nm).
In contrast to FN30kDa, 2–5FNI titration into SfbI-UR12345 showed one broad transition that could be fitted to a one-site model (stoichiometry of 5) (Table 1 and Fig. 5, 20). The apparent Kd value of the indistinguishable sites was only very weakly enhanced in comparison with SfbI-4 binding (188 nm). This is in contrast to the significantly lowered Kd values observed for binding of all other FN fragments to the SfbI-2/4 site in two- or five-site constructs (e.g. 4 nm for FN30kDa and SfbI-UR12345). This observation is the first experimental evidence for an involvement of FN-FN interactions in multiple binding to FnBPs.
Cryptic Sites in FN100kDa Are Exposed upon Binding to SfbI
Streptococci and staphylococci bind to the N-terminal region of FN to establish a link to host cell integrins. The major integrin-binding site in FN, the RGD motif, is located in the 10FNIII module, i.e. remote from the bacterial binding site. As in fibrillogenesis, recruitment of FN to bacteria will result in RGD exposure. Indeed it has been demonstrated that an SfbI fragment “FUD,” representing SfbI-UR1 lacking six and three residues at the N and C termini, respectively, exposes epitopes of the RGD-containing 10FNIII domain in soluble FN (33). To explore the possibility that RGD exposure is linked to a conformational change at the bacterial binding site, SfbI constructs, and FN100kDa, the smallest available FN fragment containing cryptic integrin-binding sites were used in a cell migration assay.
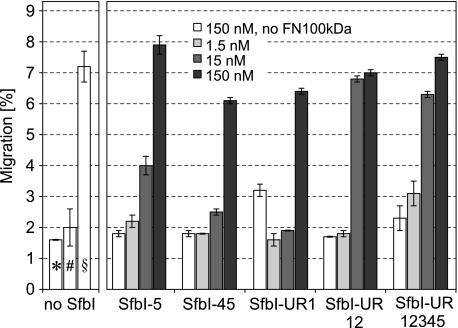
Adult skin fibroblasts were plated on to the surface of three-dimensional collagen type I gels in the presence of FN100kDa and increasing concentrations of SfbI-5, SfbI-UR1, SfbI-45, SfbI-UR12, or SfbI-UR12345. After incubation for 4 days, the number of cells that had migrated into the collagen gel was determined microscopically. At the highest concentration used (150 nm), all SfbI constructs induced migration of fibroblasts (Fig. 6) at a level comparable with the one induced by migration-stimulating factor (truncated oncofetal FN) or the very similar FN70kDa. For these proteins, it has been shown that the only active sequences for stimulating fibroblast migration into collagen gels are the IGD motifs present on 7FNI and 9FNI (12, 34). The FN100kDa-R222A mutant also stimulated cell migration (Fig. 6) (9), suggesting that the IGD sites are at least partially exposed due to a less compact conformation of this mutant in comparison with nonactive FN100kDa (9). Our data indicate that IGD sites in FN100kDa are exposed as a consequence of an interaction with SfbI. Although the results of migration assays cannot confidently be interpreted quantitatively, SfbI-UR12 and SfbI-UR12345 were the only constructs inducing the full level of migration stimulation already at a concentration of 15 nm. Assuming the affinities are similar in ITC and under conditions used in the migration assay, ∼80–100% of FN100kDa should be bound to each of the SfbI constructs at 15 nm concentration.
FIGURE 6.
Effect of increasing concentrations of SfbI constructs on the motogenic activity of FN100kDa. The number of fibroblast that had migrated into three-dimensional collagen gels was determined microscopically. The concentration of FN100kDa in all samples was 100 pg/ml (∼1 pm) except for control experiments (* and §) where FN100kDa was omitted. Neither FN100kDa alone (#) nor SfbI constructs at 150 nm concentrations in the absence of FN100kDa (white bars) induced migration, in contrast to FN100kDa-R222A at 100 pg/ml (∼1 pm) (§).
SfbI Binding May Compete with an Intramolecular Interaction in FN100kDa
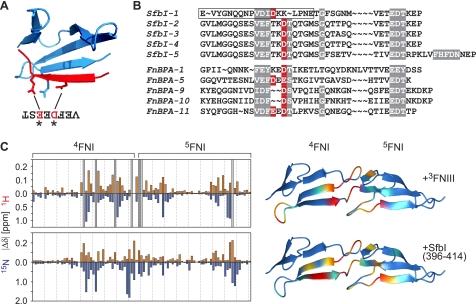
Vakonakis et al. (9) identified an intramolecular salt bridge involving residues Arg-222 (in 4FNI) and acidic side chains in 3FNIII as a determinant of a closed, inactive conformation of FN100kDa (Fig. 4C). The side chain of Arg-222 was also shown to be targeted by one or two acidic residues in the 4FNI-binding motif in bacterial peptides from FnBPA (Fig. 7A) (17). SfbI FN-binding repeats contain conserved aspartate residues in 4FNI-specific regions that are very likely to form a salt bridge with Arg-222 when bound to 4FNI (Fig. 7B).
FIGURE 7.
Competition of SfbI with intramolecular FN interactions. A, ribbon diagram showing the 4FNI-binding motif from FnBPA-5 (red) in complex with 4FNI (blue) (Protein Data Bank code 2RL0) (17). The side chains of FN R222 and two acidic residues in FnBPA are shown. B, alignment of SfbI repeats with repeats from S. aureus FnBPA. Highly conserved residues are highlighted gray. The negatively charged residues that are known or predicted to interact with Arg-222 in 4FNI are highlighted red. The peptide used for the NMR binding experiment shown in C is boxed. C, chemical shift perturbation maps for the interaction of 15N-4FNI5FNI with 3FNIII (top) (9) and the 4FNI5FNI-binding motif of SfbI-1 (bottom) (18). Red bars correspond to absolute values of amide proton chemical shift changes observed upon addition of the unlabeled binding partners to 15N-4FNI5FNI. Blue bars represent absolute 15N amide chemical shift changes. Gray bars in the 3FNIII shift map represent resonances that could not be traced in the titration experiment due to intermediate exchange, indicative of large chemical shift changes. The combined chemical shift changes (|Δδ 1HN| + |(Δδ 15N)/5|) are mapped onto secondary structure diagrams of 4FNI5FNI (Protein Data Bank code 1FBR). Small to large chemical shift changes are indicated by color gradation from blue to red (prepared with MOLMOL (45)).
To support the hypothesis of a competition of SfbI with an intramolecular FN interaction, the chemical shift perturbation map of 4FNI5FNI upon addition of a peptide representing the 4FNI5FNI-binding motif of SfbI-1 (residues 396–414) (18) was compared with a similar experiment monitoring the effect of 3FNIII addition to 15N-4FNI5FNI (9). Although the observed perturbation patterns are not identical, the overlap of binding sites identified in both experiments is obvious (Fig. 7C) and strongly suggests the possibility of a displacement of 3FNIII from its intramolecular binding site by SfbI. It should be noted that the NMR shift mapping used here only observes backbone amide group resonances. Involvement of an amino acid side chain in a binding interaction does not necessarily induce chemical shift changes in the backbone amide of the same residue. Arg-222, despite being involved in binding to 3FNIII and most likely SfbI, does not show large backbone chemical shift changes upon binding to 3FNIII or the SfbI-1 peptide.
DISCUSSION
To induce their uptake by nonprofessional phagocytes, bacteria utilizing FN-integrin interactions have to recruit FN to their surface in a density high enough to trigger integrin-mediated outside-to-inside signaling known to be exploited by many pathogens for cell invasion (1). Although it has not been demonstrated yet, an obvious assumption would be that the purpose of localized multiple binding of FN to FnBPs is to present a critical number of RGD motifs to induce integrin clustering. However, it is unclear how an interaction of bacteria with the N-terminal domain of FN results in exposure of the remote cell-binding site. FN and the large complexes formed with FnBPs are not amenable to current structural techniques. We used different FN fragments and ITC, the most accurate technique for quantitative analysis of molecular interactions, complemented by a cell migration assay reporting conformational changes in FN100kDa to gain insights into purpose and consequences of multiple FN binding to bacterial surface proteins.
Recruitment of FN fragments to a gapless array of binding sites results in significantly enhanced affinities. FN·FnBP complexes have to be stable enough to mediate tight bacterial adhesion, presumably sometimes under shear stress, but also to sustain the traction forces exerted on the bacteria during internalization. In agreement with this notion, staphylococci expressing single site low affinity FnBPs do not invade endothelial cells (24). The N-terminal module 1FNI may play a role in cooperative binding of FN to SfbI, conceivably facilitated by the flexible attachment of 1FNI to the FN molecule (32). The N-terminal domain (1–5FNI) is not only the main bacterial binding site in FN but also the only region strictly required for matrix assembly (35). It is tempting to suggest a special role for 1FNI in FN interactions required for both fibrillogenesis and bacterial invasion.
To interact with host cell integrins, the RGD sequence in FN must be accessible, which requires a conformational change in plasma FN. This activation has been shown to be triggered by various ligands (e.g. heparin and gelatin), environmental conditions (8, 36), and potentially bacteria (33). The inactive plasma form of FN is generally assumed to be defined by intramolecular interactions involving the N-terminal domain of FN. Such interactions have been identified for FN100kDa, which serves as a valuable tool to study FN conformational changes associated with the exposure of cryptic sites (9). Based on our systematic analysis of FN-SfbI interactions, the observed differences in binding enthalpy changes for FN70kDa and FN100kDa binding to multiple site constructs must result from an event not directly linked to the formation of binding interfaces with SfbI. A plausible explanation for the diminished ΔH for FN100kDa binding is the occurrence of an entropically favorable, enthalpically unfavorable process such as unfolding or, more likely, a conformational change associated with a loss of an intramolecular interface resulting in increased flexibility. As the only difference between FN70kDa and FN100kDa are three FNIII modules that do not participate in SfbI binding, such a conformational change will involve these modules. Intriguingly, in all multiple site binding experiments with FN100kDa, enthalpically diminished binding is never observed for the N-terminal binding sites, i.e. SfbI-4 in SfbI-45, SfbI-UR1 in SfbI-UR12 (Fig. 4A), and presumably SfbI-UR1 in SfbI-UR12345. An FN fragment bound to the N-terminal site cannot engage in intermolecular interaction with 1FNI from a ligand attached to a neighboring site. Cooperative FN binding to SfbI appears to rely on ligand-ligand interactions involving the N-terminal 1FNI module. The conformational change (or activation) of FN100kDa as observed by ITC and cell migration appears to be facilitated by multiple binding. Cell migration experiments support the notion of a conformational change in FN triggered by SfbI. It should be noted that stimulation of cell migration is unlikely to be of any relevance for streptococcal infections as binding to full-length FN will trigger rapid RGD-dependent uptake. In this study, detection of IGD exposure in migration assays is only used as a sensitive reporter of conformational changes.
Bacterial activation of soluble FN, in some respects, is related to FN activation leading to fibrillogenesis. However, formation of insoluble FN assemblies would be detrimental to bacterial uptake. Therefore, bacterial binding is not expected to result in the exposure of cryptic self-association sites such as the interdomain linker between the 1FNIII and 2FNIII modules that are thought to require tension induced by focal adhesions (37). Although no data supporting this assumption are available for constructs containing multiple sites, an SfbI fragment FUD largely resembling SfbI-UR1 inhibited rather than promoted the formation of FN matrix (38). Another consequence of multiple FN binding to FnBPs may be the formation of well defined supra-molecular assemblages (supplemental Fig. 3), resulting in an arrangement of RGD motifs suitable for integrin clustering and probably resembling the pattern found in the extracellular matrix. The spacing of RGD motifs is crucial for effective integrin clustering, cell adhesion, and spreading on extracellular matrix-mimicking nano-patterned substrates (39, 40). The critical RGD density and spacing required for integrin activation might be difficult to achieve with multiple copies of FnBPs containing single binding sites.
SfbI-mediated internalization of S. pyogenes involves recruitment of caveolae that act as an entry port for the bacteria (41). SfbI-coated gold particles have been shown to accumulate at caveolae, very likely reflecting the formation of integrin clusters. Caveolae have also been implicated in caveolin-1-dependent α5β1 integrin-mediated FN endocytosis (42). These findings suggest the interesting possibility that SfbI-expressing streptococci and potentially other bacterial pathogens or viruses (43), through multiple binding to FN, target this cellular recycling mechanism to achieve internalization.
Supplementary Material
Acknowledgments
We thank Dr. Ioannis Vakonakis for helpful discussions and the original chemical shift perturbation data for the 3FNIII/4FNI5FNI interaction. We thank Dr. Catherine Botting for mass spectrometry. I. R. E. thanks Prof. S. L. Schor and Dr. A. M. Schor for the kind gift of the FSF44 cell line used in these studies. The MGAS6180 genomic DNA was a kind gift by Prof. Slawomir Lukomski.
This work was supported in part by the E. P. Abraham Cephalosporin Fund (Oxford, United Kingdom).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- FnBP
- fibronectin-binding protein
- FN
- fibronectin
- FN30kDa
- FN70kDa, FN100kDa, 30-, 70-, and 100-kDa N-terminal fragments of FN
- nFNI/II/III
- nth FN type-I/II/III domains
- GBD
- fibronectin gelatin-binding domain
- SfbI-UR/1/2/3/4/5
- upstream FN-binding region, 1st, 2nd, 3rd, 4th, and 5th FN-binding repeat of SfbI
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Pizarro-Cerdá J., Cossart P. (2006) Cell 124, 715–727 [DOI] [PubMed] [Google Scholar]
- 2. Nitsche-Schmitz D. P., Rohde M., Chhatwal G. S. (2007) Thromb. Haemost. 98, 488–496 [PubMed] [Google Scholar]
- 3. Ozeri V., Rosenshine I., Mosher D. F., Fässler R., Hanski E. (1998) Mol. Microbiol. 30, 625–637 [DOI] [PubMed] [Google Scholar]
- 4. Dziewanowska K., Patti J. M., Deobald C. F., Bayles K. W., Trumble W. R., Bohach G. A. (1999) Infect. Immun. 67, 4673–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leiss M., Beckmann K., Girós A., Costell M., Fässler R. (2008) Curr. Opin. Cell Biol. 20, 502–507 [DOI] [PubMed] [Google Scholar]
- 6. Clark R. A., Wikner N. E., Doherty D. E., Norris D. A. (1988) J. Biol. Chem. 263, 12115–12123 [PubMed] [Google Scholar]
- 7. Fukai F., Ohtaki M., Fujii N., Yajima H., Ishii T., Nishizawa Y., Miyazaki K., Katayama T. (1995) Biochemistry 34, 11453–11459 [DOI] [PubMed] [Google Scholar]
- 8. Ugarova T. P., Zamarron C., Veklich Y., Bowditch R. D., Ginsberg M. H., Weisel J. W., Plow E. F. (1995) Biochemistry 34, 4457–4466 [DOI] [PubMed] [Google Scholar]
- 9. Vakonakis I., Staunton D., Ellis I. R., Sarkies P., Flanagan A., Schor A. M., Schor S. L., Campbell I. D. (2009) J. Biol. Chem. 284, 15668–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schor S. L., Ellis I., Banyard J., Schor A. M. (1999) J. Cell Sci. 112, 3879–3888 [DOI] [PubMed] [Google Scholar]
- 11. Ellis I. R., Jones S. J., Staunton D., Vakonakis I., Norman D. G., Potts J. R., Milner C. M., Meenan N. A., Raibaud S., Ohea G., Schor A. M., Schor S. L. (2010) Exp. Cell Res. 316, 2465–2476 [DOI] [PubMed] [Google Scholar]
- 12. Schor S. L., Ellis I. R., Jones S. J., Baillie R., Seneviratne K., Clausen J., Motegi K., Vojtesek B., Kankova K., Furrie E., Sales M. J., Schor A. M., Kay R. A. (2003) Cancer Res. 63, 8827–8836 [PubMed] [Google Scholar]
- 13. Talay S. R., Valentin-Weigand P., Jerlström P. G., Timmis K. N., Chhatwal G. S. (1992) Infect. Immun. 60, 3837–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz-Linek U., Höök M., Potts J. R. (2004) Mol. Microbiol. 52, 631–641 [DOI] [PubMed] [Google Scholar]
- 15. Penkett C. J., Redfield C., Jones J. A., Dodd I., Hubbard J., Smith R. A., Smith L. J., Dobson C. M. (1998) Biochemistry 37, 17054–17067 [DOI] [PubMed] [Google Scholar]
- 16. Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., Briggs J. A., Gough T. S., Höök M., Campbell I. D., Potts J. R. (2003) Nature 423, 177–181 [DOI] [PubMed] [Google Scholar]
- 17. Bingham R. J., Rudiño-Piñera E., Meenan N. A., Schwarz-Linek U., Turkenburg J. P., Höök M., Garman E. F., Potts J. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12254–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schwarz-Linek U., Pilka E. S., Pickford A. R., Kim J. H., Höök M., Campbell I. D., Potts J. R. (2004) J. Biol. Chem. 279, 39017–39025 [DOI] [PubMed] [Google Scholar]
- 19. Meenan N. A., Visai L., Valtulina V., Schwarz-Linek U., Norris N. C., Gurusiddappa S., Höök M., Speziale P., Potts J. R. (2007) J. Biol. Chem. 282, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 20. Talay S. R., Zock A., Rohde M., Molinari G., Oggioni M., Pozzi G., Guzman C. A., Chhatwal G. S. (2000) Cell. Microbiol. 2, 521–535 [DOI] [PubMed] [Google Scholar]
- 21. Atkin K. E., Brentnall A. S., Harris G., Bingham R. J., Erat M. C., Millard C. J., Schwarz-Linek U., Staunton D., Vakonakis I., Campbell I. D., Potts J. R. (2010) J. Biol. Chem. 285, 36977–36983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Natanson S., Sela S., Moses A. E., Musser J. M., Caparon M. G., Hanski E. (1995) J. Infect. Dis. 171, 871–878 [DOI] [PubMed] [Google Scholar]
- 23. Towers R. J., Fagan P. K., Talay S. R., Currie B. J., Sriprakash K. S., Walker M. J., Chhatwal G. S. (2003) J. Clin. Microbiol. 41, 5398–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edwards A. M., Potts J. R., Josefsson E., Massey R. C. (2010) PLoS Pathog. 6, e1000964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Staunton D., Millard C. J., Aricescu A. R., Campbell I. D. (2009) Methods Mol. Biol. 522, 73–99 [DOI] [PubMed] [Google Scholar]
- 26. Lobley A., Whitmore L., Wallace B. A. (2002) Bioinformatics 18, 211–212 [DOI] [PubMed] [Google Scholar]
- 27. Houtman J. C., Brown P. H., Bowden B., Yamaguchi H., Appella E., Samelson L. E., Schuck P. (2007) Protein Sci. 16, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schor S. L. (1980) J. Cell Sci. 41, 159–175 [DOI] [PubMed] [Google Scholar]
- 29. Zhao Q., Liu X., Collodi P. (2001) Exp. Cell Res. 268, 211–219 [DOI] [PubMed] [Google Scholar]
- 30. Liu X., Zhao Q., Collodi P. (2003) Matrix Biol. 22, 393–396 [DOI] [PubMed] [Google Scholar]
- 31. Cooper A., Johnson C. M., Lakey J. H., Nöllmann M. (2001) Biophys. Chem. 93, 215–230 [DOI] [PubMed] [Google Scholar]
- 32. Potts J. R., Bright J. R., Bolton D., Pickford A. R., Campbell I. D. (1999) Biochemistry 38, 8304–8312 [DOI] [PubMed] [Google Scholar]
- 33. Ensenberger M. G., Annis D. S., Mosher D. F. (2004) Biophys. Chem. 112, 201–207 [DOI] [PubMed] [Google Scholar]
- 34. Millard C. J., Ellis I. R., Pickford A. R., Schor A. M., Schor S. L., Campbell I. D. (2007) J. Biol. Chem. 282, 35530–35535 [DOI] [PubMed] [Google Scholar]
- 35. Mao Y., Schwarzbauer J. E. (2005) Matrix Biol. 24, 389–399 [DOI] [PubMed] [Google Scholar]
- 36. Wierzbicka-Patynowski I., Schwarzbauer J. E. (2003) J. Cell Sci. 116, 3269–3276 [DOI] [PubMed] [Google Scholar]
- 37. Vakonakis I., Staunton D., Rooney L. M., Campbell I. D. (2007) EMBO J. 26, 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tomasini-Johansson B. R., Kaufman N. R., Ensenberger M. G., Ozeri V., Hanski E., Mosher D. F. (2001) J. Biol. Chem. 276, 23430–23439 [DOI] [PubMed] [Google Scholar]
- 39. Arnold M., Cavalcanti-Adam E. A., Glass R., Blümmel J., Eck W., Kantlehner M., Kessler H., Spatz J. P. (2004) ChemPhysChem. 5, 383–388 [DOI] [PubMed] [Google Scholar]
- 40. Cavalcanti-Adam E. A., Volberg T., Micoulet A., Kessler H., Geiger B., Spatz J. P. (2007) Biophys. J. 92, 2964–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rohde M., Müller E., Chhatwal G. S., Talay S. R. (2003) Cell. Microbiol. 5, 323–342 [DOI] [PubMed] [Google Scholar]
- 42. Shi F., Sottile J. (2008) J. Cell Sci. 121, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beer C., Pedersen L. (2007) J. Virol. 81, 8247–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pickford A. R., Smith S. P., Staunton D., Boyd J., Campbell I. D. (2001) EMBO J. 20, 1519–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.