Abstract
The Crohn's disease and early onset sarcoidosis susceptibility protein, NOD2, coordinates innate immune signaling pathways. Because dysregulation of this coordination can lead to inflammatory disease, maintaining appropriate activation of the NOD2 signaling pathway is paramount in immunologic homeostasis. In this work, we identify the atypical tumor necrosis factor-associated factor (TRAF) family member, TRAF4, as a key negative regulator of NOD2 signaling. TRAF4 inhibits NOD2-induced NF-κB activation and directly binds to NOD2 to inhibit NOD2-induced bacterial killing. We find that two consecutive glutamate residues in NOD2 are required for interaction with TRAF4 and inhibition of NOD2 signaling because mutation of these residues abrogated both TRAF4 binding and inhibition of NOD2. This work identifies a novel negative regulator of NOD2 signaling. Additionally, it defines a TRAF4 binding motif within NOD2 involved in termination of innate immune signaling responses.
Keywords: Inflammation, Innate Immunity, Protein Kinases, Signal Transduction, Ubiquitination
Introduction
Upon intracellular exposure to bacterial products, the Crohn's disease susceptibility protein, NOD2 (nucleotide-binding oligomerization domain 2), initiates innate immune inflammatory signaling pathways to help tailor the adaptive immune system such that it can eradicate offending pathogens (1–3). Both the induction and the termination of NOD2 signaling are important to immune homeostasis and must be tightly controlled. Genetic variants of NOD2 result in dysregulated signaling and are associated with inflammatory disorders, including Crohn's disease, early onset sarcoidosis (EOS),2 and Blau syndrome (4–6). Although the signaling cascade linking NOD2 stimulation to NF-κB activation has been extensively studied, the mechanisms by which NOD2, itself, is regulated remain elusive.
Upon intracellular exposure to a breakdown product of bacterial peptidoglycan, muramyl dipeptide (MDP), NOD2 binds to the scaffolding protein kinase, RIP2 (receptor-interacting protein 2), via caspase recruitment domain interactions (1, 7). Following activation of the NOD2-RIP2 complex, the (CARD) IKK scaffolding protein IKKγ (NEMO) becomes polyubiquitinated via Lys63 linkages on Lys285 of NEMO. These ubiquitin chains on NEMO are thought to help nucleate TAK1 (TNF receptor-associated kinase 1) such that it can phosphorylate the activation loop of IKKβ and ultimately activate the NF-κB transcription factors (8–11).
Previous work from this laboratory shows that one point of coordination for NOD2-induced innate immune signaling occurs at the level of the mitogen-activated protein kinase kinase kinases (MAP3Ks). MAP3Ks are the upper tier of a sequential cascade of MAPK-activating kinases that ultimately leads to MAPK activation. Specifically, the MAP3K, MEKK4, was identified as a regulator of NOD2. Both basally and in response to MDP, MEKK4 helps the cell to maintain low levels of NF-κB activity (12). In addition to phosphorylating MAP2Ks, the MAP3K proteins act as scaffolding proteins for signaling complexes (13). This is evidenced by the fact that one atypical tumor necrosis receptor-associated factor (TRAF) family member, TRAF4, has been implicated in MEKK4-dependent signaling pathways. TRAF4 binds to MEKK4, and the TRAF4 knock-out mouse closely resembles both the kinase-inactive MEKK4 knock-in mouse and the MEKK4 knock-out mouse because all of these mice show embryonic lethality with impaired neural tube closure and skeletal malformations (14–17).
Consistent with structural characteristics of other TRAF protein family members, TRAF4 has an N-terminal RING domain followed by a series of zinc fingers. The RING domain of TRAF proteins confers E3 ubiquitin ligase capability. The C terminus contains a TRAF domain that engages in heterotypic protein-protein interactions (18). The TRAF domain of each TRAF family member recognizes unique motifs in their binding partners. Although the TRAF domains of TRAF2 and TRAF6 show similar but separate peptide binding interactions, the peptide binding motif of TRAF4 has not been defined (19–21). Unlike other TRAF proteins, TRAF4 is largely uncharacterized, with few studies examining its role in inflammatory signaling responses.
In this work, we identify TRAF4 as a novel negative regulator of NOD2 signaling. We confirm the interaction of TRAF4 with MEKK4 and find that, like MEKK4, TRAF4 negatively regulates MDP-induced NF-κB activation and killing of intracellular bacteria. Furthermore, we find that TRAF4 binds NOD2, and we map the sites of interaction between NOD2 and TRAF4 to specific regions of these two proteins. We identify two critical amino acid residues within a putative TRAF binding motif in NOD2 that are essential for both binding of TRAF4 and TRAF4 inhibition of NOD2 function. Our findings demonstrate a novel mechanism for regulation of the host response to bacterial infection.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, Immunoprecipitations, and Western Blotting
HEK293T cells were maintained in DMEM containing 5% FBS (Hyclone). RAW 264.7 macrophages and HCT116 were maintained in DMEM containing 10% FBS (Hyclone). HT-29 cells were maintained in RPMI media containing 10% FBS (Hyclone). All media contained antibiotic/antimycotic solution (Invitrogen). Stably transfected cell lines were generated by retroviral infection of the indicated cells followed by neomycin selection (300 μg/ml; InvivoGen) in their respective media. Clones (>1000) were pooled. Calcium phosphate precipitation transfections in HEK293T cells were carried out as described previously (8, 9, 12, 22). Immunoprecipitations were conducted in Cell Signaling Lysis Buffer (50 mm Tris (pH 7.5), 150 mm NaCl, 1% Triton X-100, 1 mm EGTA, 1 mm β-glycerophosphate, 1 mm PMSF, 1 mm NaVO4, 10 nm Calyculin A in the presence of protease inhibitor mixture (Sigma)). Protein G-Sepharose beads (Invitrogen) were added to lysates, and immunoprecipitates were washed five times prior to Western blotting. A high stringent lysis buffer (Cell Signaling Lysis Buffer containing 1 m NaCl and 1% SDS) was used to wash immunoprecipitates from ubiquitination assays. Western blotting was completed on nitrocellulose membranes (Bio-Rad) as described previously (8, 9, 12).
Antibodies, Plasmids, and Reagents
Myc (9E10), RIP2, Omni, and TRAF4 antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). GST, Myc (rbt), phosphotyrosine, phospho-IKKβ, phospho-IκBα, and total IκBα antibodies were purchased from Cell Signaling Technology. Anti-HA (HA-11) was purchased from Covance. MEKK4 (ms) and FLAG (rbt), as well as FLAG beads (M2) were purchased from Sigma. Anti-Salmonella antibody (ms) was purchased from AbD Serotec. Goat anti-mouse Alexa568 antibody was purchased from Invitrogen. HA-MEKK4 was a generous gift from the laboratory of Gary Johnson (University of North Carolina School of Medicine, Chapel Hill, NC). Omni-MEKK4 was generated by subcloning MEKK4 (XhoI-XbaI) into HisMax pcDNA4 vector (Invitrogen). FLAG-TRAF4, TRAF2, and TRAF3 were obtained from the laboratory of Xiaoxia Li (Lerner Research Institute, Cleveland, OH). Omni-TRAF4 was generated by subcloning FLAG-TRAF4 (BamHI-EcoRI) into HisMax pcDNA4 vector (Invitrogen). pcDNA3 ΔRING (16376) and ΔTRAF (16377) constructs were purchased from AddGene (22) and subcloned (BglII-EcoRV) into HisMax pcDNA4 vector (Invitrogen). HA-NOD2 and NOD2 deletion mutants were used as described previously (23). The HA-E279A/E280A NOD2 construct was generated by QuikChange site-directed mutagenesis (Stratagene). Point mutations were verified by sequence analysis. pMXn-FLAG-NOD2 was used as previously described (24). Luciferase assay plasmids pBVIx-Luc and pEFBOS-βgal were used as described previously (1). A 5′ FLAG-fused TRAF4 was generated by PCR amplification using Omni-TRAF4 as a template and subcloned (BamHI-NotI) into pMXn to generate retroviral FLAG-TRAF4. HA-ubiquitin, Myc-K399R NEMO, Omni-RIP2, HA-RIP2, Omni-TRAF6, and GST-IKKβ were used as described previously (8, 9, 12). MDP was obtained from Bachem.
siRNA
Four separate siRNAs were purchased from Qiagen. The sequences of these were as follows: siTRAF4-2, CACCAGCACATTCGAAAGCGA; siTRAF4-5, AAGCTGGAAGTACAAGTATTG; siTRAF4-7, CTGCAGGAGTTCCTCAGTGAA; siTRAF4-8, CCGGAGCTGGAAGTACAAGTA. TRAF4 siRNAs were numbered according to the number of oligonucleotides given by Qiagen. HCT116 cells were transfected with 5 nm siRNA using Polyfect (Qiagen) for 48 h prior to the assay, according to the manufacturer's instructions.
Recombinant Protein Production
NOD2 amino acids 260–308 were PCR-amplified from HA-NOD2 and subcloned (BamHI-NotI) into pGEX-4T vector (GE Healthcare). BL21 (LysS) bacteria were transformed with pGEX-260–308 or empty PGEX-4T vector. Exponentially growing bacteria were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Bacterial pellets were subjected to freeze/thaw cycles in a buffer containing 50 mm Tris (pH 7.5), 100 mm NaCl, I mm DTT, 50 mg/ml lysozyme, and protease inhibitor mixture (Sigma). Glutathione-Sepharose was added to the lysates following DNase treatment and centrifugation. Beads were collected and washed extensively.
Luciferase Reporter Gene Assays
HEK293T cells were plated in triplicate at 4 × 104 cells/well. Reporter plasmids and expression constructs were transfected with Polyfect (Qiagen) as described previously (25). Cells were lysed in 1× reporter lysis buffer (Promega) and assayed for luciferase and β-galactosidase activity (1). Luciferase values were normalized to β-galactosidase activity.
Salmonella Infection and Gentamycin Protection Assays
Intracellular killing of Salmonella enterica serovar typhimurium SL1344 (gift of Gabriel Nuñez, University of Michigan) was assessed by a gentamycin protection assay. Overnight cultures in LB broth were incubated at 30 °C, subjected to shaking at 180 rpm, and then diluted 1:7 and grown until A600 = 0.5. Cells were infected in triplicate at a multiplicity of infection of 1:10 for 30 min, washed with PBS, and further incubated in DMEM containing 50 μg/ml gentamycin (Sigma) for either 1 h (HEK293T, HCT116) or 3 h (HT-29). Cells were then lysed in 1% Triton, PBS for 10 min on ice. Serial dilutions were plated on LB agar in duplicate and grown overnight at 30 °C. Colonies recovered were counted, and colony-forming units (cfu)/well were calculated.
Confocal Imaging
HCT116 cells were grown on coverslips and infected with Salmonella as described above. Cells were washed with PBS and fixed in 4% paraformaldehyde in PBS. Cells were permeabilized with 1% Triton X-100 in PBS. After blocking with 3% BSA, cells were incubated with an anti-Salmonella (ms) antibody. Salmonella antibody was detected with goat anti-mouse Alexa568 secondary antibody (Invitrogen), and coverslips were mounted on slides with Vectashield containing DAPI (Vector Laboratories). Microscopy was conducted using a Leica SP5 AOBS confocal microscope and HCX PL APO λ blue 63.0 × 1.40 oil UV objective (numerical aperture 1.4). Images were acquired using LAS-AF software, and further analysis was performed using the Imaris imaging suite (Bitplane, St. Paul, MN).
Quantitative RT-PCR
Total RNA was extracted from the indicated HT-29 cell lines using the RNeasy kit (Qiagen) according to the manufacturer's instructions. DNA synthesis was performed using Qiagen's Quantitect reverse transcription kit according to the manufacturer's instructions. Real-time PCR was carried out using primers against hBD-2 (forward, 5′-ATCAGCCATGAGGGTCCTGT-3′; reverse, 5′ TTTAACCGTGGACACCAGAG-3′) and human GAPDH (forward, 5′-GACCTGACCTGCCGTCTA-3′; reverse, 5′-GTTGCTGTAGCCAAATTCGTT-3′) along with the iQ SYBR Green Supermix (Bio-Rad) and detection using a Bio-Rad iCycler. Data shown are normalized to GAPDH. hBD-2 primers were obtained from the laboratory of Dr. Alan Levine (Case Western Reserve University).
Statistics
Data are presented as means, with error bars representing S.E. values of at least three different experiments for each condition. Statistical analysis was completed using Prism 5 Software. Significance was determined by p values of <0.05 after one-way analysis of variance. p values are compared with vector.
RESULTS
TRAF4 Inhibits NOD2-induced NF-κB Activation
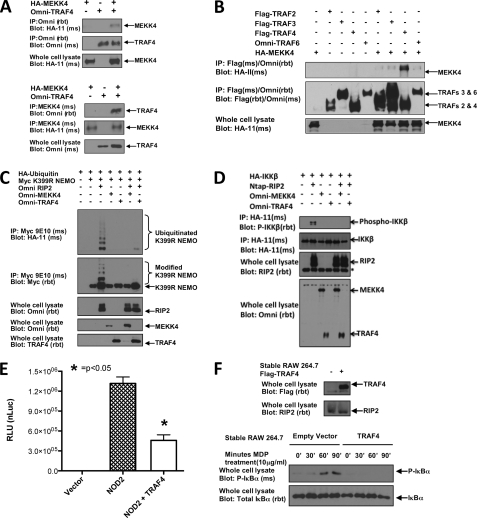
NOD2 signaling must be tightly regulated to provide a strong inflammatory response to bacterial infection and quickly down-regulated to prevent unnecessary tissue damage and inflammatory disease. However, only a handful of NOD2 regulatory proteins have been identified to date. To identify additional NOD2 regulatory proteins, we sought to characterize proteins that may be in a complex with a previously identified NOD2 regulator, MEKK4 (12, 13). One known MEKK4-interacting protein is TRAF4 (14, 15, 17). Given that TRAF proteins are key regulators of innate immune and inflammatory signaling, we wanted to confirm and further investigate MEKK4 binding to TRAF4. To this end, Omni-TRAF4 was cotransfected with HA-MEKK4 in HEK293T cells. TRAF4 was immunoprecipitated from lysates, and binding of MEKK4 was assayed by Western blotting. The reciprocal experiment was also performed in which MEKK4 was immunoprecipitated from lysates and TRAF4 binding was assayed by Western blotting. TRAF4 binding to MEKK4 was detected in the reciprocal coimmunoprecipitations (Fig. 1A). A related TRAF family member, TRAF6, is known to play a role in NOD2 signaling (9, 11). To determine if TRAF6 or other TRAF family members also bind to MEKK4, FLAG-TRAF2, FLAG-TRAF3, FLAG-TRAF4, or Omni-TRAF6 was cotransfected with HA-MEKK4 into HEK293T cells. The TRAF proteins were immunoprecipitated, and MEKK4 binding was detected by Western blotting. Of the TRAFs tested, only TRAF4 reproducibly and strongly coprecipitated with MEKK4 (Fig. 1B).
FIGURE 1.
TRAF4 mimics MEKK4 inhibition of NF-κB activation. A, reciprocal coimmunoprecipitations were conducted to confirm the previously published MEKK4-TRAF4 interaction. HA-MEKK4 was cotransfected into HEK293T cells with Omni-TRAF4. In the upper panels, the lysates were subjected to immunoprecipitation using an Omni antibody. In the bottom panels, the lysates were subjected to immunoprecipitation (IP) using a MEKK4 antibody. Both sets of experiments were immunoblotted with the indicated antibodies. TRAF4 and MEKK4 were present in reciprocal precipitations. B, to compare MEKK4 specificity for TRAF4 with other TRAF family members, HA-MEKK4 was cotransfected with FLAG-TRAF2, FLAG-TRAF3, FLAG-TRAF4, or Omni-TRAF6. Lysates were subjected to immunoprecipitation using either a FLAG or Omni antibody, and immunoblots were performed using the indicated antibodies. MEKK4 bound most robustly to TRAF4. C, HA-ubiquitin was cotransfected into HEK293T cells with Myc-K399R NEMO, Omni-RIP2, and either Omni-MEKK4 or Omni-TRAF4. Lysates were subjected to immunoprecipitation with Myc antibody under stringent washing conditions (1 m NaCl, 1% SDS). Immunoprecipitates and whole cell lysates were immunoblotted using the indicated antibodies. Both MEKK4 and TRAF4 strongly inhibited RIP2-induced NEMO ubiquitination. D, HA-IKKβ was cotransfected into HEK293T cells with NTAP-RIP2 and either Omni-MEKK4 or Omni-TRAF4. Lysates were generated and subjected to immunoprecipitation with HA-11 antibody. The precipitated HA-IKKβ was immunoblotted with a phospho-specific antibody that recognizes phosphorylated, active IKKβ. Whole cell lysates were immunoblotted with the indicated antibodies. Both MEKK4 and TRAF4 strongly inhibited RIP2-induced IKKβ activation. E, luciferase reporter gene assays were conducted in HEK293T cells transfected with NF-κB luciferase reporter plasmid, β-galactosidase, and NOD2 in the absence or presence of TRAF4. Raw luciferase values were normalized to β-galactosidase values to control for transfection efficiency (nLuc), and averages with S.E. values (error bars) are shown. Cotransfection of TRAF4 with NOD2 significantly inhibited NOD2-induced NF-κB activity. F, RAW 264.7 cells were retrovirally transduced to establish stable FLAG-TRAF4 expression. Lysates were immunoblotted with anti-FLAG for TRAF4 or anti-RIP2 to confirm TRAF4 protein expression (upper panel). The MDP response of FLAG-TRAF4-expressing cells is compared with cells transduced with an empty retroviral vector in the lower panel. Cells were treated with MDP (10 μg/ml) for 0, 30, 60, or 90 min. Lysates were collected and standardized for equal protein concentration prior to immunoblotting with anti-IκBα or anti-phospho IκBα. Empty vector-transduced cells had normal activation of NF-κB in response to MDP, whereas TRAF4-expressing cells failed to initiate NF-κB in response to MDP.
Because the mouse knock-out phenotypes so closely match one another, we hypothesized that TRAF4 might function like MEKK4 in the inhibition of the NOD2-RIP2 signaling pathway. For this reason, the effect of TRAF4 on NF-κB activation was compared with that of MEKK4. To determine if TRAF4 has a similar inhibitory effect on NF-κB activation, RIP2-induced ubiquitination of the IKK scaffolding protein NEMO (IKKγ) was assayed. RIP2 induces the Lys63 ubiquitination of NEMO at Lys285 following NOD2 activation to initiate the IKK signalosome upstream of NF-κB nuclear translocation (8). To study this, HA-ubiquitin was cotransfected with Myc-K399R NEMO and Omni-RIP2 in the presence of either Omni-MEKK4 or Omni-TRAF4. The K399R NEMO mutant was used to eliminate basal ubiquitination and more readily assay for RIP2-induced ubiquitination at Lys285. NEMO was immunoprecipitated from lysates and subjected to stringent washing (1% SDS, 1 m NaCl) to eliminate ubiquitinated binding proteins of NEMO. Immunoprecipitates were immunoblotted with an HA antibody to detect ubiquitinated NEMO. In the absence of MEKK4 and TRAF4, RIP2 induced NEMO ubiquitination. In the presence of MEKK4 or TRAF4, the RIP2 ubiquitination of NEMO was strongly attenuated (Fig. 1C). As an alternative measure of the MEKK4 and TRAF4 inhibitory effect of NF-κB activation, RIP2-induced phosphorylation of the catalytic signalosome subunit IKKβ was assayed in the absence or presence of either MEKK4 or TRAF4. HA-IKKβ was cotransfected with Omni-RIP2 and either Omni-MEKK4 or Omni-TRAF4 in HEK293T cells. HA-IKKβ was immunoprecipitated from lysates with anti-HA and immunoblotted with a phospho-specific antibody for Ser177 and Ser181 within the activation loop of IKKβ (Fig. 1D). RIP2 induced the phosphorylation of IKKβ in the absence of MEKK4 and TRAF4. The presence of MEKK4 or TRAF4 inhibited RIP2-induced phosphorylation of IKKβ (Fig. 1D).
To demonstrate the effects of TRAF4 overexpression on NOD2-dependent signaling, luciferase reporter gene assays were conducted to measure NF-κB activation (Fig. 1E). Overexpression of TRAF4 inhibited NOD2-induced NF-κB reporter activity (Fig. 1E). To further validate the negative regulation by NF-κB of TRAF4 in a more endogenous, stimulus-dependent setting, FLAG-TRAF4 was retrovirally transduced into RAW 264.7 macrophages (Fig. 1F). After confirming TRAF4 expression, cells were treated with MDP (10 μg/ml) for 0, 30, 60, or 90 min. Lysates were collected and immunoblotted for total or phosphorylated IκBα. Empty vector-transduced RAW 264.7 cells responded to MDP with increased detection of phosphorylated IκBα at 60 and 90 min post-treatment. TRAF4-expressing cells, however, failed to initiate an NF-κB response to MDP (Fig. 1F), suggesting that TRAF4 can inhibit MDP-induced NF-κB responses in a more endogenous setting. In total, these results suggest that, like MEKK4, TRAF4 can negatively regulate NOD2-RIP2-induced NF-κB activation.
NOD2 Binds to TRAF4 and Bridges TRAF4 to RIP2
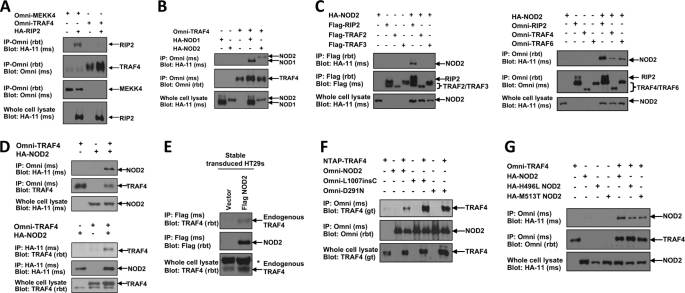
In the absence of MDP, MEKK4 is thought to maintain low levels of NOD2-induced NF-κB activation by sequestering RIP2, thereby not allowing binding to NOD2 (12). To determine the role of TRAF4 in this biochemical process, the binding of TRAF4 to RIP2 was assessed in coimmunoprecipitation assays in HEK293T cells. These assays were performed in HEK293T cells because this cell line does not express NOD2 (2) and allows the analysis of interactions between proteins involved in NOD2 signaling in the absence of NOD2 expression. Omni-MEKK4 or Omni-TRAF4 was cotransfected with HA-RIP2. MEKK4 and TRAF4 were immunoprecipitated from lysates, and RIP2 binding was assayed by Western blot (Fig. 2A). RIP2 coprecipitated with MEKK4 but not with TRAF4 (Fig. 2A). This finding suggests that, unlike MEKK4, the inhibition of NOD2 signaling by TRAF4 may not be due to direct competition for binding to RIP2.
FIGURE 2.
NOD2 binds TRAF4. A, to determine if, like MEKK4, TRAF4 also binds RIP2, Omni-MEKK4 or Omni-TRAF4 was cotransfected into HEK293T cells with HA-RIP2. Lysates were subjected to immunoprecipitations (IP) using an Omni antibody. Immunoblotting was performed with the indicated antibodies. RIP2 was present in the MEKK4 immunoprecipitate but not in the TRAF4 immunoprecipitate. B, additional components of innate immune signaling pathways were screened for TRAF4 binding. Either HA-NOD1 or HA-NOD2 was cotransfected into HEK293T cells with Omni-TRAF4. Lysates were subjected to immunoprecipitations using an Omni antibody. Immunoblots were performed using the indicated antibodies. TRAF4 coprecipitated with both NOD1 and NOD2. C, NOD2 was screened for binding to additional TRAF family member proteins. Left, FLAG-RIP2, FLAG-TRAF2, or FLAG-TRAF3 was cotransfected with HA-NOD2; right, Omni-RIP2, Omni-TRAF4, or Omni-TRAF6 was co-transfected with HA-NOD2. Lysates were subjected to immunoprecipitation, and immunoblotting was performed using the indicated antibodies. NOD2 coprecipitated with RIP2, TRAF4, and TRAF6. D, to further validate the TRAF4 interaction with NOD2, following transfection with the indicated constructs, reciprocal coimmunoprecipitations followed by immunoblotting were performed. TRAF4 coprecipitated with NOD2 in the reciprocal precipitations. E, HT-29s were retrovirally transduced to establish stable expression of FLAG-NOD2. Lysates were subjected to immunoprecipitations with a FLAG antibody. Immunoprecipitates were immunoblotted with an antibody to endogenous TRAF4. TRAF4 weakly interacts with NOD2 under these non-stimulated conditions. *, nonspecific band. F, Crohn's disease-associated NOD2 mutant alleles were tested for TRAF4 binding. NTAP-TRAF4 was cotransfected into HEK293T cells with Omni-NOD2, Omni-L1007insC, or Omni-D291N. Omni-NOD2 mutants were immunoprecipitated from lysates, and immunoprecipitates were immunoblotted with the indicated antibodies. TRAF4 coprecipitated with each of the NOD2 mutants. G, early onset sarcoidosis-associated NOD2 mutants were screened for TRAF4 binding. Omni-TRAF4 was cotransfected with HA-NOD2, HA-H496L NOD2, or HA-M513T NOD2. TRAF4 was immunoprecipitated from lysates with an Omni antibody, and immunoblots were performed with the indicated antibodies. Each of the NOD2 mutants coprecipitated with TRAF4.
These findings also indicate the presence of a separate TRAF4-containing complex that inhibits NF-κB activation. For this reason, other components of innate immune signaling pathways were screened for TRAF4 binding. Omni-TRAF4 was cotransfected with HA-NOD1 or HA-NOD2 in HEK293T cells. Omni-TRAF4 was immunoprecipitated from lysates, and NOD1 or NOD2 binding was detected by Western blot. TRAF4 was found to bind to both of these Nod-Like Receptor (NLR) proteins (Fig. 2B).
Given the association of altered NOD2 function with inflammatory disease, we decided to focus our studies on the effects of TRAF4 interaction with NOD2. Additional TRAF family member proteins were screened to determine the specificity of the NOD2 interaction with TRAF4 (Fig. 2C). HA-NOD2 was cotransfected with FLAG-TRAF2, FLAG TRAF3, or FLAG-RIP2 into HEK293T cells with HA-NOD2. In a separate experiment, Omni-TRAF4, Omni-TRAF6, or Omni-RIP2 was cotransfected into HEK293T cells. RIP2 binding was used as a positive control for each experiment (Fig. 2C). Neither TRAF2 nor TRAF3 was detected with precipitated NOD2 (Fig. 2C). However, NOD2 precipitated with both TRAF4 and TRAF6 (Fig. 2C). TRAF6 binding to NOD2 is consistent with previously published data (9, 11). To further confirm the TRAF4 interaction with NOD2, reciprocal immunoprecipitations were conducted in HEK293T cells cotransfected with Omni-TRAF4 and HA-NOD2. Binding was detected in these reciprocal coimmunoprecipitations (Fig. 2D). Because we have not found an antibody that reliably immunoprecipitates endogenous NOD2, HT-29 cells were then retrovirally transduced with FLAG-NOD2, and after neomycin selection, clones (>1000) were pooled. Stably expressing NOD2 HT-29s were used to verify TRAF4 binding in a more endogenous setting. FLAG-NOD2 was immunoprecipitated from lysates, and Western blotting was conducted to detect endogenous TRAF4 binding. NOD2 could weakly bind TRAF4 in the uninduced state (Fig. 2E). Crohn's disease or EOS/Blau syndrome-associated NOD2 mutants were also screened for TRAF4 binding. NTAP-TRAF4 was cotransfected into HEK293T cells with Omni-NOD2 or one of the Crohn's disease-associated NOD2 mutants, Omni-L1007insC or Omni-D291N. NOD2, L1007inC, or D291N NOD2 was immunoprecipitated via their Omni tag and immunoblotted with TRAF4 antibody to detect binding. TRAF4 coprecipitated with the wild type as well as the Crohn's disease-associated NOD2 proteins (Fig. 2F). In addition, Omni-TRAF4 was cotransfected into HEK293T cells with HA-NOD2 or one of the EOS/Blau syndrome mutants, Omni-H496L NOD2 or Omni-M513T NOD2 (26). Omni-TRAF4 was immunoprecipitated from lysates and immunoblotted with HA to detect NOD2 binding. Wild type NOD2 as well as each of the EOS/Blau syndrome-associated NOD2 proteins coprecipitated with TRAF4 (Fig. 2G).
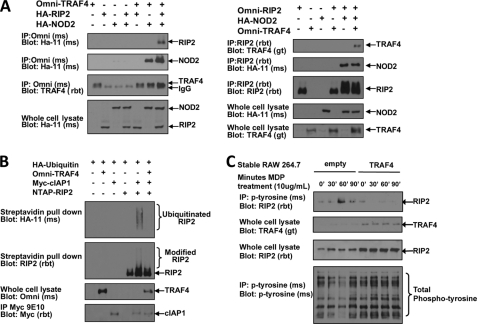
In order to investigate the molecular mechanisms of TRAF4 inhibition of NOD2 signaling, we first characterized the components of the TRAF4-NOD2 complex. In the absence of NOD2 expression, we do not detect an interaction between TRAF4 and RIP2 (Fig. 2D). Given that RIP2 binds NOD2 upon either NOD2 overexpression or MDP stimulation, TRAF4 binding to RIP2 was tested in the presence of NOD2. Reciprocal immunoprecipitations were performed. First, Omni-TRAF4 was cotransfected into HEK293T cells with HA-RIP2 alone or with HA-RIP2 and HA-NOD2. TRAF4 was immunoprecipitated from lysates and Western blotted for RIP2 and NOD2 binding. As shown previously, NOD2 coprecipitated with TRAF4. This interaction was independent of the presence of RIP2. When cotransfected in the absence of NOD2, RIP2 did not coprecipitate with TRAF4 (Fig. 3A, left). However, when NOD2 was cotransfected, RIP2 coprecipitated with TRAF4. This was observed in a reciprocal coimmunoprecipitation because TRAF4 only coprecipitated with RIP2 in the presence of NOD2 (Fig. 3A, right).
FIGURE 3.
TRAF4 inhibits RIP2 activation. A, to determine if TRAF4 is present in the NOD2-RIP2 complex, TRAF4 binding to RIP2 was assayed in the presence of NOD2. Left, Omni-TRAF4 was co-transfected with HA-RIP2 alone or with HA-RIP2 and HA-NOD2; right, Omni-RIP2 was cotransfected with Omni-TRAF4 alone or with Omni-TRAF4 and HA-NOD2. Lysates were subjected to immunoprecipitation (IP), and immunoblotting was performed using the indicated antibodies. NOD2 co-precipitated with both RIP2 and TRAF4, whereas RIP2 and TRAF4 only coprecipitated when NOD2 was present. B, the effect of TRAF4 on cIAP1-induced ubiquitination of RIP2 was measured to determine if TRAF4 affected the activation state of RIP2. HA-ubiquitin was cotransfected with NTAP-RIP2 and Myc-cIAP1 alone or with cIAP1 and Omni-TRAF4. NTAP-RIP2 was precipitated using streptavidin-agarose under stringent washing conditions. Precipitates and whole cell lysates were immunoblotted with the indicated antibodies. cIAP1 induced ubiquitination of RIP2; however, cotransfection of TRAF4 with cIAP1 inhibited the ubiquitination of RIP2. C, the effect of TRAF4 on another marker of RIP2 activation, tyrosine autophosphorylation, was assayed for. RAW 264.7 macrophages and RAW 264.7 macrophages that stably overexpress TRAF4 were treated with MDP for 0, 30, 60, or 90 min. Lysates were collected and subjected to immunoprecipitation with a phosphotyrosine antibody. Precipitates were immunoblotted with a RIP2 antibody. RIP2 is precipitated with the phosphotyrosine antibody most strongly at 60 min in the RAW 264.7 cells but not in the TRAF4-expressing 264.7 macrophages.
The above findings are consistent with a model in which TRAF4 binds to and inhibits an active NOD2-RIP2 complex. To test this model and to determine if TRAF4 inhibited the next proximal signaling events, we investigated the effect of TRAF4 on molecular markers of an active NOD2-RIP2 complex. One such marker is the cIAP1-induced ubiquitination of RIP2 following NOD2 activation (27). HA-ubiquitin was cotransfected into HEK293T cells with NTAP-RIP2 and Myc-cIAP1 in the absence or presence of Omni-TRAF4 (Fig. 3B). NTAP-RIP2 was precipitated from lysates with streptavidin-agarose and immunoblotted with HA to detect ubiquitinated RIP2. cIAP1 induced ubiquitination of RIP2. Cotransfection of TRAF4 with cIAP1 inhibited ubiquitination of RIP2 (Fig. 3B). Additionally, our laboratory has recently shown that RIP2 autophosphorylates on a tyrosine residue following NOD2 activation (28). To determine if TRAF4 also inhibited this marker of RIP2 activation, tyrosine phosphorylation of RIP2 was compared in RAW 264.7 to RAW 264.7 macrophages that had stable overexpression of TRAF4 (Fig. 3C). Cells were treated with MDP (10 μg/ml) for 0, 30, 60, or 90 min. Lysates were collected and subjected to immunoprecipitation using a phosphotyrosine antibody. Precipitates were immunoblotted with RIP2 antibody to detect tyrosine-phosphorylated RIP2. RIP2 tyrosine phosphorylation was strongly induced in the RAW 264.7 cells at 60 min, whereas the TRAF4-expressing RAW macrophages remained unresponsive. Collectively, these results indicate that TRAF4 is recruited to the active NOD2-RIP2 signaling complex through interactions with NOD2.
TRAF4 Binding to NOD2 Inhibits NOD2-induced Bacterial Killing
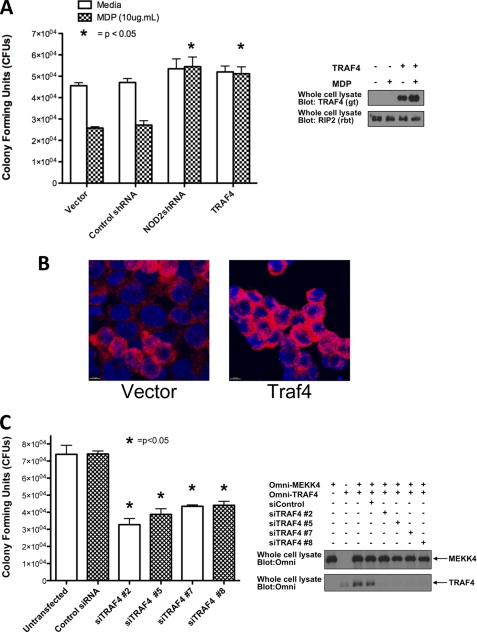
It has been shown that NOD2 plays an important role in the clearance of the intracellular bacterium, Salmonella typhimurium (25, 29, 30). In light of this, the role of TRAF4 in NOD2-induced bacterial killing was examined by studying the effect of TRAF4 on NOD2-induced Salmonella killing in gentamycin protection assays. HCT116 cells, a human epithelial colorectal carcinoma cell line that expresses endogenous NOD2 (30), were transfected with a vector control plasmid DNA, control shRNA, NOD2 shRNA, or TRAF4 plasmid DNA. This NOD2 shRNA has been shown previously to inhibit endogenous NOD2 expression in this cell type (25). MDP induced bacterial killing in the vector- and control shRNA-containing cells. NOD2 shRNA prevented the MDP-induced bacterial killing, confirming that MDP is acting through a NOD2-dependent process. TRAF4 overexpression (expression levels shown in the right panels of Fig. 4A) inhibited bacterial killing to the same extent as the NOD2 shRNA (Fig. 4A). This is consistent with TRAF4 having an inhibitory effect on NOD2 signaling. To confirm these findings in an independent assay, HCT116 cells were grown on coverslips and infected with Salmonella. Confocal immunofluorescence using a Salmonella primary antibody and an Alexa568 secondary antibody showed that the TRAF4-overexpressing cells have increased intracellular Salmonella (Fig. 4B), independently supporting the cfu data.
FIGURE 4.
TRAF4 inhibits NOD2-induced Salmonella killing. A, gentamycin protection assays were performed to determine the effect of TRAF4 on NOD2-induced Salmonella killing. HCT116 cells, a cell line that endogenously expresses NOD2, were transfected with an empty vector, TRAF4, control shRNA, or shRNA targeting endogenous NOD2. TRAF4 expression is shown in the panel on the right. After transfection, cells were treated with either media or MDP (10 μg/ml) and infected with Salmonella at a multiplicity of infection of 10:1. cfu/well were calculated from duplicate measures of triplicate wells. Average cfu/well with S.D. values (error bars) are shown. NOD2 shRNA-transfected cells and TRAF4-transfected cells had no MDP-induced Salmonella killing. B, to independently verify these findings, empty vector- or TRAF4-transfected HCT116 cells were grown on coverslips and infected with Salmonella. Fixed cells were permeabilized before incubation with an anti-Salmonella antibody with detection by an Alexa568 secondary antibody (red) and nuclei staining with DAPI (blue). Confocal imaging shows increased bacterial load in the cytosol of the TRAF4-transfected cells. Scale bars, 10 μm. C, Salmonella killing was increased when TRAF4 expression was inhibited. TRAF4 expression was inhibited by four different siRNAs in HCT116 cells, and gentamycin protection assays were performed. The panel on the right shows the efficacy of knockdown by Western blot of overexpressed TRAF4 in HEK293T cells. Omni-MEKK4 was included as a transfection efficiency and specificity control.
The ability of NOD2 to induce Salmonella killing in HCT116 cells in which TRAF4 expression was inhibited was also tested (Fig. 4C). Four separate siRNAs targeting TRAF4 were transfected individually into HCT116 cells, and gentamycin protection assays were conducted. Knockdown of TRAF4 enhances basal Salmonella killing (Fig. 4C). Knockdown of transfected TRAF4 in HEK293T cells is shown in the right panel. Omni-MEKK4 was included as a control for transfection efficiency and siRNA specificity (Fig. 4C). These results demonstrate that modulation of TRAF4 expression alters the antibacterial function of NOD2.
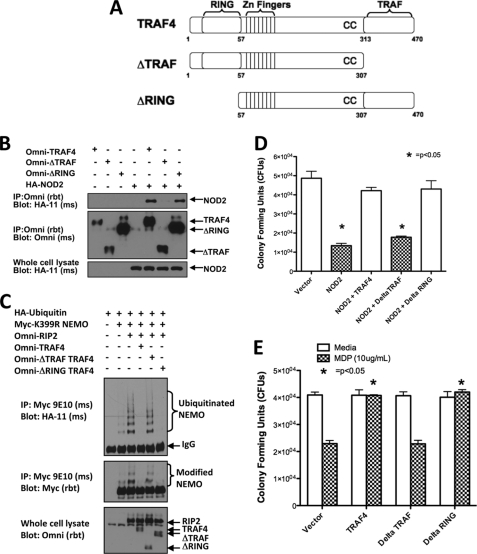
The mechanism of TRAF4 inhibition of NOD2-induced signaling could be due to a direct physical effect of TRAF4 binding to NOD2. To test this, deletion mutants of TRAF4 (Fig. 5A) were tested for their ability to interact with NOD2 and also to inhibit NOD2 function. Omni-TRAF4 mutants lacking either the TRAF domain (ΔTRAF) or RING domain (ΔRING) were cotransfected into HEK293T cells with HA-NOD2. Full-length TRAF4 and TRAF4 mutants were immunoprecipitated from lysates, and NOD2 binding was detected by Western blotting. NOD2 coprecipitated with full-length TRAF4 and the ΔRING TRAF4 but failed to coprecipitate with the ΔTRAF TRAF4 mutant (Fig. 5B). TRAF4 binding to NOD2 through the TRAF domain of TRAF4 is consistent with other TRAF family member proteins forming heterotypic protein-protein interactions via their TRAF domains (18).
FIGURE 5.
TRAF4 binding to NOD2 and inhibition of NOD2-induced Salmonella killing is TRAF domain-dependent. A, the depicted TRAF4 mutants were used to map the regions of TRAF4 required for NOD2 binding. The ΔTRAF mutant is lacking the C-terminal TRAF domain, and the ΔRING mutant is lacking the N-terminal RING domain. B, to map the regions of TRAF4 required for NOD2 binding, Omni-TRAF4, Omni-ΔTRAF TRAF4, and/or Omni-ΔRING TRAF4 were cotransfected into HEK293T cells with HA-NOD2. Lysates were subjected to immunoprecipitation (IP) with an Omni antibody, and immunoblotting was performed using the indicated antibodies. HA-NOD2 binding to the full-length TRAF4 and the ΔRING TRAF4 was detected but was not detected using ΔTRAF TRAF4. C, NEMO ubiquitination was measured to assess the effect of these TRAF4 deletion mutants on RIP2 activity. After transfection with the indicated constructs, immunoprecipitation of NEMO was performed under stringent washing conditions. Immunoblotting was then performed using the indicated antibodies. RIP2-induced NEMO ubiquitination was inhibited by full-length TRAF4 and ΔRING TRAF4 but not by ΔTRAF TRAF4. D, gentamycin protection assays were performed to determine if TRAF4 binding was required for NOD2-induced Salmonella killing. Expression of NOD2 induced Salmonella killing in HEK293T cells. Only ΔTRAF TRAF4 did not have an inhibitory effect on NOD2. E, gentamycin protection assays were then performed in NOD2-expressing HCT116 cells transfected with either empty vector, TRAF4, ΔTRAF TRAF4, or ΔRING TRAF4. MDP induced Salmonella killing in vector-transfected cells. MDP-induced killing was inhibited with the transfection of either TRAF4 or the ΔRING TRAF4 but not with ΔTRAF TRAF4. Error bars, S.E.
The deletion mutants were tested for their ability to inhibit NOD2 signaling and NOD2-dependent bacterial killing. First, their molecular effect on NOD2 signaling was tested by assessing their ability to inhibit RIP2-stimulated NEMO ubiquitination. HA-ubiquitin was cotransfected with Myc-K399R NEMO, Omni-RIP2, and either full-length Omni-TRAF4, Omni-ΔTRAF TRAF4, or Omni-ΔRING TRAF4 (Fig. 5C). Myc-K399R NEMO was immunoprecipitated from lysates via the Myc tag and immunoblotted with HA antibody to detect ubiquitinated NEMO. As shown previously (Fig. 1C), TRAF4 inhibited RIP2 induced ubiquitination of NEMO. The ΔRING TRAF4 mutant, which retains NOD2 binding, inhibited RIP2-induced ubiquitination of NEMO, whereas the ΔTRAF TRAF4 mutant, which does not bind NOD2, did not inhibit NEMO ubiquitination (Fig. 5C).
Next, the TRAF4 mutants were tested for their ability to inhibit NOD2-dependent bacterial killing. As shown previously (25), expression of NOD2 alone induced bacterial killing in HEK293T cells (Fig. 5D). TRAF4, ΔTRAF TRAF4, or ΔRING TRAF4 was then cotransfected into HEK293T cells with HA-NOD2. Similar to the results in the NEMO ubiquitination assay, expression of either TRAF4 or the ΔRING TRAF4 inhibited the NOD2-induced Salmonella killing. In contrast, cotransfection of ΔTRAF TRAF4 had no effect on NOD2-induced bacterial killing (Fig. 5D), implying that the effect of TRAF4 on NOD2 may be a direct effect dependent on their binding. To further verify this finding, HCT116 cells, which express endogenous NOD2, were transfected with TRAF4, ΔTRAF TRAF4, or ΔRING TRAF4 and then either exposed or not exposed to MDP (to activate endogenous NOD2). MDP exposure induced bacterial killing in cells transfected with vector. TRAF4 and ΔRING TRAF4 expression prevented MDP-induced killing, whereas expression of ΔTRAF TRAF4 did not (Fig. 5E). In total, these experiments suggest that TRAF4 binding to NOD2 is required for the TRAF4 inhibitory effect on NOD2-dependent Salmonella killing.
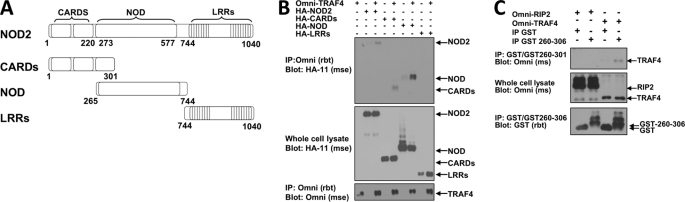
In reciprocal experiments, NOD2 mutants were used to map the regions of NOD2 required for TRAF4 binding (Fig. 6A). Each of these mutants was cotransfected with Omni-TRAF4 (Fig. 6B). Full-length HA-NOD2 and each of the HA-tagged NOD2 mutants were immunoprecipitated from lysates. TRAF4 binding was then detected by Western blotting. TRAF4 binding was detected with full-length NOD2, the NOD2 CARDs, and the NOD2 NOD domain. The Leucine-rich repeats (LRRs) of NOD2 were not able to bind TRAF4 (Fig. 6B). The CARDs NOD2 and NOD NOD2 mutants were compared to identify overlapping regions that might confer binding in both. A 36-amino acid region spanning amino acids 265–301 is present in both mutants (Fig. 6A). To determine if amino acids 265–301 of NOD2 were sufficient for TRAF4 binding, a bacterial recombinant GST-NOD2(260–306) protein containing GST linked to the 36 amino acids was generated. Because recombinant proteins produced in bacteria are often contaminated with bacterial cell wall components (i.e. LPS and MDP), we specifically performed these in vitro assays using RIP2 and TRAF4 purified from HEK293T cells, which do not express NOD2 or other NLR family members activated by MDP, to reduce the likelihood that an activated NLR would alter the results of this assay. This binding of TRAF4 to the peptide was specific because RIP2 did not bind GST-NOD2(260–306), which is consistent with previous studies demonstrating that RIP2 binds the CARDs of NOD2 (1, 31) and TRAF4 did not bind the GST peptide alone (Fig. 6C). These assays demonstrate a direct, physical interaction of TRAF4 with a small region of NOD2 composed of amino acids 260–306.
FIGURE 6.
TRAF4 binds to amino acids 260–301 of NOD2. A, the depicted NOD2 mutants were used to map the regions of NOD2 required for TRAF4 binding. There is a 36-amino acid overlap between the CARDs-only mutant and the NOD-only mutant. B, each of these mutants was cotransfected with Omni-TRAF4 in HEK293T cells. Lysates were subjected to immunoprecipitation (IP) with an Omni antibody, and immunoprecipitates were immunoblotted with the indicated antibodies. TRAF4 binding was detected with full-length NOD2 and with the CARD and NOD mutants but not with the LRR mutant, suggesting that an overlapping region consisting of amino acids 265–301 of NOD2 was responsible for TRAF4 binding. C, a bacterial recombinant GST-fused peptide of NOD2 amino acids 260–306 was generated. This purified GST-NOD2(260–306) (GST-260–306) was incubated with lysates from cells transfected with Omni-TRAF4 or Omni-RIP2 followed by precipitation using glutathione beads. TRAF4 binding was detected to the GST-NOD2(260–306) but not the GST alone. RIP2 did not precipitate with the GST-NOD2(260–306) because this polypeptide lacks the CARD domains of NOD2.
Two Amino Acids in NOD2 Are Critical for Binding TRAF4 and Allowing Negative Regulation of NOD2 Signaling
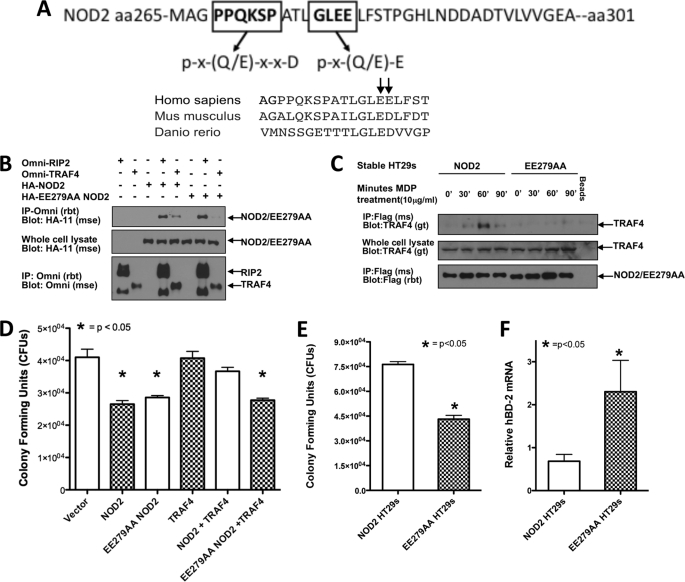
Having narrowed down TRAF4 binding to a 36-amino acid-spanning region of NOD2, the primary amino acid sequence was scrutinized for potential TRAF binding sites (Fig. 7A). Two putative TRAF binding sites were identified. One resembled the minor TRAF2 binding motif of PX(Q/E)XXD, whereas another resembled the TRAF2 major binding motif, (P/S/A/T)X(Q/E)E (19, 21). The minor binding motif present in NOD2 is partially conserved in the mouse, but its conservation is lost in zebrafish. The major binding motif, however, is conserved in both mouse and zebrafish (Fig. 7A). NOD2 has a general lack of conservation between species (32); thus, the conservation in this region between species signified to us that this could be an important TRAF binding site. Due to the similarity of this site to the TRAF2 major binding motif, glutamate residues at 279 and 280 were focused on as potential TRAF4 binding residues (Fig. 7A) and were mutated to alanine residues (Fig. 7B). The E279A/E280A NOD2 mutant was first tested for TRAF4 binding. HA-E279A/E280A NOD2 was cotransfected with either Omni-RIP2 or Omni-TRAF4 (Fig. 7B). RIP2 binding was used as a positive control to ensure that these mutations in NOD2 did not affect global NOD2 protein structure or protein trafficking. RIP2 and TRAF4 were immunoprecipitated from cell lysates, and NOD2 and E279A/E280A NOD2 binding was determined by Western blotting. NOD2 coprecipitated with both TRAF4 and RIP2. Although the E279A/E280A NOD2 mutant maintained RIP2 binding, TRAF4 binding was lost (Fig. 7B), demonstrating the requirement of this motif in interaction of NOD2 and TRAF4.
FIGURE 7.
E279A/E280A NOD2 abrogates TRAF4 binding and inhibition of NOD2-induced Salmonella killing. A, primary amino acid sequence 260–301 of NOD2 was analyzed for potential TRAF binding sites. Shown in boldface type are residues consistent with the published TRAF2 minor and major binding motifs. Evolutionary conservation of NOD2 Glu279 and Glu280 and the surrounding sequence is also shown. B, a NOD2 mutant containing point mutations E279A and E280A (EE279AA) was generated, and co-transfections followed by immunoprecipitations (IP) were performed to determine if this region was required for TRAF4 binding. NOD2 coprecipitated with TRAF4, whereas the E279A/E280A NOD2 did not. RIP2 coprecipitated with both NOD2 and E279A/E280A NOD2. C, HT-29s were retrovirally transduced to establish stable expression of FLAG-E279A/E280A NOD2 and FLAG-NOD2. After selection, cells were treated with MDP (10 μg/ml) for 0, 30, 60, and 90 min. Protein concentrations were standardized and subjected to immunoprecipitations with a FLAG antibody. Immunoblotting was performed with the indicated antibodies. TRAF4 binding to NOD2 but not E279A/E280A NOD2 is strongly induced upon MDP treatment. D, gentamycin protection assays were performed in HEK293T cells cotransfected with Omni-TRAF4 and either HA-NOD2 or HA-E279A/E280A NOD2. Expression of both NOD2 and E279A/E280A NOD2 induced Salmonella killing. Cotransfection of TRAF4 inhibited NOD2-induced Salmonella killing but had no effect on E279A/E280A NOD2-induced Salmonella killing. E, to confirm findings from C and D, basal Salmonella killing in stable NOD2 and E279A/E280A NOD2 cell lines was compared. The E279A/E280A NOD2-stable cells have significantly increased Salmonella killing compared with the NOD2-expressing cells. F, quantitative RT-PCR was performed on isolated RNA from control, NOD2-expressing HT-29s or E279A/E280A NOD2 HT-29s to compare basal levels of the NOD2-inducible gene, hBD-2. hBD-2 expression is increased basally in the E279A/E280A NOD2 HT-29s compared with NOD2 HT-29s. Data are normalized to GAPDH. Error bars, S.E.
To further show the requirement of glutamate residues at 279 and 280, the effect of MDP on endogenous TRAF4 binding to E279A/E280A NOD2 was also examined. HT-29 cells were transduced with an E279A/E280A NOD2 retrovirus for stable expression of the mutant NOD2. E279A/E280A NOD2 cells were treated with MDP for 0, 30, 60, and 90 min, and their responses were compared with those of HT-29 cells stably transduced with wild type NOD2 (Fig. 7C). FLAG-NOD2 and FLAG-E279A/E280A NOD2 were immunoprecipitated from lysates, and endogenous TRAF4 binding was detected by Western blotting. Although some basal binding was present, MDP stimulation transiently increased TRAF4 binding (peak binding was detected at 60 min poststimulation). TRAF4 binding to the E279A/E280A NOD2 mutant was not detected, further supporting the requirement of these residues in the binding of TRAF4 to NOD2 (Fig. 7C).
In light of the previous data showing that TRAF4 binding inhibits NOD2-induced Salmonella killing (Fig. 5, D and E), we would predict that the E279A/E280A NOD2 mutant should be resistant to inhibition by overexpression of TRAF4. HEK293T cells were cotransfected with either NOD2 or E279A/E280A NOD2 in the absence or presence of TRAF4 and infected with Salmonella. Expression of both wild type NOD2 and the E279A/E280A NOD2 mutant increased Salmonella killing (Fig. 7D). Overexpression of TRAF4 inhibited wild type NOD2-induced bacterial killing but had no effect on the killing stimulated by the E279A/E280A NOD2 mutant, demonstrating that the inhibition of NOD2 function by TRAF4 is dependent on interaction with specific residues in NOD2. There was not increased basal killing of Salmonella killing by the E279A/E280A NOD2 in this overexpression system; endogenous TRAF4 expression is probably too low to have an appreciable effect on the overexpressed NOD2 proteins.
For this reason, the above findings were confirmed in additional assays performed in HT-29 cell lines stably expressing wild type NOD2 and E279A/E280A NOD2. In gentamycin protection assays, E279A/E280A NOD2-expressing cells had significantly increased Salmonella killing when compared with wild type NOD2-expressing cells. This finding is consistent with the data from the experiments with TRAF4 expression knocked down by siRNA, leading to a constitutively active NOD2 (Fig. 4C). In support of the loss of TRAF4 interaction leading to increased NOD2 activity, we examined the expression of human β-defensin-2 (hBD-2) by quantitative RT-PCR. hBD-2 is a NOD2-induced antimicrobial peptide produced by epithelial cells (33). The amount of hBD-2 transcript amplified was normalized to GAPDH levels, and levels of HBD-2 expression were compared between the two cell lines. The E279A/E280A NOD2 cells had significantly increased basal expression of hBD-2 (Fig. 7F). Taken together, these results demonstrate that TRAF4 has an inhibitory effect on NOD2 signaling and bacterial killing dependent on a physical interaction between the TRAF domain of TRAF4 and glutamate residues 279 and 280 of NOD2. Disruption of this interaction results in ligand-independent activation of NOD2, illustrating the importance of this interaction in maintaining innate immune system homeostasis.
DISCUSSION
Downstream signaling events linking NOD2 stimulation to NF-κB activation have been extensively studied. Less well studied are the negative feedback mechanisms that aim to limit the NF-κB signaling pathway after initial NOD2 activation. Many of the negative regulators of NOD2 signaling dampen NF-κB activation downstream of NOD2. For example, the deubiquitinases CYLD and A20 as well as nuclear factor κB inhibitor (IκBα) are among the first genes transcribed upon NOD2 and NF-κB activation, and all help terminate signaling downstream of NOD2 (8, 34, 35). Despite these findings, studies investigating the mechanisms by which NOD2 is directly regulated are less prominent. In this work, we have identified TRAF4 as a novel negative regulator and binding protein of NOD2. Binding of NOD2 to TRAF4 is observed in reciprocal coimmunoprecipitations of overexpressed protein as well as the coprecipitation of endogenous TRAF4 with stably expressed FLAG-NOD2 (Figs. 1–3). The TRAF domain of TRAF family members generally confers heterotypic protein-protein interactions. Consistent with this, NOD2 binding is lost with the expression of a TRAF4 mutant lacking the TRAF domain (ΔTRAF) (Fig. 5). Unable to bind, the ΔTRAF TRAF4 also is unable to inhibit NOD2 signaling (Fig. 5). Given that TRAF4 binding is required for its inhibition of NOD2 signaling, we considered that TRAF4 affected recruitment of the NOD2 effector protein, RIP2. The presence of TRAF4 did not affect NOD2 binding to RIP2 but did inhibit the next proximal signaling events, cIAP1 ubiquitination of RIP2 and RIP2 tyrosine autophosphorylation.
Deletion mutants of NOD2 revealed a 36-amino acid region required for TRAF4 binding. This was confirmed with the generation and use of a bacterial recombinant GST-fused peptide of this region, GST-NOD2(260–305) (Fig. 6).
The TRAF domains of each TRAF family member recognize and bind a distinct motif in their binding partner proteins (18). Although binding motifs for TRAF2 and TRAF6 have been characterized, a TRAF4 binding motif has not been described (19–21, 36). Sequence comparison by BLAST analysis reveals that the TRAF domain of TRAF4 is more similar to TRAF2 than TRAF6. Thus, we considered that the binding motif of TRAF4 might resemble that of TRAF2. We examined the 36-amino acid region of NOD2 for the presence of potential binding sites and identified two imperfect TRAF2 binding sites. When conservation of these sites was compared, we found that the binding site resembling the major TRAF2 binding site, (P/S/A/T)X(Q/E)E (19, 21), was conserved in both mouse and zebrafish. For this reason, we generated NOD2 point mutants with glutamate residues 279 and 280 replaced with alanine residues. TRAF4 binding to the E279A/E280A NOD2 mutant was severely diminished, and TRAF4 could no longer function to inhibit E279A/E280A NOD2 (Fig. 7).
In addition to the activation of inflammatory pathways, a role for NOD2 has been documented in maintaining an appropriate intestinal flora. NOD2-deficient mice have increased commensal bacteria and a decreased ability to prevent colonization of pathogenic bacteria (37). NOD2 has been reported as an antibacterial factor in epithelial cells and Paneth cells, whose antibacterial responses include defensin secretion (25, 29, 30, 33). The role of TRAF4 in NOD2-induced bacterial killing was examined by observing the effect of TRAF4 on NOD2-induced Salmonella killing in gentamycin protection assays. We find that TRAF4 binding to NOD2 inhibits the ability of NOD2 to kill Salmonella and that this inhibition is lost when mutants are used that abrogate TRAF4 binding to NOD2 (Fig. 4). This is not the first time TRAF4 has been shown to inhibit an innate immune signaling pathway. TRAF4 binds to TRAF6 and TRIF to dampen TLR-mediated NF-κB and IFNβ activation (38). In addition, TRAF4 has been shown to affect reactive oxygen species generation. The exact role of TRAF4 in reactive oxygen species generation remains elusive; however, three laboratories have independently shown TRAF4 binding the cytosolic NADPH oxidase complex subunit p47phox (38–40). It is possible that decreases in NOD2-induced bacterial killing by TRAF4 are linked to its role in mediating a reactive oxygen species response. This hypothesis is further supported by the recently published data indicating that NOD2 interacts with DUOX2 to mediate NOD2-dependent generation of reactive oxygen species in epithelial cells (41) and would be an interesting avenue for future investigation.
In summary, this study identified a novel NOD2-binding protein and regulator of NOD2 signaling. This finding provides insight that allows further dissection of NOD2 signaling pathways to elucidate how dysregulated NOD2 signaling leads to disease. Having mapped the specific regions of binding provides opportunities to manipulate the system for further research or future drug targeting techniques. Last, having identified a TRAF4 binding motif allows for identification of additional TRAF4-binding proteins within innate immune signaling pathways to elucidate the function of this understudied TRAF family member.
Acknowledgments
We thank George Dubyak (Case Western Reserve University) and George Stark and Xiaoxia Li (Cleveland Clinic Foundation) for critical comments on the manuscript. We also thank Alex Huang (Case Western Reserve University) and members of the Huang laboratory for use of and assistance with the confocal microscope.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant R01GM86550-01 (to D. W. A.), NIH Cell and Molecular Biology Training Grant 5T32GM008056-28 (to J. M. M.), and NIH Research Grant R01DK082437 (to C. M.). This work was also supported by Burroughs Wellcome Career Award for Biomedical Scientists 10061206.01 (to D. W. A.) and the generosity of Gerald and Nancy Goldberg (to C. M.).
- EOS
- early onset sarcoidosis
- MDP
- muramyl dipeptide
- CARD
- caspase activation recruitment domain
- NF-κB
- nuclear factor κB
- TRAF
- tumor necrosis factor-associated factor
- NEMO
- NF-κB essential modifier
- hBD-2
- human β-defensin 2
- cfu
- colony-forming unit(s)
- rbt
- rabbit
- ms
- mouse
- gt
- goat.
REFERENCES
- 1. Inohara N., Koseki T., Lin J., del Peso L., Lucas P. C., Chen F. F., Ogura Y., Núñez G. (2000) J. Biol. Chem. 275, 27823–27831 [DOI] [PubMed] [Google Scholar]
- 2. Inohara N., Ogura Y., Fontalba A., Gutierrez O., Pons F., Crespo J., Fukase K., Inamura S., Kusumoto S., Hashimoto M., Foster S. J., Moran A. P., Fernandez-Luna J. L., Nuñez G. (2003) J. Biol. Chem. 278, 5509–5512 [DOI] [PubMed] [Google Scholar]
- 3. Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 4. Fritz J. H., Ferrero R. L., Philpott D. J., Girardin S. E. (2006) Nat. Immunol. 7, 1250–1257 [DOI] [PubMed] [Google Scholar]
- 5. Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 6. Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 7. Strober W., Murray P. J., Kitani A., Watanabe T. (2006) Nat. Rev. Immunol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 8. Abbott D. W., Wilkins A., Asara J. M., Cantley L. C. (2004) Curr. Biol. 14, 2217–2227 [DOI] [PubMed] [Google Scholar]
- 9. Abbott D. W., Yang Y., Hutti J. E., Madhavarapu S., Kelliher M. A., Cantley L. C. (2007) Mol. Cell. Biol. 27, 6012–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Windheim M., Lang C., Peggie M., Plater L. A., Cohen P. (2007) Biochem. J. 404, 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang Y., Yin C., Pandey A., Abbott D., Sassetti C., Kelliher M. A. (2007) J. Biol. Chem. 282, 36223–36229 [DOI] [PubMed] [Google Scholar]
- 12. Clark N. M., Marinis J. M., Cobb B. A., Abbott D. W. (2008) Curr. Biol. 18, 1402–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kolch W. (2005) Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 14. Abell A. N., Johnson G. L. (2005) J. Biol. Chem. 280, 35793–35796 [DOI] [PubMed] [Google Scholar]
- 15. Abell A. N., Rivera-Perez J. A., Cuevas B. D., Uhlik M. T., Sather S., Johnson N. L., Minton S. K., Lauder J. M., Winter-Vann A. M., Nakamura K., Magnuson T., Vaillancourt R. R., Heasley L. E., Johnson G. L. (2005) Mol. Cell. Biol. 25, 8948–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chi H., Sarkisian M. R., Rakic P., Flavell R. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Régnier C. H., Masson R., Kedinger V., Textoris J., Stoll I., Chenard M. P., Dierich A., Tomasetto C., Rio M. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5585–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chung J. Y., Park Y. C., Ye H., Wu H. (2002) J. Cell Sci. 115, 679–688 [DOI] [PubMed] [Google Scholar]
- 19. Park Y. C., Burkitt V., Villa A. R., Tong L., Wu H. (1999) Nature 398, 533–538 [DOI] [PubMed] [Google Scholar]
- 20. Ye H., Arron J. R., Lamothe B., Cirilli M., Kobayashi T., Shevde N. K., Segal D., Dzivenu O. K., Vologodskaia M., Yim M., Du K., Singh S., Pike J. W., Darnay B. G., Choi Y., Wu H. (2002) Nature 418, 443–447 [DOI] [PubMed] [Google Scholar]
- 21. Ye H., Park Y. C., Kreishman M., Kieff E., Wu H. (1999) Mol. Cell 4, 321–330 [DOI] [PubMed] [Google Scholar]
- 22. Tao M., Scacheri P. C., Marinis J. M., Harhaj E. W., Matesic L. E., Abbott D. W. (2009) Curr. Biol. 19, 1255–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 24. McDonald C., Chen F. F., Ollendorff V., Ogura Y., Marchetto S., Lécine P., Borg J. P., Nuñez G. (2005) J. Biol. Chem. 280, 40301–40309 [DOI] [PubMed] [Google Scholar]
- 25. Homer C. R., Richmond A. L., Rebert N. A., Achkar J. P., McDonald C. (2010) Gastroenterology 139, 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanazawa N., Okafuji I., Kambe N., Nishikomori R., Nakata-Hizume M., Nagai S., Fuji A., Yuasa T., Manki A., Sakurai Y., Nakajima M., Kobayashi H., Fujiwara I., Tsutsumi H., Utani A., Nishigori C., Heike T., Nakahata T., Miyachi Y. (2005) Blood 105, 1195–1197 [DOI] [PubMed] [Google Scholar]
- 27. Bertrand M. J., Doiron K., Labbé K., Korneluk R. G., Barker P. A., Saleh M. (2009) Immunity 30, 789–801 [DOI] [PubMed] [Google Scholar]
- 28. Tigno-Aranjuez J. T., Asara J. M., Abbott D. W. (2010) Genes Dev. 23, 2666–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hisamatsu T., Suzuki M., Reinecker H. C., Nadeau W. J., McCormick B. A., Podolsky D. K. (2003) Gastroenterology 124, 993–1000 [DOI] [PubMed] [Google Scholar]
- 30. Yamamoto-Furusho J. K., Barnich N., Hisamatsu T., Podolsky D. K. (2010) Inflamm. Bowel Dis. 16, 736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kobayashi K., Inohara N., Hernandez L. D., Galán J. E., Núñez G., Janeway C. A., Medzhitov R., Flavell R. A. (2002) Nature 416, 194–199 [DOI] [PubMed] [Google Scholar]
- 32. Ogura Y., Saab L., Chen F. F., Benito A., Inohara N., Nuñez G. (2003) Genomics 81, 369–377 [DOI] [PubMed] [Google Scholar]
- 33. Voss E., Wehkamp J., Wehkamp K., Stange E. F., Schröder J. M., Harder J. (2006) J. Biol. Chem. 281, 2005–2011 [DOI] [PubMed] [Google Scholar]
- 34. Hitotsumatsu O., Ahmad R. C., Tavares R., Wang M., Philpott D., Turer E. E., Lee B. L., Shiffin N., Advincula R., Malynn B. A., Werts C., Ma A. (2008) Immunity 28, 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hutti J. E., Turk B. E., Asara J. M., Ma A., Cantley L. C., Abbott D. W. (2007) Mol. Cell. Biol. 27, 7451–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chung J. Y., Lu M., Yin Q., Lin S. C., Wu H. (2007) Adv. Exp. Med. Biol. 597, 122–130 [DOI] [PubMed] [Google Scholar]
- 37. Petnicki-Ocwieja T., Hrncir T., Liu Y. J., Biswas A., Hudcovic T., Tlaskalova-Hogenova H., Kobayashi K. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15813–15818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takeshita F., Ishii K. J., Kobiyama K., Kojima Y., Coban C., Sasaki S., Ishii N., Klinman D. M., Okuda K., Akira S., Suzuki K. (2005) Eur. J. Immunol. 35, 2477–2485 [DOI] [PubMed] [Google Scholar]
- 39. Li J. M., Fan L. M., Christie M. R., Shah A. M. (2005) Mol. Cell. Biol. 25, 2320–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Y. C., Wu R. F., Gu Y., Yang Y. S., Yang M. C., Nwariaku F. E., Terada L. S. (2002) J. Biol. Chem. 277, 28051–28057 [DOI] [PubMed] [Google Scholar]
- 41. Lipinski S., Till A., Sina C., Arlt A., Grasberger H., Schreiber S., Rosenstiel P. (2009) J. Cell Sci. 122, 3522–3530 [DOI] [PubMed] [Google Scholar]