Abstract
Protein arginine methylation plays a critical role in differential gene expression through modulating protein-protein and protein-DNA/RNA interactions. Although numerous proteins undergo arginine methylation, only limited information is available on how protein arginine methyltransferases (PRMTs) identify their substrates. The human PRMT5 complex consists of PRMT5, WD45/MEP50 (WD repeat domain 45/methylosome protein 50), and pICln and catalyzes the symmetrical arginine dimethylation of its substrate proteins. pICln recruits the spliceosomal Sm proteins to the PRMT5 complex for methylation, which allows their subsequent loading onto snRNA to form small nuclear ribonucleoproteins. To understand how the PRMT5 complex is regulated, we investigated its biochemical composition and identified RioK1 as a novel, stoichiometric component of the PRMT5 complex. We show that RioK1 and pICln bind to PRMT5 in a mutually exclusive fashion. This results in a PRMT5-WD45/MEP50 core structure that either associates with pICln or RioK1 in distinct complexes. Furthermore, we show that RioK1 functions in analogy to pICln as an adapter protein by recruiting the RNA-binding protein nucleolin to the PRMT5 complex for its symmetrical methylation. The exclusive interaction of PRMT5 with either pICln or RioK1 thus provides the first mechanistic insight into how a methyltransferase can distinguish between its substrate proteins.
Keywords: Adaptor Proteins, Protein Methylation, Protein-Protein Interactions, Ribonuclear Protein (RNP), RNA Binding Protein, PRMT5, RioK1, U SnRNP, Nucleolin
Introduction
Posttranslational modifications regulate the localization, stability, and catalytic activity of proteins. In recent years, protein arginine methylation has emerged as a common theme to modulate protein-protein and/or protein-nucleic acid interactions (1). The enzymes catalyzing this posttranslational modification, protein arginine methyltransferases (PRMTs),4 have accordingly been implicated in the regulation of diverse processes ranging from DNA damage repair and transcriptional regulation to RNA splicing (2, 3). So far, nine PRMTs are known in humans and are classified into two major types based on substrate and reaction product specificity (4). Type I and II PRMTs catalyze the formation of monomethylarginines, but only type I PRMTs catalyze the formation of asymmetric dimethylarginines (1). Type II PRMTs, on the other hand, catalyze the formation of symmetric dimethylarginines and encompass PRMT5 (5), PRMT7 (6), and PRMT9 (7).
Most insight has been gained into the function of PRMT5, whose substrate proteins include myelin basic protein (8), histones (9), and the spliceosomal Sm proteins (10, 11). PRMT5 fulfils its role in methylation of Sm proteins within a trimeric complex, termed the PRMT5 complex, containing PRMT5, WD45/MEP50, and pICln (chloride channel nucleotide sensitive 1A) (10, 12, 13). Although newly synthesized Sm proteins can be spontaneously incorporated into U small nuclear ribonucleoproteins in vitro (14), this process depends on the cooperate action of the PRMT5 complex and the SMN (survival of motor neuron) complex in vivo (13, 15). The PRMT5 complex symmetrically dimethylates the Sm proteins B/B′, D1, and D3 within an arginine/glycine-rich “RG-box” (arginine and glycine rich protein region) motive (10, 11, 16), which enhances their affinity for the SMN complex (17, 18). Subsequently, the SMN complex, composed of SMN and Gemins2–8 (components of gems number 2–8), facilitates the loading of methylated Sm proteins onto snRNA, resulting in the formation of the small nuclear ribonucleoprotein core (13, 15, 19–22).
Even though the role of PRMT5 in small nuclear ribonucleoprotein biogenesis is relatively well understood, the roles of WD45/MEP50 and pICln are only beginning to emerge. WD45/MEP50 associates with various PRMT5 substrates (23), but its functional role within the PRMT5 complex remains unclear. The other component of the PRMT5 complex, pICln, originally described as component of ion channels (24), directly binds Sm proteins (10, 11, 25) and most likely acts as an Sm chaperone (26). Furthermore, silencing of pICln expression has been reported to be essential for motor neuron outgrowth in zebrafish, resembling spinal muscular atrophy, which is the phenotypic manifestation of reduced SMN protein levels in humans (27, 28).
Here, we have investigated the composition of the PRMT5 complex at a biochemical level. We identified the Rio domain-containing protein RioK1 as a novel component of the PRMT5 complex, which interacts directly with PRMT5 in a stoichiometric manner. Interestingly, RioK1 and pICln bind to the N terminus of PRMT5 in a mutually exclusive fashion. Our data thus redefine the PRMT5 complex into a core complex consisting of PRMT5 and WD45/MEP50, which either interacts with pICln or RioK1. Although pICln recruits Sm proteins, RioK1 recruits nucleolin for its symmetrical methylation to the PRMT5 complex. The mutually exclusive interaction of two adapter proteins with PRMT5 thus provides the first mechanistic hint at how a methyltransferase can distinguish between its substrate proteins.
EXPERIMENTAL PROCEDURES
cDNA Constructs
Plasmids encoding full-length cDNAs corresponding to the open reading frames of PRMT5, WD45/MEP50, and pICln have been described previously (11). The full-length open reading frames of RioK1, RioK2, RioK3, and nucleolin were amplified by PCR using the following primers: RioK1, 5′-CGGACGTCGACATATGGACTACCGGCGGCTTCTCATG-3′ and 5′-CTGATGCGGCCGCCTATTTGCCTTTTTTCGTCTTGGC-3′; RioK2, 5′-GTTGGATCCATGGGGAAAGTGAATG-3′ and 5′-CATCTCGAGTTATTCTCCCCAAAAG-3′; RioK3, 5′-GTTGGATCCATGGATCTGGTAGGAG-3′ and 5′-CATCTCGAGCTATTCATCATATAG-3′; and nucleolin, 5′-AGGAATTCATGGTGAAGCTCGCG-3′ and 5′-CTGCTCGAGCTATTCAAACTTCG-3′ from a human brain cDNA library and subcloned into pEGEX6P-1 (GE Healthcare) and pHA vector (an N-terminal HA tag containing derivate of pCDNA3.1, Invitrogen). Truncations of PRMT5 (aa 1–291, 5′-GGATGGATCCATGGCGGCGATGGCG-3′ and 5′-GCACTCTCGAGACGGTTCTGGCTTAAG-3′; aa 295–637, 5′-CATGGGATCCAATGCCTATGAACTC-3′ and 5′-GCATCTCGAGCTAGAGGCCAATGGTATATG-3′), RioK1 (aa 1–120, 5′-GTAGAATTCATGGACTACCGGCG-3′ and 5′-CATCTCGAGTCAATTAATTTTATTCTC-3′; aa 121–242, 5′-GATGCGGCCGCAATTTAGATAAGC-3′ and 5′-CATCTCGAGTCACATTTTCCTAGGGT-3′; and aa 1–242, 5′-CGGACGTCGACATATGGACTACCGGCGGCTTCTCATG-3′ and 5′-CGTGCGGCCGCTCACATTTCCTAGGG-3′), and nucleolin (aa 648–710, 5′-CAGGAATTCATGGGTGAAGGTGGCTTC-3′ and 5′-CTGCTCGAGCTATTCAAACTTCG-3′) were generated by PCR and subcloned into pEGEX6P-1 and pHA, respectively.
RNA Interference
Cellular levels of RioK1 in HEK293T cells were reduced by transfection of a mixture of two double-stranded 21-nt-long siRNAs (sequences, 5′-GAGAAGGAUGACAUUCUGUTT-3′ and 5′-ACAGAAUGUCAUCCUUCUCTT-3′). Both specific siRNAs as well as a scrambled siRNA control were purchased from IBA Nucleic Acids Synthesis, Göttingen, Germany) and transfected with OligofectamineTM (Invitrogen) following the protocol of the manufacturer. Silencing of RioK1 was assayed by Western blotting of cell extracts 8, 24, 48, and 72 h after transfection using RioK1 specific antibodies.
Liquid Chromatography-Mass Spectrometry Analysis
Sample preparation was performed according to a modified protocol (29, 30). Nano-LC-MS/MS analyses were accomplished on an LTQ XL (Thermo Scientific, Dreieich, Germany) coupled to an Ultimate 3000 nano-HPLC system (Dionex, Idstein, Germany). Peptides were preconcentrated in 0.1% TFA on a 100-μm inner diameter custom-made RP trapping column (HydroRP, 2-cm length, 4-μm particle size) (31) and subsequently separated on a custom-made 75-μm inner diameter RP column (HydroRP, 15-cm length, 2-μm particle size) by applying a 40-min binary gradient (solvent A, 0.1% TFA; solvent B, 0.1% TFA and 84% acetonitrile) ranging from 5 to 50% of solvent B at a flow rate of 250 nl/min. MS acquisition as well as data processing and analysis were performed as described elsewhere (32).
Recombinant Proteins and in Vitro Protein Binding Assays
Expression and purification of GST-tagged fusion proteins were carried out as described previously (33). [35S]methionine-labeled proteins were produced using the TNT-T7 Quick Coupled Transcription/Translation system (Promega, Mannheim, Germany). In vitro translated proteins were incubated with ∼2 μg purified GST-fusion proteins, immobilized on a glutathione-Sepharose resin, and allowed to bind in lysis buffer (50 mm Tris/HCl, pH 7.5, 200 mm NaCl, 0.01% IGEPAL CA-630, 1 mm dithiothreitol, 5 mm EDTA, 5 mm EGTA, 1 mg/ml bovine serum albumin) at 4 °C for 1 h. After washing the resin three times with lysis buffer, bound proteins were eluted by boiling in 2× SDS sample buffer, resolved by SDS-PAGE, and analyzed by Coomassie staining. Labeled proteins were detected by autoradiography of the dried gel.
Preparation of Cell Extracts, Antibodies, and Immunoprecipitations
All cell lines were obtained from the ATCC and cultured in appropriate media. Total cell extract was prepared as described previously (11). Cytoplasmic and nuclear extracts were prepared following a protocol of Dignam et al. (34). Antibodies against PRMT5 and WD45/MEP50 were raised by injection of recombinant full-length human proteins into rabbits. Antibodies against RioK1 were raised by injection of recombinant RioK1 comprising aa 1–242. Antibodies were affinity-purified on columns with the respective covalently linked antigen. The anti-SmB/D1/D3 antibody Y12 was kindly provided by J. Steitz (Yale University). SmB/B′ antibody was kindly provided by R. Lührmann (Max Planck Institute of Biophysical Chemistry), and PRP4 (pre-mRNA processing protein 4) antibody was a kind gift of B. Laggerbauer (Biocenter of the University of Würzburg). Antibodies recognizing pICln and unrip (unr interacting protein) have been described previously (33). SYM10 antibody and nucleolin antibody were purchased from Millipore (Schwalbach, Germany). Immunoprecipitations were carried out in HeLa or HEK293T extract, using antibodies covalently linked to protein A-Sepharose. Precipitated complexes were washed extensively with buffer containing 50 mm Tris/HCl, pH 7.5, 200 mm NaCl, 0.01% IGEPAL CA-630, 5 mm EDTA, and 5 mm EGTA and eluted by boiling in SDS sample buffer. Samples were resolved by SDS-PAGE and further analyzed by silver staining, Coomassie staining, or Western blotting.
Glycerol Gradient Centrifugation and Analytical Size Exclusion Chromatography
250 ml of HeLa extract was layered on a 10–30% glycerol gradient and centrifuged for 16 h in a SW60Ti rotor (4 °C at 24,000 rpm, Beckman Coulter Optima L-80 XP). Gradients were automatically fractionated from bottom to the top and analyzed by SDS-PAGE and Western blotting. Analytical size exclusion chromatography was achieved by fractionating HeLa extract on a Superdex 200 HR 10/300 gel filtration column (GE Healthcare) after incubation with purified recombinant pICln or RioK1 protein (2 h at 4 °C). Obtained fractions were separated by SDS-PAGE and further analyzed by Western blotting. For calibration of the column, a molecular weight standard from GE Healthcare was used.
Immunofluorescence Microscopy
HeLa cells grown on cover slides in DMEM and 10% FCS were washed once in PBS, fixed with 3% paraformaldehyde for 7 min, and permeabilized in 0.2% Triton X-100/PBS for 5 min on ice. After blocking with 2% BSA in PBS, cells were incubated for 1 h with affinity-purified polyclonal rabbit anti-RioK1 antibody. Following staining with rhodamine-conjugated secondary anti-rabbit antibodies, cells were analyzed with a 63× oil immersion lens on a Zeiss Axiovert 200 m microscope. HeLa cells transfected with an N-terminal GFP-RioK1 fusion were directly analyzed 48-h posttransfection without fixation.
In Vitro Methylation Assay
Purified recombinant substrate proteins were incubated with the PRMT5 complex (immunoprecipitated by anti-WD45/MEP50 antibody from HeLa extract) in 30 μl PBS and 2 μl [3H]S-adenosyl methionine for 2 h at 37 °C. The reaction was stopped by boiling in 2× SDS-sample buffer and analyzed by Coomassie staining and radiography of the dried gel.
Recombinant Protein Pulldown Assay, Competition Assay
For recombinant precipitation assays from HeLa extracts, purified GST-tagged RioK1, and truncations thereof, RioK2, RioK3, pICln, and GST (∼10 μg) were covalently cross-linked to glutathione-Sepharose by dimethyl pimelimidate dihydrochloride and incubated with HeLa extract for 1.5 h. The matrix was washed extensively, and bound proteins were eluted by boiling in 2× SDS sample buffer and analyzed by Coomassie staining.
To analyze the competition of pICln and RioK1, the PRMT5 complex was immunoprecipitated from HeLa extract by anti-WD45/MEP50 antibody. Identical amounts of the precipitated complex immobilized to protein A-Sepharose were incubated with increasing amounts of recombinant pICln or RioK1 (0.1 to 10 μg) for 1 h at 4 °C on a head-over-tail rotator. The matrix was washed extensively in incubation buffer (1× PBS, 0.01% IGEPAL), and bound proteins were analyzed by SDS-PAGE and Western blotting.
RESULTS
RioK1 Is a Novel Stoichiometric Component of Human PRMT5 Complex
To identify novel regulators of the PRMT5 complex, we performed immunoprecipitations of PRMT5 from HeLa lysate (Fig. 1A). Among the well characterized components of the PRMT5 complex, pICln, WD45/MEP50, and PRMT5 itself, we identified a novel protein, RioK1, of ∼90 kDa in size via mass spectrometry. To analyze whether RioK1 is a constitutive component of the human PRMT5 complex, we generated polyclonal antibodies against all components of this complex, including RioK1. Immunoprecipitations with these antibodies performed from HeLa extract robustly pulled down PRMT5 and WD45/MEP50. Interestingly, although PRMT5 and WD45/MEP50 were always present in stoichiometric amounts in all immunoprecipitations, pICln and RioK1 were only clearly enriched in the corresponding immunoprecipitations (Fig. 1B). This finding indicates that only a part of the methyltransferase complex might be associated with pICln and/or RioK1. Furthermore, only immunoprecipitations of pICln but not RioK1 co-precipitated notable amounts of Sm proteins (Fig. 1B, lane 4). To further analyze protein composition of the complex, immunoprecipitations of Fig. 1B were analyzed by Western blotting. Although pICln and RioK1 were clearly enriched in their corresponding precipitations, all complex components could be detected (Fig. 1C).
FIGURE 1.
RioK1 is a novel stoichiometric component of the human PRMT5 complex. A, the PRMT5 complex was immunoprecipitated from HeLa extract using anti-PRMT5 antibodies covalently coupled to protein A-Sepharose, resolved via SDS-PAGE, and visualized by silver staining. Normal rabbit serum (NRS) coupled to protein A-Sepharose was used as control (lane 2). The asterisk-marked band at ∼90 kDa was excised and identified as RioK1 by mass spectrometry (lane 3). HC and LC represent the heavy and light chain of co-eluted antibodies. Lane 1 shows protein standard (M). B, the PRMT5 complex was immunoprecipitated from HeLa extract using antibodies directed against PRMT5 (lane 2), WD45/MEP50 (lane 3), pICln (lane 4), RioK1 (lane 5), or normal rabbit serum (NRS) as control (lane 6). The precipitated complex components were analyzed by SDS-PAGE and silver staining. Note that protein content was normalized to PRMT5, explaining the different intensities of co-precipitated pICln in A (lane 3) and B (lane 2). C, the PRMT5 complex was immunoprecipitated similarly to B, resolved by SDS-PAGE, and analyzed by Western blotting using the indicated antibodies. D, HeLa total cell lysate (TCL) was separated by gel filtration chromatography, and fractions were analyzed by Western blotting. E, total protein extracts were generated from MCF7 (breast cancer cell line), HeLa (cervix carcinoma cell line), Kelly (neuroblastoma cell line), NEC8 (testicular tumor cell line), HEK293T (embryonic kidney cell line), MelHo (melanoma cell line), and HS68 (primary foreskin fibroblast cell line) and normalized to β-actin (lowest panel). Expression of RioK1 and the PRMT5 complex components PRMT5, WD45/MEP50, and pICln were analyzed by immunoblotting with specific antibodies as indicated. F, to analyze whether RioK1 is part of the PRMT5 complex in different cell lines, RioK1 was immunoprecipitated from MCF7, HEK293T, and HS68 extracts. Co-precipitation of PRMT5 complex components was assessed by Western blotting with specific antibodies as indicated (lanes 3–5). Normal rabbit serum was used as control (lanes 6–8).
To strengthen that RioK1 is part of the human PRMT5 complex, we fractionated HeLa cytosolic extract by size exclusion chromatography and assayed the obtained fractions by immunoblotting. As shown in Fig. 1D, RioK1 and pICln partly co-migrated with the core components of the PRMT5 complex including PRMT5 and WD45/MEP50 in the so called 20 S complex at 700 to 400 kDa (10). Yet, RioK1 was separated in a more diffuse pattern, ranging from 700 to 200 kDa, indicating its participation in complexes other than the PRMT5 complex. Similar results were obtained when cell lysates were fractioned using glycerol gradients (data not shown). pICln, in addition, associates with the SMN complex (700 kDa to 1 MDa) and with Sm proteins in the so called 6 S complex (10).
To analyze whether RioK1 is a ubiquitous protein, we examined its expression in different cell lines. RioK1 was present in all analyzed immortalized cell lines, although at different amounts (Fig. 1E). The expression in HS68 primary fibroblasts was low but detectable after longer exposure of the gel (data not shown). Next, we were interested whether RioK1 indeed participates in the PRMT5 complex in the different cell lines. To this end, we immunoprecipitated RioK1 from MCF7, HEK293T, and HS68 extract with specific antibodies and analyzed for co-precipitation of PRMT5 (Fig. 1F). In all of the cell lines tested, PRMT5 co-immunoprecipitated with RioK1, indicating that RioK1 is a ubiquitous component of the PRMT5 complex.
RioK1 Is an Exclusively Cytoplasmic Protein
To analyze the subcellular localization of endogenous RioK1, we assayed its localization in comparison with the other PRMT5 complex components by immunofluorescence in HeLa cells. Although RioK1 was predominantly cytoplasmic, PRMT5 and WD45/MEP50 were evenly distributed between cytoplasm and nucleus. For pICln, in contrast, we observed a predominantly nuclear localization (Fig. 2A). A cytoplasmic localization of RioK1 was not only observed for the endogenous protein but also after overexpression (Fig. 2B). To biochemically analyze the distribution of the components of the PRMT5 complex, HeLa cells were biochemically separated into a nuclear and a cytoplasmic fraction and analyzed by Western blotting (Fig. 2C). PRMT5 and WD45/MEP50 were present in the cytoplasm and nucleus but were clearly enriched in the cytoplasmic fraction, whereas the Sm adapter protein pICln showed a slight enrichment in the nucleus. In stark contrast and in line with immunofluorescence analysis, RioK1 was exclusively present in the cytoplasmic fraction. Unrip, the pre-mRNA processing factor PRP4, and Sm proteins were used as controls and confirmed the proper separation of cellular compartments. From these data, we conclude that RioK1 is the only component of the PRMT5 complex that localizes exclusively to the cytoplasm.
FIGURE 2.
RioK1 is an exclusively cytoplasmic protein. A, HeLa cells were grown on coverslips, fixed, permeabilized, and stained with antibodies against PRMT5, WD45/MEP50, pICln, and RioK1. DAPI was used to visualize DNA in the nucleus (upper panel). Cytoplasmic localization of RioK1 was confirmed by merge of DAPI and rhodamin channels. B, GFP-RioK1 was overexpressed in HeLa cells for 48 h and compared with GFP expression for control (lower panel). C, HeLa cells were fractionated into total cell lysate (TCL, lane 1), cytoplasmic extract (CPE, lane 2), and nuclear extract (NE, lane 3). The fractions were analyzed by Western blotting using the indicated antibodies.
RioK1 Directly Interacts with PRMT5 Complex via its N Terminus
We next investigated which component of the PRMT5 complex binds RioK1. To this end, in vitro translated, [35S]methionine-labeled RioK1 was incubated with the GST-tagged fusion proteins of PRMT5, WD45/MEP50, and pICln in an interaction assay. RioK1 interacted exclusively with PRMT5, but not with WD45/MEP50 or pICln (Fig. 3A). GST-tagged RioK1, immobilized on glutathione-Sepharose and in vitro translated PRMT5 and truncations thereof, were utilized in interaction assays to assess the interacting regions of PRMT5 and RioK1. This revealed that the N-terminal part of PRMT5 (aa 1–291) interacts with RioK1 (Fig. 3B). Further analysis with truncated versions of RioK1 (aa 1–120, 1–242, 121–242, and 227–568) indicated that this interaction is probably mediated via amino acids 1–242 of RioK1, where aa 1–120 contribute to the major interaction compared with aa 121–242 (Fig. 3C, compare Fig. 3C, lanes 3, 4, 5, and 6).
FIGURE 3.
RioK1 directly interacts with PRMT5 via its very N-terminal region. A, in an interaction assay, in vitro translated, radioactively [35S]methionine-labeled RioK1 was incubated with GST-tagged PRMT5 (lane 2), WD45/MEP50 (lane 3), pICln (lane 4), and GST (lane 5) bound to GSH-Sepharose. After extensive washing, the precipitated proteins were eluted with sample buffer, resolved by SDS-PAGE, and visualized by Coomassie staining. The in vitro translated protein was visualized by autoradiography of the dried gel. For comparison, 10% of in vitro translated RioK1 was loaded (lane 6). B, GST-tagged truncations of RioK1 comprising aa 1–242 (lanes 2–4), aa 227–568 (lanes 5–7), aa 1–120 (lanes 8–10), or GST (lanes 11–13) bound to GSH-Sepharose were incubated with in vitro translated, [35S]methionine-labeled full-length (fl), N-terminal (nt; aa 1–291), or C-terminal (ct; aa 295–637) versions of PRMT5 in an interaction assay. For comparison, 10% of in vitro translated proteins were loaded (lanes 14–16). The interaction assay was analyzed as described in A. C, GST-PRMT5 (lanes 2–6) and GST (lanes 7–11) bound to GSH-Sepharose were incubated with an in vitro translated, [35S]methionine-labeled version of RioK, including full-length RioK1, aa 1–242, aa 227–568, aa 1–120, and aa 121–242. For comparison, 10% of in vitro translated RioK1 was loaded (lanes 12–16). D, GST-tagged pICln (lanes 2 and 3), RioK1 (lanes 4–6), GST alone (lanes 7 and 8), RioK2 (lanes 9 and 10), or RioK3 (lanes 11 and 12), covalently cross-linked to GSH-Sepharose were incubated with or without HeLa extract in a pulldown assay. As a control, non-cross-linked protein was analyzed by SDS-PAGE. After extensive washing, the precipitated proteins were eluted from the matrix by sample buffer, resolved by SDS-PAGE, visualized via Coomassie staining, and analyzed by mass spectrometry. Note, that lanes containing non-cross-linked, recombinant proteins exhibit degradation products of these proteins.
The region aa 1–242 comprises the so called Rio domain of RioK1 (Fig. 4E). We were therefore interested whether the other two members of the Rio family are likewise able to bind to the PRMT5 complex. To this end, we performed pulldown assays from HeLa lysates using recombinant GST-tagged Rio proteins (Fig. 3D). Of the Rio proteins, only GST-RioK1 was able to precipitate the PRMT5 complex, but not RioK2 or RioK3. The identity of the precipitated proteins was ensured by mass spectrometry, which did not yield any PRMT5 peptides in the pulldowns with GST-RioK2 or GST-RioK3 (Fig. 3D and data not shown). Instead, GST-RioK2 associated mainly with proteins of the 40 S ribosome as has been described previously (35–40), whereas no particular protein family was enriched in the GST-RioK3 precipitations. Taken together, only RioK1 is capable to bind to the PRMT5 complex.
FIGURE 4.
RioK1 and pICln compete for binding to PRMT5. A, GST-tagged RioK1 (lane 2) and pICln (lane 3) or GST alone (lane 4), covalently cross-linked to GSH-Sepharose were incubated with HeLa extract in a pulldown assay. After extensive washing, the precipitated proteins were eluted from the matrix by sample buffer, resolved by SDS-PAGE, and visualized via Coomassie staining. Intervening lanes have been spliced out. Lane 1 shows protein standard (M). B, in a competition assay, the PRMT5 complex was immunoprecipitated with anti-WD45/MEP50 antibodies from HeLa cell extract. The purified complex was incubated with increasing amounts (0.1 to 10 μg) of GST-tagged RioK1 (aa 1–242, left panel) or pICln (right panel). The complex composition was afterward analyzed by Western blotting using the indicated antibodies. C, HeLa cell extract was incubated with GST (20 μg, upper panel), GST-RioK1 (20 μg, middle panel), or GST-pICln (<20 μg, lower panel) and was afterward fractionated by size exclusion chromatography. The collected fractions were analyzed by Western blotting with the indicated antibodies. D, GST-tagged pICln (lanes 2–4) or GST (lanes 5–7) immobilized on glutathione-Sepharose were incubated with in vitro translated, [35S]methionine-labeled, full-length PRMT5 or truncations thereof in an interaction assay. After extensive washing, the precipitated proteins were eluted from the matrix in loading buffer, resolved by SDS-PAGE, Coomassie stained, and visualized by autoradiography. For comparison, 10% of in vitro translated proteins were loaded (lanes 8–10). E, schematic overview of the interacting domains of PRMT5, RioK1, and pICln. Interactions are indicated by arrows. Interacting domains are colored black. fl, full length; nt, N-terminal; ct, C-terminal; TCL, total cell lysate.
RioK1 and pICln Compete for Binding to PRMT5
To elucidate the role of RioK1 in the composition of the PRMT5 complex, pulldowns from HeLa extract using recombinant GST-RioK1 and GST-pICln were performed. In both cases, PRMT5 and WD45/MEP50 were robustly precipitated (Fig. 4A). Yet, RioK1 did not precipitate pICln and vice versa, as deduced from Coomassie staining (Fig. 4A) and mass spectrometric analysis (data not shown). This finding indicated that RioK1 and pICln might bind to the same site in PRMT5 and thus compete for binding. To address this issue, we performed in vitro competition assays using PRMT5 complex immunopurified from HeLa cell extract with anti-WD45/MEP50 antibodies. The immobilized complex was then incubated with rising amounts of purified recombinant pICln and N-terminal RioK1 protein, respectively (Fig. 4B and supplemental Fig. S1). Recombinant pICln and RioK1 efficiently displaced their endogenous counterparts from the PRMT5 complex, indicating that recombinant pICln and RioK1 were correctly folded and specifically interacted with PRMT5. Indeed, recombinant pICln displaced endogenous RioK1 from the PRMT5 complex, whereas recombinant RioK1 correspondingly displaced endogenous pICln. GST alone, on the other hand, did not displace either endogenous RioK1 or pICln from the PRMT5 complex (supplemental Fig. S1). Importantly, the 1:1 stoichiometry of PRMT5:WD45/MEP50 was not disturbed by either treatment.
To investigate the competition between RioK1 and pICln in a carrier-free situation, we incubated HeLa extract with recombinant, purified RioK1 or pICln, followed by gel filtration chromatography (Fig. 4C). Western blotting of the fractions revealed that, upon addition of recombinant RioK1 to the extract, pICln was excluded from the high molecular mass PRMT5 complex migrating in the range of 700–400 kDa. In turn, when the extract was incubated with recombinant pICln, endogenous RioK1 no longer co-migrated with the PRMT5 complex. Mock-treated (GST) extract was separated and analyzed as control.
The previous data raised the possibility that RioK1 and pICln may compete for the same binding site in PRMT5. To this end, we investigated whether pICln binds to the N terminus of PRMT5, just as RioK1. Interaction assays utilizing GST-tagged pICln and in vitro translated PRMT5 and truncations thereof revealed that pICln bound to the N terminus of PRMT5 (Fig. 4D). These findings further substantiated that both proteins, RioK1 and pICln, utilize the same binding site within the PRMT5 protein (Fig. 4E).
RioK1 Interacts with PRMT5 Methylation Substrate Nucleolin
As shown above, RioK1 competes with pICln for binding to PRMT5. We thus asked whether this differential interaction might influence methylation of Sm substrate proteins, which are recruited to PRMT5 by pICln (10, 11). To assess whether PRMT5 methylation activity is influenced by varying amounts of RioK1 or pICln, the PRMT5 complex was immunoprecipitated with antibodies directed against the individual components of the complex. The anti-RioK1 and anti-pICln immunoprecipitations were thus enriched for RioK1 and pICln, respectively, allowing for the investigation of the influence of these proteins on PRMT5 methylation activity. Equal amounts of the methyltransferase were incubated with recombinant, purified SmD1 substrate protein and [3H]-labeled S-adenosyl methionine (Fig. 5A). In this set of experiments, no difference in methylation activity was detectable in regard to Sm protein methylation. Furthermore, similar data were obtained by in vitro methylation assays with the PRMT5 complex, precipitated from HeLa cells overexpressing pICln (data not shown).
FIGURE 5.
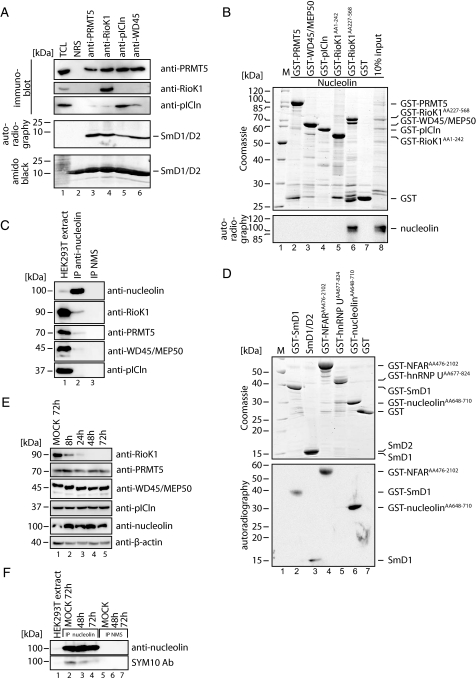
RioK1 interacts with the PRMT5 methylation substrate nucleolin. A, the PRMT5 complex was immunoprecipitated (IP) from HeLa lysate with the indicated antibodies and incubated with recombinant SmD1/D2 in a methylation assay with [3H]S-adenosyl methionine at 37 °C for 2 h. The reaction was stopped by the addition of sample buffer, and the eluted proteins were resolved by SDS-PAGE and Western blotting. HeLa total cell lysate (TCL) was used as control. Amido Black staining of the PVDF membrane was used to visualize equal loading of SmD1/D2. The identity of the precipitated proteins was ensured via immunoblotting. The methylated species of SmD1/D2 were visualized via autoradiography. Normal rabbit serum (NRS) was used as control for immunoprecipitation. B, GST-tagged members of the PRMT5 complex were incubated with in vitro translated, [35S]methionine-labeled nucleolin in an interaction assay (lanes 2–7). The precipitated proteins were visualized by autoradiography and Coomassie staining. For comparison, 10% of in vitro translated nucleolin was loaded (lane 8). Note that bands visible in the Coomassie staining below the expected molecular mass of the recombinant, GST-tagged proteins represent degradation products of these proteins. Lane 1 shows protein standard (M). C, nucleolin was specifically immunoprecipitated from HEK293T extract, and the precipitates were analyzed for the presence of PRMT5 complex components by immunoblotting with the respective specific antibodies (lane 2). Normal mouse serum (NMS) was used for control immunoprecipitation (lane 3). Lane 1 shows total extract of HEK293T. D, in an in vitro methylation assay of GST-SmD1 (lane 2), SmD1/D2 heterodimer (lane 3), the RG-boxes of NFAR (aa 476–2102, lane 4), hnRNP U (aa 677–824, lane 5), nucleolin (aa 648–710, lane 6), or GST (lane 7) were incubated with [3H]S-adenosyl methionine. The reaction was stopped by the addition of sample buffer and resolved by SDS-PAGE. The methylated proteins were visualized by autoradiography. Note that bands visible in the Coomassie staining below the expected molecular weight of the recombinant, GST-tagged proteins represent degradation products of these proteins. E, HEK293T cells were treated with siRNAs against RioK1 and harvested at the indicated time points and analyzed by immunoblotting for indicated proteins. F, cell extracts obtained in E were used for antinucleolin immunoprecipitations and analyzed for the presence of symmetrically methylated nucleolin by immunoblotting with SYM10 antibody.
If, as the previous experiment suggests, RioK1 does not function as a direct regulator of Sm-protein methylation, it may function as an adapter molecule, recruiting substrate proteins to the methylosome that are distinct from those recruited by pICln. To address this possibility, we carried out anti-RioK1 immunoprecipitations and GST-RioK1 pulldowns from HeLa extract followed by mass spectrometry (supplemental Fig. S2 and data not shown). Both approaches identified a very similar array of proteins as candidate interaction partners of RioK1 including mainly RG-box-containing, methylated proteins like hnRNPs, histones, or nucleolin (for a detailed list of identified proteins refer to supplemental Table 1).
Nucleolin is involved in the synthesis and maturation of ribosomes, whereas arginine methylation of nucleolin is regarded as a prerequisite for its interaction with RNA (41–44). To prove the specificity of the protein-protein interactions identified in the immunoprecipitation and GST pulldown assays, we carried out in vitro interaction assays of [35S]methionine-labeled, in vitro translated nucleolin with GST-tagged components of the PRMT5 complex (Fig. 5B). Nucleolin interacted only with the C terminus of RioK1, suggesting that RioK1 recruits nucleolin to the PRMT5 complex.
To confirm the interaction between nucleolin and the PRMT5 complex in cells, we performed anti-nucleolin immunoprecipitations from HEK293T cell extracts (Fig. 5C). Indeed, RioK1 co-immunoprecpitated with nucleolin. Interestingly, although PRMT5 and WD45/MEP50 were likewise present in the anti-nucleolin immunoprecipitations, we were not able to detect pICln. This data strongly indicated that the RioK1-containing PRMT5 complex can associate with nucleolin.
To investigate whether nucleolin is a PRMT5 substrate, an in vitro methylation assay was performed with PRMT5 complex immunoprecipitated from HeLa cells with the RG-box of nucleolin (44) as substrate (Fig. 5D). The known PRMT5 substrates SmD1 (10, 11) and NFAR5 were used as positive controls. hnRNPU was used as negative control to ensure specificity of methylation, as hnRNPU is asymmetrically methylated by PRMT1 (45, 46). Compared with SmD1 and NFAR, nucleolin was by far the most efficient PRMT5 substrate. Taken together, these data indicate that RioK1 recruits nucleolin to the PRMT5 complex for methylation.
To elucidate whether RioK1 is required for symmetrical arginine methylation of nucleolin in cells, we analyzed the methylation status of nucleolin after transfection of HEK293T cells with siRNA oligonucleotides targeting RioK1. Although RioK1 protein levels were already severely reduced 8 h after siRNA transfection, PRMT5, WD45/MEP50, and pICln protein levels were not affected by the knockdown of RioK1 (Fig. 5E). We then subjected extracts 48 and 72 h after transfection with RioK1 siRNAs to immunoprecipitation with anti-nucleolin antibody. Although the total amount of nucleolin was not affected by RioK1 siRNA treatment, symmetrically methylated nucleolin remarkable declined concomitantly with decreasing RioK1 protein levels, as detected by Western blotting with the methylation-specific antibody SYM10 (Fig. 5F). This finding clearly indicates that RioK1 specifically promotes the symmetrical methylation of nucleolin by recruiting it to the PRMT5 complex.
DISCUSSION
The PRMT5 complex has been described to consist of PRMT5, WD45/MEP50, and pICln in a 1:1:1 stoichiometry (10–12). Here, we have used immunoprecipitation and in vitro binding experiments to study the composition and regulation of the PRMT5 complex.
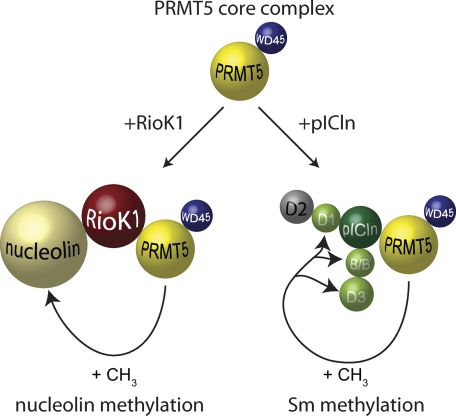
We identified RioK1 as a novel component of the human PRMT5 complex. Interestingly, RioK1 and pICln both bind to PRMT5, utilizing the same binding site within the methyltransferase. The competitive binding of RioK1 and pICln and our detailed analysis of the PRMT5 complex by precipitating all complex components with the respective antibodies revealed the existence of different PRMT5 complexes in vivo. A PRMT5 core-complex that was precipitated by PRMT5 and WD45/MP50 antibody encompasses WD45/MEP50 and PRMT5 (Fig. 6). This core complex then binds pICln or RioK1 to form distinct subcomplexes. Importantly, both trimeric complexes (PRMT5-WD45/MEP50-RioK1 and PRMT5-WD45/MEP50-pICln) are composed of a 1:1:1 stoichiometry and capable to recruit specific substrate proteins (Fig. 6).
FIGURE 6.
Model of PRMT5 substrate recognition through RioK1 and pICln. Schematic view of the mutually exclusive interaction of PRMT5 with RioK1 or pICln, which results in either the recruitment of nucleolin by RioK1 or Sm proteins by pICln for subsequent methylation by the PRMT5 complex.
RioK1 might therefore function to recruit substrate proteins to the PRMT5 complex, analogous to pICln (10, 11). Indeed, RioK1 binds PRMT5 with its N terminus, which leaves its C terminus free to serve as a recruitment platform for a variety of predicted and validated protein arginine methylation substrates which have been identified as RioK1-interacting proteins in this study (see supplemental Table 1 for details). RioK1 mainly interacts with RG-box containing, methylated proteins such as hnRNPs, histones, or ribosomal proteins, e.g. the very recently identified new PRMT5 substrate protein RPS10 (47), suggesting RioK1 to be a more general adapter protein than pICln.
Nucleolin is recruited to the PRMT5 complex by RioK1 for methylation in this manner as well. This also explains how PRMT5 regulates the localization of nucleolin in vivo as described previously (48). Association of nucleolin with the exclusively cytoplasmic RioK1 could trap nucleolin in the cytoplasm and concomitantly facilitate its methylation by PRMT5. Interestingly, nucleolin is known to be required for early rRNA processing (41–43), and methylation of its RG-box modulates its interaction with nucleic acids (44). This implicates a role for the RioK1-PRMT5 complex in rRNA processing and goes in line with the ascribed function of its yeast homolog, Rio1p in rRNA processing (49).
Mutually exclusive binding of pICln and RioK1 to PRMT5 should have immediate consequences for the methylation of its substrates. Oscillations in the ratio of RioK1-bound to pICln-bound PRMT5 should result in altered methylation of Sm proteins. Even though this is not the case in vitro, recruitment of distinct substrates to the different PRMT5 complexes might well occur in vivo, especially considering the important role of pICln as chaperone of Sm proteins (26). Indeed, excess pICln inhibits the methylation activity of PRMT5 toward histones and interestingly, lack of pICln leads to the SMN disease phenotype (27). A corollary of this model is that mechanisms exist to alter RioK1 and/or pICln protein levels or change their affinity for PRMT5 and thereby impart spatial and temporal control over substrate specificity of the PRMT5 complex. Indeed, yeast Rio1p is degraded at the G1/S transition of the cell cycle in a casein kinase 2 (CK2)-dependent manner (50). This, together with the fact that human RioK1 is a CK2 substrate in vitro5 suggests that a similar mechanism may control RioK1 protein levels in human cells.
In summary, we present RioK1 as a novel, stoichiometric component of the human PRMT5 complex. Importantly, RioK1 and pICln associate with PRMT5 in a mutually exclusive fashion, which allows for the recruitment of distinct methylation substrates such as nucleolin and Sm proteins, respectively. The work presented here not only redefines the PRMT5 complex, but also raises interesting new questions. Future research should focus on how RioK1 influences methylation of e.g. nucleolin, histones, or Sm proteins in vivo and how the interaction of RioK1 and pICln with PRMT5 is controlled by posttranslational modification.
Supplementary Material
Acknowledgments
We thank Dr. R. Lührmann (Max Planck Institute for Biophysical Chemistry), Dr. J. Steitz (Yale University), and Dr. Bernhard Laggerbauer (Biocenter of the University of Würzburg) for the generous gift of antibodies. We are indebted to E. Dinkl for excellent technical help and M. Reinhart for programming and help with evaluation of mass spectrometry data. We are grateful to H. J. Gross und H. Beier for very stimulating discussions.
This work was supported by grants of the German Research Foundation (Deutsche Forschungsgemeinschaft Fi573/4-1) and families of spinal muscle atrophy (FSMA).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
G. Guderian, C. Peter, J. Wiesner, A. Sickmann, K. Schulze-Osthoff, U. Fischer, and M. Grimmler, unpublished data.
- PRMT
- protein arginine methyltransferase
- aa
- amino acids.
REFERENCES
- 1. Gary J. D., Clarke S. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61, 65–131 [DOI] [PubMed] [Google Scholar]
- 2. Bedford M. T., Richard S. (2005) Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 3. Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bachand F. (2007) Eukaryot. Cell 6, 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Branscombe T. L., Frankel A., Lee J. H., Cook J. R., Yang Z., Pestka S., Clarke S. (2001) J. Biol. Chem. 276, 32971–32976 [DOI] [PubMed] [Google Scholar]
- 6. Miranda T. B., Miranda M., Frankel A., Clarke S. (2004) J. Biol. Chem. 279, 22902–22907 [DOI] [PubMed] [Google Scholar]
- 7. Cook J. R., Lee J. H., Yang Z. H., Krause C. D., Herth N., Hoffmann R., Pestka S. (2006) Biochem. Biophys. Res. Commun. 342, 472–481 [DOI] [PubMed] [Google Scholar]
- 8. Ghosh S. K., Paik W. K., Kim S. (1988) J. Biol. Chem. 263, 19024–19033 [PubMed] [Google Scholar]
- 9. Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., Sif S. (2004) Mol. Cell Biol. 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friesen W. J., Paushkin S., Wyce A., Massenet S., Pesiridis G. S., Van Duyne G., Rappsilber J., Mann M., Dreyfuss G. (2001) Mol. Cell Biol. 21, 8289–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meister G., Eggert C., Bühler D., Brahms H., Kambach C., Fischer U. (2001) Curr. Biol. 11, 1990–1994 [DOI] [PubMed] [Google Scholar]
- 12. Friesen W. J., Wyce A., Paushkin S., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) J. Biol. Chem. 277, 8243–8247 [DOI] [PubMed] [Google Scholar]
- 13. Meister G., Bühler D., Pillai R., Lottspeich F., Fischer U. (2001) Nat. Cell Biol. 3, 945–949 [DOI] [PubMed] [Google Scholar]
- 14. Raker V. A., Plessel G., Lührmann R. (1996) EMBO J. 15, 2256–2269 [PMC free article] [PubMed] [Google Scholar]
- 15. Pellizzoni L., Yong J., Dreyfuss G. (2002) Science 298, 1775–1779 [DOI] [PubMed] [Google Scholar]
- 16. Kambach C., Walke S., Young R., Avis J. M., de la Fortelle E., Raker V. A., Lührmann R., Li J., Nagai K. (1999) Cell 96, 375–387 [DOI] [PubMed] [Google Scholar]
- 17. Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R. (2001) RNA 7, 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friesen W. J., Dreyfuss G. (2000) J. Biol. Chem. 275, 26370–26375 [DOI] [PubMed] [Google Scholar]
- 19. Gubitz A. K., Feng W., Dreyfuss G. (2004) Exp. Cell Res. 296, 51–56 [DOI] [PubMed] [Google Scholar]
- 20. Pellizzoni L. (2007) EMBO Rep. 8, 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Neuenkirchen N., Chari A., Fischer U. (2008) FEBS Lett. 582, 1997–2003 [DOI] [PubMed] [Google Scholar]
- 22. Meister G., Fischer U. (2002) EMBO J. 21, 5853–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Furuno K., Masatsugu T., Sonoda M., Sasazuki T., Yamamoto K. (2006) Biochem. Biophys. Res. Commun. 345, 1051–1058 [DOI] [PubMed] [Google Scholar]
- 24. Paulmichl M., Li Y., Wickman K., Ackerman M., Peralta E., Clapham D. (1992) Nature 356, 238–241 [DOI] [PubMed] [Google Scholar]
- 25. Pu W. T., Krapivinsky G. B., Krapivinsky L., Clapham D. E. (1999) Mol. Cell Biol. 19, 4113–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chari A., Golas M. M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., Fischer U. (2008) Cell 135, 497–509 [DOI] [PubMed] [Google Scholar]
- 27. Winkler C., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U. (2005) Genes Dev. 19, 2320–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. (2008) Cell 133, 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 30. Winkler C., Denker K., Wortelkamp S., Sickmann A. (2007) Electrophoresis 28, 2095–2099 [DOI] [PubMed] [Google Scholar]
- 31. Mitulović G., Smoluch M., Chervet J. P., Steinmacher I., Kungl A., Mechtler K. (2003) Anal. Bioanal. Chem. 376, 946–951 [DOI] [PubMed] [Google Scholar]
- 32. Kroiss M., Schultz J., Wiesner J., Chari A., Sickmann A., Fischer U. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10045–10050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grimmler M., Otter S., Peter C., Müller F., Chari A., Fischer U. (2005) Hum. Mol. Genet 14, 3099–3111 [DOI] [PubMed] [Google Scholar]
- 34. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geerlings T. H., Faber A. W., Bister M. D., Vos J. C., Raué H. A. (2003) J. Biol. Chem. 278, 22537–22545 [DOI] [PubMed] [Google Scholar]
- 36. Léger-Silvestre I., Milkereit P., Ferreira-Cerca S., Saveanu C., Rousselle J. C., Choesmel V., Guinefoleau C., Gas N., Gleizes P. E. (2004) EMBO J. 23, 2336–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rouquette J., Choesmel V., Gleizes P. E. (2005) EMBO J. 24, 2862–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schäfer T., Strauss D., Petfalski E., Tollervey D., Hurt E. (2003) EMBO J. 22, 1370–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vanrobays E., Gelugne J. P., Gleizes P. E., Caizergues-Ferrer M. (2003) Mol. Cell Biol. 23, 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zemp I., Wild T., O'Donohue M. F., Wandrey F., Widmann B., Gleizes P. E., Kutay U. (2009) J. Cell Biol. 185, 1167–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Allain F. H., Bouvet P., Dieckmann T., Feigon J. (2000) EMBO J. 19, 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ginisty H., Amalric F., Bouvet P. (1998) EMBO J. 17, 1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ginisty H., Serin G., Ghisolfi-Nieto L., Roger B., Libante V., Amalric F., Bouvet P. (2000) J. Biol. Chem. 275, 18845–18850 [DOI] [PubMed] [Google Scholar]
- 44. Raman B., Guarnaccia C., Nadassy K., Zakhariev S., Pintar A., Zanuttin F., Frigyes D., Acatrinei C., Vindigni A., Pongor G., Pongor S. (2001) Nucleic Acids Res. 29, 3377–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Herrmann F., Bossert M., Schwander A., Akgün E., Fackelmayer F. O. (2004) J. Biol. Chem. 279, 48774–48779 [DOI] [PubMed] [Google Scholar]
- 46. Tang J., Kao P. N., Herschman H. R. (2000) J. Biol. Chem. 275, 19866–19876 [DOI] [PubMed] [Google Scholar]
- 47. Ren J., Wang Y., Liang Y., Zhang Y., Bao S., Xu Z. (2010) J. Biol. Chem. 285, 12695–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teng Y., Girvan A. C., Casson L. K., Pierce W. M., Jr., Qian M., Thomas S. D., Bates P. J. (2007) Cancer Res. 67, 10491–10500 [DOI] [PubMed] [Google Scholar]
- 49. Vanrobays E., Gleizes P. E., Bousquet-Antonelli C., Noaillac-Depeyre J., Caizergues-Ferrer M., Gélugne J. P. (2001) EMBO J. 20, 4204–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Angermayr M., Hochleitner E., Lottspeich F., Bandlow W. (2007) FEBS J 274, 4654–4667 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.