Abstract
Recoding by adenosine-to-inosine RNA editing plays an important role in diversifying proteins involved in neurotransmission. We have previously shown that the Gabra-3 transcript, coding for the α3 subunit of the GABAA receptor is edited in mouse, causing an isoleucine to methionine (I/M) change. Here we show that this editing event is evolutionarily conserved from human to chicken. Analyzing recombinant GABAA receptor subunits expressed in HEK293 cells, our results suggest that editing at the I/M site in α3 has functional consequences on receptor expression. We demonstrate that I/M editing reduces the cell surface and the total number of α3 subunits. The reduction in cell surface levels is independent of the subunit combination as it is observed for α3 in combination with either the β2 or the β3 subunit. Further, an amino acid substitution at the corresponding I/M site in the α1 subunit has a similar effect on cell surface presentation, indicating the importance of this site for receptor trafficking. We show that the I/M editing during brain development is inversely related to the α3 protein abundance. Our results suggest that editing controls trafficking of α3-containing receptors and may therefore facilitate the switch of subunit compositions during development as well as the subcellular distribution of α subunits in the adult brain.
Keywords: Brain, Cell Surface Receptor, Double-stranded RNA, GABA Receptors, Receptor Modification, RNA Editing, Trafficking
Introduction
Adenosine to inosine (A-to-I)2 RNA editing is a mechanism used in the mammalian nervous system to provide alterations in the protein sequence by co-transcriptional modification of single nucleotides. This modification is catalyzed by adenosine deaminases that act on RNA (ADAR1 and ADAR2) that can selectively modify adenosine to inosine residues within double stranded pre-mRNAs. Within mRNA transcripts, inosine is read as guanosine by the translation machinery. Therefore, this mechanism has the potential to change the amino acid sequence and thereby the function of the protein. Several gene products encoding proteins involved in neurotransmission have been shown to be A-to-I edited, including ligand- and voltage-gated ion channels as well as a G-protein-coupled receptor and thereby creating diverse isoforms of proteins essential for balanced neuronal kinetics (reviewed in Ref. 1).
One of the most well studied substrates for editing in the brain is the transcript coding for the AMPA glutamate receptor (GluA). AMPA receptors consist of four subunits (GluA1–GluA4) in different combinations. Changing a codon for glutamine to arginine in GluA2 is essential to the organism and required for a normal brain development (2, 3).
We have previously found that the mouse Gabra-3 transcript, coding for the α3 subunit of the GABAA receptor undergoes site-selective A-to-I editing causing an isoleucine to methionine (I/M) change in the third transmembrane region (TM3) (4). The chloride-permeable (GABAA) receptors are the main mediators of fast inhibitory neurotransmission in the mammalian central nervous system (reviewed in Ref. 5). These heteropentameric ligand-gated chloride ion channels can be formed from at least 16 different subunits: α1–6, β1–3, γ1–3, δ, ϵ, θ, and π (6, 7). The functional diversity of GABAA receptors is based on subunit heterogeneity. Individual neurons express a range of subunits resulting in the formation of a large variety of structurally and functionally different receptor subtypes (7–9).
In the adult brain, the most common GABAA receptor is composed of two α1, two β2, and one γ2 subunit (7–9). However, subunit composition is different during embryogenesis and changes throughout development (9, 10). The major change occurs among the α subunits, where α1 expression increases with age and peaks in the adult animal, whereas the α3 subunit is the predominant subunit during early development but declines with age (11, 12). Receptors containing the α3 subunit are characterized by slow activation, desensitization, and deactivation and by low GABA sensitivity (13–16). These unique kinetic properties have been shown to play an important role in the maturation of developing neurons (17). Receptors containing the α3 subunit represent ∼10–15% of the total GABAA receptors in the adult brain (18).
The purpose of this study was to analyze whether editing at the I/M site influences the subcellular distribution of α3 containing GABAA receptors. We show that the developmental increase in editing at the I/M site is inversely related to the α3 protein expression. By using transiently expressed GABAA receptor subunits, we illustrate that I/M editing of α3 reduces cell surface and total numbers of this subunit. We hypothesize that A-to-I editing decreases the amount of α3 subunit containing GABAA receptors, possibly in favor of α1 containing receptors that is more abundant in the adult brain.
EXPERIMENTAL PROCEDURES
Generation of Recombinant GABAA Receptor Subunits
pRK5 plasmids containing cDNAs encoding rat α3(M) (the edited form), α1, β2, β3, and γ2L subunits were kind gifts from Professor Hartmut Lüddens (University of Mainz, Mainz, Germany). Expression vector pRK5-rα3(I) (unedited) was generated by replacing guanosine with adenosine at the I/M site of pRK5-rα3(M) (edited) using QuikChange site-directed mutagenesis (Stratagene). An adenosine-to-guanosine change at the same position as the I/M site of α3 was carried out on the Gabra-1 expression vector pPK5-rα1 by site-directed mutagenesis. This mutation of rα1(I315M) mimics an edited gabra-1. The enhanced GFP-tagged γ2L subunit expression vector was constructed by inserting the enhanced GFP (Clontech) into the large intracellular loop of γ2L between TM3 and TM4 at the EcoRV restriction site. The complete coding strand of the variant of the red fluorescent protein from Discosoma sp. (DsRed-Mono; Clontech) was inserted into the second MscI site in the large intracellular loop between TM3 and TM4 of the resultant protein of the expression vectors for α3(I) and α3(M), respectively. All of the fusion constructs were sequenced, functionally tested by patch clamp, and shown to resemble the characteristics of wild-type subunits (data not shown).
Transfection of Recombinant GABAA Receptor Subunits
HEK293 cells were grown at 37 °C in 5% CO2, 95% air in DMEM (Invitrogen) containing 10% FBS (Invitrogen) and 1% (100 units/ml) of penicillin and streptomycin (Invitrogen). Transfections using calcium phosphate or Lipofectamine 2000 (Invitrogen) with the GABAA subunit-encoded constructs were carried out in a 1:1:1 ratio or in the following ratio: 1.2 α3(I) or (M):10 β2:0.5 γ2L as described in Ref. 19. The cells were transfected with a total of 10 μg of (plasmid) DNA using calcium phosphate or with a total of 4 μg of plasmid DNA when Lipofectamine 2000 was used.
In Vitro Translation
In vitro transcripts were made using the MEGAscript (Ambion) from the SP6 promoter on the pRK5-rα3(I) and pRK5-rα3(M) plasmids. In vitro translation was made using radiolabeled [35S]methionine and rabbit retic lysate (Ambion) according to the protocol by the manufacturer.
Confocal Microscopy and Image Analysis
To analyze cell surface localized receptors, HEK293 cells were transfected with expression vectors for α3(I) or α3(M) as well as α1(WT) or α1(I315M) together with β2 or β3 and γ2 in 35-mm 6-well plates with coverslips. The cells were fixed in 4% paraformaldehyde and blocked in 3% BSA, 48 h after transfection. To determine the total protein expression, the cells were permeabilized with 0.5% Triton X-100 prior to blocking in BSA and incubated with primary antibodies followed by fluorescence-coupled secondary antibodies. Mounting medium (Vectashield; Vector Laboratories) was added, and the coverslip was mounted and analyzed. The primary antibodies were directed against α3 (Chemicon International) or α1 (Abcam) subunits, both antibodies recognizing the extracellular N terminus of the α subunits. The secondary antibody was a goat anti-rabbit TRITC (DakoCytomation). In the fusion fluorescent protein experiments, the cells were fixed with 4% paraformaldehyde and mounted.
To analyze the internalization of α3(I) and α3(M), the cells were transfected as in the experiments above and incubated with the anti-α3 antibody in buffer A (25 mm HEPES, pH 7.4, 119 mm NaCl, 5 mm KCL, 2 mm CaCl2, 2 mm MgCl2, 30 mm glucose) containing 3% BSA for 30 min at 4 °C. The cells were washed extensively with ice-cold buffer A and incubated for 60 min at 37 °C to allow internalization. Control cells were left at 4 °C, a condition that prevents internalization. The cells were then fixed for 10 min (4% paraformaldehyde containing 4% sucrose). The cells were rinsed with PBS and incubated with Alexa 488-conjugated secondary antibody (Invitrogen) for 30 min at room temperature. After washing with PBS, the cells were permeabilized for 10 min with 0.5% Triton X-100 in buffer A, followed by staining of internalized receptors with Alexa 555-conjugated secondary antibody (Invitrogen) for 30 min at room temperature. After washing, the cells were mounted and analyzed. The cells were imaged using a Zeiss LSM 510 META microscope with 40×, 63×, or 100× objectives. The confocal microscope settings were identical for all experiments, and all of the images presented in the figures were unprocessed. For each experiment single-blind tests were performed, and 100 cells from triplicate experiments were randomly chosen for quantification of fluorescence intensity using the ImageJ software (NIH Image). The background fluorescence was subtracted from the images.
Western Blot Analysis
Cell extracts from transfected HEK293 cells were obtained with Lysis-M (Roche Applied Science) containing a protease inhibitor mixture (Roche Applied Science). Crude protein extracts of whole mouse brain from E15, E19, P2, P7, P14, P21, and adult after P21 were obtained by homogenizing the brain in Lysis-M (Roche Applied Science) containing a protease inhibitor mixture. The samples were diluted 1:1 with Laemmli sample buffer (Bio-Rad), and 20 μg of total proteins from the transfected cells or 30 μg of proteins from whole brain were loaded onto a 10% SDS-PAGE and transferred to polyvinylidine fluoride membranes (Bio-Rad). After blocking in 5% (w/v) nonfat dry milk or 5% BSA (w/v) in Tris-buffered saline-Tween, the membranes were incubated with a polyclonal GABAA α3 antibody (Chemicon International) or polyclonal GABAA α1 antibody (Abcam). An actin antibody (Sigma) was used as an internal loading control. As secondary antibody, a horseradish peroxidase-coupled swine anti-rabbit antibody was used (DakoCytomation). The antibodies were detected using ECL Plus according to the manufacturer's instructions (GE Healthcare) and developed in a CCD camera (LAS 1000).
Cell Surface Biotinylation
HEK293 cells were transfected with α3(I) or α3(M) together with β2 and γ2L. An expression vector with a FLAG tag was used as a transfection control. After 48 h, the live cells were washed with PBS and incubated with 1.5 mg/ml sulfosuccinimidyl-2-(biotinamido) ethyl-1,3-dithiopropionate (sulfo-NHS-SS-biotin) using cell surface protein isolation (Pierce) for 30 min at 4 °C. The sulfo-NHS-SS-biotin was quenched, and the cells were lysed and incubated with immobilized strepavidin according to the manufacturer's description. Biotinylated proteins were eluted from the streptavidin by incubation with Laemmli sample buffer (Bio-Rad) at room temperature. The intracellular proteins and the cell surface biotinylated proteins were analyzed by Western blotting as described above. As a loading control of the biotinylated proteins, an anti-transferrin antibody (Biogenesis) was used. To determine the transfection efficiency between the different transfections, an anti-FLAG antibody (Sigma) was used on the intracellular protein lysates (data not shown).
Proteasome and Lysosome Inhibition
Transfections were performed as described above. After 24 h, the cells were treated with or without (controls) 250 μm epoxomicin (Biomol International) and/or 50 μm leupeptin (Sigma). After 16–20 h of incubation, the cells were prepared for image analysis with quantification of the fluorescence intensity as described above. The transfected epoxomicin-treated cells were also assessed by Western blotting as described above.
Analysis of Gabra-3 RNA Editing
Total RNA from adult whole brain was isolated from human, mouse, chicken, and frog (Xenopus laevis) using TRIzol (Invitrogen). From each sample 3 μg of total RNA was reverse transcribed by Superscript III RT (Invitrogen). The cDNAs from adult whole brain of dog, rabbit, and rat were purchased from Biochain. Primers specific for Gabra-3 of the different species were used in the PCR using Taq polymerase (Invitrogen). The PCR products were gel-purified and sequenced (Eurofins MWG Operon). For editing determination of Gabra-3 during development, the 454 sequencing method (Roche Applied Science) was used (20). Whole brains from NMRI mice were collected at five different developmental days: E15, E19, P2, P7, and P21 as described in Ref. 21. Total RNA was isolated as described above. The numbers of sequences collected for each developmental day were: E15 (592 sequences), E19 (901 sequences), P2 (971 sequences), P7 (68 sequences), and P21 (638 sequences). DIALIGN 2 was used to create multiple sequence alignments (22). The sequences of primers used in PCR amplification are available from the authors upon request.
Statistical Analysis
For comparison of fluorescence intensity between treatments, two-way analysis of variance analysis was performed, with the experimental day as a blocking factor. Separate analyses were done for each experiment. The data used were the experimental day averages of the cells log-transformed fluorescence intensity. The estimates of treatment differences together with 95% confidence intervals (CI) were back-transformed and expressed as ratios. All of the intensity measurements are shown as relative expression normalized to 1.
RESULTS
Editing of Gabra-3 Is Evolutionarily Conserved
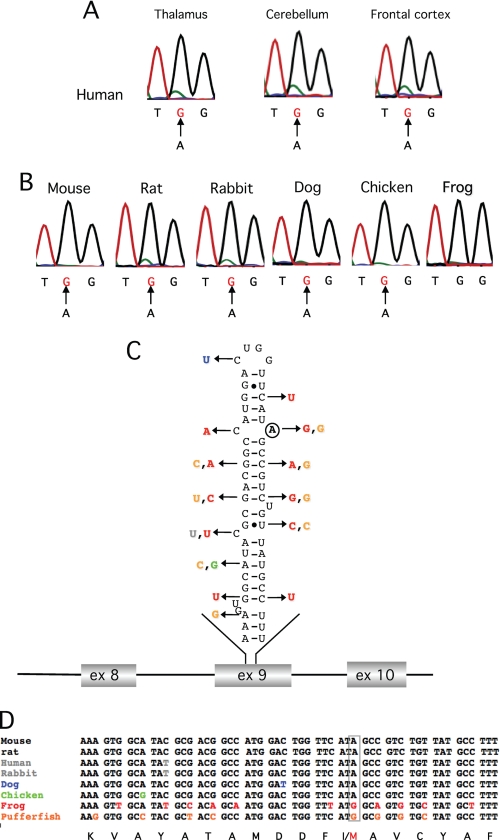
We sought to determine whether the RNA editing event in the Gabra-3 transcript coding for the GABAA receptor subunit α3 is phylogenetically conserved. Thus, cDNAs from the brain of adult rat, mouse, human, rabbit, dog, chicken, and frog were used as templates for PCR in the region of the putative I/M site in exon 9 of Gabra-3. The extent of editing at the I/M site was determined by direct DNA sequencing. In human brain samples from thalamus, cerebellum, and frontal cortex, the Gabra-3 transcripts were extensively edited, shown as an adenosine to guanosine (A to G) change in the chromatogram at the I/M site (Fig. 1A). A minor adenosine peak (green) indicates that few unedited Gabra-3 transcripts are present in these tissues. In the other species, total brain samples were analyzed. Editing was close to 100% in all species except pipid frog (Fig. 1B), which permanently codes for a methionine at this site because of a genomically encoded guanosine (Fig. 1D). Interestingly, both the structure and the sequence of the putative stem-loop required for editing within the coding sequence of exon 9 are conserved in all species with an edited Gabra-3 transcript (Fig. 1, C and D). The only variation is found in the transcript from chicken, where an A:U base pair is changed to a G:U base pair, upstream of the edited site, presumably with little effect on the structure. Taken together, the Gabra-3 transcript is efficiently edited in all species analyzed that have an adenosine at the I/M site.
FIGURE 1.
Conserved editing of the Gabra-3 transcript. A, frequency of editing at the I/M site in Gabra-3 in the thalamus, cerebellum, and frontal cortex of the human brain as indicated. The arrows indicate the A to G change as a result of A-to-I editing. B, editing frequency at the I/M site in Gabra-3 in total brain from mouse, rat, rabbit, dog, and chicken. Pipid frog has a genome encoded G at the I/M site and is therefore not edited. C, the predicted stem-loop structure of the Gabra-3 transcript at the I/M site in mouse (mGabra-3). A circle indicates the edited adenosine. The arrows indicate nucleotide substitutions in Gabra-3 of other species as shown in D. D, sequence alignment of the gabra-3 gene, showing the part of exon 9 that forms the putative stem loop in different species. The site of editing is boxed in gray. The amino acid sequence is shown at the bottom with the I/M site indicated.
Editing at the I/M Site of Gabra-3 Is Developmentally Regulated
We and others have previously shown that editing at the I/M site of Gabra-3 increases with age (4, 21, 23). It is also known that the α3 protein decreases during brain development (12). We wanted to investigate whether there is a correlation between Gabra-3 editing and the α3 protein levels. For a thorough investigation of editing frequency, we used our previously published data analyzing A-to-I editing within single Gabra-3 transcripts by the 454 sequencing technology (21). The advantage of 454 sequencing is its accuracy in determining the editing frequency, because exceedingly few edited transcripts are detected. We extended our previous 454 sequencing analysis during mouse development with an additional time point at P7. Thereby the sequence of over 3100 transcripts at five different developmental stages was analyzed, making the determination of editing levels highly accurate.
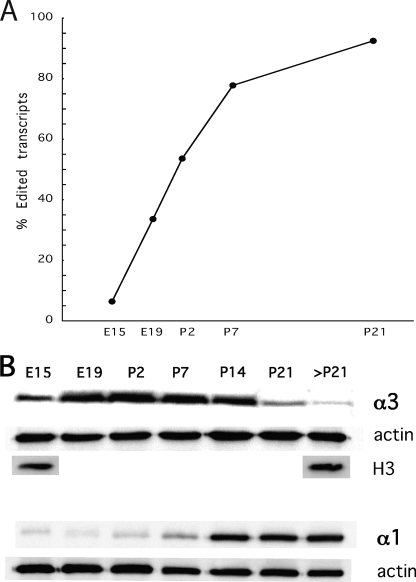
In all, RNA was analyzed from mouse brain at E15 and E19 as well as P2, P7, and P21. At E15, only 6.4% of the sequences analyzed had a G at the I/M site of Gabra-3 (Fig. 2A). The editing frequency increased throughout development, where 36% of the transcripts were edited at E19. After birth, editing increased gradually, reaching 54% at P2 and 78% at P7. Finally, 92% of the transcripts were edited in the mouse brain at P21.
FIGURE 2.
Editing of the Gabra-3 transcript and the protein levels of α3 and α1 during mouse brain development. A, the editing frequency at the I/M site of the Gabra-3 transcript was determined by 454 amplicon sequencing of RT-PCR products from whole-mouse brain at E15, E19, P2, P7, and P21. B, Western blot analysis on protein levels of α3 and α1 subunits during developmental day E15, E19, P2, P7, P14, P21, and P21 and after. Actin was detected and used as a loading control for both α3 and α1. To ensure that actin correlates to the same number of cells, histone H3 was detected and used as a loading control for α3.
To relate the editing frequencies to the amount of α3 protein, we performed Western blot analysis with an α3 antibody on total brain extracts, from developmental days E15, E19, P2, P7, P14, P21, and adult after P21. The amount of α3 protein was low at E15 but increased rapidly to the highest level between P2 and P7. At P14, the amount of α3 protein is reduced compared with P7 and in older mice α3 decreases even further (Fig. 2B). The total amount of α3 protein was compared with the α1 protein levels. The α1 protein concentration was low during early development and increased when the α3 protein level was decreasing, reaching the highest level in the adult brain (Fig. 2B). In summary, these results show that the accumulation of edited transcripts throughout development is inversely related to the reduction in α3 protein and related to the increase in α1 protein.
Gabra-3 Editing Affects α3 Cell Surface Presentation
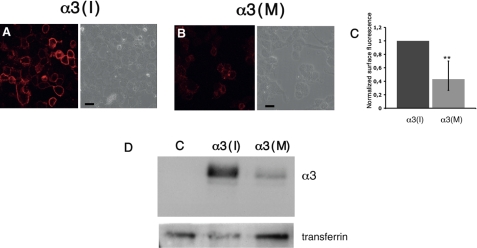
The isoleucine to methionine change after editing is located in the third of four transmembrane regions of the α3 subunit, in a region that is important for trafficking of GABAA α subunits (24, 25). To address cellular changes in trafficking or stability of the GABAA receptor, resulting from A-to-I editing, we first asked whether the event changed the surface distribution of the receptor. Expression vectors for the unedited α3(I) or edited α3(M), β2, and γ2L subunits of GABAA were co-transfected into HEK293 cells. To visualize the cell surface distribution of α3-containing receptors, nonpermeabilized cells were incubated with an α3 antibody recognizing the extracellular N-terminal of the protein. By confocal microscopy, the intensity of a TRITC-conjugated secondary antibody was analyzed. Fluorescence intensity was measured as described under “Experimental Procedures.” As seen in Fig. 3A (left panel), α3(I) was clearly expressed on the cell surface after transfection. Interestingly, the presence of α3 on the cell surface was vastly decreased after the amino acid change (Fig. 3B, left panel). The cell surface level of α3(M), quantified as fluorescence intensity, was reduced by almost 60% compared with α3(I) (n = 3, p < 0.01; Fig. 3C).
FIGURE 3.
Confocal microscopy showing cell surface presented α3-containing GABAA receptors. HEK293 cells were transfected with expression vectors for α3(I) or (M), β2, and γ2L subunits in the ratio 1.2:10:0.5. A, cell surface distributed α3(I) visualized in nonpermeabilized cells using an α3 antibody and a TRITC-conjugated secondary antibody. The phase contrast image of the cells is shown on the right. B, cell surface distribution of the α3(M) subunits as in A. The scale bars represent 10 μm. C, the relative reduction in fluorescence intensity of α3(M) from the experiments in A and B as described under “Experimental Procedures” with the rate of α3(I) normalized to 1 (n = 3). **, p < 0.01. The values reported are the means ± 95% CI. D, HEK293 cells transfected with: empty vector, β2, and γ2L (lane C); α3(I), β2, and γ2L (lane α3(I)); and α3(M), β2, and γ2L (lane α3(M)). The transfected cells were incubated with sulfo-NHS-SS-biotin, and the biotinylated proteins were analyzed by Western blot. The cell surface bound transferrin receptors was detected and used as a loading control.
To confirm that the low fluorescence intensity in cells expressing the α3(M)-containing receptor was due to a reduction in surface presentation, we performed cell surface biotinylation experiments. HEK293 cells were transfected with the GABAA subunit expression vectors as above and incubated with sulfo-NHS-SS-biotin. The biotinylated proteins were isolated as described under “Experimental Procedures.” The amount of membrane-bound α3(I) and α3(M) was determined by Western blot using the α3 antibody (Fig. 3D). The difference in surface distribution between α3(I) and (M) is remarkable. The change from Ile to Met in TM3 greatly reduces the number of α3 subunits in the membrane. Taken together, these results reveal that editing at the I/M site has a negative effect on the cell surface numbers of the α3 subunit.
Editing Affects the Presence of α3 Subunits
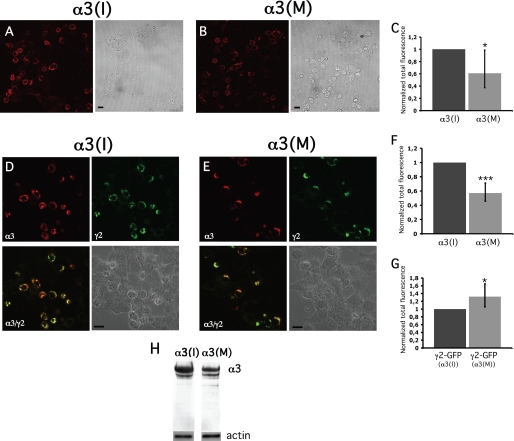
We wanted to investigate whether editing at the I/M site could serve as a regulator of the α3 protein expression. To investigate whether I/M editing has an effect on α3 protein stability, the transiently transfected cells were permeabilized prior to exposure of the α3 antibody. Quantification indicated a reduction in the total level of α3(M) compared with α3(I), although not as distinct as the reduction in cell surface presentation. (Fig. 4A, B and C). To ensure that the observed reduction in α3(M) expression was not due to a decrease in translation efficiency, the proteins were translated in vitro. As shown in supplemental Fig. S3, no difference in translation efficiency was observed when α3(I) and α3(M) were translated using retic lysate.
FIGURE 4.
Confocal microscopy showing total expression of the α3 subunits. HEK293 cells were transfected with expression vectors for α3 (I) or (M), β2, and γ2L subunits, ratio 1.2:10:0.5. A, total expression of the α3(I) subunit visualized in permeabilized cells using an α3antibody and a TRITC-conjugated secondary antibody. A phase contrast image is shown on the right. B, total expression of α3(M) subunit, visualized as in A. The scale bars represent 10 μm. C, the relative total level of α3(M) averaged to 0.61 when α3(I) was set to 1 (n = 3). D, total expression of α3(I)-DsRed (top left); γ2L-GFP (top right); and the merged pictures of α3-DsRed and γ2L-GFP (bottom left). Bottom right, the phase contrast image of the cells. The scale bar represents 10 μm. E, total expression of the α3(M)-DsRed and as in D. F, the relative fluorescence intensity of α3(M)-DsRed averaged to 0.57 when α3(I)-DsRed was normalized to 1 (n = 3). G, the relative fluorescence intensity of γ2L-GFP from the transfections in D and E. *, p < 0.05; ***, p < 0.001. The error bars represent 95% CI. H, Western blot analysis of whole cell lysates from transfections in D and E using an α3 antibody. Actin was detected and used as a loading control (lower panels).
In heterologous expression systems, α and β subunits alone can form receptors that are transported to the cell surface (26). To confirm that α3 assembles with the γ2L subunit, red and green fluorescent protein reporters were fused to α3 (α3-DsRed) and γ2L (γ2L-GFP), respectively, and transfected as above together with the β2 subunit. The α3(I)-DsRed protein co-localized with γ2L-GFP, indicating that the receptors contain all three subunit forms (Fig. 4D). The α3(M)-DsRed protein also co-localized with γ2L, although the intensity from α3(M)-DsRed-expressing cells was diminished compared with cells expressing α3(I)-DsRed (Fig. 4, E and F). This is in line with the results from the immunostaining of permeabilized cells.
To determine the transfection efficiency, cells were transfected with α3(I)-DsRed or α3(M)-DsRed together with the β2 and γ2L-GFP subunits and thereafter quantifying the fluorescent intensity of γ2L-GFP. This revealed that the reduced protein level of α3(M)-DsRed was not due to a lower transfection efficiency because the average amount of γ2L-GFP was similar or even slightly higher when co-transfected with α3(M)-DsRed than with α3(I)-DsRed or empty vector (Fig. 4G and supplemental Fig. S2).
Immunoblotting was used as a second method to determine the effect of editing on the expression of α3-DsRed subunits (Fig. 4H). Indeed, the concentration of α3(M) was reduced compared with α3(I) in line with the results from the immunocytochemical experiments. In conclusion, our results indicate that the recoded amino acid at the I/M site reduced the total concentration of α3 protein.
The Diminished α3(M) Surface Localization Is Independent of Subunit Composition
Our results on receptor presentation are based on transfection of vectors expressing specific GABAA subunits. It is therefore possible that the effect we observe is influenced by the subunit ratio and/or subunit combinations used in the transfections. In the experiments presented hitherto, the ratio between α3, β2, and γ2L has been 1.2:10:0.5 according to previously established conditions for receptor assembly (19). Recently the ratio (1:1:1) between the subunits α3, β3, and γ2L have been used to study the electrophysiological effects of α3 I/M editing (23, 27). To ensure that the effect that we are observing on surface presentation after editing also persists during these conditions, receptor subunit transfections were done using the 1:1:1 ratio between α3:β2:γ2L as well as the α3:β3:γ2L receptor subunits. The transfected cells were immunostained as in the cell surface experiments above. The different ratio or the replacement of β2 with β3 did not affect the reduced α3(M) surface distribution (supplemental Fig. S1). However, we observe a stronger signal at the surface for both the unedited and edited α3 in the α3:β3:γ2L transfection compared with α3:β2:γ2L, indicating that the former subunit combination assembled more efficiently. Taken together, these results clearly demonstrate that the observed reduction in α3(M) surface levels compared with α3(I) is independent of the subunit combination.
The Reduced α3(M) Surface Presentation Is Independent of Proteasomal Degradation
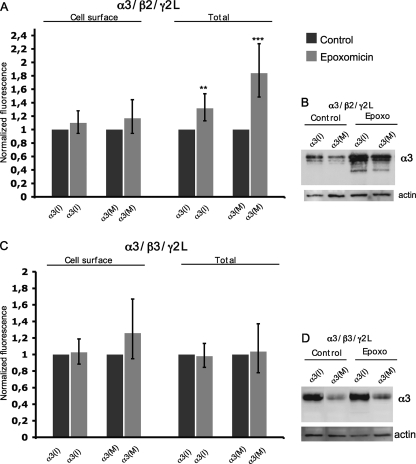
GABAA receptor subunits assemble in the endoplasmic reticulum (ER) via defined pathways (28). Misfolded and unassembled α1 subunits have been shown to be subjected to ER-associated degradation (29). Our initial results did not reveal whether the decrease in surface numbers of α3 after editing is due to facilitated protein degradation or alterations in receptor trafficking. To address this, we have analyzed whether editing at the I/M site triggers degradation of the α3 subunit by ER-associated degradation and the proteasome. Transfected cells expressing α3, β2, and γ2L subunits were treated with the proteasome inhibitor epoxomicin prior to the immunostaining. Cell surface presentation and the total levels of α3 in epoxomicin-treated cells were compared with untreated transfected cells and quantified using confocal microscopy. Total protein levels were also analyzed by Western blot. Quantification of both α3(I) and α3(M) revealed comparable surface levels in epoxomicin-treated as in control experiments (Fig. 5A), displaying reduced levels of α3(M) compared with α3(I) as seen in previous cell surface experiments (Fig. 3, A–C). This indicates that the reduced surface level of α3 after editing is not caused by proteasomal degradation.
FIGURE 5.
In assembly with β2 and γ2L, α3 is a proteasome substrate. Relative fluorescence intensity and Western blot analysis of α3 levels in control and epoxomicin-treated cells. A, cell surface and total α3 levels in cells transfected with α3(I) or α3(M), β2, and γ2L subunits, ratio (1:1:1) in untreated and epoxomicin-treated cells. The rate of epoxomicin-treated cells was normalized to untreated cells (n = 3). The values are represented as the means with 95% CI. Only minor effects on stabilization of both α3(I) and α3(M) were observed in epoxomicin-treated cells. An increase of the average relative intensity of α3(I) and α3(M) is seen when the total α3 expression was measured in epoxomicin-treated compared with untreated cells. B, Western blot analysis of whole cell lysates from the same cells as in A using an α3 antibody. C, the cell surface and total level of α3 in cells transfected with α3(I) or α3(M), β3, and γ2L subunits (n = 3). The relative cell surface intensity of α3(I) in epoxomicin-treated cells normalized to untreated cells is presented as indicated on the left. The relative intensity of the total α3(I) expression in epoxomicin-treated cells normalized to untreated cells is presented for α3(I) and α3(M) as indicated on the right. **, p < 0.01; ***, p < 0.001. The error bars represent 95% CI. D, Western blot analysis of whole cell lysates from the experiments represented in C using an α3 antibody. Actin was detected and used as a loading control.
To analyze total protein expression, transfected cells were permeabilized prior to immunostaining. When normalized to untreated cells, a stabilization of α3(I) was seen in epoxomicin-treated cells with the average intensity of 1.3 (n = 3, p = 0.005; Fig. 5A). An even more prominent stabilization of α3(M) was observed, with an average of 1.8 compared with the untreated cells normalized to 1 (n = 3; p = 0.0004; Fig. 5A), suggesting that α3(M) may be targeted by the proteasome to a larger extent than the α3(I) subunit. In Western blot analysis, a stabilization of both α3(I) and α3(M) in epoxomicin-treated cells compared with untreated cells was seen (Fig. 5B). Moreover, a facilitated degradation of α3(M) compared with α3(I) indicates that there is an additional degradation pathway that might affect receptor cell surface presentation (Fig. 5, A and B).
It has been demonstrated that the β3 subunit is important for the expression of the α3 subunit (30, 31). We wanted to investigate whether α3(M) is targeted for proteasome degradation in the presence of the β3 subunit. The cells were transfected with α3(I) or α3(M), β3, and γ2L and treated as in the above proteasome inhibition experiments. Consistent with the β2 experiment, we observe no stabilization of α3(M) on the surface after proteasome inhibition (Fig. 5C). Interestingly, confocal and Western blot analysis revealed that the total levels of both α3(I) and α3(M) were independent of the proteasome showing no stabilization after inhibition (Fig. 5, C and D). Thus, this result confirms our cell surface studies showing that α3 is more stable when assembled with β3 than with β2. Taken together, the reduced cell surface levels of the α3 subunit upon editing could not be explained by facilitated proteasomal degradation.
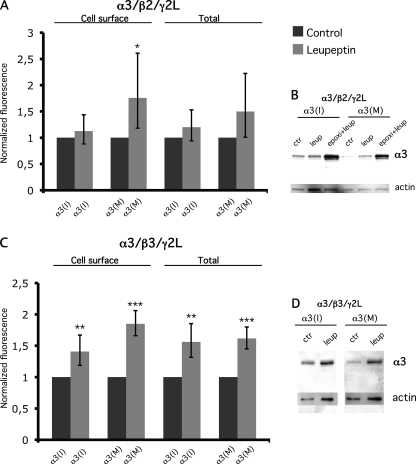
Blocking of the Lysosomal Pathway Increase α3(M) Surface Levels
To further explore the possibility of an increased protein degradation after I/M editing, we analyzed processing through the lysosomal pathway. To analyze the possible role of this pathway on the surface and total protein levels, we used the lysosomal protease inhibitor leupeptin. The cells were transfected as described above and treated with leupeptin for 16 h prior to immunostaining and Western blot. After lysosome inhibition, an increased surface level was seen for α3(M) but not α3(I) when assembled with the β2 subunit, indicating that the α3(M) subunit is less stable on the cell surface (Fig. 6). Only a minor increase in the total expression of α3(I) and α3(M) could be observed by immunostaining and Western blot after lysosome inhibition. To investigate whether the α3(M) subunit could be further stabilized, both the proteasome and the lysosome pathway was inhibited. A further increased stabilization could be observed for both α3(I) and α3(M) after the double inhibition using Western blot. The stabilization was more prominent for α3(M) than α3(I). This result indicates that the α3 subunit is targeted by both the proteasome and the lysosome when associated with the β2 subunit and that the α3(M) subunit is more susceptible for degradation. Interestingly, when α3 (I or M) was assembled with β3, an even more prominent stabilization of both α3(I) and α3(M) could be observed after lysosome inhibition at both the cell surface and total protein levels. It is noteworthy that the largest effect on stabilization was seen for α3(M) at the cell surface, indicating that the α3(M)-containing receptors are more susceptible for lysosomal degradation than those containing α3(I), affecting the cell surface presentation.
FIGURE 6.
Inhibition of the lysosomal pathway increase the surface levels of the α3(M) subunit. Relative fluorescence intensity and Western blot analysis of α3 in control and leupeptin-treated cells. A, cell surface and total α3 levels in control and leupeptin-treated cells transfected with α3(I) or α3(M), β2, and γ2L subunits, ratio (1:1:1). The rate of epoxomicin-treated cells was normalized to untreated cells (n = 3). The values are represented as the means with 95% CI. A more prominent stabilization of cell surface α3(M) was observed compared with α3(I) in leupeptin-treated cells. When total expression was measured, the average relative intensity of both α3(I) and α3(M) was increased in leupeptin-treated cells compared with controls, although the stabilization was stronger for the α3(M) subunit. B, Western blot analysis of whole cell lysates from the same cells as in A and experiments where the proteasome and lysosome were inhibited. ctr, control; epoxi, epoxomicin; leup, leupeptin. C, the cell surface and total level of α3 in cells transfected with α3(I) or α3(M), β2, and γ2L subunits (n = 3). The relative cell surface intensity of α3 in leupeptin-treated cells normalized to untreated cells is presented as indicated on the left. A stabilization of both α3(I) and α3(M) was observed. The relative intensity of the total α3 levels in leupeptin-treated cells normalized to untreated cells as indicated on the right. *, p < 0.05; **, p < 0.01; ***, p < 0.001. The error bars represent 95% CI. D, Western blot analysis of whole cell lysates from the experiments represented in C using an α3 antibody. Actin was detected and used as a loading control.
Increased Internalization of the α3(M) Subunit
We wanted to analyze whether the facilitated degradation of the α3(M) subunit was due to an altered surface stability. To identify receptor internalization, surface α3(I) or α3(M) subunits were prebound by the anti-α3 antibody at 4 °C and thereafter incubated at 37 °C, whereas controls were left at 4 °C to prevent internalization. Our results indicate that α3(I) and α3(M) internalize efficiently to an intracellular compartment. Internalization efficiency was measured by comparing cell surface α3 intensity on cells left at 4 and 37 °C using confocal microscopy. Our results indicate that the α3(M) subunit may internalize at a faster rate than α3(I), suggesting that the α3(M) subunit is less stable on the cell surface (supplemental Fig. S3).
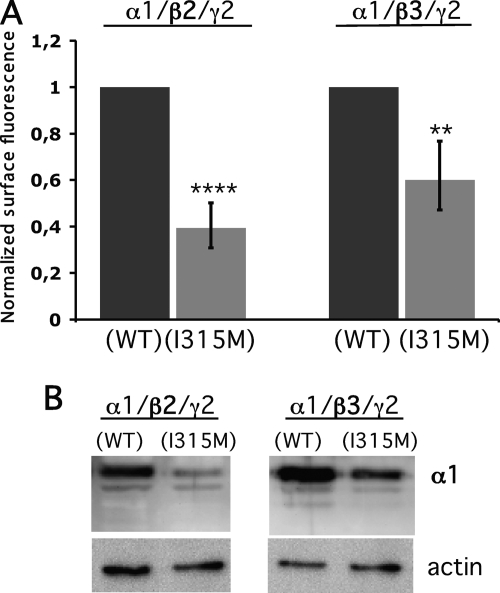
The Ile-to-Met Change Affects the α1 Cell Surface Distribution
The α3 subunit is one of six different subtypes of GABAA receptor α subunits. However, the Gabra-3 transcript coding for α3 is the only α subunit that is subjected to editing. The I/M site in α3 is located in a region that is highly conserved between the α subunits. All six α subtypes code for an Ile at this site in their genomes. To determine whether this site plays a general role in controlling receptor trafficking, we made the Ile-to-Met residue change in the rat α1 subunit, α1(I315M) to resemble an edited I/M site in α3. The cells were transfected with α1(WT) or α1(I315M), β2, and γ2L. In a separate experimental setup, β2 was replaced by β3. Cell surface-presented α1 was identified in nonpermeabilized cells using an α1 antibody. Confocal microscopy analysis revealed that the surface presentation of the mutated α1(I315M) in assembly with β2 was reduced >60% compared with α1(WT)-expressing cells (n = 3, p < 0.001; Fig. 7A). In assembly with β3, a 40% reduction of the α1(I315M) compared with the α1(WT) was observed (p < 0.01). Western blot analysis of the transfected cells, using the same α1 antibody, revealed that the total protein concentration of α1(I315M) was also vastly reduced compared with wild-type α1 (Fig. 7B). In summary, this supports the assumption that this residue in the α subunits is of general importance for the trafficking of GABAA receptors.
FIGURE 7.
Cell surface presentation of α1 is affected by a mutation mimicking the edited I/M site. Relative fluorescence intensity and Western blot analysis of wild-type α1(WT) or mutated α1(I315M) expression in HEK293 cells. A, average surface intensity of α1 when co-transfected with either β2 and γ2L or β3 and γ2L expression vectors in a 1:1:1 ratio. The relative cell surface intensity of α1(I315M) is presented when normalized to wild-type α1 (n = 3). **, p < 0.01; ***, p < 0.001. The error bars represent 95% CI. B, Western blot analysis of whole cell lysates from experiments represented in A. An α1 antibody was used to detect the total protein concentration. Actin was detected and used as a loading control.
DISCUSSION
The Nucleotide Sequence Is Evolutionarily Conserved in the Vicinity of the Edited Site
Here we demonstrate that the RNA editing event, giving rise to an isoleucine-to-methionine change in the α3 subunit of the GABAA receptor occurs in all species from chicken to human. In these species the nucleotide sequence in the vicinity of the I/M site is exceptionally conserved. Further, we show that the pipid frog does not edit the Gabra-3 transcript but rather encodes a guanosine (G) in its genome and thus a permanent methionine in the protein. Comparison of the genetic code of other species also revealed that the pufferfish (Tetradon nigroviridis) has a guanosine at the I/M site. A similar editing pattern has previously been observed in GluA2 at the Q/R site where the hagfish encodes an Arg codon in the genome (32, 33). In the gabra-3 sequence surrounding the G at the I/M site from frog and pufferfish, the nucleotides at the third wobble position in the genetic code are frequently different from the other species (Fig. 1D). Interestingly, the differences also give rise to a less stable Gabra-3 RNA stem-loop structure in the unedited compared with the edited species (Fig. 1C). This observation can be explained by the lack of selection pressure to preserve the nucleotide sequence required for editing. Similarly, editing at the R/G site of GluA2 has been shown to be conserved from man to bird with only minor phylogenetic variations in the sequence of the hairpin forming the substrate for editing, whereas the sequence of Tetradon with a guanosine at the R/G site is frequently different (34, 35). Recently, further examples have been identified in which A-to-I editing sites genomically encode guanosine in distantly related species (36). Our result further supports the use of this feature as a bioinformatic tool to search for novel sites of A-to-I editing.
The Effects of Developmental Regulation of I/M Editing in α3
We know from our previous studies and others that editing at the I/M site in Gabra-3 is developmentally regulated (4, 21, 23). In a previous work (21) and in the present study, we show by 454 high throughput sequencing that the editing frequency increase from 6.4% at embryonic day 15 to 92% at postnatal day 21. The number of edited transcripts at E15 is one-third of what was previously reported by Rula et al. (23). This might be due to the use of different mouse strains or the difficulty in determining lower editing levels with high accuracy using methods other than deep sequencing. According to their analysis, editing reaches a maximum at P7, whereas we show a continual increase until P21. The subunit combination of GABAA receptors change during brain development (9, 10, 37). This switch of subunits, resulting in a faster inhibitory signaling, is required for the maturation of developing neurons. Subunit α3 is one of the early subunits expressed during embryogenesis. These early receptors containing α3 activate and deactivate more slowly than receptors composed of other α subunits. Mechanisms by which the NMDA receptor activity controls the subunit switch have been suggested (38). However, the detailed mechanism underlying the developmental change is unclear. It has been shown that the α1 subunit increases at the expense of α3 during development (12, 38, 39). In mice this switch is initiated after P6, and the peak is near P14. In agreement with the subunit switch, we show that the majority of the transcripts coding for α3 are edited at P6 with a continuing increase until P21. It is known that the level of Gabra-3 transcripts decreases during the mammalian brain development (9, 23). Here we demonstrate that RNA editing of the Gabra-3 transcript increases concurrent with the decrease in α3 protein levels during development. We speculate that α3 editing is involved in the switch of the subunits. In our system we found that α3 editing alters the stability of the subunit, supporting the theory that the diminished α3 protein levels seen during development are partly regulated by editing. We suggest that RNA editing reduces the number of α3-containing receptors and thereby allows for an increase in α1-containing receptors.
The Relation between Editing of Gabra-3 and the Expression of α3
The GABAA subunits are synthesized, N-glycosylated, and assembled within the ER, where they associate with proteins required for assembly and transport to the plasma membrane (40–43). Receptors on the cell surface undergo endocytosis, a mechanism that rapidly modifies the number of receptors at the synapses (44). The internalized receptors are then either reinserted into the membrane or targeted for degradation (45).
We found that the editing event of the α3 transcripts result in GABAA receptors with altered subcellular distribution. Transient transfections of α3, β2, and γ2L subunits as well as α3, β3, and γ2L revealed a substantial decrease of cell surface-exposed α3(M) compared with α3(I) (Fig. 3 and supplemental Fig. S1). However, proteasome inhibition experiments revealed that the diminished α3(M) on cell surface is not due to facilitated ER-associated degradation (Fig. 5). Our results indicate that the α3(M) internalize more efficiently than α3(I). The reduced α3(M) surface level is due to an enhanced lysosomal degradation. These results indicate that α3(I) is more stable and therefore recycles to the cell surface to a higher extent than α3(M). However we cannot exclude the possibility that the reduced level of membrane-bound α3(M) also is due to less efficient assembly compared with the α3(I) subunit. Previous electrophysiological studies done using whole cell recordings demonstrate that the edited α3 produce receptors with smaller amplitudes, slower activation, and faster deactivation compared with receptors assembled with the unedited α3 (23, 27). Interestingly, the 2-fold current amplitude differences observed in these studies in cells expressing the edited α3 compared with unedited α3 could be due to fewer receptors on the cell surface. However, we cannot exclude the possibility that editing at the I/M site might have multiple functions. Even so, the reduced current amplitude seen for the α3(M)-containing receptors could be at least partly explained by reduced cell surface expression of the α3(M) subunit.
The TM3 Segment in the α Subunits Is Important for Trafficking of GABAA Receptors
As mentioned above, our results suggest that the amino acid change from Ile to Met in TM3 of the α3 subunit alters trafficking of the GABAA receptor. We found that this effect on cell surface presentation was not specific for the α3 subunit. A mutation in the α1 subunit (I315M) at the position corresponding to the α3 I/M site display reduced cell surface abundance similar to what was observed for the α3(M) subunit. These findings support the preceding evidence that the TM3 segment in α subunits is important for receptor trafficking. It has previously been shown that the mutation A322D in the α1 subunit reduces the cell surface and the total amount of α1 (24, 29). This mutation in α1 is situated only 5 amino acids downstream of the corresponding I/M site in the α3 subunit. The edited residue in α3 is located close to the extracellular transmembrane 2/3 linker. The change from isoleucine to methionine maintains a highly hydrophobic nature of the region but might affect the shape of the subunit and influence conformational stability of the receptor and/or the binding of GABAA receptor-interacting proteins. It is also possible that this amino acid change affects the interaction between the α and γ subunits similar to what was seen for the A322D mutation (24).
RNA editing has previously been shown to affect receptor assembly. In addition to channel gating, Q/R as well as R/G editing of GluA2 influences the assembly of the AMPA receptor by reducing self-assembly of GluA2 homotetramers (46, 47). Therefore, editing at both sites shifts assembly of GluA2 to preferred heteromerization. Because R/G editing of GluA2 is developmentally regulated in a similar way as α3, subunit heteromerization may be regulated with developmental progression (21, 48). Further, RNA editing has been shown to reduce the ability to tetramerize in the squid Kv potassium channel (49). Thus, RNA editing is frequently used to control receptor composition in neural receptors and thereby expand the diversity within the complex nervous system.
In summary, our results suggest an important role for RNA editing of the α3 subunit in balancing the spatial and temporal distribution of GABAA receptor subunits. This balance is also important during neurogenesis to achieve the proper subunit compositions required for essential changes in synaptic transmission.
Supplementary Material
Acknowledgments
We thank Dr. Hartmut Lüddens and his laboratory for reagents and fruitful discussions. We thank Dr. Patrick Young for help on the proteasomal work and for critically reading the manuscript. We are grateful to Dr. Jan-Olov Persson and Hicham Loukili for help on the statistical analysis as well as the 454 sequencing facility at the Royal Institute of Technology Genome Center (Stockholm, Sweden).
This work was supported by the Swedish Research Council.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- A-to-I
- adenosine to inosine
- I/M
- isoleucine to methionine
- TM
- transmembrane region
- En
- embryonic day n
- Pn
- postnatal day n
- CI
- confidence interval(s)
- TRITC
- tetramethylrhodamine isothiocyanate
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Jepson J. E., Reenan R. A. (2008) Biochim. Biophys. Acta 1779, 459–470 [DOI] [PubMed] [Google Scholar]
- 2. Brusa R., Zimmermann F., Koh D. S., Feldmeyer D., Gass P., Seeburg P. H., Sprengel R. (1995) Science 270, 1677–1680 [DOI] [PubMed] [Google Scholar]
- 3. Feldmeyer D., Kask K., Brusa R., Kornau H. C., Kolhekar R., Rozov A., Burnashev N., Jensen V., Hvalby O., Sprengel R., Seeburg P. H. (1999) Nat. Neurosci. 2, 57–64 [DOI] [PubMed] [Google Scholar]
- 4. Ohlson J., Pedersen J. S., Haussler D., Ohman M. (2007) RNA 13, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Farrant M., Kaila K. (2007) Prog. Brain Res. 160, 59–87 [DOI] [PubMed] [Google Scholar]
- 6. Baumann S. W., Baur R., Sigel E. (2001) J. Biol. Chem. 276, 36275–36280 [DOI] [PubMed] [Google Scholar]
- 7. Farrar S. J., Whiting P. J., Bonnert T. P., McKernan R. M. (1999) J. Biol. Chem. 274, 10100–10104 [DOI] [PubMed] [Google Scholar]
- 8. Benke D., Mertens S., Trzeciak A., Gillessen D., Mohler H. (1991) J. Biol. Chem. 266, 4478–4483 [PubMed] [Google Scholar]
- 9. Laurie D. J., Wisden W., Seeburg P. H. (1992) J. Neurosci. 12, 4151–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fritschy J. M., Paysan J., Enna A., Mohler H. (1994) J. Neurosci. 14, 5302–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bosman L. W., Rosahl T. W., Brussaard A. B. (2002) J. Physiol. 545, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hutcheon B., Fritschy J. M., Poulter M. O. (2004) Eur. J. Neurosci. 19, 2475–2487 [DOI] [PubMed] [Google Scholar]
- 13. Böhme I., Rabe H., Lüddens H. (2004) J. Biol. Chem. 279, 35193–35200 [DOI] [PubMed] [Google Scholar]
- 14. Barberis A., Mozrzymas J. W., Ortinski P. I., Vicini S. (2007) Eur. J. Neurosci. 25, 2726–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gingrich K. J., Roberts W. A., Kass R. S. (1995) J. Physiol. 489, 529–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Verdoorn T. A. (1994) Mol. Pharmacol. 45, 475–480 [PubMed] [Google Scholar]
- 17. Ortinski P. I., Lu C., Takagaki K., Fu Z., Vicini S. (2004) J. Neurophysiol. 92, 1718–1727 [DOI] [PubMed] [Google Scholar]
- 18. McKernan R. M., Whiting P. J. (1996) Trends Neurosci. 19, 139–143 [DOI] [PubMed] [Google Scholar]
- 19. Korpi E. R., Lüddens H. (1993) Mol. Pharmacol. 44, 87–92 [PubMed] [Google Scholar]
- 20. Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z., Dewell S. B., Du L., Fierro J. M., Gomes X. V., Godwin B. C., He W., Helgesen S., Ho C. H., Irzyk G. P., Jando S. C., Alenquer M. L., Jarvie T. P., Jirage K. B., Kim J. B., Knight J. R., Lanza J. R., Leamon J. H., Lefkowitz S. M., Lei M., Li J., Lohman K. L., Lu H., Makhijani V. B., McDade K. E., McKenna M. P., Myers E. W., Nickerson E., Nobile J. R., Plant R., Puc B. P., Ronan M. T., Roth G. T., Sarkis G. J., Simons J. F., Simpson J. W., Srinivasan M., Tartaro K. R., Tomasz A., Vogt K. A., Volkmer G. A., Wang S. H., Wang Y., Weiner M. P., Yu P., Begley R. F., Rothberg J. M. (2005) Nature 437, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wahlstedt H., Daniel C., Ensterö M., Ohman M. (2009) Genome Res. 19, 978–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morgenstern B. (1999) Bioinformatics 15, 211–218 [DOI] [PubMed] [Google Scholar]
- 23. Rula E. Y., Lagrange A. H., Jacobs M. M., Hu N., Macdonald R. L., Emeson R. B. (2008) J. Neurosci. 28, 6196–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gallagher M. J., Song L., Arain F., Macdonald R. L. (2004) J. Neurosci. 24, 5570–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guzmán J. N., Hernández A., Galarraga E., Tapia D., Laville A., Vergara R., Aceves J., Bargas J. (2003) J. Neurosci. 23, 8931–8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pritchett D. B., Sontheimer H., Gorman C. M., Kettenmann H., Seeburg P. H., Schofield P. R. (1988) Science 242, 1306–1308 [DOI] [PubMed] [Google Scholar]
- 27. Nimmich M. L., Heidelberg L. S., Fisher J. L. (2009) Neurosci. Res. 63, 288–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Connolly C. N., Krishek B. J., McDonald B. J., Smart T. G., Moss S. J. (1996) J. Biol. Chem. 271, 89–96 [DOI] [PubMed] [Google Scholar]
- 29. Gallagher M. J., Shen W., Song L., Macdonald R. L. (2005) J. Biol. Chem. 280, 37995–38004 [DOI] [PubMed] [Google Scholar]
- 30. Homanics G. E., DeLorey T. M., Firestone L. L., Quinlan J. J., Handforth A., Harrison N. L., Krasowski M. D., Rick C. E., Korpi E. R., Mäkelä R., Brilliant M. H., Hagiwara N., Ferguson C., Snyder K., Olsen R. W. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4143–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramadan E., Fu Z., Losi G., Homanics G. E., Neale J. H., Vicini S. (2003) J. Neurophysiol. 89, 128–134 [DOI] [PubMed] [Google Scholar]
- 32. Kung S. S., Chen Y. C., Lin W. H., Chen C. C., Chow W. Y. (2001) FEBS Lett. 509, 277–281 [DOI] [PubMed] [Google Scholar]
- 33. Wu Y. M., Kung S. S., Chen J., Chow W. Y. (1996) DNA Cell Biol. 15, 717–725 [DOI] [PubMed] [Google Scholar]
- 34. Aruscavage P. J., Bass B. L. (2000) RNA 6, 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ensterö M., Åkerborg O., Lundin D., Wang B., Furey T. S., Öhman M., Lagergren J. (2010) BMC Bioinformatics 11, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tian N., Wu X., Zhang Y., Jin Y. (2008) RNA 14, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Araki T., Kiyama H., Tohyama M. (1992) Neuroscience 51, 583–591 [DOI] [PubMed] [Google Scholar]
- 38. Henneberger C., Jüttner R., Schmidt S. A., Walter J., Meier J. C., Rothe T., Grantyn R. (2005) Eur. J. Neurosci. 21, 431–440 [DOI] [PubMed] [Google Scholar]
- 39. Liu Q., Wong-Riley M. T. (2006) Brain Res. 1098, 129–138 [DOI] [PubMed] [Google Scholar]
- 40. Baumann S. W., Baur R., Sigel E. (2002) J. Biol. Chem. 277, 46020–46025 [DOI] [PubMed] [Google Scholar]
- 41. Connolly C. N., Kittler J. T., Thomas P., Uren J. M., Brandon N. J., Smart T. G., Moss S. J. (1999) J. Biol. Chem. 274, 36565–36572 [DOI] [PubMed] [Google Scholar]
- 42. Gorrie G. H., Vallis Y., Stephenson A., Whitfield J., Browning B., Smart T. G., Moss S. J. (1997) J. Neurosci. 17, 6587–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wisden W., Laurie D. J., Monyer H., Seeburg P. H. (1992) J. Neurosci. 12, 1040–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kittler J. T., Delmas P., Jovanovic J. N., Brown D. A., Smart T. G., Moss S. J. (2000) J. Neurosci. 20, 7972–7977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kittler J. T., Thomas P., Tretter V., Bogdanov Y. D., Haucke V., Smart T. G., Moss S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Greger I. H., Khatri L., Kong X., Ziff E. B. (2003) Neuron 40, 763–774 [DOI] [PubMed] [Google Scholar]
- 47. Greger I. H., Khatri L., Ziff E. B. (2002) Neuron 34, 759–772 [DOI] [PubMed] [Google Scholar]
- 48. Lomeli H., Mosbacher J., Melcher T., Höger T., Geiger J. R., Kuner T., Monyer H., Higuchi M., Bach A., Seeburg P. H. (1994) Science 266, 1709–1713 [DOI] [PubMed] [Google Scholar]
- 49. Rosenthal J. J., Bezanilla F. (2002) Neuron 34, 743–757 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.