Abstract
The hereditary bone disorder osteogenesis imperfecta is often caused by missense mutations in type I collagen that change one Gly residue to a larger residue and that break the typical (Gly-Xaa-Yaa)n sequence pattern. Site-directed mutagenesis in a recombinant bacterial collagen system was used to explore the effects of the Gly mutation position and of the identity of the residue replacing Gly in a homogeneous collagen molecular population. Homotrimeric bacterial collagen proteins with a Gly-to-Arg or Gly-to-Ser replacement formed stable triple-helix molecules with a reproducible 2 °C decrease in stability. All Gly replacements led to a significant delay in triple-helix folding, but a more dramatic delay was observed when the mutation was located near the N terminus of the triple-helix domain. This highly disruptive mutation, close to the globular N-terminal trimerization domain where folding is initiated, is likely to interfere with triple-helix nucleation. A positional effect of mutations was also suggested by trypsin sensitivity for a Gly-to-Arg replacement close to the triple-helix N terminus but not for the same replacement near the center of the molecule. The significant impact of the location of a mutation on triple-helix folding and conformation could relate to the severe consequences of mutations located near the C terminus of type I and type III collagens, where trimerization occurs and triple-helix folding is initiated.
Keywords: Bacteria, Collagen, Protein Conformation, Protein Folding, Protein Stability, Mutation, Osteogenesis Imperfecta
Introduction
The close packing of the three chains in the collagen triple-helix structure requires that the smallest amino acid, Gly, occupies every third position and leads to the regular (Gly-Xaa-Yaa)n amino acid sequence pattern (1–3). The presence of missense mutations in collagen that change one Gly residue to a larger residue leads to hereditary connective tissue disorders (4). The most well characterized collagen disease is osteogenesis imperfecta (OI),2 a dominant disorder caused by mutations in the α1 or α2 chain of type I collagen, the major collagen in bone (5–7). Single-base substitutions in a Gly codon can lead to its replacement by one of eight bulkier residues, and most of these substituted residues are well represented in the more than 600 missense mutations in type I collagen reported to result in OI (7).
All OI cases are characterized by bone fragility and increased susceptibility to fracture, but the phenotype can vary from very mild to perinatal lethal (6, 7). In general, clinical severity is milder for mutations in the α2(I) chain compared with the α1(I) chain, and mutations of Gly to Arg, Val, Asp, and Glu are found more often in lethal cases than mutations to Ser, Cys, and Ala (7). A small degree of triple-helix destabilization has been reported for OI collagens expressed in fibroblasts (8). Delayed folding also appears to be a consequence of a Gly substitution (5, 9, 10). Type I collagen folding is initiated by trimerization of globular C-propeptides, followed by triple-helix nucleation and C-to-N-terminal triple-helix propagation (11). During collagen biosynthesis, post-translational modification of Pro to Hyp and of Lys to Hyl and glycosylation of Hyl take place only on unfolded collagen chains. The observation of increased post-translational modification N-terminal to the mutation in OI collagens suggests delayed folding at the mutation site (10). Abnormal folding is also supported by the decreased rate of triple-helix formation and collagen secretion in cultured OI fibroblasts and by increased degradation in the endoplasmic reticulum for an OI mouse model (9, 12).
The location of a type I collagen OI mutation with respect to the C-propeptide trimerization domain may affect the degree of folding delay and could be a factor in determining clinical severity (5, 6, 10, 13). Early studies noted that the clinical consequences of an identical Gly replacement depended on its location within the α1(I) chain. For instance, Starman et al. (13) reported that a Gly-to-Cys mutation at residue 94 of the triple helix led to a very mild form of OI, whereas Gly-to-Cys mutations at residues 526 and 718 led to severe and lethal forms of the disease, respectively. The local sequences around these different mutation sites vary and represent another possible factor in clinical severity (14).
Peptides have been excellent models for studying the impact of Gly substitutions (15, 16), but it is not possible to make significant changes in location in these relatively short peptides to assess the effect of position. A recombinant bacterial collagen is utilized here, in which a Gly mutation can be introduced at any location within the triple helix while controlling the sequence surrounding it. A construct based on the Scl2 protein from Streptococcus pyogenes (group A streptococcus serotype M28) was designed to contain the N-terminal globular domain (denoted as V), which is required for trimerization and refolding, followed by the collagen-like triple-helix domain that consists of a (Gly-Xaa-Yaa)79 sequence (denoted as CL) (17). Previous studies have shown that recombinant VCL expressed in Escherichia coli can be obtained in high yields and forms a stable triple helix with a melting temperature (Tm) of 36–38 °C, close to the stability found in human collagens (17–20). This stability is achieved in the absence of Hyp because bacteria do not appear to contain prolyl hydroxylases to carry out this post-translational modification. Here, site-directed mutagenesis was used to introduce a Gly-to-Arg or Gly-to-Ser mutation at a site near the middle or near the N terminus of the triple helix. All mutations led to small decreases in stability. Mutations near the center of the triple helix resulted in a significant delay in folding, whereas a mutation near the N terminus led to a far more dramatic decrease in the folding rate, together with significant local loosening of the triple-helix structure.
EXPERIMENTAL PROCEDURES
Construction of pColdIII-VCL with Gly Substitutions
pColdIII-VCL was constructed using pColdIII-163, encoding the p163 polypeptide based on Scl2.28 as described previously (20). A seven-residue thrombin/trypsin cleavage site was introduced between the V and CL domains to facilitate enzymatic isolation of the collagen domain (20). Versions of pColdIII-VCL with Gly substitutions were constructed by site-directed mutagenesis at residue 199 near the middle of the CL triple-helix domain and at residue 109, which is five triplets from the N-terminal end of the CL domain (see Fig. 1). The substitution of Gly at residue 199 with Arg and Ser was performed by PCR; the codon GGC (Gly) was changed to CGT (Arg) or AGC (Ser) at the position of the 199th amino acid in the VCL sequence to create VCL(G199R) or VCL(G199S), respectively. A Gly-to-Arg mutation was also made near the N terminus at Gly-109. The original sequence around Gly-109 is GPTGPA, but to better match the local environment of Gly-199, the codon for Thr-108 (ACC) was changed to ATG (Met), creating GPMGPA (protein denoted as VCL(T108M)). The substitution of Gly at residue 109 with Arg in VCL(T108M) was performed by PCR with the codon GGA (Gly) changed to CGA (Arg), yielding VCL(T108M,G109R). The mutations were confirmed by DNA sequencing.
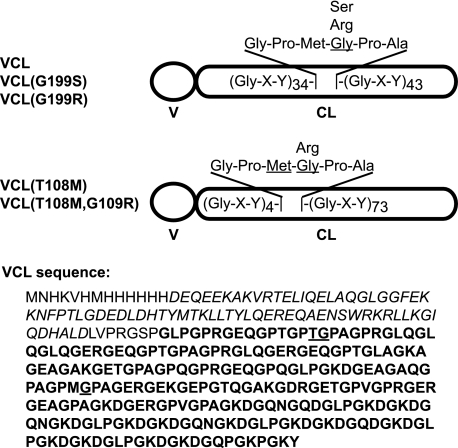
FIGURE 1.
Schematic illustration of the design of VCL constructs with Gly substitutions at different positions. The VCL amino acid sequence is shown below, with the trimerization V domain in italics, the triple-helix domain in boldface, and the Gly replacement locations underlined.
Expression of Bacterial Collagen with Gly Substitutions
Wild-type VCL, VCL(G199R), VCL(G199S), VCL(T108M), and VCL(T108M,G109R) were all expressed in the E. coli BL21 strain under the conditions described previously (20). Cells were harvested by centrifugation after overnight induction, and after cell disruption using a French press, the supernatant fraction with protein was obtained and analyzed by SDS-PAGE (20).
Purification of Bacterial Collagen VCL
The supernatant fraction was purified on a nickel-Sepharose resin column as described previously (20), and the purity was confirmed by SDS-PAGE. The purified proteins were dialyzed against PBS (pH 7.0). Protein concentrations were determined using an extinction coefficient at 275 nm of ϵ = 9579 m−1 cm−1.
Trypsin Digestion
VCL proteins in PBS (pH 7.0) were digested with trypsin at 25 °C for 5 min at a ratio of 1 mg of protein to 1 μg of trypsin as described previously (20). After digestion, 50 mm PMSF was used to inactivate trypsin, the material was centrifuged at 3000 rpm for 10 min, and the digestion products were analyzed by SDS-PAGE and MALDI-TOF mass spectrometry. CL triple-helix domains were obtained as described previously (20) and checked by SDS-PAGE.
Circular Dichroism Spectroscopy
CD spectra and thermal transitions were determined on an Aviv Model 62DS spectrophotometer as described previously (21). From a curve obtained at a heating rate of 0.1 °C/min, the Tm was defined as the temperature at which the fraction folded was equal to 0.5. For folding studies, samples were denatured at 55 °C for 20 min and then immediately transferred to CD cell holders pre-equilibrated at 0 °C. Previous studies established that both the N-terminal V domain and the collagen triple helix are fully denatured at this temperature. The dead time was on the order of 40 s. The ellipticity at 220 nm was recorded with a time constant of 2 s and a time interval of 10 s. The half-time of refolding (t½) was defined as the time for the fraction folded to reach 0.5.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) data were measured on a Nano-DSC II Model 6100 scanning calorimeter (Calorimetry Sciences Corp.) with a 1 °C/min heating rate as described previously (21).
Dynamic Light Scattering
Dynamic light scattering measurements were performed in a 12-μl quartz cuvette at 5 °C using a DynaPro Titan instrument (Wyatt Technology Corp., Santa Barbara, CA) equipped with a temperature controller. Before measurement, the samples were filtered through a 0.1-μm Whatman Anotop filter.
RESULTS
Recombinant Bacterial Collagens Designed to Include Gly Substitutions
Gly substitutions were introduced into a control protein that consisted of a portion of the collagen-like protein Scl2.28 from S. pyogenes. The control protein, referred to as VCL, contained an N-terminal globular trimerization domain (V) and a triple-helix domain of sequence (Gly-Xaa-Yaa)79 (Fig. 1). A Gly-to-Arg substitution was introduced at amino acid position 199, GPMGPA → GPMRPA, and the expressed protein was denoted as VCL(G199R). This mutation was located near the middle of the triple helix, with 43 Gly-Xaa-Yaa triplets C-terminal to the mutation and 35 Gly-Xaa-Yaa triplets N-terminal to the mutation (Fig. 1). To examine the effect of a different residue replacing Gly at the same position, a homologous protein with a Gly-to-Ser substitution was constructed and denoted as VCL(G199S).
To investigate the effect of the position of the mutation within the triple helix, an additional construct was made with a Gly-to-Arg substitution near the N terminus of the triple helix at amino acid position 109, with five Gly-Xaa-Yaa triplets N-terminal to the mutation and 73 triplets C-terminal to the mutation (Fig. 1). The original sequence around Gly-109 is GPTGPA. The immediate environment around a mutation site may be an important factor, so the Thr N-terminal to Gly-109 was changed to Met to create a GPMGPA local sequence comparable with that seen for Gly-199. The construct with only a Thr-to-Met substitution at position 108 was expressed as a control and denoted as VCL(T108M), whereas the mutant construct was designated as VCL(T108M,G109R).
Triple-helix Formation and Resistance to Trypsin
The well established resistance of triple-helix collagen to digestion by enzymes such as trypsin was used to probe the triple-helix nature of the constructs based on VCL (22). Treatment of VCL, VCL(G199S), VCL(G199R), and VCL(T108M) proteins with trypsin at room temperature led to a single SDS-PAGE band (Fig. 2), and the molecular weight as determined by mass spectroscopy is consistent with the mass expected for the full-length CL triple-helix domain: 22,813 observed versus 22,840 predicted for CL; 22,910 observed versus 22,939 predicted for CL(G199R); 22,840 observed versus 22,870 predicted for CL(G199S); and 22,955 observed versus 22,870 predicted for CL(T108M).
FIGURE 2.
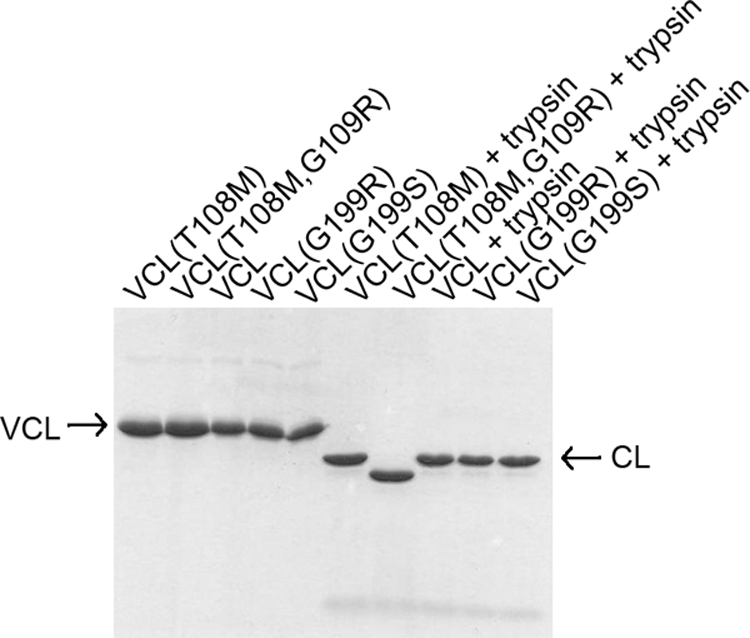
SDS-PAGE of VCL proteins as purified and after trypsin digestion. The lower band after trypsin digestion shows that the V domains were cleaved by trypsin, leaving the CL triple-helix domain intact. The band of VCL(T108M,G109R) after trypsin digestion is lower than the bands of the other control and mutant proteins, indicating that the mutation allowed partial cleavage of the CL domain; mass spectroscopy indicated that cleavage occurred at Arg-114.
Trypsin digestion of the protein with a Gly-to-Arg mutation near the N terminus, VCL(T108M,G109R), showed a band that ran faster than the trypsin products of all other constructs on SDS-PAGE (Fig. 2). Mass spectrometry gave a value of 20,683, indicating a trypsin cleavage site at Arg-114 in the seventh triplet in from the beginning of the triple-helix (Gly-Xaa-Yaa)n domain. The G109R substitution near the N terminus of the triple helix apparently leads to local unwinding near the mutation site or unwinding of the N-terminal residues of the triple helix up to residue 114.
CD and DSC Studies of Conformation and Stability of Bacterial Collagen with Gly Substitutions
CD spectra of proteins with Gly substitutions, VCL(G199R), VCL(G199S), and VCL(T108M,G109R), were very similar to those of the controls, with the characteristic triple-helix maximum at 220 nm and a minimum at 198 nm (Table 1), indicating that Gly substitutions did not result in any major perturbation of the overall conformation. The CD spectra were recorded for collagen domains of the control and mutant proteins isolated from the full-length proteins by trypsin digestion: CL, CL(G199S), and CL(G199R). The Rpn (ratio of the intensity of the positive peak near 220 nm over the intensity of the negative peak near 198 nm) values of the collagen domains were assessed to determine triple-helix content (23). The control and mutant collagen domains all had Rpn values of 0.13, consistent with fully folded triple helices.
TABLE 1.
Physical properties of bacterial collagen protein with Gly replacements (PBS at pH 7.0)
| Protein | MRE220 | MRE198 |
Tm |
t½ | |
|---|---|---|---|---|---|
| CD | DSC | ||||
| degrees cm2 dmol−1 | degrees cm2 dmol−1 | °C | min | ||
| VCL | 1340 | −34,400 | 38.5 | 37.3 | 10 |
| VCL(G199R) | 1320 | −36,700 | 36.5 | 34.4 | 55 |
| VCL(G199S) | 1275 | −38,600 | 37.0 | 34.5 | 50 |
| VCL(T108M) | 1350 | a | 38.3 | 37.4 | 10 |
| VCL(T108M,G109R) | 1250 | a | 36.7 | 36.6 | >1000 |
a The low wavelength data at 198 nm were not calculated due to increased light scattering.
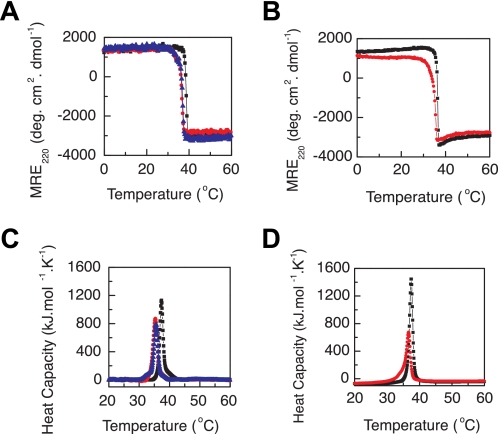
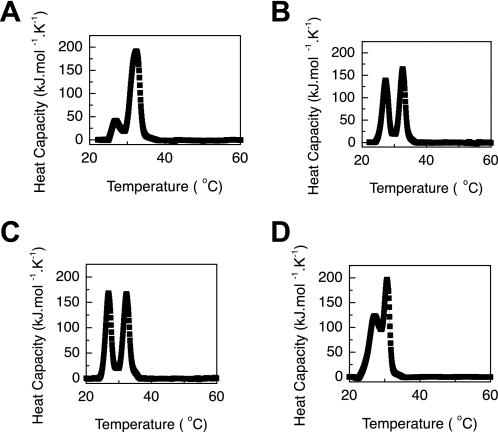
Gly substitutions at position 199 in the VCL(G199R) and VCL(G199S) proteins and at position 109 in the VCL(T108M,G109R) protein resulted in a small but reproducible decrease in thermal stability (ΔTm) of 1.5∼2.0 °C as measured by CD (Fig. 3, A and B, and Table 1). DSC thermal transitions showed values similar to those observed by CD, with a 1–3 °C decrease as a result of the Gly substitutions (Fig. 3, C and D). The purified collagen domains with mutations G199R and G199S also showed similar small decreases in thermal stability (Fig. 4).
FIGURE 3.
Thermal transition of VCL proteins by CD (A and B) and DSC (C and D) showing that Gly-to-Arg and Gly-to-Ser mutations at different positions lead to small but significant decreases in stability (PBS at pH 7). A, CD thermal transitions for VCL (black), VCL(G199R) (red), and VCL(G199S) (blue) (0.35 mg/ml). B, CD thermal transitions for VCL(T108M) (black) and VCL(T108M,G109R) (red) (1.0 mg/ml). C, DSC profiles of VCL (black), VCL(G199R) (red), and VCL(G199S) (blue) (0.35 mg/ml). D, DSC profiles of VCL(T108M) (black) and VCL(T108M,G109R) (red) (0.5 mg/ml). deg, degrees.
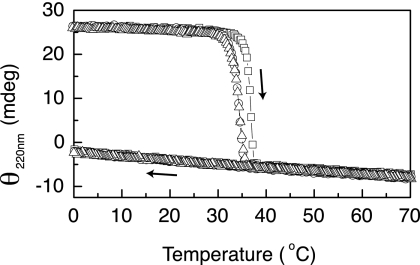
FIGURE 4.
CD thermal transitions of the collagen CL domains of control and mutant proteins. The sharp cooperative transitions observed with increasing temperature (right arrow) show the same small decrease in Tm values as a result of mutations as seen for VCL proteins. Cooling at the same rate (left arrow) led to only a small linear increase in ellipticity, suggesting no correct refolding in the absence of the V domain. Squares, CL; circles, CL(G199R); triangles, CL(G199S). (PBS at pH 7.0, 0.35 mg/ml). The observed raw ellipticity values are plotted versus temperature, rather than mean residue ellipticity (MRE), because of the difficulty in obtaining a reliable value of the concentration for the collagen domains, which contain no aromatic residues. Various estimates of the concentration through weighing and absorbance at 214 nm are consistent with estimates obtained by subtracting the V domain from the VCL spectrum and suggest that MRE220 ∼ 7500 degrees cm2 dmol−1 for the CL domains. mdeg, millidegrees.
Gly Mutations Delay in Vitro Refolding
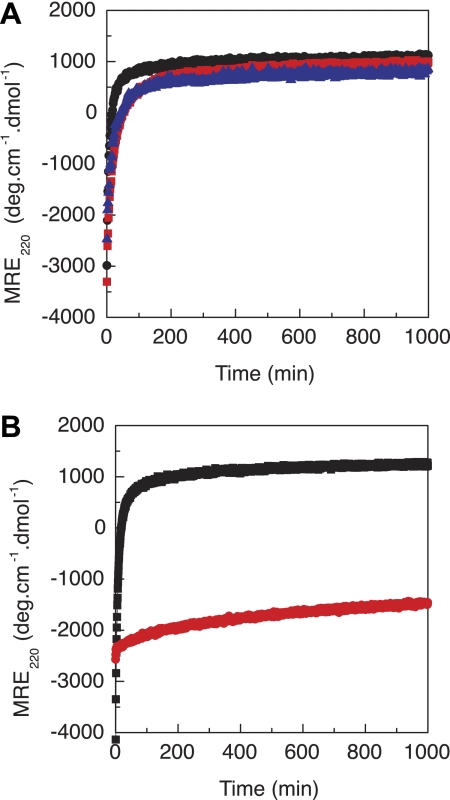
To monitor refolding, recombinant bacterial collagen proteins were denatured at 55 °C for 20 min (PBS, pH 7, 0.35 mg/ml) and quickly cooled to 0 °C, at which the recovery of the characteristic triple-helix CD maximum at 220 nm was monitored as a function of time. The replacement of Gly-199 with Arg or Ser resulted in a significant delay in folding time (Fig. 5A and Table 1), increasing from t½ = 10 min for VCL to t½ = 55 min for VCL(G199R) and t½ = 50 min for VCL(G199S). Replacement of Gly with Arg near the N terminus in the VCL(T108M,G109R) mutant led to a much more dramatic decrease in the rate of folding compared with mutations near the middle of the triple helix (Fig. 5B). The folding was extremely slow and did not reach 50% folding within the experiment time of 16 h, so t½ could not be calculated. In the absence of the trimerization V domain, all isolated CL domains showed little refolding after heat denaturation, although some signal that may represent misfolded species was regained after 1 week (see reverse transitions in Fig. 4).
FIGURE 5.
Monitoring the refolding of VCL proteins by CD, plotted as MRE220versus time. A, VCL (black), VCL(G199R) (red), and VCL(G199S) (blue) (PBS at pH 7.0, 0.35 mg/ml). B, VCL(T108M) (black) and VCL(T108M,G109R) (red) (PBS at pH 7.0, 0.5 mg/ml). Samples were denatured at 55 °C for 20 min and then rapidly cooled to 0 °C, at which the CD signal at 220 nm was measured. The half-time of refolding (t½) is defined as the time for the fraction folded to reach 0.5. The data are expressed as MRE220 values versus time rather than as fraction folded because the MRE220 value is a combination of decreasing values during the rapid refolding of the α-helix V domain and increasing values during the slower refolding of the triple helix, which presumably occurs after the V domain is folded. deg, degrees.
Effect of Acidic pH on VCL with Mutations
The VCL protein has a very high content of charged residues, with a pI of 6.10, and the properties of the bacterial collagen constructs in 0.1 m acetic acid solutions (pH 2.9) were investigated. The CD spectra of VCL, VCL(G199R), and VCL(G199S) in acetic acid all showed similar spectra with maxima at 220 nm and minima at 198 nm (data not shown), which means that the bacterial collagen has a typical triple-helix conformation at low pH that is not affected by the Gly substitutions. However, a significant decrease in triple-helix stability was observed, as well as changes in domain structure and/or cooperativity at acid pH. DSC of the control VCL protein in acid showed a large transition peak at 32 °C and a smaller transition near 27.0 °C. The homologous proteins with Gly substituted with Arg and Ser at residue 199 also showed two peaks at the same positions, but the intensity of the lower temperature transition was substantially increased to give two almost equal peaks (Fig. 6), whereas the VCL(T108M,G109R) construct was more similar to the control in having a dominant peak at the higher temperature. After the VCL and VCL(G199R) proteins were heated at 55 °C for 20 min in acetic acid buffer, no refolding occurred upon cooling. This is consistent with the previous observation that the V domain is unfolded and unable to trimerize at acid pH (24).
FIGURE 6.
DSC profiles of samples in 0.1 m acetic acid showing decreased thermal stability and multiple thermal transitions at acid pH. A, VCL; B, VCL(G199R); C, VCL(G199S); D, VCL(T108M,G109R).
DISCUSSION
OI collagens synthesized by fibroblasts always represent a mixture of molecular species, containing zero, one, or two mutant chains, and it is not feasible to purify a single molecular species containing mutations in amounts needed for study. The recombinant bacterial collagen system makes it possible to obtain homogeneous proteins containing a Gly mutation at a specific site within a defined triple-helix environment. Sufficient amounts of protein can be obtained from this expression system to permit in vitro folding studies and other methods of biophysical characterization. In addition, the length of the bacterial collagen domain and the presence of the natural trimerization domain in the VCL protein system make its folding mechanism similar to that of native type I collagen, unlike model peptides that were previously used.
The observed decrease of ∼2 °C in thermal stability (ΔTm) for the bacterial triple helix with a Gly substitution is similar to ΔTm values reported by the Leikin laboratory for fibroblast OI collagens with a mutation in just one chain (8). Their DSC studies on 28 OI collagens showed that replacement of Gly with Ser, Cys, Arg, Val, Asp, or Glu in one α1(I) chain decreases the Tm value by an average of 2.3 °C (ranging from 0.8 to 4.6 °C). The similarity of the ΔTm values for the homotrimeric recombinant bacterial system containing Gly mutations in all three chains to the ΔTm values seen for OI type I heterotrimers containing one or two chains with mutations suggests that the stability decrease is not significantly affected by the number of chains with mutations. This is consistent with studies on model peptides that showed that the first Gly-to-Ser replacement leads to a major decrease in stability, whereas Gly-to-Ser mutations in the second and third chains cause significantly less additional destabilization (25).
In addition, the degree of destabilization does not appear to be significantly affected by the different amino acid sequence/composition of the bacterial collagen protein compared with animal collagens. Although the overall composition of the bacterial CL domain is more highly charged and acidic than that of the human type I collagen (19), the local amino acid sequence environments near the sites selected for mutations are not very different from those found in some sites of OI mutations in human collagens. For instance, the immediate sequence around position 199, GPAGPMGPAGERGEK, is similar to that surrounding the G626S mutation site in the α1(I) chain, which leads to a mild form of OI, GPPGPAGPAGERGEQ (7).
The replacement of Gly with Ser at residue 199 in bacterial collagen showed the same ∼2 °C stability decrease seen for the Gly-to-Arg mutation. Although studies of model peptides have shown that the degree of destabilization depends on the identity of the residue replacing Gly (26, 27), Makareeva et al. (8) observed that the degree of destabilization of mutant collagens obtained from fibroblasts of patients with OI did not correlate with the identity of the residue replacing Gly. The lack of effect of the identity of the residue replacing Gly in bacterial collagen as well as OI collagens supports the concept that destabilizing effects of mutations are magnified in peptide models because of their shorter length (typically ∼10–15 tripeptides). Although the bacterial collagen triple helix is only 79 triplets long, compared with 338 triplets in type I collagen, it appears to be long enough to be a good model for this feature.
The in vitro refolding of the bacterial VCL triple helix has been shown to require the N-terminal globular trimerization V domain (19, 24), a feature reminiscent of the requirement for C-terminal C-propeptide trimerization prior to type I collagen folding (11). By analogy to type I collagen, it is probable that trimerization at the N-terminal V domain holds the three chains in proximity and facilitates nucleation of the triple helix, which is followed by directional propagation of the triple helix. Preliminary studies indicate that the isolated V domain refolds very rapidly, within 1 min.3 The control VCL protein folds more slowly than the isolated V domain (t½ = 10 min), and more than one kinetic step can be observed. Possible slow steps include nucleation of the triple helix and propagation of the triple helix limited by cis,trans-proline isomerization. Because trimerization occurs at the N-terminal V domain, nucleation of the CL triple helix is likely to occur near the N terminus of the (Gly-Xaa-Yaa)79 sequence, where there is an unusual stretch of Pro-containing triplets (GSPGLPGPRGEQGPTGPTGPAGPR).
Gly mutations near the center of the CL domain lead to slower folding rates (t½ = 50–55 min), whereas a Gly mutation near the N terminus hinders the folding so dramatically that t½ cannot be obtained. All Gly mutations successfully model the folding delay seen for OI type I collagens (9, 10). The observation that a Gly-to-Arg mutation introduced five tripeptides from the N-terminal trimerization V domain results in a much greater decrease in folding rate than seen for a mutation near the center of the triple helix suggests that it interferes with the nucleation of the triple helix. Similar interference with triple-helix nucleation of Gly mutations located near the trimerization domain in fibril-forming collagens could explain their severe clinical outcome. For instance, examination of mutations near the C terminus of the α1 chain of type I collagen, where nucleation of the triple helix occurs, shows a preponderance of lethal versus non-lethal OI cases (28). In addition, Gly mutations near the C terminus of type III collagen, where disulfide bonding and triple-helix nucleation take place, were observed to have particularly severe consequences in terms of collagen loss, secretion, and fibril structure (29). Interference of Gly mutations near the trimerization domain with triple-helix nucleation could result in dramatic folding delays such as seen in the bacterial collagen model. An extreme folding delay could affect collagen secretion and degradation and thus relate to their severe phenotype.
Acknowledgments
We acknowledge the helpful discussions of Drs. Jean Baum, Jianxi Xiao, Marion Gordon, and Sergey Leikin and the technical support of Teresita Silva.
This work was supported, in whole or in part, by National Institutes of Health Grant GM60048 (to B. B.) and Training Grant T32 GM008360 (to E. H.). This work was also supported by a China Scholarship Council fellowship (to H. C.).
E. Hwang and Z. Yu, unpublished data.
- OI
- osteogenesis imperfecta
- DSC
- differential scanning calorimetry.
REFERENCES
- 1. Ramachandran G. N. (1967) Treatise on Collagen, Academic Press, New York [Google Scholar]
- 2. Rich A., Crick F. H. (1961) J. Mol. Biol. 3, 483–506 [DOI] [PubMed] [Google Scholar]
- 3. Brodsky B., Persikov A. V. (2005) Adv. Protein Chem. 70, 301–339 [DOI] [PubMed] [Google Scholar]
- 4. Myllyharju J., Kivirikko K. I. (2004) Trends Genet. 20, 33–43 [DOI] [PubMed] [Google Scholar]
- 5. Byers P. H. (2001) Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 151–7; discussion 157–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Byers P. H., Cole W. G. (2002) Connective Tissue and Its Hereditable Disorders: Molecular, Genetic and Medical Aspects (Royce P. M., Steinmann B. eds) 2nd Ed. pp. 385–430, Wiley-Liss, New York [Google Scholar]
- 7. Marini J. C., Forlino A., Cabral W. A., Barnes A. M., San Antonio J. D., Milgrom S., Hyland J. C., Körkkö J., Prockop D. J., De Paepe A., Coucke P., Symoens S., Glorieux F. H., Roughley P. J., Lund A. M., Kuurila-Svahn K., Hartikka H., Cohn D. H., Krakow D., Mottes M., Schwarze U., Chen D., Yang K., Kuslich C., Troendle J., Dalgleish R., Byers P. H. (2007) Hum. Mutat. 28, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makareeva E., Mertz E. L., Kuznetsova N. V., Sutter M. B., DeRidder A. M., Cabral W. A., Barnes A. M., McBride D. J., Marini J. C., Leikin S. (2008) J. Biol. Chem. 283, 4787–4798 [DOI] [PubMed] [Google Scholar]
- 9. Raghunath M., Bruckner P., Steinmann B. (1994) J. Mol. Biol. 236, 940–949 [DOI] [PubMed] [Google Scholar]
- 10. Bonadio J., Byers P. H. (1985) Nature 316, 363–366 [DOI] [PubMed] [Google Scholar]
- 11. Engel J., Prockop D. J. (1991) Annu. Rev. Biophys. Biophys. Chem. 20, 137–152 [DOI] [PubMed] [Google Scholar]
- 12. Forlino A., Kuznetsova N. V., Marini J. C., Leikin S. (2007) Matrix Biol. 26, 604–614 [DOI] [PubMed] [Google Scholar]
- 13. Starman B. J., Eyre D., Charbonneau H., Harrylock M., Weis M. A., Weiss L., Graham J. M., Jr., Byers P. H. (1989) J. Clin. Invest. 84, 1206–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bächinger H. P., Morris N. P., Davis J. M. (1993) Am. J. Med. Genet 45, 152–162 [DOI] [PubMed] [Google Scholar]
- 15. Baum J., Brodsky B. (1999) Curr. Opin. Struct. Biol. 9, 122–128 [DOI] [PubMed] [Google Scholar]
- 16. Bhate M., Wang X., Baum J., Brodsky B. (2002) Biochemistry 41, 6539–6547 [DOI] [PubMed] [Google Scholar]
- 17. Xu Y., Keene D. R., Bujnicki J. M., Höök M., Lukomski S. (2002) J. Biol. Chem. 277, 27312–27318 [DOI] [PubMed] [Google Scholar]
- 18. Lukomski S., Nakashima K., Abdi I., Cipriano V. J., Ireland R. M., Reid S. D., Adams G. G., Musser J. M. (2000) Infect. Immun. 68, 6542–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mohs A., Silva T., Yoshida T., Amin R., Lukomski S., Inouye M., Brodsky B. (2007) J. Biol. Chem. 282, 29757–29765 [DOI] [PubMed] [Google Scholar]
- 20. Yoshizumi A., Yu Z., Silva T., Thiagarajan G., Ramshaw J. A., Inouye M., Brodsky B. (2009) Protein Sci. 18, 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Persikov A. V., Xu Y., Brodsky B. (2004) Protein Sci. 13, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bruckner P., Prockop D. J. (1981) Anal. Biochem. 110, 360–368 [DOI] [PubMed] [Google Scholar]
- 23. Feng Y., Melacini G., Goodman M. (1997) Biochemistry 36, 8716–8724 [DOI] [PubMed] [Google Scholar]
- 24. Yu Z., Mirochnitchenko O., Xu C., Yoshizumi A., Brodsky B., Inouye M. (2010) Protein Sci. 19, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gauba V., Hartgerink J. D. (2008) J. Am. Chem. Soc. 130, 7509–7515 [DOI] [PubMed] [Google Scholar]
- 26. Beck K., Chan V. C., Shenoy N., Kirkpatrick A., Ramshaw J. A., Brodsky B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bryan M. A., Cheng H., Brodsky B. (March 18, 2010) Biopolymers 10.1002/bip.21432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Byers P. H. (1995) Connect. Tissue Res. 31, 257–259 [DOI] [PubMed] [Google Scholar]
- 29. Smith L. T., Schwarze U., Goldstein J., Byers P. H. (1997) J. Invest. Dermatol. 108, 241–247 [DOI] [PubMed] [Google Scholar]