Abstract
The miR-200 family plays a crucial role in epithelial to mesenchymal transition via controlling cell migration and polarity. We hypothesized that miR-200b, one miR-200 family member, could regulate angiogenic responses via modulating endothelial cell migration. Delivery of the miR-200b mimic in human microvascular endothelial cells (HMECs) suppressed the angiogenic response, whereas miR-200b-depleted HMECs exhibited elevated angiogenesis in vitro, as evidenced by Matrigel® tube formation and cell migration. Using in silico studies, miR target reporter assay, and Western blot analysis revealed that v-ets erythroblastosis virus E26 oncogene homolog 1 (Ets-1), a crucial angiogenesis-related transcription factor, serves as a novel direct target of miR-200b. Knocking down endogenous Ets-1 simulated an anti-angiogenic response of the miR-200b mimic-transfected cells. Certain Ets-1-associated genes, namely matrix metalloproteinase 1 and vascular endothelial growth factor receptor 2, were negatively regulated by miR-200b. Overexpression of Ets-1 rescued miR-200b-dependent impairment in angiogenic response and suppression of Ets-1-associated gene expression. Both hypoxia as well as HIF-1α stabilization inhibited miR-200b expression and elevated Ets-1 expression. Experiments to identify how miR-200b modulates angiogenesis under a low oxygen environment illustrated that hypoxia-induced miR-200b down-regulation de-repressed Ets-1 expression to promote angiogenesis. This study provides the first evidence that hypoxia-sensitive miR-200b is involved in induction of angiogenesis via directly targeting Ets-1 in HMECs.
Keywords: Endothelium, Hypoxia, Metalloprotease, Oxidation Reduction, Oxygen Radicals
Introduction
Micro-RNAs (miRs)2 are small non-coding RNAs consisting of ∼22 nucleotide base pairs. These small nucleic acids could regulate gene expression by binding to the 3′ untranslated region (3′ UTR) of mRNA, resulting in either translational repression or transcript degradation. Around 30% of the genes in the whole genome are subjected to regulation by miRs (1). Because of the short sequence requirement for binding the target 3′ UTR, a single miR could interact with a wide range of target transcripts, which could substantially alter gene expression and dictate cell fate such as proliferation, apoptosis, and cell migration.
Angiogenesis, the formation of vessels from the existing vascular structure, is a crucial biological response in wound healing, menstrual cycle, tumor aggression, and diabetic retinopathy. Endothelial sprouting, which requires extracellular matrix remodeling and endothelial cell migration, is the pre-requisite process of angiogenic response. Such biological response relies on highly coordinated gene expression in a temporal and spatial manner. Increasing evidence revealed that miRs play a pivotal role in the angiogenic process. Our group and others reported that dicer, a ribonuclease III catalyzing miR maturation, is involved in the angiogenic process in human endothelial cells (2–4). Endothelial-specific dicer knock-out mice exhibited an impaired angiogenic response (5). Certain miRs such as miR-126 and miR-296 have been shown to exert pro-angiogenic effects, whereas other reports indicated that miR-130a, -221, and -222 inhibited angiogenesis (6). Recently, the miR-200 family has been shown to arrest cell migration and modulate epithelial-mesenchymal transition in a wide range of epithelial cancer cells (7–12). We hypothesized that miR-200b, one member in miR-200 family, regulates endothelial cell migration and angiogenic responses. In this study, we present the first evidence demonstrating that miR-200b is hypoxia-inducible and down-regulates v-ets erythroblastosis virus E26 oncogene homolog 1 (Ets-1), a novel direct target of miR-200b, subsequently promoting angiogenic response of HMEC.
EXPERIMENTAL PROCEDURES
Cells, Cell Culture, and Hypoxic Treatment
Human dermal microvascular endothelial cells (HMECs) were cultured in a humidified chamber (37 °C, 20% O2 and 5% CO2) in MCDB-131 medium supplemented with 10% FBS, 10 mm l-glutamine, and 100 IU/ml of penicillin, 0.1 mg/ml of streptomycin (Invitrogen), as described previously (3). Primary adult human dermal microvascular endothelial cells were cultured at 37 °C (20% O2 and 5% CO2) in EBM-2 medium (Lonza) supplemented with EGM-2MV single quotes (Lonza) as described by the manufacturer. HEK-293 cells were grown at standard cell culture conditions (37 °C, 20% O2 and 5% CO2) with DMEM supplemented with 10% FBS and 100 IU/ml of penicillin, 0.1 mg/ml of streptomycin. For hypoxic treatment, cells were seeded on a 35-mm dish at 0.5 × 106 cells/plate 1 day before hypoxic treatment. Medium was refreshed and incubated at either normoxic (20% O2) or hypoxic (1% O2) in a chamber with the same humidity and temperature as indicated. For the miR-200b mimic hypoxia study, cells were first transfected with control or miR-200b mimic for 48 h, followed by versenization and plated on a 35-mm dish. The cells were allowed to settle for 4 h and subjected to hypoxic treatment after refreshing the medium, as described above. After 24 h, cells were either lysed or plated on Matrigel® pre-coated plates for further analysis.
Transfection of miR Mimic, miR Inhibitors, siRNA, or Ets-1 Expressing Plasmid
One day before transfection, cells were seeded in a 12-well plate at 0.17 × 106 cells/well (∼70% confluence). DharmaFECTTM 1 transfection reagent (Dharmacon RNA Technologies) was employed to transfect cells with miR-200b mimic (50 nm), miR-200b inhibitor (100 nm), or siRNA smart pool for human Ets-1, ZEB1, and ZEB2 (100 nm), as indicated. Non-targeting miR mimic, miR inhibitors, or siRNA were transfected in the cells to serve as negative controls, respectively. All the miR mimics, inhibitors, and siRNA were obtained from Dharmacon RNA Technologies. The corresponding sequences of siRNA are shown in Table 1. Unless specified, the cells were either harvested directly or seeded on regular or Matrigel pre-coated plates for further analysis after a 72-h transfection (3). For Ets-1 overexpression studies, an expressing vector encoding Ets-1 (Ets-1 pcDNA) (13), a generous gift from Dr. Michael Ostrowski, The Ohio State University, was transfected in HMECs at the indicated concentrations using LipofectamineTM LTX/plus reagent (Invitrogen) according to the manufacturer's protocol. Empty vector (pcDNA) served as the negative control. 48 h after transfection, cells were either lysed or further transfected with control or miR-200b mimics as described above for further analysis.
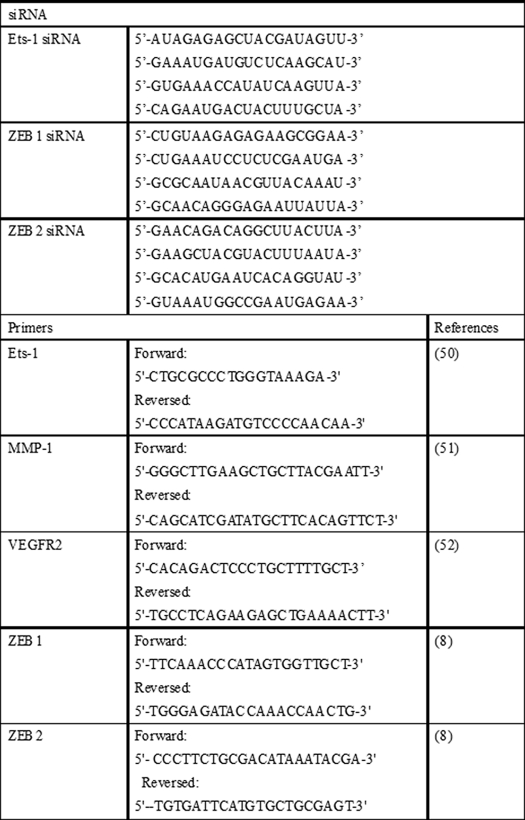
TABLE 1.
siRNA and primer used in the study
RNA Isolation and Quantitative Real Time PCR
Total RNA was extracted using the miRVana miRNA Isolation Kit according to the manufacturer's protocol (Ambion). For determination of miR expression, specific TaqMan assays for miRs and the TaqMan Micro-RNA Reverse Transcription Kit were employed, followed by semi-quantitative real time PCR using the Universal PCR Master Mix (Applied Biosystems) (3, 14). For gene expression studies, total cDNA synthesis was achieved using the SuperScriptTM III First Strand Synthesis System (Invitrogen). The transcription levels of Ets-1, MMP-1, VEGFR2, ZEB1, and ZEB2 were assessed by real time PCR using SYBR Green-I (Applied Biosystems). The corresponding sequences of primers were shown in Table 1. miR-16 and β-actin served as loading controls as described previously (14–16).
Infection of Adenovirus Encoding Oxygen-insensitive Hypoxia-inducing Factor (HIF)-α
HMECs were infected with adenovirus encoding oxygen-insensitive HIF-1α as described previously (14, 17). After a 48-h infection, the cells were harvested for protein, RNA, or cell lysate by a HIF-dependent reporter luciferase assay.
miR Target Reporter Luciferase Assay
HEK-293 cells were transfected with 100 ng of pLuc-Ets1–3′UTR plasmid (Signosis) or a control construct using Lipofectamine LTX/Plus reagent (Invitrogen) according to the manufacturer's protocol. The construct was designed based on the sequence of miR-200b binding sites and a total of 200 bp (starting from 500–700 of Ets-1 3′ UTR) were cloned in the 3′ UTR of the pLuc plasmid (for details, see supplementary data). Normalization was achieved by co-transfection with Renilla plasmid (10 ng). Cells were lysed after 48 h, and luciferase activity was determined using the dual-luciferase reporter assay system (Promega). Data are presented as ratio of firefly to Renilla luciferase assay.
In Vitro Angiogenesis and Cell Migration Assay
In vitro angiogenesis and cell migration were assessed by the tube formation ability on Matrigel and scratch wound assay on monolayer cells, respectively, as described previously (3). For the in vitro angiogenesis assay, cells were plated out after transfection or treatment and seeded on a Matrigel pre-coated 4-well plates at 5 × 104 cells/well. The angiogenic property was assessed 8 h after seeding as described previously (18), and the tube length was measured using the AxioVision Rel 4.6 software (Zeiss). For the cell migration assay, cells were seeded at the confluent monolayer on a 4-well plate. After seeding for 4 h, the in vitro wound was induced by scratching gently using a 10-μl pipette tip. Cell migratory distance was measured 4 h post-wounding using AxioVision Rel 4.6 software (Zeiss). Cell migration was expressed in percentage of wound closure relative to 0 h.
Western Blot Analysis and Immunocytochemistry
Western blot and immunocytochemistry was performed using polyclonal rabbit antibody against Ets-1 (Santa Cruz biotechnology) as described previously (3). Briefly, the cells were harvested in lysis buffer containing 10 mm Tris, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% deoxycholic acid, 0.1% SDS, and 5 mm EDTA. Cell lysates were subjected to SDS-PAGE, transblotted to PVDF membrane (Amersham Biosciences), blocked, and incubated with primary antibody against Ets-1 at a 1:5,000 concentration overnight at 4 °C. β-Actin (Sigma, 1:10,000) serves as a loading control. The signal was visualized using the corresponding secondary antibody against rabbit (Amersham Biosciences, 1:3,000) and ECL PlusTM Western blotting Detection Reagents (Amersham Biosciences). For immunocytochemistry, cells were fixed with paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with normal goat serum, and incubated with primary antibody against Ets-1 at a 1:500 concentration overnight. The signal was visualized using Alexa Fluor® 568 dye-conjugated antibody against rabbit (Invitrogen, 1:200), together with DAPI. Images were captured by microscope using AxioVision Rel 4.6 (Zeiss).
Statistical Analyses
Data reported represent mean ± S.E. of at least 3 independent experiments. Difference between two means was tested by Student's t test, whereas one-way analysis of variance was employed to compare three groups or more. p < 0.05 was considered statistically significance.
RESULTS
miR-200b Arrests Angiogenic Response of HMECs
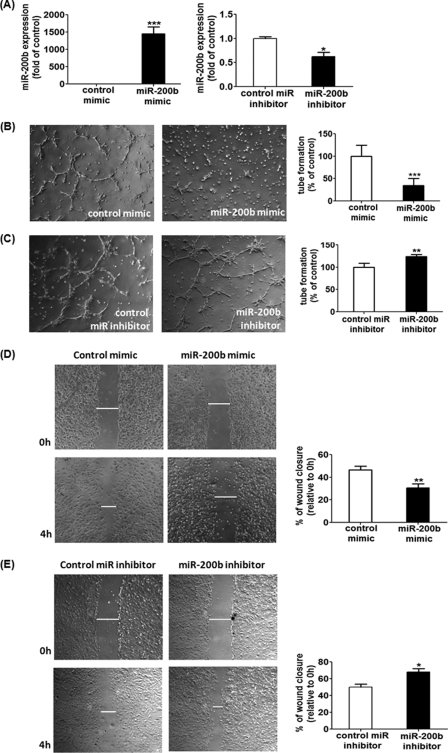
To elucidate the specific role of miR-200b to angiogenic response of endothelial cells, HMECs were transiently transfected with miR-200b mimic or mR-200b inhibitor to up- or down-regulate the endogenous miR-200b level, respectively (Fig. 1A), and subsequently plated on a Matrigel-coated plate to study the tube formation ability. Delivery of 50 nm miR-200b mimic significantly blunted the length of tube, compared with the mimic-transfected control HMECs (Fig. 1B). The anti-angiogenic effects of miR-200b mimic were also observed at 0.5 and 5 nm concentrations (supplemental Fig. S1, A and B). On the contrary, inhibition of endogenous miR-200b by transfection of the miR-200b inhibitor enhanced the tube formation ability on Matrigel, compared with inhibitor-transfected control cells (Fig. 1C). One of the crucial factors in determining angiogenic property is cell migration, which was assessed by the in vitro scratch wound closure assay. Control HMECs exhibited 50% wound closure 4 h post-wounding, whereas such migration was clearly compromised in the miR-200b mimic-delivered cells, attaining ∼30% wound closure (Fig. 1D). Consistently, miR-200b depleted HMECs showed a significantly elevated cell migration (65% closure), compared with control transfected cells (Fig. 1E). To test that miR-200b exerts a general anti-angiogenic effect on endothelial cells, similar experiments were performed in primary adult human dermal microvascular endothelial cells. Delivery of miR-200b mimic to primary endothelial cells significantly attenuated the tube formation ability on Matrigel (supplemental Fig. S2, A and B), which is comparable with the effect on HMECs.
FIGURE 1.
miR-200b exhibited anti-angiogenic effects in HMECs. A, real time PCR analysis of miR-200b expression after transfection of miR-200b mimic (left) or miR-200b inhibitor (right). Results are mean ± S.E. *** indicates p < 0.001; * represents p < 0.05 compared with control. Matrigel tube formation visualized by phase-contrast microscopy at 8 h after miR-200b mimic delivery (B) or down-regulation (C). Representative image of at least 3 independent experiments. Quantification of the length of tube formation (% of control) of the miR-200b mimic or miR-200b inhibitor-transfected cells. Results are mean ± S.E. *** indicates p < 0.001; ** represents p < 0.01 compared with control. Representative images of the in vitro scratch wound closure assay were visualized by phase-contrast microscopy at 0 and 4 h post-wounding in miR-200b mimic-delivered (D) or miR-200b-depleted (E) HMECs. Cell migration was assessed by quantification of scratch wound closure (relative to 0 h). Results are mean ± S.E. ** indicates p < 0.01; * represents p < 0.05 compared with control.
Ets-1 Is a Novel Direct Target of MiR-200b
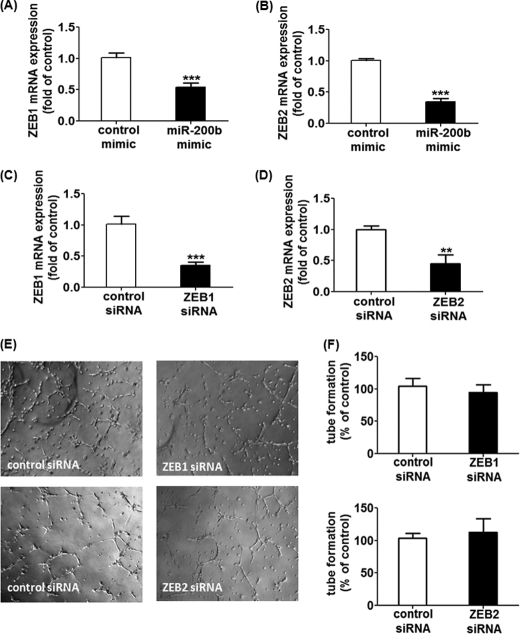
Previous studies revealed that miR-200b negatively regulates epithelial-mesenchymal transition via modulating the expression of its direct target zinc finger E-box-binding homeobox 1 (ZEB1) and ZEB2 (7–10). On top of that, ZEB1 has been reported to control endothelial cell migration and attachment, thus regulating tumor angiogenesis (19, 20). In this regard, we tested the hypothesis whether miR-200b arrests angiogenesis via direct targeting of ZEB1 and ZEB2. miR-200b mimic delivery significantly suppressed the expression of both ZEB1 and ZEB2 (Fig. 2, A and B). However, down-regulation of ZEB1 or ZEB2 alone using specific siRNA failed to simulate the anti-angiogenic response in miR-200b mimic-delivered HMECs (Fig. 2, C–F), indicating that ZEB1 and ZEB2 do not mediate the anti-angiogenic response demonstrated by miR-200b.
FIGURE 2.
ZEB1 or ZEB2 do not serve as the mediators in miR-200b-associated antiangiogenic response. Real time PCR analysis of ZEB1 and ZEB2 expression after transfection of miR-200b mimic (A and B), ZEB1 siRNA (C), or ZEB2 (D). Results are mean ± S.E. ** represents p < 0.01; *** indicates p < 0.001 compared with corresponding control. E, Matrigel tube formation visualized by phase-contrast microscopy at 8 h after transfection of either ZEB1 siRNA or ZEB2 siRNA. Representative image of at least 3 independent experiments are shown. F, quantification of the length of tube formation (% of control) of cells with ZEB1 or ZEB2 knockdown is shown. Results are mean ± S.E.
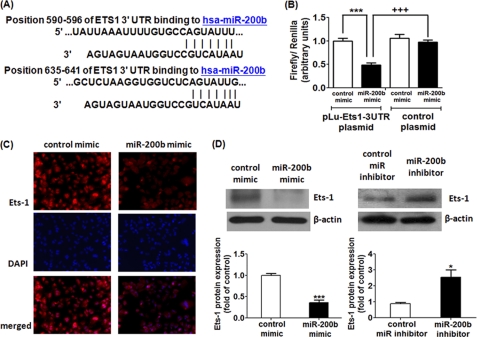
Next, in silico studies were performed to understand possible miR-200b targets that may be responsible for its anti-angiogenic function using data base resources including Targetscan (21), MiRanda (21), Pictar (22), miRBase Target Data base (23), and miRDB (24). Bioinformatic analysis predicted that the seed sequence of miR-200b may bind to the Ets-1 3′ UTR in two different sites at 590–596 and 635–641 (Fig. 3A and supplemental text). To validate the direct binding between miR-200b and the Ets-1 3′ UTR region, we performed miR target reporter luciferase assay using the pLuc-Ets1–3′UTR plasmid in HEK-293 cells. Approximately 50% reduction of luciferase activity from the pLuc-Ets1–3′UTR plasmid was observed in miR-200b mimic-delivered cells compared with control mimic-transfected cells (Fig. 3B). To confirm that repression in luciferase activity was specific for the selected region of the Ets-1 3′ UTR, a similar reporter assay was carried out using the construct without the insert of the selected Ets-1 3′ UTR sequence (control plasmid). As expected, deletion of the Ets-1 3′ UTR sequence from the construct abolished the inhibitory effects of miR-200b on luciferase activity (Fig. 3B). These results establish that miR-200b interact with the Ets-1 3′ UTR to exert translational repression.
FIGURE 3.
Ets-1 serves as a novel direct target of miR-200b. A, in silico study revealing two possible binding sites in Ets-1 3′ UTR for miR-200b as predicted by Targetscan, Pictar, MiRanda, MiRBase Target Data base, and miRDB. B, miR target reporter luciferase assay after the miR-200b mimic delivery in HEK-293 cells. Open and solid bars represent control mimic and miR-200b mimic-delivered cells, respectively. Results were normalized with data obtained from an assay with Renilla luciferase and expressed as mean ± S.E. *** indicates p < 0.001 compared with control mimic-transfected cells; +++ represents p < 0.001 compared with control plasmid-transfected cells. C, representative diagram showing Ets-1 immunoreactivity after miR-200b mimic delivery from three independent experiments. Nuclear counterstain with DAPI and the corresponding merged image are shown in the lower panels. D, Western blot analysis of Ets-1 protein expression in miR-200b mimic-delivered (left) and -depleted (right) HMECs. β-Actin serves a loading control. Representative blot from three independent experiments is shown. Quantification of the band intensity relative to control. Results are mean ± S.E. *** indicates p < 0.001; * represents p < 0.05 compared with control.
Ets-1 Is Negatively Regulated by MiR-200b in HMECs
To determine whether miR-200b regulates Ets-1 expression in HMECs, Western blot and immunocytochemistry were performed to determine the protein expression level. Nearly 70% reduction of Ets-1 protein was observed in miR-200b mimic-delivered HMECs. Consistently, miR-200b inhibitor significantly up-regulated the Ets-1 protein by 2.5-fold (Fig. 3D). These results were further confirmed by immunocytochemistry. In the presence miR-200b mimic, HMECs exhibited blunted Ets-1 immunoreactivity compared with control mimic-transfected cells (Fig. 3C). The specificity of the antibody was confirmed by similar experiments with the omission of primary antibody (supplemental Fig. S3). Similarly, primary endothelial cells with the miR-200b mimic delivered exhibited attenuated Ets-1 expression (supplemental Fig. S2C). It was thus identified that Ets-1 is negatively regulated by miR-200b in endothelial cells.
Ets-1 Down-regulation Simulates the Anti-angiogenic Effects of miR-200b
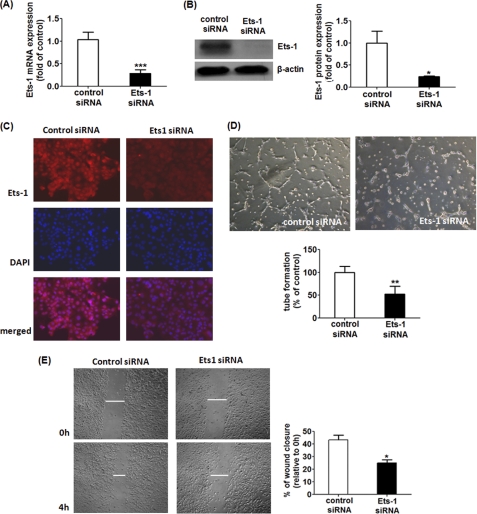
If miR-200b regulates the angiogenic response via modulating Ets-1, knocking-down Ets-1 alone should mimic, or at least partially simulate, the effects of miR-200b on angiogenesis. Using specific siRNA targeting Ets-1, endogenous Ets-1 expression could be significantly down-regulated in the extent similar to that observed in miR-200b mimic delivery (Fig. 4, A–C). Down-regulation of Ets-1 compromised Matrigel tube formation as well as cell migration, compared with control siRNA-transfected HMECs (Fig. 4, D and E).
FIGURE 4.
Down-regulation of Ets-1 simulated the effects of miR-200b mimic in angiogenic response and cell migration. A, real time PCR analysis of Ets-1 mRNA after transfection of Ets-1 siRNA. Results are mean ± S.E. *** indicates p < 0.001 compared with control siRNA. B, Western blot analysis of Ets-1 protein expression after transfection of Ets-1 siRNA; β-actin serves a loading control. Representative blot from three independent experiments. Quantification of band intensity relative to control. Results are mean ± S.E. * represent p < 0.05 compared with control siRNA. C, representative diagram showing Ets-1 immunoreactivity after knocking down Ets-1 from three independent experiments. Nuclear counterstain with DAPI and the corresponding merged image are shown in the lower panels. D, Matrigel tube formation visualized by phase-contrast microscopy at 8 h after Ets-1 down-regulation. Representative image of at least three independent experiments. Quantification of the length of tube formation (% of control) of Ets-1 siRNA-transfected cells. Results are mean ± S.E. ** indicates p < 0.01 compared with control. E, representative image of a in vitro scratch wound closure assay visualized by phase-contrast microscopy at 0 and 4 h post-wounding in Ets-1 knock-down cells. Cell migration of Ets-1-depleted cells was assessed by quantification of scratch wound closure (relative to 0 h). Results are mean ± S.E. * represent p < 0.05 compared with control.
MiR-200b Regulates Ets-1-associated Genes
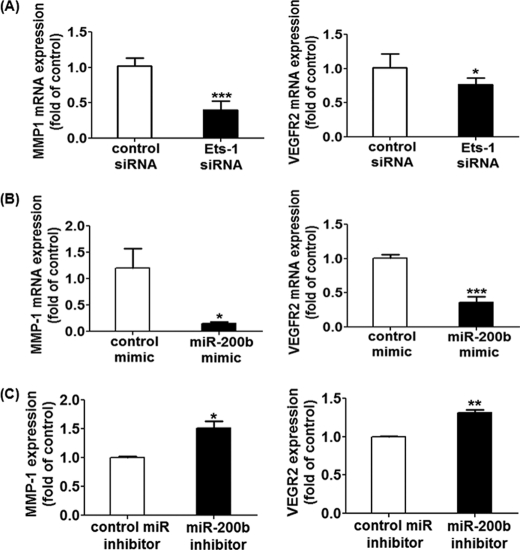
Ets-1 is the transcription factor that controls the expression of genes relevant to the regulation of angiogenesis. Of note, matrix metalloproteinase-1 (MMP-1) and vascular endothelial growth factor receptor 2 (VEGFR2) are the Ets-1-associated mediators for extracellular matrix remodeling and endothelial migration. Knocking-down Ets-1 in HMECs significantly inhibited MMP-1 and VEGFR2 transcript expression (Fig. 5A). Interestingly, up-regulation of miR-200b by miR mimic delivery simulated the effects of Ets-1 siRNA, inhibiting the mRNA expression of MMP-1 and VEGFR2 in both HMECs (Fig. 5B) and primary endothelial cells (supplemental Fig. S2D). Consistently, suppression of endogenous miR-200b induced MMP-1 and VEGFR2 expressions (Fig. 5C).
FIGURE 5.
Expression of Ets-1 related genes in Ets-1 knock-down, miR-200b mimic-delivered, and miR-200b-depleted HMECs. Real time PCR analysis of MMP-1 and VEGFR2 expression after transfection of Ets-1 siRNA (A), miR-200b mimic (B), or miR-200b inhibitor (C). Results are mean ± S.E. *** indicates p < 0.001. ** indicates p < 0.01; * represents p < 0.05 compared with corresponding control.
Overexpression of Ets-1 Abolished the miR-200b-dependent Anti-angiogenic Effect and Depletion in Ets-1-associated Gene Expression
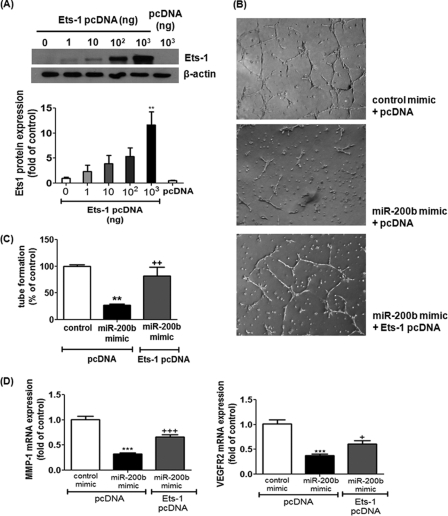
To test whether Ets-1 serves as a downstream mediator of miR-200b signaling, HMECs were subjected to Ets-1 overexpression, followed by miR-200b mimic delivery. Transfection of the expression vector encoding Ets-1 (Ets-1 pcDNA), but not empty vector (pcDNA), dose-dependently increased Ets-1 protein expression in HMECs (Fig. 6A). Overexpression of Ets-1 significantly reversed miR-200b-associated impairment in Matrigel tube formation (Fig. 6, B and C). Furthermore, miR-200b-induced MMP-1 and VEGFR2 down-regulation was significantly rescued by overexpression of Ets-1 (Fig. 6D).
FIGURE 6.
Ets-1 overexpression reversed miR-200b mimic-mediated anti-angiogenic effects and its associated down-regulation of MMP-1 and VEGFR2. A, Western blot analysis of Ets-1 protein expression after transient transfection of various amounts of expressing plasmid encoding Ets-1 (Ets-1 pcDNA) in HMECs; β-actin serves a loading control. Representative blot from three independent experiments is shown. Quantification of the band intensity relative to control. Results are mean ± S.E. ** represent p < 0.01 compared with control empty vector (pcDNA). B, Matrigel tube formation visualized by phase-contrast microscopy at 8 h after delivery of control or miR-200b mimic in the presence or absence of Ets-1 pcDNA. Representative image of at least three independent experiments is shown. Quantification of the length of tube formation (% of control). Results are mean ± S.E. ** indicates p < 0.01 compared with control mimic + pcDNA; ++ indicates p < 0.01 compared with miR-200b mimic + pcDNA. C, real time PCR analysis of MMP-1 and VEGFR2 expression after delivery of control or miR-200b mimic in the presence or absence of Ets-1 pcDNA. Results are mean ± S.E. *** indicates p < 0.001 compared with control mimic + pcDNA. +++ indicates p < 0.001; + represents p < 0.05 compared with miR-200b mimic + pcDNA.
Hypoxia-induced Ets-1 Up-regulation and Angiogenic Response Depend on MiR-200b Expression
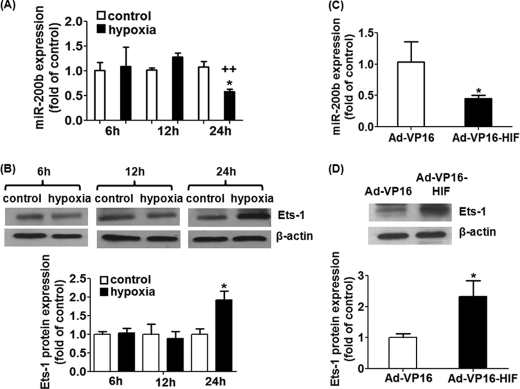
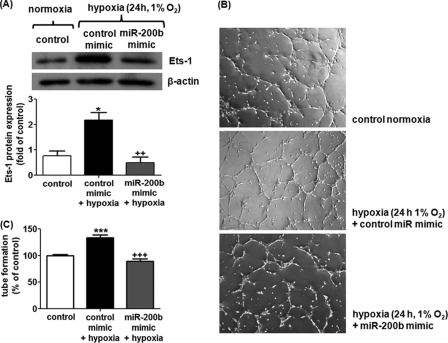
Hypoxia is a well known physiological stimulus that drives angiogenesis in a wound setting. We questioned whether there is any association between miR-200b expression and low oxygen tension. Treatment of HMECs with hypoxia (1% O2) for 24 h, but not 6 or 12 h, significantly inhibited endogenous miR-200b expression, attaining 40% depletion (Fig. 7A). Intriguingly, the expression pattern of miR-200b negatively correlated with that of the Ets-1 protein, implying that miR-200b is associated with Ets-1 down-regulation under hypoxic conditions (Fig. 7B). To elucidate which particular mechanism is involved in miR-200b down-regulation under hypoxia, we tested the significance of HIF stabilization. HIF-1α stabilization was achieved by infection of HMECs with adenovirus carrying an oxygen-insensitive HIF-1α (Ad-VP16-HIF), resulting in HIF-1α overexpression under normoxic conditions as described previously (14). The HIF transactivation activity under hypoxia or Ad-VP16-HIF infection was confirmed by reporter luciferase assay after infection of adenovirus encoding HIF responsive element-dependent luciferase (supplemental Fig. S4). Such forced stabilization of HIF-1α induced 55% depletion of miR-200b expression, compared with control adenovirus (Ad-VP16)-infected HMECs (Fig. 7C). Similarly, HIF-stabilized HMECs exhibited nearly 2-fold up-regulation of Ets-1 protein (Fig. 7D). These results demonstrate that HIF-1α stabilization is responsible for regulation of miR-200b and Ets-1 under hypoxic conditions. To further examine the role of miR-200b in hypoxia-induced Ets-1 up-regulation and angiogenesis, HMECs were subjected to miR-200b mimic delivery, followed by hypoxic treatment. Treatment of the miR-200b mimic significantly blunted hypoxia-induced Ets-1 up-regulation (Fig. 8A). This result was in good agreement with the in vitro angiogenesis observation. Hypoxia pre-conditioning promoted tube formation in Matrigel, which was significantly compromised by the miR-200b mimic delivery (Fig. 8, B and C).
FIGURE 7.
Down-regulation of miR-200b and up-regulation of Ets-1 was observed upon hypoxia or HIF-1α stabilization by infection of adenovirus encoding oxygen-insensitive HIF-1α (Ad-VP16-HIF). A and C, real time PCR analysis of miR-200b expression in hypoxia (1% O2 for 24 h)-treated HMECs or HIF-stabilized HMECs. Results are mean ± S.E. * represent p < 0.05 compared with corresponding control; ++ indicates p < 0.01 compared with the 12-h hypoxia. B and D, Western blot analysis of Ets-1 protein expression in hypoxia (1% O2 for 24 h)-treated HMECs or HIF-stabilized HMECs; β-actin serves a loading control. Representative blot from three independent experiments. Quantification of the band intensity relative to control. Results are mean ± S.E. * represent p < 0.05 compare with corresponding control or control virus (Ad-VP16)-infected cells, respectively.
FIGURE 8.
Hypoxia-induced Ets-1 up-regulation and angiogenesis were abrogated by miR-200b mimic delivery in HMEC. A, Western blot analysis of Ets-1 in HMECs treated with hypoxia (1% O2 for 24 h) with or without miR-200b mimic. β-Actin serves a loading control. Representative blot from three independent experiments is shown. Quantification of the band intensity relative to control. Results are mean ± S.E. * represents p < 0.05 compared with control; ++ indicates p < 0.01 compared with co-treatment of hypoxia and control mimic. B, Matrigel tube formation visualized by phase-contrast microscopy at 8 h after hypoxia pre-conditioning treatment for 24 h in the presence or absence of miR-200b mimic (B and C). Representative image of at least three independent experiments. Quantification of the length of tube formation (% of control) of hypoxia-preconditioned HMECs in the presence or absence of miR-200b mimic. Results are mean ± S.E. *** indicates p < 0.001; +++ indicates p < 0.001 compared with co-treatment of hypoxia and control mimic.
DISCUSSION
The miR-200 family is a cluster of micro-RNAs consisting of five members: miR-200a, miR-200b, miR-200c, miR-429, and miR141, which lies in two different loci in the genome. miR-200a, miR-200b, and miR-429 are clustered in chromosome 1p36, whereas miR-200c and miR-141 are located in chromosome 12p13 (25). The miR-200 family is highly conserved among species (26) and co-regulated upon certain extracellular stimuli such as transforming growth factor (TGF)-β1 and platelet-derived growth factor (PDGF) (8, 27). Loss of the miR-200 family, with the concomitant elevation of expression of the epithelial-mesenchymal transition inducer ZEB1/2, has been shown to be highly correlated with the mesenchymal phenotype (8). Recent findings revealed that the individual member of the miR-200 family simulates the effects of the function of the cluster. Overexpression of miR-200c, one of homologues of miR-200b, alone is sufficient to regulate ZEB1 and restore E-cadherin in breast cancer cells (28, 29). These are supported by in vivo studies indicating that overexpression of miR-200c alone attenuated the invasiveness of breast cancer cells (7) and impaired mammary outgrowth (11). In this study, we employed both the up-regulation and down-regulation approach to dissect the role of miR-200b on the angiogenic response of endothelial cells. Similar to epithelial cells, miR-200b mitigated endothelial cell migration, and more importantly, it suppressed the tube formation ability of endothelial cells, establishing that miR-200b serves as an anti-angiogenic miR under physiological conditions.
Ets-1 is a key transcription factor that is known to support angiogenesis. Certain pro-angiogenic stimulus such as VEGF (30, 31), angiotensin II (32, 33), and fibroblast growth factor (FGF) (34) induce Ets-1 expression. In vitro scratch wound on the monolayer of confluent human endothelial cells up-regulated Ets-1 expression specifically in migrating edges, whereas it returned back to the basal level after wound closure completion (35). Antisense against Ets-1 inhibited basal (36) and VEGF-induced (30) endothelial cell invasion in Matrigel. Suppression of endogenous Ets-1 in murine endothelial cells impaired cell spreading, F-actin formation, and vinculin assembly on the vitronectin-coated surface (36). On the other hand, overexpression of Ets-1 exaggerated endothelial cell invasion in Matrigel, with concomitant up-regulation of MMP-1, MMP-3, MMP-9, and integrin β3 (36). The pro-angiogenic effects of Ets-1 were further supported by a number of in vivo studies. Ets-1 up-regulation was reported in balloon catheter injury in rat aortic endothelium (35) and murine proliferative retinopathy (31), which require angiogenesis for the remodeling process. Injection of adenovirus encoding dominant-negative Ets-1 significantly attenuated retinal neovascularization (31). These observations are in line with our data indicating that endogenous Ets-1 is a key mediator of cell migration and Matrigel tube formation in HMECs. Although the regulatory mechanism of Ets-1 in angiogenesis is well documented, understanding the post-transcriptional control of Ets-1 remains poorly developed (37–40). Here, we reported a novel post-transcriptional regulation of Ets-1 by miR-200b: binding to the 3′ UTR of Ets-1 mRNA to induce translational repression. This hypothesis is further strengthened by the inverse relationship between miR-200b and Ets-1-related downstream mediators including MMP-1 and VEGFR2. The promoter regions of MMP-1 (41) and VEGFR2 (42, 43) contain Ets binding sites, and mutation of Ets binding sites in the 5′ flanking region in MMP-1 or VEGFR2 gene significantly compromises basal or stimulus-induced promoter transactivation activity (41, 43). Furthermore, overexpression of Ets-1 reversed the phenotypic changes caused by miR-200b mimics, further supporting the notion that miR-200b inhibited the angiogenic response via silencing of Ets-1. One interesting finding in the present study is that down-regulation of transcripts of MMP-1 and VEFGR2 by miR-200b up-regulation is much more prominent than by knocking down Ets-1 alone. Overexpression of Ets-1 did not completely reverse miR-200b-associated MMP-1 and VEGFR2 down-regulation. These observations indicate that miR-200b, apart from targeting Ets-1, might silence other target proteins involved in transcription of the indicated genes.
Most studies addressing the miR-200 family have focused on regulation of ZEB1 and ZEB2, subsequently controlling cluster of gene expression including E-cadherin and vitmentin (8, 10). Previous findings from another group indicated that ZEB1 negatively regulates angiogenesis in macrovascular endothelial cells (19, 20). Knocking down ZEB1 exaggerated tube formation and Matrigel invasion in human umbilical vascular endothelial cells (19, 20). Aorta isolated from ZEB1 knock-out mice exhibited enhanced sprouting on Matrigel compared with the wild type littermates (20). In contrast, we did not observe any change in Matrigel tube formation in response to silencing ZEB1 or its homologue ZEB2. The discrepancy could be explained by the difference in properties of endothelial cells derived from microvessels (HMECs) and macrovessels (human umbilical vascular endothelial cells). Heterogeneity between microvascular and macrovascular endothelial cells in terms of morphology, antigen presentation, biological response, and functional characteristics have been characterized (44). Microvascular and macrovascular endothelial cells have different contractile response and MMP expression profile upon stimulation (45–47). Thus, it is not surprising that ZEB1 is critical in regulation of the macrovascular angiogenic response, but not microvascular angiogenesis.
Hypoxia is broadly recognized as a general cue that drives angiogenesis. Our results demonstrate that miR-200b is involved in enabling the hypoxia-induced angiogenic response. Findings of this study support the direct involvement of HIF stabilization in repressing miR-200b expression. Furthermore, hypoxia-induced aggressiveness of Matrigel tube formation as well as Ets-1 up-regulation was rescued by delivery of the miR-200b mimic. These findings consolidate the notion that under hypoxic conditions miR-200b down-regulation is required to relieve Ets-1 repression resulting in successful angiogenic outcomes. Several possible mechanisms have been proposed for down-regulation of miRs under a low oxygen environment. Inhibition of transcription of specific miRs under conditions of hypoxia represents a key player. Hypoxia, for instance, down-regulates expression of the miR-17-92 cluster in colonic cancer cells (48). Low oxygen tension induces recruitment of p53 to the promoter of miR-17-92, thus repressing its transcription (48). The expression of the pri-miR-17-92 cluster negatively correlates with the p53 status in colorectal cancer, further supporting the notion of transcription control under hypoxia (48). Another possible mechanism by which hypoxia may lower miR abundance is by accelerating miR degradation. In cardiac myocytes, hypoxia suppressed the expression of mature miR-199a, whereas its passenger strand miR-199a*, which was derived from the same stem-loop precursor, remained unaffected (49). This finding suggested that hypoxia might selectively destabilize the guide strand (miR-199a) but not the passenger strand (miR-199a*). Whether miR-200b is regulated through a similar mechanism remains unclear. Further investigation is required to elucidate the underlying mechanism of how hypoxia induces miR-200b down-regulation.
Taken together, our results indicate that hypoxia-sensitive miR-200b is crucial in inducing angiogenesis via Ets-1 direct targeting. These findings not only provide a novel regulation of angiogenesis in the context of Ets-1 but also open up a new avenue for addressing the post-transcriptional control of Ets-1, which could potentially be applied in different scenarios including wound healing, other ischemic disorders, and tumor biology.
Supplementary Material
Acknowledgment
We thank Dr. Michael Ostrowski for providing the Ets-1 cDNA clone as a generous gift.
This work was supported, in whole or in part, by National Institutes of Health Grants GM077185 and GM069589 (to C. K. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental data and Figs. S1–S4.
- miR
- micro-RNA
- HMEC
- human microvascular endothelial cell
- Ets-1
- v-ets erythroblastosis virus E26 oncogene homolog 1
- HIF
- hypoxia-inducing factor
- ZEB
- zinc finger E-box-binding homeobox
- MMP-1
- matrix metalloproteinase-1
- VEGFR
- vascular endothelial growth factor receptor.
REFERENCES
- 1. Rajewsky N. (2006) Nat. Genet. 38, (suppl.) S8–13 [DOI] [PubMed] [Google Scholar]
- 2. Kuehbacher A., Urbich C., Zeiher A. M., Dimmeler S. (2007) Circ. Res. 101, 59–68 [DOI] [PubMed] [Google Scholar]
- 3. Shilo S., Roy S., Khanna S., Sen C. K. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 471–477 [DOI] [PubMed] [Google Scholar]
- 4. Suárez Y., Fernández-Hernando C., Pober J. S., Sessa W. C. (2007) Circ. Res. 100, 1164–1173 [DOI] [PubMed] [Google Scholar]
- 5. Suárez Y., Fernández-Hernando C., Yu J., Gerber S. A., Harrison K. D., Pober J. S., Iruela-Arispe M. L., Merkenschlager M., Sessa W. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14082–14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sen C. K., Gordillo G. M., Khanna S., Roy S. (2009) J. Vasc. Res. 46, 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. (2008) EMBO Rep. 9, 582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 9. Korpal M., Lee E. S., Hu G., Kang Y. (2008) J. Biol. Chem. 283, 14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park S. M., Gaur A. B., Lengyel E., Peter M. E. (2008) Genes Dev. 22, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S. P., Chiao E., Dirbas F. M., Somlo G., Pera R. A., Lao K., Clarke M. F. (2009) Cell 138, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wellner U., Schubert J., Burk U. C., Schmalhofer O., Zhu F., Sonntag A., Waldvogel B., Vannier C., Darling D., zur Hausen A., Brunton V. G., Morton J., Sansom O., Schüler J., Stemmler M. P., Herzberger C., Hopt U., Keck T., Brabletz S., Brabletz T. (2009) Nat. Cell Biol. 11, 1487–1495 [DOI] [PubMed] [Google Scholar]
- 13. Yang B. S., Hauser C. A., Henkel G., Colman M. S., Van Beveren C., Stacey K. J., Hume D. A., Maki R. A., Ostrowski M. C. (1996) Mol. Cell. Biol. 16, 538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Biswas S., Roy S., Banerjee J., Hussain S. R., Khanna S., Meenakshisundaram G., Kuppusamy P., Friedman A., Sen C. K. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6976–6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fasanaro P., D'Alessandra Y., Di Stefano V., Melchionna R., Romani S., Pompilio G., Capogrossi M. C., Martelli F. (2008) J. Biol. Chem. 283, 15878–15883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roy S., Khanna S., Hussain S. R., Biswas S., Azad A., Rink C., Gnyawali S., Shilo S., Nuovo G. J., Sen C. K. (2009) Cardiovasc. Res. 82, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khanna S., Roy S., Maurer M., Ratan R. R., Sen C. K. (2006) Free Radic. Biol. Med. 40, 2147–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ono K., Ishihara M., Ishikawa K., Ozeki Y., Deguchi H., Sato M., Hashimoto H., Saito Y., Yura H., Kurita A., Maehara T. (2002) Br. J. Cancer 86, 1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inuzuka T., Tsuda M., Kawaguchi H., Ohba Y. (2009) Biochem. Biophys. Res. Commun. 379, 510–513 [DOI] [PubMed] [Google Scholar]
- 20. Inuzuka T., Tsuda M., Tanaka S., Kawaguchi H., Higashi Y., Ohba Y. (2009) Cancer Res. 69, 1678–1684 [DOI] [PubMed] [Google Scholar]
- 21. Hsu P. W., Lin L. Z., Hsu S. D., Hsu J. B., Huang H. D. (2007) Nucleic Acids Res. 35, D381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 23. Griffiths-Jones S., Grocock R. J., van Dongen S., Bateman A., Enright A. J. (2006) Nucleic Acids Res. 34, D140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X. (2008) RNA 14, 1012–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bracken C. P., Gregory P. A., Kolesnikoff N., Bert A. G., Wang J., Shannon M. F., Goodall G. J. (2008) Cancer Res. 68, 7846–7854 [DOI] [PubMed] [Google Scholar]
- 26. Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A., Brownstein M. J., Tuschl T., Margalit H. (2005) Nucleic Acids Res. 33, 2697–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kong D., Li Y., Wang Z., Banerjee S., Ahmad A., Kim H. R., Sarkar F. H. (2009) Stem Cells 27, 1712–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hurteau G. J., Carlson J. A., Roos E., Brock G. J. (2009) Cell Cycle 8, 2064–2069 [DOI] [PubMed] [Google Scholar]
- 29. Hurteau G. J., Carlson J. A., Spivack S. D., Brock G. J. (2007) Cancer Res. 67, 7972–7976 [DOI] [PubMed] [Google Scholar]
- 30. Chen Z., Fisher R. J., Riggs C. W., Rhim J. S., Lautenberger J. A. (1997) Cancer Res. 57, 2013–2019 [PubMed] [Google Scholar]
- 31. Watanabe D., Takagi H., Suzuma K., Suzuma I., Oh H., Ohashi H., Kemmochi S., Uemura A., Ojima T., Suganami E., Miyamoto N., Sato Y., Honda Y. (2004) Am. J. Pathol. 164, 1827–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ni W., Zhan Y., He H., Maynard E., Balschi J. A., Oettgen P. (2007) Circ. Res. 101, 985–994 [DOI] [PubMed] [Google Scholar]
- 33. Pearse D. D., Tian R. X., Nigro J., Iorgulescu J. B., Puzis L., Jaimes E. A. (2008) Am. J. Physiol. Renal Physiol. 294, F1094–1100 [DOI] [PubMed] [Google Scholar]
- 34. Forough R., Weylie B., Collins C., Parker J. L., Zhu J., Barhoumi R., Watson D. K. (2006) J. Vasc. Res. 43, 327–337 [DOI] [PubMed] [Google Scholar]
- 35. Tanaka K., Oda N., Iwasaka C., Abe M., Sato Y. (1998) J. Cell. Physiol. 176, 235–244 [DOI] [PubMed] [Google Scholar]
- 36. Oda N., Abe M., Sato Y. (1999) J. Cell. Physiol. 178, 121–132 [DOI] [PubMed] [Google Scholar]
- 37. Du C., Liu C., Kang J., Zhao G., Ye Z., Huang S., Li Z., Wu Z., Pei G. (2009) Nat. Immunol. 10, 1252–1259 [DOI] [PubMed] [Google Scholar]
- 38. Romania P., Lulli V., Pelosi E., Biffoni M., Peschle C., Marziali G. (2008) Br. J. Haematol. 143, 570–580 [DOI] [PubMed] [Google Scholar]
- 39. Ji Z., Degerny C., Vintonenko N., Deheuninck J., Foveau B., Leroy C., Coll J., Tulasne D., Baert J. L., Fafeur V. (2007) Oncogene 26, 395–406 [DOI] [PubMed] [Google Scholar]
- 40. Nishida T., Terashima M., Fukami K., Yamada Y. (2007) Biochem. J. 405, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jinnin M., Ihn H., Mimura Y., Asano Y., Yamane K., Tamaki K. (2005) Nucleic Acids Res. 33, 3540–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kappel A., Schlaeger T. M., Flamme I., Orkin S. H., Risau W., Breier G. (2000) Blood 96, 3078–3085 [PubMed] [Google Scholar]
- 43. Elvert G., Kappel A., Heidenreich R., Englmeier U., Lanz S., Acker T., Rauter M., Plate K., Sieweke M., Breier G., Flamme I. (2003) J. Biol. Chem. 278, 7520–7530 [DOI] [PubMed] [Google Scholar]
- 44. Lang I., Pabst M. A., Hiden U., Blaschitz A., Dohr G., Hahn T., Desoye G. (2003) Eur. J. Cell Biol. 82, 163–173 [DOI] [PubMed] [Google Scholar]
- 45. Sachidanandam K., Harris A., Hutchinson J., Ergul A. (2006) Exp. Biol. Med. 231, 1016–1021 [PubMed] [Google Scholar]
- 46. Glassberg M. K., Nolop K. B., Jackowski J. T., Abraham W. M., Wanner A., Ryan U. S. (1992) J. Appl. Physiol. 72, 1681–1686 [DOI] [PubMed] [Google Scholar]
- 47. Jackson C. J., Nguyen M. (1997) Int. J. Biochem. Cell Biol. 29, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 48. Yan H. L., Xue G., Mei Q., Wang Y. Z., Ding F. X., Liu M. F., Lu M. H., Tang Y., Yu H. Y., Sun S. H. (2009) EMBO J. 28, 2719–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rane S., He M., Sayed D., Vashistha H., Malhotra A., Sadoshima J., Vatner D. E., Vatner S. F., Abdellatif M. (2009) Circ. Res. 104, 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murakami Y., Yamagoe S., Noguchi K., Takebe Y., Takahashi N., Uehara Y., Fukazawa H. (2006) J. Biol. Chem. 281, 28113–28121 [DOI] [PubMed] [Google Scholar]
- 51. Roy S., Shah H., Rink C., Khanna S., Bagchi D., Bagchi M., Sen C. K. (2007) DNA Cell Biol. 26, 627–639 [DOI] [PubMed] [Google Scholar]
- 52. Olszewska-Pazdrak B., Hein T. W., Olszewska P., Carney D. H. (2009) Am. J. Physiol. Cell Physiol. 296, C1162–1170 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.