Abstract
The KCNQ family of potassium channels underlie a repolarizing K+ current in the heart and the M-current in neurones. The assembly of KCNQ1 with KCNE1 generates the delayed rectifier current IKs in the heart. Characteristically these channels are regulated via Gq/11-coupled receptors and the inhibition seen after phospholipase C activation is now thought to occur from membrane phosphatidylinositol (4,5)-bisphosphate (PIP2) depletion. It is not clear how KCNQ1 recognizes PIP2 and specifically which residues in the channel complex are important. Using biochemical techniques we identify a cluster of basic residues namely, Lys-354, Lys-358, Arg-360, and Lys-362, in the proximal C terminus as being involved in binding anionic phospholipids. The mutation of specific residues in combination, to alanine leads to the loss of binding to phosphoinositides. Functionally, the introduction of these mutations into KCNQ1 leads to shifts in the voltage dependence of channel activation toward depolarized potentials and reductions in current density. Additionally, the biophysical effects of the charge neutralizing mutations, which disrupt phosphoinositide binding, mirror the effects we see on channel function when we deplete cellular PIP2 levels through activation of a Gq/11-coupled receptor. Conversely, the addition of diC8-PIP2 to the wild-type channel, but not a PIP2 binding-deficient mutant, acts to shift the voltage dependence of channel activation toward hyperpolarized potentials and increase current density. In conclusion, we use a combined biochemical and functional approach to identify a cluster of basic residues important for the binding and action of anionic phospholipids on the KCNQ1/KCNE1 complex.
Keywords: Heart, Ion Channels, Phosphatidylinositol Signaling, Plasma Membrane, Potassium Channels
Introduction
There are five members of the KCNQ family of voltage-gated potassium channels (KCNQ1–5). KCNQ1 together with KCNE1 constitutes an important repolarizing current in mammalian ventricular myocytes that has characteristically slow activation kinetics and is known as IKs (1–3). The channel is of great medical importance as mutations in the genes are a cause of hereditary sudden arrhythmic death as part of the long QT syndrome. Long QT syndrome is characterized by prolongation of the QT interval on the ECG and this predisposes the individual to torsade-de-pointes and subsequent sudden death due to ventricular fibrillation (4, 5). About half of all cases of hereditary long QT are associated with mutations in the genes encoding the pore forming KCNQ1 and auxiliary subunit KCNE1 (LQT1 and LQT5, respectively) (6). In addition, KCNQ1 (together with KCNE3) underlies a time independent current in gastric epithelial cells (7). KCNQ2, KCNQ3, and KCNQ5 are the molecular correlates of the M-current in neurones, and mutations in KCNQ2 can lead to hereditary epilepsy (8–12). KCNQ4 is present in the inner ear and defects in the channel once again can result in hereditary deafness (13).
A characteristic property of these channels is that they are inhibited after agonist activation of Gq/11-coupled receptors, in particular the M1 muscarinic receptor (9, 14, 15). This is well recognized in neurons where inhibition of the current can have significant effects on excitability (16). However, inhibition of IKs after activation of the α1 Gq/11-coupled receptors also occurs and may influence the cardiac action potential (17). The second messenger accounting for receptor-mediated regulation remained mysterious for a number of years but it is now thought that the major mechanism by which this occurs is through depletion of the membrane anionic phospholipid phosphatidylinositol (4,5)-bisphosphate (PIP2)3 (18–21). It is clear that anionic phospholipids can regulate a range of channels and transporters including inwardly rectifying K+ channels and the Na+/Ca2+ exchanger (22). In the inwardly rectifying family of K+ channels the regulation occurs by direct interaction with a binding site on the channel. The residues underlying this are well defined by both biochemical and functional studies (23–26). Despite the obvious physiological significance of the regulation there is much less consensus as regards to the site of action on the KCNQ family of channels and for KCNQ1 only speculation as to the exact binding site (27). In this study, we describe a combined biochemical and functional approach to identify a cluster of basic residues in the proximal C terminus of KCNQ1 as being of critical importance.
EXPERIMENTAL PROCEDURES
Molecular Biology
The cytoplasmic C terminus of KCNQ1 (amino acids 353 to 676) was cloned into the pmalc2x vector (New England Biolabs) using standard cloning techniques. The MBPKCNQ1C protein was expressed and purified as previously described (26, 28). Site-directed mutagenesis was carried out using the QuikChange mutagenesis kit as per the manufacturer's instructions (Agilent Technologies).
Protein-Lipid Overlay Assay
PIP strips (Invitrogen) were used as previously described (26, 28). PIP arrays (Invitrogen) were also used following the same method as for the PIP strips. Protein concentration used varied between 1 and 5 μg/ml for the PIP strips to demonstrate that the amount of protein was not a limiting factor in the binding of the phosphoinositides. 1 μg/ml of protein was used for the PIP arrays.
Liposome Preparation
PIP2 lipids were obtained from Avanti Polar Lipids and PE, PS, and PC were obtained from Sigma. All were resuspended in 2:1, chloroform:methanol. A total of 1.5 mm lipids were added in the ratio of PC:PE:PS:PIP2 (40:30:20:10). The lipids were dried using a N2 stream before resuspension in HBS-N (10 mm HEPES, pH 7.4, 500 mm NaCl). Lipids were incubated at 37 °C for 45 min before subjecting them to 7 freeze/thaw cycles. Sonication for 5 min at 40 °C was followed by extrusion through a mini extruder (100 nm filter, Avanti Polar Lipids).
Surface Plasmon Resonance
Measurements were made using a Biacore 3000 and a L1 chip (GE Healthcare). The L1 chip was prepared using 40 mm octyl glucoside (2 × 10 μl injections at a flow rate of 10 μl/min). The chip was loaded with liposomes at a flow rate of 5 μl/min for a total of 15 min. An injection of 10 mm NaOH (20 μl) at a flow rate of 100 μl/min was used to remove any unbound lipids. Protein was washed over the surface of the chip at a flow rate of 30 μl/min. First BSA (120 μl, 0.1 mg/ml) was used to reduce nonspecific binding (wait 15 min) before the desired protein (120 μl) was washed over the chip surface. After a 15 min wait, a NaOH (20 μl, flow 100 μl/min) injection was used to remove protein bound to the lipids allowing the chip surface to be regenerated.
Electrophysiology
Currents produced by KCNQ1 and KCNE1 were recorded from transfected CHO-K1 cells or CHO cells stably expressing the human M1 muscarinic acetylcholine receptor (CHO-hM1) (15), 2 days after transfection. 500 ng of each vector was transfected using Lipofectamine (Invitrogen) as per the manufacturer's instructions. CHO-K1 cells and CHO-hM1 cells were cultured as previously described (15, 29). Whole cell voltage-clamp recording was carried out at room temperature using an Axopatch 200B amplifier (Axon Instruments). Transfected cells were identified by epifluorescence. The pipette solution for recordings made in CHO-K1 cells contained (mm): 150 KCl, 5 EGTA, 10 HEPES, 2 MgCl2, 1 CaCl2, and 5 (Na)2ATP (pH 7.2 with KOH). For recordings made in CHO-hM1 cells, the pipette solution was supplemented with extra CaCl2 to give 100 nm “free” Ca2+ (1.97 mm CaCl2). We included 100 nm free Ca2+ in this intracellular pipette solution because it has been shown that phospholipase C requires at least resting levels of calcium to retain activity (30). The extracellular solution for both cell lines contained (mm): 150 NaCl, 5 KCl, 10 HEPES, 2 MgCl2, and 1 CaCl2 (pH 7.4 with NaOH). Currents were recorded at 2 min, after break-in, unless stated otherwise, by holding the cell at a voltage of −80 mV followed by stepped depolarizations from −80 to +80 mV for 6 s in 10 mV increments, followed by a repolarizing pulse back to −20 mV for 2 s (to measure tail currents) and back to −80 mV. Series resistance was at least 70% compensated using the amplifier circuitry. Pipette resistance, when filled with intracellular solution, was ∼1.5–2.5 megaohms, and pipette capacitance was reduced by coating the tip with a parafilm oil suspension. Data were analyzed using Clampfit and Microcal Origin software (23, 31). Current-voltage relationships were determined by normalizing the maximal current densities at the end of each pulse potential to cell capacitance (nA or pA/pF). Peak tail current density (PTCD) was determined by normalizing the maximal current densities of the peak tail currents in response to each pulse potential to cell capacitance (pA/pF). The voltage dependence of channel activation (steady-state activation) (V0.5, indicates the potential at which the activation is half-maximal) was determined by fitting the normalized amplitude of the peak tail currents (y/ymax) versus test potential (Vt) with a Boltzmann function (y/ymax = 1/(1 + exp[(V0.5 − Vt)/k])) (k indicates the slope factor). Voltage-dependent rates of activation (activation t½) were determined by calculating the time taken to reach half-maximal current density, after subtracting any instantaneous current, during the 6 s depolarization step at a range of voltages from +30 to +80 mV. To determine channel deactivation time constants (deactivation tau (τ)) the cells were repolarized to −20 mV for 2 s following a 6 s pulse to +30, +40, or +50 mV and the resulting tail currents were fit to a single exponential function. To perform thermal energy calculations, single Boltzmann functions, see above for details, were fit to each of the normalized peak tail currents and the difference in free energy between closed and open states at 0 mV (ΔG0) was calculated according to ΔG0 = 0.2389 zFV50. Where V50 is the half-activation voltage (V0.5), Z is the equivalent gating charge, and F is the Faraday constant. The change in ΔG0 brought about by each mutation (ΔΔG0) was calculated as follows: ΔΔG0 = ΔG0mutant − ΔG0wild-type. Diacylglycerol C8 (diC8)-PIP2 was obtained from Echelon Biosciences and oxotremorine methiodide (Oxo-M) was obtained from Sigma.
Statistical Analysis
Data are expressed as mean ± S.E., and statistical comparisons were made using a one-way ANOVA with Dunnett's multiple comparison post hoc test or a two-way ANOVA with Bonferroni post hoc test. Statistical analysis was performed using GraphPad Prism (San Diego, CA). Data were considered to be statistically significant when p < 0.05.
RESULTS
Biochemical Demonstration of Binding of the C Terminus of KCNQ1 to Anionic Phospholipids
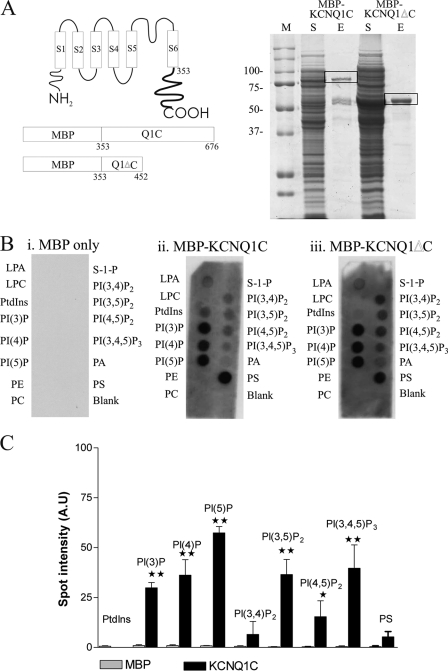
MBPKCNQ1C (79 kDa) was expressed and purified to around 70% purity, although there was some degradation of the protein to a 42-kDa fragment (possibly MBP) and also a 50-kDa product (Fig. 1A). On incubation of the protein with PIP strips, it emerged that MBPKCNQ1C has a broad specificity of lipid binding, binding to most lipids including the monophosphoinositols, diphosphoinositols, and PIP3 (Fig. 1, B and C). The MBP when expressed alone does not show any binding to the PIP strips (Fig. 1, B and C). This indicates that the binding observed comes directly from the KCNQ1C portion of the protein. To determine the region of KCNQ1 that may be involved in this lipid binding, the C terminus was truncated to create a protein consisting of the first 100 amino acids proximal to the membrane domains (amino acids 353 to 452, as shown in Fig. 1A). Purification of this protein shows that it is around 90% pure (Fig. 1A). Comparison of various lipids such as phosphatidylinositol 4-phosphate (PI4P), PIP2, and PIP3 show that there is little difference between the full-length and the truncated version (Fig. 1B). This indicates that the lipid-binding region is located within this proximal C-terminal region of KCNQ1.
FIGURE 1.
Purification and lipid binding of MBP, MBPKCNQ1C, and MBPKCNQ1ΔC. A, schematic diagram showing the topology of the KCNQ1 channel. The proposed amino acid at the start of the C-terminal domain is indicated. The constructs used and the amino acids of KCNQ1 used to create them are also shown. A 10% SDS-PAGE gel was run with the soluble fraction from the purification (S) and 2 μg of eluted protein (E) for both MBPKCNQ1C (79 kDa) and MBPKCNQ1ΔC proteins (50 kDa). The Bio-Rad broad range prestained marker is shown and the marker sizes in kilodaltons are indicated. B, PIP strips that have been incubated overnight with 1 μg/ml of the desired protein as indicated and binding detected using an anti-MBP antiserum (1:1000) followed by anti-rabbit antibody (1:5000) from the ECL kit (GE Healthcare). (i) indicates binding observed with MBP, whereas (ii) and (iii) are MBPKCNQ1C and MBPKCNQ1ΔC, respectively. C, the relative binding of MBPKCNQ1C in comparison to MBP for a selection of the phosphoinositols present. MBPKCNQ1C has a broad specificity of binding. **, p < 0.01 and *, p < 0.05 as determined using one-way ANOVA followed by Dunnett's post test (n = 3). Data are presented as mean ± S.E.
Residues Responsible for the Binding
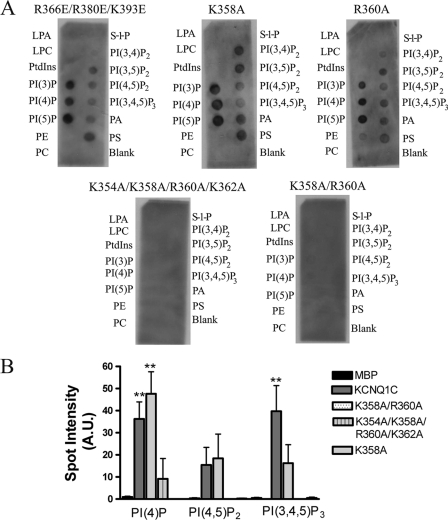
To determine the residues involved in lipid binding the mutagenesis of suitable arginine and lysine residues within the full-length MBPKCNQ1C was undertaken. We targeted positively charged residues within the region contained in the deletion mutation and located close to the last transmembrane domain (residues Lys-354 to Lys-393) as proposed by Loussouarn et al. (27) based on homology modeling. In particular they proposed by homology with the inward rectifiers that the most proximal C-terminal basic residues might be important. As a potential control we mutated three basic residues (in the full-length MBPKCNQ1C), located within MBPKCNQ1ΔC and at the C-terminal end of this proposed region, Arg-366, Arg-380, and Lys-393 to glutamate and observed no difference in binding whether these mutations were introduced singly or in combinations (Fig. 2A and not shown). In contrast, targeting the most proximal basic residues was more revealing. Mutation of all 4 residues together (Lys-354, Lys-358, Arg-360, and Lys-362) to alanine caused loss of binding to all of the phosphoinositols. In addition, Lys-358 and Arg-360 seemed to be particularly important as mutation of these two residues to alanine together led to a comparable loss of binding (Fig. 2, A and B). In contrast, the mutation of Lys-354 and Lys-358 together to alanine qualitatively retained binding (not shown). The mutation of the Lys-354, Lys-358, Arg-360, and Lys-362 alone, to alanine, also did not obviously alter the lipid binding profile of the protein (Fig. 2, A and B, and not shown).
FIGURE 2.
Lipid binding of MBPKCNQ1C mutant proteins. A, a representative PIP strip for representative mutants as indicated. B, the relative spot intensities for PI(4)P, PI(4,5)P2, and PI(3,4,5)P3 for a variety of the mutants in comparison to the full-length MBPKCNQ1C and MBP. Statistical significance between MBP and MBPKCNQ1C mutants for different phosphoinositols is indicated as ** p < 0.01, determined using one-way ANOVA followed by Dunnett's post test (n = 3). Data are presented as mean ± S.E.
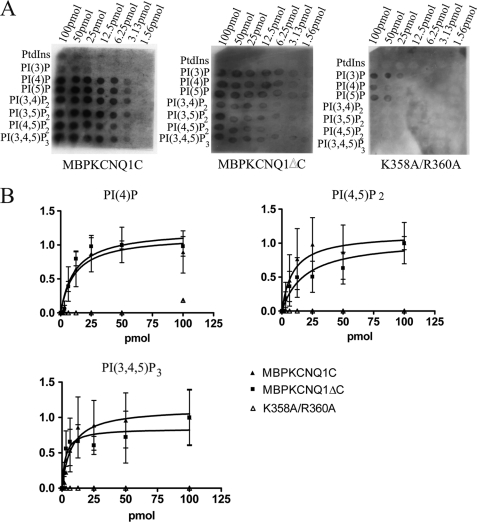
To further examine the relative change in affinity, PIP arrays were incubated with the MBPKCNQ1C, MBPKCNQ1ΔC, and MBPKCNQ1C K358A/R360A fusion proteins. The binding of MBPKCNQ1C and MBPKCNQ1ΔC saturated and there was no obvious difference in the affinity of the two proteins. In contrast, very little binding of MBPKCNQ1C K358A/R360A was detectable particularly to the di- and triphosphatidylinositol species (Fig. 3, A and B).
FIGURE 3.
PIP arrays of MBPKCNQ1C, MBPKCNQ1ΔC and MBPKCNQ1C K358A/R360A. A, shows representative PIP arrays for MBPKCNQ1C, MBPKCNQ1ΔC, and MBPKCNQ1C K358A/R360A. Spot intensities are from left to right, 100, 50, 25, 12.5, 6.25, 3.13, and 1.56 pmol. B, shows plots of mean spot intensity versus lipid concentration for MBPKCNQ1C (filled triangle), MBPKCNQ1ΔC (filled square), and MBPKCNQ1C K358A/R360A (open triangle) for (i) PI(4)P, (ii) PI(4,5)P2, and (iii) PI(3,4,5)P3. All values were normalized and n = 3. Data are presented as mean ± S.E.
An Alternative Biochemical Approach
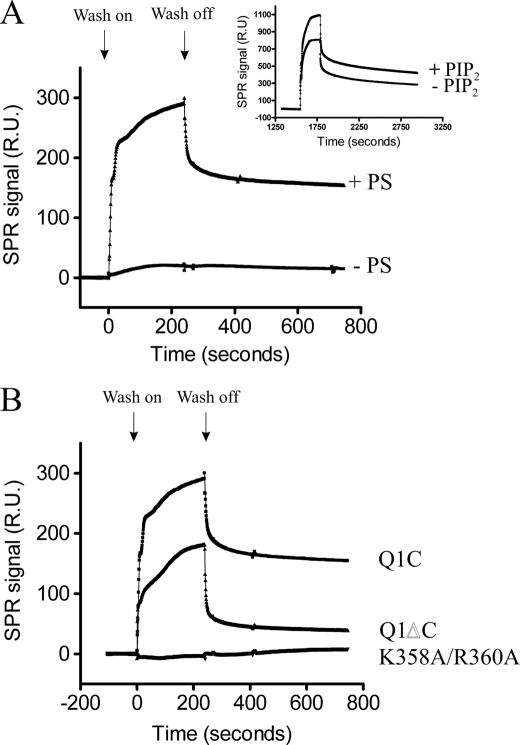
To show that this effect is reproducible using a different type of assay, we used surface plasmon resonance. The L1 chip was chosen on the basis that it is easier to manipulate and because a bilayer is formed. Once liposomes were made and loaded onto the chip first, 1 and 10 μm MBP were washed over the chip. No binding was observed with the control protein indicating that any binding that is observed is coming from the KCNQ1 C terminus (not shown). Then various concentrations of MBPKCNQ1C were washed over the chip. Interestingly, the presence of PS was required for binding of MBPKCNQ1C to occur. Liposomes that only contained PC/PE ± PIP2 showed no binding of MBPKCNQ1C even at 5 μm concentration. In the presence of PC/PE/PS ± PIP2 binding occurred (Fig. 4A). Although there is binding occurring to PC/PE/PS liposomes, the presence of PIP2 increases the ability of MBPKCNQ1C to bind (Fig. 4A, inset). A response of around 250–300 response units (RU) was observed for 1 μm MBPKCNQ1C. Increasing or decreasing the protein concentration used results in a subsequent increase or decrease in the RU. This indicates that this method is able to detect binding of the protein to PIP2. When 1 μm MBPKCNQ1ΔC was washed over the chip surface binding of around 150–200 RU was observed, not too dissimilar to the full-length C terminus (Fig. 4B). When 1 μm MBPKCNQ1C K358A/R360A was used, binding was reduced to 0–10 RU (Fig. 4B). This was repeated on three separate occasions with similar results and validates the observations seen using the protein-lipid overlay assay.
FIGURE 4.
Surface plasmon resonance experiments. A, shows a representative trace for binding of 1 μm MBPKCNQ1C to liposomes containing either PC/PE/PS/PIP2 or PC/PE/PIP2. The traces show the subtraction of the flow cell containing PIP2 minus the flow cell without PIP2 (inset in the figure are the raw traces that are subtracted). The presence of PS is required for binding of MBPKCNQ1C. B, this shows a representative trace of 1 μm of each MBPKCNQ1C, MBPKCNQ1ΔC, and MBPKCNQ1C K358A/R360A. These were repeated 3 times giving similar results each time. No binding is observed with MBPKCNQ1C K358A/R360A.
What Are the Effects of These Mutations on Channel Function?
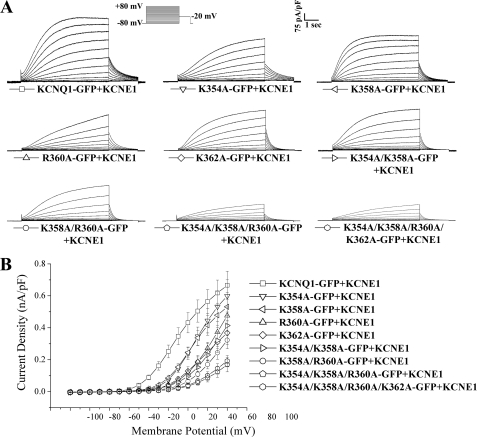
We next sought to investigate the consequences of mutating the residues identified as important for binding phosphoinositides on the electrophysiological properties of the channel complex. We introduced these alanine mutations singly or in combination into KCNQ1-GFP and co-expressed them with KCNE1 in CHO-K1 cells to replicate the molecular basis of the cardiac current. We have previously shown that KCNQ1-GFP behaves in an essentially identical fashion to KCNQ1 from a functional point of view (31). We used whole cell patching to measure and characterize the currents. All the mutants generated current but this was significantly (p < 0.05) reduced compared with control KCNQ1-GFP/KCNE1 and this reduction became more prominent as additional mutations were introduced such that the triple and quadruple mutant expressed only small amounts of current even at quite depolarized potentials (Fig. 5, A and B, and Table 1). Of the double mutations, K358A/R360A led to the most prominent effects on current density at a given potential (Fig. 5B and Table 1).
FIGURE 5.
The electrophysiological characterization of the effect of charge neutralizing mutations in a region involved in phosphoinositide binding on KCNQ1-GFP. A, representative whole cell recordings of KCNQ1-GFP and mutants as indicated co-expressed with KCNE1. Currents were normalized to cell capacitance. The voltage protocol used is shown in the inset. B, mean current-voltage relationships. Current-voltage relationships were determined by normalizing the maximal current densities at the end of each pulse potential to cell capacitance (nA/pF). Data are presented as mean ± S.E. The number of cells analyzed are indicated in Table 1.
TABLE 1.
Analysis of the effects on IKs channel function of charge neutralizing mutations, the intracellular application of diC8-PIP2, and stimulation of the M1 muscarinic 1 receptor
The following represent: E1; KCNE1. n = number of cells analyzed; V0.5 represents the voltage, in mV, at which the channel is half-maximally activated; Z, equivalent gating charge. ΔG0 and ΔΔG0 were calculated as described under “Experimental Procedures.” Data represent mean ± S.E.
| Channel | n | Current density (+40 mV) | PTCD (+40 mV) | V0.5 | Slope factor | Activation t½ (+40 mV) | Deactivation τ (+40 mV) | Z | ΔG0 | ΔΔG0 |
|---|---|---|---|---|---|---|---|---|---|---|
| pA/pF | mV | ms | kcal/mol | |||||||
| CHO-K1 | ||||||||||
| KCNQ1-GFP + E1 | 15 | 431.3 ± 63.4 | 215.9 ± 33.4 | 13.3 ± 1.2 | 12.3 ± 0.5 | 1336.7 ± 100.6 | 950.2 ± 52.6 | 2.0 ± 0.2 | 0.6 ± 0.1 | |
| K354A-GFP + E1 | 8 | 239.2 ± 39.6a | 119.3 ± 21.3a | 37.9 ± 1.6b | 12.5 ± 0.1 | 2122.8 ± 257.0b | 792.3 ± 38.5a | 2.2 ± 0.3 | 1.9 ± 0.1b | 1.2 |
| K358A-GFP + E1 | 8 | 243.9 ± 32.8a | 113.7 ± 15.4a | 31.1 ± 1.5a | 12.4 ± 0.7 | 1151.8 ± 89.4 | 393.7 ± 15.1b | 2.2 ± 0.2 | 1.6 ± 0.1b | 1.0 |
| R360A-GFP + E1 | 8 | 116.7 ± 22.6b | 54.6 ± 11.6b | 57.3 ± 5.4b | 16.0 ± 2.2 | 2550.0 ± 222.1b | 502.8 ± 45.0b | 1.9 ± 0.2 | 2.6 ± 0.3b | 1.9 |
| K362A-GFP + E1 | 8 | 128.7 ± 27.0b | 63.4 ± 13.6b | 45.0 ± 2.2b | 14.2 ± 0.8 | 1747.3 ± 185.0 | 442.6 ± 38.5b | 1.6 ± 0.1 | 1.7 ± 0.1b | 1.1 |
| K354A/K358A-GFP + E1 | 8 | 96.5 ± 19.4b | 40.3 ± 9.2b | 74.4 ± 10.0b | 20.9 ± 2.5b | 1164.8 ± 159.4 | 348.8 ± 37.8b | 1.7 ± 0.2 | 2.5 ± 0.4b | 1.9 |
| K358A/R360A-GFP + E1 | 10 | 64.7 ± 10.2b | 26.9 ± 3.6b | 72.0 ± 5.3b | 18.5 ± 1.7b | 2098.2 ± 162.3b | 338.0 ± 25.8b | 1.6 ± 0.1 | 2.5 ± 0.2b | 1.9 |
| K354A/K358A/R360A-GFP + E1 | 8 | 34.3 ± 9.3b | 12.0 ± 5.1b | 74.6 ± 6.3b | 14.5 ± 2.0 | NDc | ND | 1.7 ± 0.3 | 3.4 ± 0.3b | 2.8 |
| K354A/K358A/R360A/K362A-GFP + E1 | 5 | 39.7 ± 12.3b | 12.8 ± 4.2b | 67.6 ± 4.4b | 13.3 ± 1.7 | ND | ND | 1.7 ± 0.2 | 3.1 ± 0.3b | 2.5 |
| KCNQ1-GFP+E1+ diC8-PIP2 | 10 | 767.9 ± 96.2d | 341.8 ± 45.6e | -4.0 ± 1.1d | 8.5 ± 0.6 | 843.2 ± 68.2d | 974.2 ± 44.0 | 3.8 ± 0.7d | −0.5 ± 0.3d | −1.1 |
| K358A/R360A-GFP + E1 + diC8-PIP2 | 11 | 71.2 ± 14.8 | 31.0 ± 7.7 | 67.2 ± 2.5 | 15.3 ± 0.7 | 2058.2 ± 91.0 | 341.7 ± 25.6 | 1.4 ± 0.1 | 2.6 ± 0.2 | 2.0 |
| CHO-hM1 | ||||||||||
| KCNQ1-GFP + E1 | 8 | 383.9 ± 44.2 | 160.8 ± 19.1 | −13.9 ± 1.4 | 9.8 ± 0.9 | 936.0 ± 81.9 | 1509.3 ± 130.7 | 2.6 ± 0.1 | −0.8 ± 0.2 | |
| KCNQ1-GFP + E1 + Oxo-M | 8 | 167.3 ± 28.8d | 76.3 ± 14.4d | 19.0 ± 2.1d | 16.7 ± 1.12e | 1325.0 ± 118.4e | 665.4 ± 60.5d | 1.4 ± 0.1d | 0.7 ± 0.2d | 1.5 |
| K358A/R360A-GFP + E1 | 8 | 99.3 ± 20.7 | 44.7 ± 9.3 | 54.9 ± 5.1 | 22.2 ± 2.49 | 1386.8 ± 210.6 | 421.7 ± 37.2 | 1.5 ± 0.1 | 1.7 ± 0.3 | 2.5 |
| K358A/R360A-GFP + E1 + Oxo-M | 8 | 44.2 ± 7.2 | 14.5 ± 3.3e | 69.4 ± 3.8e | 18.7 ± 1.29 | 1507.8 ± 116.4 | 313.5 ± 17.9e | 1.4 ± 0.1 | 2.2 ± 0.1 | 3.0 |
a p < 0.05 compared to KCNQ1-GFP/KCNE1.
b p < 0.01 compared to KCNQ1-GFP/KCNE1. Statistical comparisons between the wild-type channel (KCNQ1-GFP/KCNE1) and mutants were performed using a one-way ANOVA with a Dunnett's multiple comparison post hoc test. For statistical comparison between treated and untreated groups, for current density, PTCD, activation and deactivation values, (e.g. KCNQ1-GFP/KCNE1 +/− diC8-PIP2 or KCNQ1-GFP/KCNE1 +/− Oxo-M) a two-way ANOVA with a Bonferroni post hoc test was performed. For statistical comparison between treated and untreated groups, for V0.5, slope factor, Z and ΔG0 values, (e.g. KCNQ1-GFP/KCNE1 +/− diC8-PIP2 or KCNQ1-GFP/KCNE1 +/− Oxo-M) a one-way ANOVA with a Dunnett's multiple comparison post hoc test was performed. For experimental details, please refer to Figs. 5–8.
c ND, not determined.
d p < 0.01 diC8-PIP2 (25 μm) or Oxo-M (10 μm)-treated group compared to untreated group.
e p < 0.05 diC8-PIP2 (25 μm) or Oxo-M (10 μm)-treated group compared to untreated group.
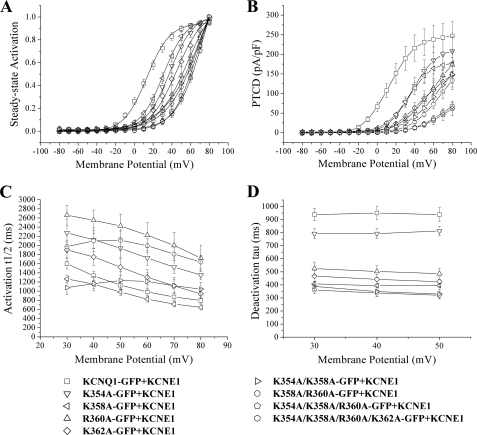
This behavior could arise from a rightward shift in the steady-state activation curve and/or an intrinsic reduction in current with maximal activation. We investigated these possibilities by constructing voltage-dependent activation (steady-state activation) curves (Fig. 6A) and by measuring peak tail current density (PTCD) (Fig. 6B). All the mutations and combinations of mutations led to a rightward shift of the former. The change was particularly prominent for R360A, a shift of ∼44 mV, of the single mutants, and K358A/R360A of the double mutants (Fig. 6A and Table 1). In addition, there was also a significant (p < 0.05) reduction in PTCD, after a depolarization step to +40 mV, and this was particularly pronounced for the K354A/K358A/R360A and K354A/K358A/R360A/K362A mutants. Of the single mutants R360A and K362A reduced PTCD, after a depolarization step to +40 mV, by ∼70–75%, and the double mutants reduced PTCD by ∼80–90% (Fig. 6B and Table 1). We investigated the kinetic basis for the change in voltage-dependent gating. We measured activation t½ over a range of voltages and deactivation Tau at +30, +40, and +50 mV (Fig. 6, C and D, and Table 1). In general, all of the mutations and combinations thereof significantly (p < 0.05) accelerated the rate of deactivation (Fig. 6D and Table 1). The effects on activation were more selective with only K354A, R360A, and K358A/R360A showing a significant (p < 0.05) slowing (Fig. 6C and Table 1).
FIGURE 6.
Detailed examination of the electrophysiological properties of the currents generated by charge neutralizing mutations in a region involved in phosphoinositide binding on KCNQ1-GFP. A, normalized voltage-dependent activation (steady-state activation) curves. The steady-state activation curves were determined by fitting the normalized amplitude of the peak tail currents versus test potential with a Boltzmann function (solid lines). B, PTCD was determined by normalizing the maximal peak tail current densities in response to each pulse potential to cell capacitance (pA/pF). C and D, activation and deactivation kinetics with voltage. The rates of activation (activation t½) and deactivation (deactivation τ) were determined as described under “Experimental Procedures.” Data are presented as mean ± S.E. The number of cells analyzed are indicated in Table 1.
The Functional Effects of PIP2
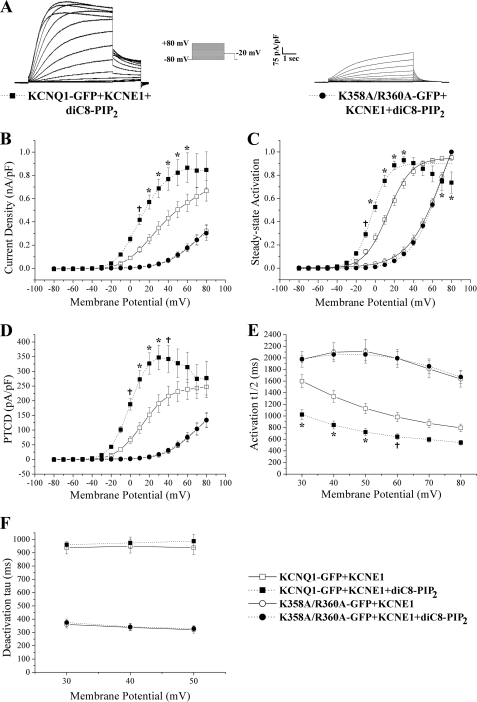
It has been reported that the application of PIP2 or a water soluble derivative (diC8-PIP2) to KCNQ1/KCNE1 results in a leftward shift of the steady-state activation curve and a slowing of deactivation at a given potential (27, 32). We investigated the effects of diC8-PIP2 in our hands on the wild-type channel complex (KCNQ1-GFP/KCNE1) and on the K358A/R360A mutant. We included 25 μm diC8-PIP2 in the patch pipette in whole cell recordings and performed our standard pulse protocol. For the control channel (KCNQ1-GFP/KCNE1) the inclusion of the phospholipid led to a significant (p < 0.05) shift in a hyperpolarized direction of the steady-state activation curve, an increase in current density, and an increase in peak tail current density (Fig. 7, A–D, and Table 1). The enhancement of steady-state gating appeared to occur because of an acceleration of the activation rate and not deactivation rate (Fig. 7, E and F, and Table 1). In contrast, the inclusion of 25 μm diC8-PIP2 in the patch pipette did not act to significantly (p > 0.05) alter any of the properties of the current generated by the K358A/R360A mutant when expressed with KCNE1 (Fig. 7, A–F, and Table 1). The inclusion of diC8-PIP2 with KCNQ1-GFP/KCNE1 also seemed to promote a time-dependent inactivation of current at very non-physiological, depolarized potentials. This can be seen in the trace shown in Fig. 7A and is reflected in the shape of the steady-state activation curve displayed in Fig. 7C. We believe this is not a technical artifact and are investigating it further.
FIGURE 7.
The electrophysiological effects of the intracellular addition of diC8-PIP2 on the wild-type channel and a PIP binding site mutant. A, representative whole cell recordings of KCNQ1-GFP and mutants as indicated co-expressed with KCNE1 in the presence of 25 μm diC8-PIP2. Currents were normalized to cell capacitance. The voltage protocol used is shown in the inset. B, mean current-voltage relationships. Current-voltage relationships were determined by normalizing the maximal current densities at the end of each pulse potential to cell capacitance (nA/pF). C, normalized voltage-dependent activation (steady-state activation) curves. The steady-state activation curves were determined by fitting the normalized amplitude of the peak tail currents versus test potential with a Boltzmann function (solid or dashed lines). D, PTCD was determined by normalizing the maximal peak tail current in response to each pulse potential to cell capacitance (pA/pF). E and F, activation and deactivation kinetics with voltage. The rates of activation (activation t½) and deactivation (deactivation τ) were determined as described under “Experimental Procedures.” Data are presented as mean ± S.E. The number of cells analyzed are indicated in Table 1. For statistical comparison between diC8-PIP2 (25 μm) treated and untreated groups (e.g. KCNQ1-GFP/KCNE1 ± diC8-PIP2), a two-way ANOVA with a Bonferroni post hoc test was performed. †, p < 0.05 diC8-PIP2-treated (25 μm) group compared with untreated group. *, p < 0.01 diC8-PIP2-treated (25 μm) group compared with untreated group.
In an effort to obtain a single reading that highlights the efficacy of the energy perturbation produced by each mutation we calculated the free energy difference between closed and open states at 0 mV (ΔG0) for each mutation and then calculated the difference in ΔG0 between wild-type (KCNQ1-GFP/KCNE1) and mutant channels (ΔΔG0). All of the mutations acted to cause significant (p < 0.05) differences in ΔG0 (Table 1). In particular, the single mutant R360A and double mutant K358A/R360A acted to cause the strongest relative stabilization of the closed state (Table 1). This is also reflected in the ΔΔG0 values, which show that of the single mutants R360A acts to stabilize the closed state most strongly (Table 1). The addition of diC8-PIP2 to the wild-type but not K358A/R360A channels acted to significantly (p < 0.05) shift the ΔG0 toward a negative ΔG0. This indicates that the addition of exogenous PIP2 to KCNQ1-GFP/KCNE1 acts to favor a relative stabilization of the open state (Table 1).
Stimulation of the Muscarinic 1 Acetylcholine Receptor (hM1) Acts to Modulate IKs Channel Function
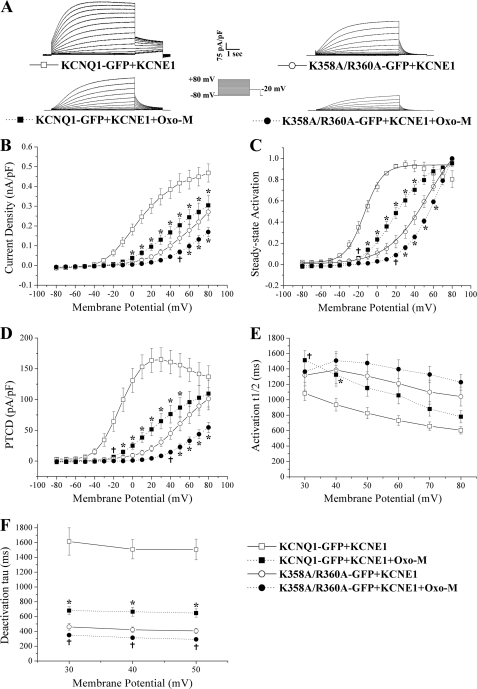
As stated earlier it has been reported that the application of diC8-PIP2 to KCNQ1/KCNE1 results in a leftward shift of the steady-state activation curve and a slowing of the rate of deactivation at a given potential (27, 32). In our hands the intracellular application of diC8-PIP2 to KCNQ1-GFP/KCNE1 resulted in a significant shift (p < 0.05) in the steady-state activation curve. It did not, however, significantly (p > 0.05) alter the rate of deactivation and instead appeared to significantly (p < 0.05) alter the rate of activation at a given potential. To attempt to reconcile these differences we investigated the effect that depletion of PIP2 has on channel function. To do this we decided to express both the wild-type channel and the double mutant, K358A/R360A, in a CHO cell line that stably expresses the Gαq-coupled human M1 muscarinic acetylcholine receptor (15). The activation of this receptor by Oxo-M has previously been shown to result in rapid depletion of the membrane located at PIP2 through activation of a PIP2-specific phospholipase C (30, 33). Additionally, it has also been shown, in this cell line (CHO-hM1), that M1 receptor stimulation leads to closure of the KCNQ1/KCNE1 channel (15). When we expressed the wild-type channel and the double mutant in this cell line we initially noticed that the V0.5 values were shifted in a hyperpolarized direction by ∼20 mV (Fig. 8C and Table 1), in comparison to the values seen in CHO-K1 cells. This difference did not appear to be due to the extra calcium included in the pipette solution as wild-type currents recorded in this cell line using the pipette solution with lower calcium also displayed leftward shifted V0.5 values (data not shown). We do not know why the channel has different V0.5 values in different cell types but have also previously found that the V0.5 value of this channel when expressed in HEK-293 cells is, in contrast, shifted in a depolarized direction (34).
FIGURE 8.
Effects of muscarinic 1 receptor (hM1) activation on the currents generated by the wild-type channel and a PIP2 binding site mutant. Currents were recorded in a CHO cell line stably expressing the human muscarinic 1 receptor (CHO-hM1). After the whole cell configuration was achieved 10 μm Oxo-M was either applied or not in the extracellular solution using a gravity based perfusion system for 5 min before and throughout recording. A, representative whole cell recordings of KCNQ1-GFP and K358A/R360A-GFP as indicated co-expressed with KCNE1 in the presence or absence of 10 μm Oxo-M. Currents were normalized to cell capacitance. The voltage protocol used is shown in the inset. B, mean current-voltage relationships. Current-voltage relationships were determined by normalizing the maximal current densities at the end of each pulse potential to cell capacitance (nA/pF). C, normalized voltage-dependent activation (steady-state activation) curves. The steady-state activation curves were determined by fitting the normalized amplitude of the peak tail currents versus test potential with a Boltzmann function (solid or dashed lines). D, PTCD was determined by normalizing the maximal peak tail current in response to each pulse potential to cell capacitance (pA/pF). E and F, activation and deactivation kinetics with voltage. The rates of activation (activation t½) and deactivation (deactivation τ) were determined as described under “Experimental Procedures.” Data are presented as mean ± S.E. The number of cells analyzed are indicated in Table 1. For statistical comparison between Oxo-M-treated (10 μm) and untreated groups (e.g. KCNQ1-GFP/KCNE1 ± Oxo-M), a two-way ANOVA with a Bonferroni post hoc test was performed. †, p < 0.05 Oxo-M-treated (10 μm) group compared with untreated group. *, p < 0.01 Oxo-M-treated (10 μm) group compared with untreated group.
Upon comparison of the currents produced by the wild-type channel in the CHO-hM1 cell line in the presence or absence of the M1 muscarinic receptor stimulation by Oxo-M (10 μm) we noted various changes in the biophysical characteristics. We found receptor stimulation acted to significantly (p < 0.05) shift the V0.5 toward a more depolarized potential (a shift of ∼35 mV) (Fig. 8C and Table 1). Application of Oxo-M to the wild-type channel also resulted in significantly reduced (p < 0.05) current density and peak tail current density values at a given potential (Fig. 8, B and D, and Table 1). M1 receptor stimulation also significantly (p < 0.05) affected the kinetics of wild-type channel activation and deactivation. Oxo-M acted to significantly (p < 0.05) slow the rate of channel activation and dramatically increase the rate of channel deactivation (Fig. 8, E and F, and Table 1). Interestingly, when we investigated the effects of Oxo-M (10 μm) application on the currents produced by the double mutant we found that it acted to (p < 0.05) shift, although not as dramatically as that seen for the wild-type channel, the V0.5 toward a more depolarized potential (a shift of ∼14 mV) (Fig. 8C and Table 1). Oxo-M also acted to significantly (p < 0.05) reduce current density and peak tail current density values, at given potentials (Fig. 8, B and D, and Table 1). Given the extreme rightward shift of the voltage activation curves these data should be interpreted with caution, as we cannot be certain that we are maximally activating the channels at the examined test potentials. In comparison to the effects seen for the wild-type channel, the effects on the deactivation and activation kinetics of the double mutant were less marked but M1 receptor stimulation did act to significantly (p < 0.05) alter the rate of channel deactivation, but not activation (Fig. 8, E and F, and Table 1).
M1 receptor stimulation acted to significantly (p < 0.05) shift the ΔG0 of the wild-type channel, but not the double mutant, toward a positive ΔG0. This indicates that M1 receptor stimulation acts, in an opposite manner to the addition of diC8-PIP2, to favor a relative stabilization of the closed state (Table 1).
DISCUSSION
Our major novel finding is that we have identified a cluster of basic residues in the C terminus of KCNQ1 for which we provide both biochemical and functional data showing their importance in binding anionic phospholipids. Of the positively charged residues in this cluster Lys-358 and Arg-360 seem to be particularly important. It is worth reviewing some of the findings to justify this statement. We use two biochemical techniques to directly assess the binding of purified protein to phospholipids. The full-length C terminus binds to anionic phospholipids well and this interaction is maintained if the C terminus is truncated to the proximal ∼100 amino acids. We focused on the basic residues that are located close to the final transmembrane domain (Lys-354, Lys-358, Arg-360, and Lys-362). The binding results show that no single mutation is sufficient but that at least two residues must be mutated before binding is lost with the combined deletion of Lys-358 and Arg-360 to alanine resulting in substantial loss of binding. The binding assays we have used are more qualitative than quantitative, however, it is possible to extract ΔΔGibbs free energy estimations from the shift in voltage dependence of activation (V0.5) that each mutant causes compared with control (Table 1). As single mutations these calculated energies reveal that R360A contributes most strongly to stabilizing the relative closed state. The double, triple, and quadruple mutants act to further stabilize the relative closed state.
The best characterized PIP2 binding site is that in the inwardly rectifying family of potassium channels. There is a wealth of data supporting the interaction of anionic phospholipids with a highly conserved basic motif in the proximal C terminus of the channel (23, 25, 26, 35). For example, in Kir3.1 these residues are KKR (amino acids 188–190). Structural studies of a chimera containing transmembrane domains from a bacterial inward rectifier with the N and C terminus of Kir3.1 were very revealing. The KKR residues formed a membrane facing charge cluster that could potentially electrostatically interact with the inositol head group (36). A number of other residues have been identified as being important and a few are present at this interfacial region. However, a majority lie within the fold of the C terminus and may have allosteric effects on the central binding site (24, 36). The inward rectifiers can also be activated quite efficiently by other phosphoinositides apart from PIP2 and this would suggest a “loose” electrostatic interaction with the membrane rather than a heavily structured binding site that is present, for example, in the interaction of PIP2 with the transcription factor tubby (37). Therefore, a cluster of basic residues as a phosphoinositide binding site has strong precedent in the inward rectifier family and homology modeling would support our hypothesis for the location of the binding site we have identified in KCNQ1. Indeed such a homology was initially proposed but there was no experimental data supporting the argument (27) and subsequent studies did not isolate these residues (32). It has been suggested, particularly for the Kir family, that PIP2 regulation is the final common pathway through which all channel regulatory influences converge (38, 39) perhaps by influencing closed open transitions at the helix-bundle crossing.
The residues we have identified in KCNQ1 are in a similar position relative to a potential helix bundle crossing if orientated with the crystal structure of the Kv1.2/2.1 channel chimera in mind (40). However, it does appear that the biophysical mechanisms of KCNQ1 regulation by PIP2 are perhaps more complex. Our data are compatible with PIP2 primarily modulating the equilibrium of voltage-dependent activation. At a kinetic level, this at first appears to be due to a slowing of the closure of the channel from the open state as evidenced by the effects of charge neutralizing mutations and PIP2 depletion, via M1 receptor activation, on the rate of channel deactivation. However, in contrast, the exogenous addition of PIP2 to membranes, where PIP2 has not been depleted, causes changes in activation (i.e. closed open transitions) but limited effects on deactivation (i.e. open closed transitions). There are discrepancies between our data and the literature that may explain the above observations. For example, Loussouarn et al. (27) observed primarily changes in deactivation but not activation kinetics with the addition of PIP2. Are there any technical reasons why this might occur? Interestingly, in studies investigating the actions of PIP2 addition on Kv11.1 (hERG) channel activation and deactivation kinetics conflicting findings have also been reported (41–43). In the whole cell configuration Bian et al. (41) showed that application of 10 μmol/liter of PIP2 in the pipette solution led to an increase in current and acted to accelerate the rate of hERG channel activation but not deactivation. In comparison, when Rodriguez et al. (43) analyzed the biophysical effects of PIP2 addition on hERG, using the inside-out macropatch configuration they found that PIP2 acted to slow the rate of hERG deactivation and had no effect on activation. These findings have led to the suggestion that the reported differences in the biophysical effects of PIP2 on the hERG channel are due to differences in the configuration of the experimental setup (whole cell compared with inside-out macropatches) (43). It is well known that a number of channels and transporters are run down in the inside-out configuration, generally this occurs less prominently in the whole cell configuration and an important factor is the relative instability of membrane concentrations of PIP2 in the former configuration (44). In the case of the effects of PIP2 addition on the KCNQ1/KCNE1 current, it is important to note that the changes in deactivation but not activation kinetics described by Loussouarn et al. (27) were seen in inside-out macropatches.
We also noticed when analyzing the effects of the sequential addition of charge neutralizing mutations on the voltage dependence of activation of the channel that the curves appear to converge on a limiting rightward (depolarizing) shift. This convergence may reflect the existence of gating mechanisms that are PIP2 independent. Experimentally this concept is supported by the convergence of the curves for the triple and quadruple mutants and the residual shift in the K358A/R360A mutant after M1 receptor activation. Importantly in the latter, the curve converges to a distribution that is similar to that seen for the wild-type channel after M1 receptor activation.
In addition to characterizing the effects on the wild-type channel, we also investigated the effects of M1 receptor activation on the double mutant (K358A/R360A) that has disrupted phosphoinositide binding. M1 receptor activation acted to shift the mutants voltage dependence of activation toward more depolarized potentials and reduce current density and peak tail current density. We feel that the changes seen in peak tail current, upon M1 receptor activation, for this mutant should be interpreted cautiously. The pronounced rightward shifted voltage dependence of activation means it is unlikely that the channel is maximally activated even at the most depolarized potentials in the voltage step prior to tail current measurement. Experimentally there are technical limitations to the observations we can make due to frequent loss of the seal upon prolonged and pronounced depolarizations (test potentials above +80 mV). Even so, this result is intriguing and could have various explanations. The first is that there may be some residual PIP2 binding to this charge cluster and we do show evidence that other residues are involved. Second it is possible that additional PIP2 binding sites exist on the channel. Alternatively, it is also possible that stimulation of the M1 receptor acts to stimulate multiple pathways that not only act to deplete PIP2 but cause changes in other signaling molecules that directly or indirectly alter the biophysical properties of the channel. For example, the production of diacylglycerol may contribute to M channel closure through the activation of protein kinase C and channel phosphorylation (45). These are all areas for investigation in future experimental work.
The comparisons with the modulation of KCNQ2 by PIP2 are intriguing. Shapiro and colleagues (46, 47) in a meticulous series of functional studies have identified a region distinct from ours in the C terminus of KCNQ2. An alignment of the relevant channels shows that this domain is relatively conserved in KCNQ3 but is quite different in KCNQ1 (Fig. 9). In contrast, three of the four residues we have identified are conserved in KCNQ2/3 with Lys-358 replaced by glutamine (Fig. 9). The equivalent histidine residue to His-361 in KCNQ1 has been identified as being of importance in the PIP2 regulation of KCNQ2 (18). It is interesting that this residue sits adjacent to the cluster of basic amino acids we have identified suggesting it could well have a modulatory role. There are two possible explanations for these varying data sets. The first is that the residues identified in our study form a core binding for PIP2 but that other regions can also modulate channel activity allosterically. Alternatively, the binding site is different in other members of the KCNQ family with other regions contributing. These are interesting questions for future investigation. It is worth mentioning that the mode of regulation is different for KCNQ2/KCNQ3: the depletion of PIP2 as occurs after Gq/11-coupled receptor activation results in a reduction in current without a prominent shift in the voltage dependence of gating (48, 49). We see some effects on KCNQ1/KCNE1 current density in the tail current measurements with the various manipulations of PIP2 and binding site mutants but the shift in voltage dependence is much more important. A general issue is that functional studies in combination with site-directed mutagenesis to determine binding sites can be complicated by functional antagonism and other effects (50). However, we have in addition directly shown in vitro binding with purified channel domains and this overcomes some of these problems. It is still possible that these residues may not be directly involved because of conformational effects on a binding site and this could only be unambiguously determined by crystallization of the full-length channel with PIP2. However, the comparisons with the inward rectifiers are striking and a crystal structure is known here (36, 51). Furthermore, mutations known to affect KCNQ1 function in the long QT syndrome can in cases be functionally ameliorated by the addition of PIP2 (32). This contrasts with the effects of diC8-PIP2 addition on the K358A/R360A mutant.
FIGURE 9.
Alignment of human KCNQ1, KCNQ2, and KCNQ3. The highlighted residues in bold in KCNQ1 are those studied in this article. Histidine 361 is underlined and the region identified by Shapiro and colleagues (46, 47) in KCNQ2 is also highlighted. The extent of the deletion mutation in KCNQ1 is indicated by italics.
The C-terminal fusion proteins we generated had a wide binding specificity that would fit well with the proposed electrostatic binding model. It is conceivable these residues might also be important in recognizing phosphoinositides, other than PIP2, for example, in vesicle trafficking. It has been proposed that increased PtdIns(3,5)P2 promotes exocytosis of KCNQ1/KCNE1 containing vesicles and thus increases the current amplitude (52). Therefore, the reduction in current amplitude we see with the mutants could have additional explanations. Interestingly, the presence of PS in the liposomes used for surface plasmon resonance was required for binding of the KCNQ1 fusion proteins and without PS no binding of the full-length KCNQ1 was possible. PS has previously been shown to be required for the membrane recruitment of the C2 domain of PKCα (53). PIP2 and PtdIns(3,4,5)P3 make up only a fraction of the negative charge in the membrane and PS is a predominantly negatively charged lipid that has been shown to be involved in recruiting proteins to this location (54). We expressed the fusion proteins in the absence of calmodulin for the sake of simplicity. It has been reported that calmodulin is an essential protein component for the assembly of the channel and it significantly improves the yield and stability of the KCNQ1 C-terminal domain when co-expressed in bacteria (55, 56). However, we found using maltose-binding protein as a fusion partner for the C-terminal domain that this yielded significant quantities of soluble and stable protein.
The physiological significance of the regulation of KCNQ2/3 by muscarinic receptors via PIP2 in central neurones is well established (57). In a similar manner, inhibition of KCNQ1/KCNE1 via receptor activation is observed after heterologous expression and angiotensin II is known to inhibit IKs in guinea pig ventricular cells (15, 17, 58). There have also been reports that a number of mutations in the long QT syndrome impair channel-PIP2 interaction but these mutations are widely distributed throughout the protein and we think that they most likely act in an allosteric fashion (32, 60). There is one mutation reported in the residues we have identified (R360G) and this leads to a severe loss of function (59). We also made this mutant and confirmed these findings (data not shown). It would be interesting to investigate the effects of this mutation further. In summary, we have defined an anionic phospholipid binding site in KCNQ1 using a combination of biochemical and functional techniques. This work could provide a platform to explore the role of PIP2 in the biology of this important family of potassium channels.
Acknowledgment
We thank Professor David A. Brown (UCL, UK) for the kind gift of the CHO-hM1 cell line and for discussing and reading the manuscript.
This work was supported by the British Heart Foundation.
- PIP2
- phosphatidylinositol (4,5)-bisphosphate
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- PC
- phosphatidylcholine
- MBP
- maltose-binding protein
- PTCD
- peak tail current density
- Oxo-M
- oxotremorine methiodide
- RU
- response units
- ANOVA
- analysis of variance
- diC8
- diacylglycerol C8.
REFERENCES
- 1. Sanguinetti M. C., Jurkiewicz N. K. (1990) J. Gen. Physiol. 96, 195–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barhanin J., Lesage F., Guillemare E., Fink M., Lazdunski M., Romey G. (1996) Nature 384, 78–80 [DOI] [PubMed] [Google Scholar]
- 3. Sanguinetti M. C., Curran M. E., Zou A., Shen J., Spector P. S., Atkinson D. L., Keating M. T. (1996) Nature 384, 80–83 [DOI] [PubMed] [Google Scholar]
- 4. Moss A. J., Kass R. S. (2005) J. Clin. Invest. 115, 2018–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nerbonne J. M., Kass R. S. (2005) Physiol. Rev. 85, 1205–1253 [DOI] [PubMed] [Google Scholar]
- 6. Splawski I., Shen J., Timothy K. W., Lehmann M. H., Priori S., Robinson J. L., Moss A. J., Schwartz P. J., Towbin J. A., Vincent G. M., Keating M. T. (2000) Circulation 102, 1178–1185 [DOI] [PubMed] [Google Scholar]
- 7. Schroeder B. C., Waldegger S., Fehr S., Bleich M., Warth R., Greger R., Jentsch T. J. (2000) Nature 403, 196–199 [DOI] [PubMed] [Google Scholar]
- 8. Wang H. S., Pan Z., Shi W., Brown B. S., Wymore R. S., Cohen I. S., Dixon J. E., McKinnon D. (1998) Science 282, 1890–1893 [DOI] [PubMed] [Google Scholar]
- 9. Shapiro M. S., Roche J. P., Kaftan E. J., Cruzblanca H., Mackie K., Hille B. (2000) J. Neurosci. 20, 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schroeder B. C., Kubisch C., Stein V., Jentsch T. J. (1998) Nature 396, 687–690 [DOI] [PubMed] [Google Scholar]
- 11. Singh N. A., Charlier C., Stauffer D., DuPont B. R., Leach R. J., Melis R., Ronen G. M., Bjerre I., Quattlebaum T., Murphy J. V., McHarg M. L., Gagnon D., Rosales T. O., Peiffer A., Anderson V. E., Leppert M. (1998) Nat. Genet. 18, 25–29 [DOI] [PubMed] [Google Scholar]
- 12. Schroeder B. C., Hechenberger M., Weinreich F., Kubisch C., Jentsch T. J. (2000) J. Biol. Chem. 275, 24089–24095 [DOI] [PubMed] [Google Scholar]
- 13. Kubisch C., Schroeder B. C., Friedrich T., Lütjohann B., El Amraoui A., Marlin S., Petit C., Jentsch T. J. (1999) Cell 96, 437–446 [DOI] [PubMed] [Google Scholar]
- 14. Brown D. A., Adams P. R. (1980) Nature 283, 673–676 [DOI] [PubMed] [Google Scholar]
- 15. Selyanko A. A., Hadley J. K., Wood I. C., Abogadie F. C., Jentsch T. J., Brown D. A. (2000) J. Physiol. 522, 349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown D. A., Hughes S. A., Marsh S. J., Tinker A. (2007) J. Physiol. 582, 917–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Matavel A., Lopes C. M. (2009) J. Mol. Cell Cardiol. 46, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang H., Craciun L. C., Mirshahi T., Rohács T., Lopes C. M., Jin T., Logothetis D. E. (2003) Neuron 37, 963–975 [DOI] [PubMed] [Google Scholar]
- 19. Suh B. C., Inoue T., Meyer T., Hille B. (2006) Science 314, 1454–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suh B. C., Hille B. (2005) Curr. Opin. Neurobiol. 15, 370–378 [DOI] [PubMed] [Google Scholar]
- 21. Winks J. S., Hughes S., Filippov A. K., Tatulian L., Abogadie F. C., Brown D. A., Marsh S. J. (2005) J. Neurosci. 25, 3400–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hilgemann D. W. (2007) J. Physiol. 582, 903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang C. L., Feng S., Hilgemann D. W. (1998) Nature 391, 803–806 [DOI] [PubMed] [Google Scholar]
- 24. Zhang H., He C., Yan X., Mirshahi T., Logothetis D. E. (1999) Nat. Cell Biol. 1, 183–188 [DOI] [PubMed] [Google Scholar]
- 25. Shyng S. L., Cukras C. A., Harwood J., Nichols C. G. (2000) J. Gen. Physiol. 116, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomas A. M., Brown S. G., Leaney J. L., Tinker A. (2006) J. Membr. Biol. 211, 43–53 [DOI] [PubMed] [Google Scholar]
- 27. Loussouarn G., Park K. H., Bellocq C., Baró I., Charpentier F., Escande D. (2003) EMBO J. 22, 5412–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomas A. M., Tinker A. (2008) Methods Mol. Biol. 491, 103–111 [DOI] [PubMed] [Google Scholar]
- 29. Harmer S. C., Wilson A. J., Aldridge R., Tinker A. (2010) Am. J. Physiol. Cell Physiol. 298, C263–C273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horowitz L. F., Hirdes W., Suh B. C., Hilgemann D. W., Mackie K., Hille B. (2005) J. Gen. Physiol. 126, 243–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilson A. J., Quinn K. V., Graves F. M., Bitner-Glindzicz M., Tinker A. (2005) Cardiovasc. Res. 67, 476–486 [DOI] [PubMed] [Google Scholar]
- 32. Park K. H., Piron J., Dahimene S., Mérot J., Baró I., Escande D., Loussouarn G. (2005) Circ. Res. 96, 730–739 [DOI] [PubMed] [Google Scholar]
- 33. Suh B. C., Hille B. (2002) Neuron 35, 507–520 [DOI] [PubMed] [Google Scholar]
- 34. Mashanov G. I., Nobles M., Harmer S. C., Molloy J. E., Tinker A. (2010) J. Biol. Chem. 285, 3664–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fan Z., Makielski J. C. (1997) J. Biol. Chem. 272, 5388–5395 [DOI] [PubMed] [Google Scholar]
- 36. Nishida M., Cadene M., Chait B. T., MacKinnon R. (2007) EMBO J. 26, 4005–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Santagata S., Boggon T. J., Baird C. L., Gomez C. A., Zhao J., Shan W. S., Myszka D. G., Shapiro L. (2001) Science 292, 2041–2050 [DOI] [PubMed] [Google Scholar]
- 38. Rapedius M., Fowler P. W., Shang L., Sansom M. S., Tucker S. J., Baukrowitz T. (2007) Neuron 55, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guy-David L., Reuveny E. (2007) Neuron 55, 537–538 [DOI] [PubMed] [Google Scholar]
- 40. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 41. Bian J., Cui J., McDonald T. V. (2001) Circ. Res. 89, 1168–1176 [DOI] [PubMed] [Google Scholar]
- 42. Bian J. S., Kagan A., McDonald T. V. (2004) Am. J. Physiol. Heart Circ. Physiol. 287, H2154–H2163 [DOI] [PubMed] [Google Scholar]
- 43. Rodriguez N., Amarouch M. Y., Montnach J., Piron J., Labro A. J., Charpentier F., Mérot J., Baró I., Loussouarn G. (2010) Biophys. J. 99, 1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hilgemann D. W., Feng S., Nasuhoglu C. (2001) Sci. STKE, RE19. [DOI] [PubMed] [Google Scholar]
- 45. Hoshi N., Zhang J. S., Omaki M., Takeuchi T., Yokoyama S., Wanaverbecq N., Langeberg L. K., Yoneda Y., Scott J. D., Brown D. A., Higashida H. (2003) Nat. Neurosci. 6, 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hernandez C. C., Zaika O., Shapiro M. S. (2008) J. Gen. Physiol. 132, 361–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hernandez C. C., Falkenburger B., Shapiro M. S. (2009) J. Gen. Physiol. 134, 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suh B. C., Horowitz L. F., Hirdes W., Mackie K., Hille B. (2004) J. Gen. Physiol. 123, 663–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gamper N., Shapiro M. S. (2007) Nat. Rev. Neurosci. 8, 921–934 [DOI] [PubMed] [Google Scholar]
- 50. Colquhoun D. (1998) Br. J. Pharmacol. 125, 924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nishida M., MacKinnon R. (2002) Cell 111, 957–965 [DOI] [PubMed] [Google Scholar]
- 52. Seebohm G., Strutz-Seebohm N., Birkin R., Dell G., Bucci C., Spinosa M. R., Baltaev R., Mack A. F., Korniychuk G., Choudhury A., Marks D., Pagano R. E., Attali B., Pfeufer A., Kass R. S., Sanguinetti M. C., Tavare J. M., Lang F. (2007) Circ. Res. 100, 686–692 [DOI] [PubMed] [Google Scholar]
- 53. Manna D., Bhardwaj N., Vora M. S., Stahelin R. V., Lu H., Cho W. (2008) J. Biol. Chem. 283, 26047–26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., Grinstein S. (2008) Science 319, 210–213 [DOI] [PubMed] [Google Scholar]
- 55. Ghosh S., Nunziato D. A., Pitt G. S. (2006) Circ. Res. 98, 1048–1054 [DOI] [PubMed] [Google Scholar]
- 56. Shamgar L., Ma L., Schmitt N., Haitin Y., Peretz A., Wiener R., Hirsch J., Pongs O., Attali B. (2006) Circ. Res. 98, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 57. Marrion N. V. (1997) Annu. Rev. Physiol. 59, 483–504 [DOI] [PubMed] [Google Scholar]
- 58. Daleau P., Turgeon J. (1994) Pflugers Arch. 427, 553–555 [DOI] [PubMed] [Google Scholar]
- 59. Yang T., Chung S. K., Zhang W., Mullins J. G., McCulley C. H., Crawford J., MacCormick J., Eddy C. A., Shelling A. N., French J. K., Yang P., Skinner J. R., Roden D. M., Rees M. I. (2009) Circ. Arrhythm. Electrophysiol. 2, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Matavel A., Medei E., Lopes C. M. (2010) Channels 4, 3–11 [DOI] [PubMed] [Google Scholar]