Abstract
One of the major characteristics of tumors is their ability to evade immunosurveillance through altering the properties and functions of host stromal and/or immune cells. CCL5 has been shown to play important roles in T cell proliferation, IFN-γ, and IL-2 production, which promotes the differentiation and proliferation of Th1 cells important for immune defense against intracellular infection. In this study we found that tumor-bearing mice were more susceptible to bacterial infection and showed reduced CCL5 levels in serum during endotoxic shock. Our data further demonstrated that the soluble factors secreted by mammary gland tumor cells but not normal mammary gland epithelial cells inhibited CCL5 expression in macrophages in response to LPS, but not to TNF-α stimulation. The inhibitory effect of tumor-secreted molecules on LPS-induced CCL5 expression was regulated at the post-transcriptional level. Blocking PGE2 synthesis by NS398 or through the use of PGE2 receptor antagonists AH-6809 (EP2 antagonist) and AH-23848 (EP4 antagonist) completely reversed the inhibitory effect of tumor-conditioned medium (TCM) on LPS-induced CCL5 expression. Moreover, PGE2 and the cAMP analog forskolin could mimic tumor-mediated CCL5 inhibition, and the inhibitory effects of TCM, PGE2, and cAMP analog on LPS-induced CCL5 expression could be completely reversed by the PKA inhibitor H89. Furthermore, blocking PGE2 synthesis in vivo led to partial recovery of CCL5 production during endotoxic shock. Taken together, our data indicate that PGE2 secreted from breast cancer cells suppresses CCL5 secretion in LPS-activated macrophages through a cAMP/PKA signaling pathway, which may result in suppression of host immune responses against subsequent bacterial infection.
Keywords: Breast Cancer, Chemokines, Innate Immunity, Lipopolysaccharide (LPS), Signal Transduction
Introduction
Solid tumors consist of both tumor and stromal cells surrounded by an extracellular matrix. The interactions between tumor cells and host cells through cytokines, chemokines, lipid molecules, and other factors create a unique microenvironment, which is favorable for tumor progression. Tumor-mediated immune suppression is a general phenomenon in cancer patients, which can be mediated via multiple mechanisms such as tumor-secreted IL-10, TGF-β, and PGE2; induction of immunosuppressive regulatory T cells or through the inhibition of CD4+ Th1 and CD8+ effector T cells (1).
Chemokines are small soluble molecules that are best known for their potent abilities to induce cellular migration, particularly by leukocytes, during inflammation and infection (2–4). Migration of leukocytes to the sites of infection is a critical step for initiating a proper immune response against invading pathogens. Regulated upon Activation, Normal T cell Expressed and Secreted (RANTES, CCL5) is a member of the CC chemokine family which also includes monocyte chemoattractant protein (MCP)-1, MCP-2, MCP-3, and macrophage inhibitory protein (MIP)-1α, MIP-1β, and I-309. CCL5 was originally discovered by subtractive hybridization in T cells but not in B cells and is expressed in platelets, macrophages, tubular epithelium, synovial fibroblasts, as well as select tumor cells (5). CCL5 plays an essential role in inflammation by recruiting T cells, macrophages, dendritic cells, eosinophils, NK cells, mast cells, and basophils to the sites of inflammation and infection (6, 7). It has been reported that CCL5 could activate macrophages to kill Trypanosoma cruzi through the induction of nitric oxide (8, 9) and promote macrophage recruitment into the lungs following endotoxemia (10). CCL5 levels inversely correlated with the APACHE (Acute Physiology And Chronic Health Evaluation) II score predicting poor outcomes (11). Besides its role as a potent chemoattractant, CCL5 plays an important role in T cell responses by facilitating the differentiation and proliferation of Th1 cells, inducing CD8 T cell responses (12–14), and enhancing type 1 immune responses through working synergistically with IFN-γ to activate macrophages, NK cells and T cells (15). Therefore, production of CCL5 is important for inducing proper immune responses against infection.
4T1 is a tumor cell line isolated from a single spontaneously arising mammary tumor from a Balb/BfC3H mouse (murine mammary tumor virus, MMTV+) (16). It is an excellent model for breast cancer research because 4T1 tumor development is well characterized both oncologically and immunologically (17). Tumor growth and metastatic spread of 4T1 cells closely mimics stage IV breast cancer (18). 4T1 tumors spontaneously metastasize to a variety of target organs, including the lungs, heart, bone, brain and liver (19). Because of the unique characteristics of 4T1 tumor, this model has been widely used to investigate important issues such as immunotherapy (20), metastasis (21), anti-angiogenesis therapy (8), and multiple chemotherapy treatments (22, 23). 4T1 tumors are poorly immunogenic in that immunization with irradiated 4T1 cells provides only slight delays in tumor growth which are not sufficient to protect the animal (24). Ours and others work demonstrated that administration of IL-12 to 4T1 tumor-bearing mice resulted in a substantial reduction in tumor size and decreased spontaneous metastases to the lungs, and significantly prolonged their survival time (25, 26). In addition, CCL5 has been detected in many clinical specimens of breast cancers (27, 28) and the presence of a high level of CCL5 in the plasma is a poor prognostic factor in inflammatory breast cancer (29, 30). 4T1 tumors also express high levels of CCL5 and blocking CCL5 signaling pathway with Met-RANTES in mice suppressed 4T1 mammary tumor growth (31). Taken together, human and mouse studies indicate that CCL5 produced from tumor cells plays an important role in breast cancer progression. However, little is known about how tumors affect host CCL5 production during infection.
It has been recognized for a long time that cancer patients have a high risk of infection (32, 33). Because CCL5 belongs to type I chemokine important for Th1 response and induction of CCL5 during endotoxemia correlates with protection (11), we decided to investigate if the presence of tumor could have any negative effect on host CCL5 production during infection. In this study, we found that tumor-bearing mice produced less CCL5 in vivo during septic shock compared with tumor-free mice. Soluble molecules secreted by tumors inhibited CCL5 protein production and mRNA expression in macrophages stimulated with LPS. The inhibitory effect of tumor-secreted molecule on CCL5 expression was regulated at the post-transcriptional level. Moreover, we found that tumor-secreted PGE2 mediated this inhibitory effect through a cAMP/PKA signaling pathway.
MATERIALS AND METHODS
Mice
6∼8-week-old female Balb/c mice (obtained from The Jackson Laboratories, Bar Harbor, ME) were housed in cages with filter tops in a laminar flow hood, fed food, and filtered water ad libitum at Saint Louis University Animal Facilities in accordance with the principles of Animal Care (National Institutes of Health publication number 85-23, revised 1985). For inducing endotoxic shock, mice were injected intraperitoneally (intraperitoneal) with lipopolysaccharide from Escherichia coli 0217:B8 (Sigma-Aldrich) (25 mg/kg). Four hours later, mice were sacrificed for collection of serum and for isolation of spleens.
Cells
The murine macrophage cell line RAW264.7 and 4T1 mammary gland tumor cells were obtained from the American Type Culture Collection (ATCC), and maintained in RPMI 1640 supplemented with 2 mm glutamine, 100 units/ml of penicillin and streptomycin and 10% FBS (Sigma, endotoxin NMT, 10.0 EU/ml). Normal mammary gland epithelial cell line, CommaD Sego cells isolated from mammary glands of female Balb/c mice (34), were kindly provided by Dr. Daniel Medina (Baylor College of Medicine), and maintained in DMEM supplemented with 5 ml HEPES buffer (per 500 ml), insulin (10 μg/ml), EGF (5 ng/ml), and 2% FBS. Mouse peritoneal macrophages were obtained by lavage 3 days after injection of sterile 3% thioglycolate broth (1 ml of intraperitoneal per mouse). Cells were washed and resuspended in RPMI containing 10% FBS and standard supplements. Macrophages were plated in 24-well tissue culture plates (1 × 106 cells/well). After 2 h of incubation to allow for adherence of macrophages, monolayers were washed three times to remove nonadherent cells, and incubated with RPMI containing 10% FBS and standard supplements. The next day, different amounts of conditioned medium collected from 4T1, B4, and 168 mammary tumor cells were added for different times as indicated in the text. Both B4 and 168 tumor cells were kindly provided by Dr. Robert Kurt at Lafayatte College (35). Tumor-conditioned media (TCMs)2 were prepared by inoculation of log-growth phase of tumor cells (4T1, B4, and 168 cells) into a T175 flask at a concentration of 0.6 × 106/ml with freshly prepared complete culture medium. 72 h later, the culture supernatants were collected, filtered through a 0.45-μm filter, aliquoted in small tubes, and stored at −20 °C for future use.
Tumor Implantation and NS398 Treatment
1 × 105 4T1 tumor cells in 100 μl phosphate-buffered saline was injected subcutaneously (s.c.) into the abdominal mammary gland area on day 0. After 7 days, NS398 (10 mg/kg) dissolved in DMSO was given intraperitoneally every 3 days until 19 days after 4T1 cell injection. The same amount of DMSO was used as negative control. 19 days after 4T1 tumor cell injection, both tumor-free and tumor-bearing mice were euthanized and blood was taken by cardiac puncture, followed by collection of serum for PGE2 and CCL5 protein assay. Mouse peritoneal macrophages were obtained as described above and cultured at 1 × 106 cells/well in 24 well plates. Culture supernatants were collected to measure levels of CCL5 protein, and RNA was isolated from macrophages.
Cecal-ligation and Puncture (CLP)
CLP was performed according to the protocol published previously (36). Briefly, 19 days after 4T1 tumor cell injection, both tumor-free and tumor-bearing mice were anesthetized with pentobarbital (50 mg per kg body weight). Then, the abdomen was shaved with an electric trimmer and disinfected with alcohol prep pads. A longitudinal skin midline incision (1.5–2 cm) was made with a scalpel. Then, the cecum was located and isolated using blunt anatomical forceps, followed by ligating the cecum at an upside position. The cecum was punctured using a 26 gauge needle and placed back into the abdominal cavity, followed by closure of the abdomen by suture. Mice were returned to clean cages at the end of surgical procedures where water and food were readily available and were closely monitored every 2 h for survival.
Plasmids and Reagents
A plasmid encoding the murine CCL5 promoter that extended from −979 to +8 was generated previously (37). All plasmid DNA for transfection were prepared with Qiagen Endo-free Maxi-Prep kits (Qiagen Inc. Valencia, CA). LPS from E. coli 0217:B8 was purchased from Sigma-Aldrich. PGE2 receptor antagonists SC-51322 (EP1), AH-23848 (EP2), AH-6809 (EP4), COX2 inhibitor NS398, and PKA inhibitor H89 were purchased from Cayman Chemical (Ann Arbor, MI).
Reverse Transcription-PCR (RT-PCR)
Reverse-transcription (RT) reactions were carried out as follows: 1-μg aliquots of DNase-treated RNA were mixed with 1 μl of oligo dT primers (0.5 mg/ml), 1 μl of 10 mm dNTPs and ddH2O to equalize volumes of all samples at 12 μl. The mixture was heated at 65 °C for 5 min, quenched on ice and spun down briefly, followed by adding 8 μl of a Master Mix consisting of 4 μl of 5 × first strand buffer (Invitrogen), 2 μl of 0.1 m DTT, 1 μl of RNase inhibitor (40 units/μl, Invitrogen), and 1 μl of Superscript II (200 μl/μl, Invitrogen). The reaction was incubated at 42 °C for 60 min, then at 70 °C for 15 min, followed by 4 °C soak. To each sample (in 20 μl of total volume) 80 μl of ddH2O was added. 5 μl of diluted cDNA was used for each PCR reaction of 25 μl volume. The following primers were used for PCR amplification of mouse CCL5 cDNA: sense, GATGGACATAGAGGACACAACT; antisense, TGGG-ACGGCAGATCTGAGGG; for mouse GAPDH cDNA: sense, AACTTTGGCATTGTGGAAGG; antisense, ACACATTGGGGGTAGGAACA.
Quantitative Real-time PCR (qRT-PCR)
To determine the level of mRNA expression by quantitative real time PCR, we used a modified protocol. Briefly, cDNA samples converted from 1 μg of total RNA, were diluted and studied at several concentrations. Diluted cDNA was mixed with a pair of primers (10 μm) targeting mouse CCL5 or GAPDH cDNA sequences as described above, and with SYBR green PCR master mix (Applied Biosystems, CA) in a 15 μl volume. PCR cycling was as followed as: 2 min at 50 °C, 10 min at 95 °C for 1 cycle, followed by 40 cycles at 15 s at 95 °C, 1 min at 60 °C.
Enzyme-linked Immunosorbent Assays (ELISA)
Supernatants from murine peritoneal macrophage and RAW cell cultures were harvested 24 h after stimulation and stored at −70 °C. Mouse CCL5 was detected using the DuoSet ELISA kits (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions. The concentration of PGE2 in serum and supernatants was detected using the prostaglandin E2 EIA kit (Cayman Chemical, Ann Arbor, MI). Concentrations were calculated by regression analysis of a standard curve.
Transfection Assay
Transient transfections were performed by electroporation. Briefly, for each condition 0.4 ml of a RAW cell suspension containing 1 × 107 cells was mixed with 16 μg of total DNA (including reporter, effector, internal control and carrier DNA) and electroporated in 0.45 cm electroporation cuvettes (Gene Pulser II, Bio-Rad) at 975 microfarade and 300 V in RPMI 1640 medium without serum. The transfected cells from different cuvettes were resuspended in RPMI 1640 containing 10% FBS, 2 mm glutamine, 10 μm chloroquine, and antibiotics, added to 24-well plates, and incubated for 48 h prior to harvesting. To measure luciferase activity, cells were pelleted by centrifugation and resuspended in 100 μl of lysis buffer containing 125 mm Tris-phosphate pH 7.8, 10 mm DTT, 10 mm 1–2-diaminocyclohexane-tetraacetic acid, 50% glycerol, 5% Triton X-100. Luciferase activity was measured in cell lysates. Transfection efficiency was routinely monitored by β-galactosidase (β-gal) assay by co-transfection with 3 μg of pCMV-β-galactosidase plasmid. Variability in β-gal activity between samples was typically within 5%. Lysates were used for both luciferase and β-gal assays.
Primary Transcript Measurement
To determine primary transcript of CCL5 gene, cDNA were synthesized with random primers using 1 μg of DNase-treated RNA generated from mouse macrophages stimulated with LPS, LPS plus tumor-conditioned medium, and LPS plus PGE2. The two sets of primers used for measurement of primary transcript by qRT-PCR were: CCL5 intron 1 (sense): TAAAGAGCCCAGCATAGCTGGCAA; CCL5 exon 2 (antisense): ACGACTGCAAGATTGGA GCACTTG, for the first set; CCL5 intron 2 (sense): TGCCTCTGGTTCCAAAGGAGAAGA, CCL5 exon 3 (antisense): TCTTCTCTGGGTTGGCACACACTT, for the second set.
Statistical Analysis
Student's t test was performed wherever applicable. Standard deviation of the mean is shown unless otherwise indicated. *: p < 0.05, **: p < 0.01, ***: p < 0.001.
RESULTS
CCL5 Expression Is Reduced in Tumor-bearing Mice during Endotoxic Shock
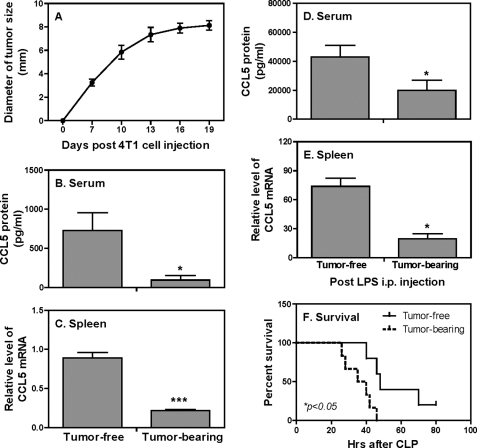
To investigate the effects of mammary gland tumor on CCL5 expression during infection, we used an endotoxic shock model by injecting LPS into 4T1 tumor-bearing mice, followed by measurement of CCL5 mRNA expression and protein production. Consistent with our previous studies (38, 39), the sizes of tumors reached about 8 mm in diameter 19 days after 4T1 tumor cell injection (Fig. 1A). At this time point, the levels of CCL5 protein in serum (Fig. 1B) and mRNA expression in spleen (Fig. 1C) in tumor-bearing mice were significantly reduced compared with tumor-free mice. To induce endotoxic shock we injected LPS into the peritoneal cavity of both tumor-free and tumor-bearing mice and collected sera and spleens for CCL5 detection 4 h later. LPS injection significantly increased CCL5 secretion (Fig. 1D) and mRNA expression (Fig. 1E) in both tumor-free and tumor-bearing mice compared with the mice without LPS treatment (Fig. 1, B and C). However, CCL5 protein secretion (Fig. 1D) and mRNA expression (Fig. 1E) in tumor-bearing mice were markedly reduced compared with tumor-free mice. To determine whether tumor-bearing mice indeed were more susceptible to infection, we employed a cecal-ligation and puncture (CLP) model to induce peritonitis with endogenous bacteria. As shown in Fig. 1F, tumor-bearing mice not only died earlier than tumor-free mice but none of them survived CLP-induced peritonitis, suggesting that the reduced CCL5 production in tumor-bearing hosts during endotoxemia may compromise host immune competence against bacterial challenge.
FIGURE 1.
CCL5 protein and mRNA expression are impaired in tumor-bearing mice during endotoxic shock. A, 1 × 105 4T1 tumor cells were injected s.c. in the abdominal mammary gland of female Balb/c mice. Tumor size in diameter (horizontal and vertical) was measured. 19 days after 4T1 cell injection, tumor-free and tumor-bearing mice were either left untreated (B and C) or injected intraperitoneal with LPS (25 mg/kg) (D and E). Four hours after LPS injection, mice were sacrificed, and serum was collected for measurement of CCL5 protein production by ELISA (B and D). Meanwhile spleens were used to extract total RNA for detection of CCL5 mRNA expression by qRT-PCR (C and E). qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the sample from the tumor-free group, which was set as 1. *, p < 0.05 and ***, p < 0.001 between two groups. Data shown represent one of three experiments with similar results and 10 mice in each group. F, 19 days after 4T1 cell injection, CLP was performed on tumor-free and tumor-bearing mice (n = 7 for each group) and survival was closely monitored in every 2 h. Data shown represent one of two experiments with similar results.
Soluble Factors Secreted by Tumor Cells Inhibit LPS-induced CCL5 Expression in Macrophages
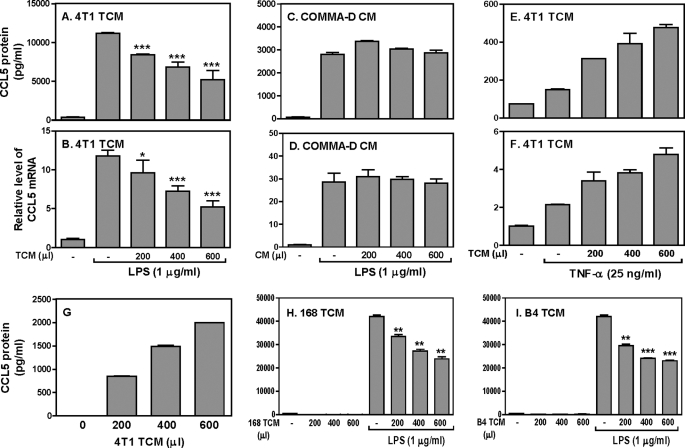
To investigate the effects of tumor on host CCL5 expression, we tested whether tumor-secreted molecules that can suppress CCL5 expression in macrophages since macrophages are a major producer of CCL5. RAW cells were stimulated with LPS and TCM collected from 4T1 mammary gland tumor cells, followed by measurement of CCL5 protein production and mRNA expression. 4T1 TCM suppressed LPS-induced CCL5 protein production (Fig. 2A) and mRNA expression (Fig. 2B) about 2-fold, which is similar to the levels of inhibition seen in tumor-bearing mice (Fig. 1, D and E). Mouse peritoneal macrophages had similar responses to TCM as the RAW cells (supplemental Fig. S1, A and B). To exclude the possibility that the soluble factors secreted from normal mammary gland epithelial cells may inhibit CCL5 expression, we collected the conditioned medium (CM) from the CommaD Bgeo cells (normal mammary gland epithelial cells isolated from Balb/c mice) and treated the macrophages with both CommaD Bgeo CM and LPS followed by measurement of CCL5 protein and mRNA expression. The results show that CommaD CM did not inhibit CCL5 protein (Fig. 2C) and mRNA (Fig. 2D) expression, indicating that the soluble factors secreted from tumor cells can suppress LPS-induced CCL5 expression. To evaluate whether the inhibitory effect is restricted to TLR4 signaling pathway, we stimulated the macrophages with TNF-α in the presence of 4T1 TCM. In contrast to LPS stimulation, 4T1 TCM enhanced TNF-α-induced CCL5 protein (Fig. 2E) and mRNA (Fig. 2F) expression in a dose-dependent manner, suggesting that soluble factors secreted from 4T1 tumor cells selectively target TLR4 signaling.
FIGURE 2.
4T1 tumor-conditioned medium inhibits LPS-induced CCL5 protein and mRNA expression in mouse macrophages. 1 × 106 RAW cells were stimulated with LPS (1 μg/ml) plus different amounts of 4T1 TCM as indicated for different times, followed by collection of culture supernatants (24 h after treatment) and total RNA (4 h after treatment) for measurement of CCL5 protein production by ELISA (A) and mRNA expression by qRT-PCR (B), respectively. 1 × 106 RAW cells were stimulated with LPS (1 μg/ml) in the absence or presence of different amounts of culture supernatants collected from the CommaD Bgeo cells as indicated to detect CCL5 protein (24 h after treatment) and mRNA expression (4 h after treatment) by ELISA (C) and qRT-PCR (D), respectively. The same number of RAW cells were stimulated with TNF-α (25 ng/ml) in the absence or presence of different amounts of 4T1 TCM as indicated to detect CCL5 protein (24 h after treatment) and mRNA expression (8 h after treatment) by ELISA (E) and qRT-PCR (F), respectively. G, different amounts of 4T1 TCM as indicated were directly used to measure the CCL5 protein levels by ELISA. Different amounts of conditioned medium collected from 168 (H) and B4 (I) tumor cells as indicated were either used directly or added to 1 × 106 RAW cells stimulated with LPS (1 μg/ml) for 24 h for measurement of CCL5 protein production in supernatants by ELISA. All data shown are means plus S.D. from three different experiments. *, p < 0.05; **, p < 0.01 between LPS-stimulated group and groups treated with LPS plus various amounts of TCM.
We noticed that 4T1 tumor cells secret a lot of CCL5 by themselves (Fig. 2G), which may affect macrophage CCL5 expression in an autocrine manner. To exclude this possibility, we cultured two different mammary gland tumor cell lines lacking of CCL5 expression, 168 and B4 cells, collected TCMs from these two types of tumor cells, and applied these TCMs to LPS-treated cells as described above. Interestingly, both TCMs collected from the 168 (Fig. 2H) and B4 (Fig. 2I) tumor cells were still able to suppress LPS-induced CCL5 expression, indicating that the inhibitory effect of tumor cells on CCL5 expression is not mediated by tumor-secreted CCL5.
The Inhibitory Effect of TCM on CCL5 Expression Is Regulated at the Post-transcriptional Level
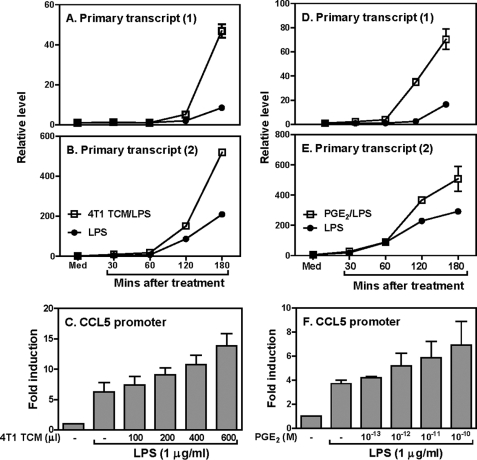
To investigate the molecular mechanisms of tumor-mediated CCL5 inhibition, we measured primary transcript rates of the CCL5 gene in response to 4T1 CM in RAW cells, with two pairs of primers corresponding to two different regions of the CCL5 gene. The use of primers specific for an intron/exon boundary region allowed us to assess the primary transcript rate of CCL5 inside the nucleus. Supplemental Fig. S2 shows a schematic of the primers used in these experiments. As shown in Fig. 3, A and B, CCL5 primary transcript was induced by LPS as early as 30 min and increased up to 3 h after LPS simulation when both primer pairs were used. To our surprise, the 4T1 TCM did not inhibit LPS-induced CCL5 primary transcript but rather increased it (Fig. 3, A and B), indicating that the inhibitory effect of tumor cells on CCL5 expression is mediated at the post-transcriptional level. To further verify the post-transcriptional inhibition of CCL5 by tumor cells, we transiently transfected the mouse CCL5 promoter into RAW cells and treated the transfected cell with different amounts of 4T1 TCM, followed by measurement of luciferase activities in cell lysates. As shown in Fig. 3C, consistent with the primary transcript results, LPS strongly activated the CCL5 promoter and 4T1 TCM further increased LPS-induced CCL5 promoter activity in a dose-dependent manner. Because our previous studies demonstrated that 4T1 tumor cells secret many molecules, including prostaglandin E2 (PGE2) that can suppress host immunity (38), we reasoned that PGE2 might mediate CCL5 inhibition by tumor cells. Therefore, we first measured the effects of PGE2 on LPS-induced CCL5 primary transcript. To our surprise, PGE2 had similar effects on CCL5 primary transcript as the 4T1 TCM in that PGE2 enhanced LPS-induced CCL5 primary transcript (Fig. 3, D and E). Furthermore, PGE2 also increased LPS-induced CCL5 promoter activity (Fig. 3F). Taken together, these data indicate that tumor-secreted molecules suppress CCL5 expression at the post-transcriptional level and PGE2 has a similar effect as the TCM on CCL5 primary transcription.
FIGURE 3.
Inhibition of LPS-induced CCL5 expression by TCM is regulated at the post-transcriptional level. CCL5 gene primary transcript was measured by qRT-PCR using PT-1 (A) and PT-2 (B) primers. Data were normalized relative to GAPDH gene expression levels in each respective sample and further normalized to the results from the untreated group (Med), which was set as 1. C, 10 × 106 RAW cells were transiently transfected with 5 μg of mouse CCL5 luciferase reporter construct as previous published (48). The transfected cells were stimulated with different amounts of 4T1 TCM as indicated for 7 h in the presence of LPS (1 μg/ml), followed by measurement of luciferase activity. 3 × 106 RAW cells were stimulated with LPS (1 μg/ml) plus 1 × 10−10 m PGE2 for different times as indicated, followed by extraction of total RNA to measure the CCL5 gene primary transcript as described above (D and E). 10 × 106 RAW cells were transiently transfected with 5 μg mouse CCL5 luciferase reporter construct as described above and the transfected cells were stimulated with different amounts of PGE2 as indicated for 7 h in the presence of LPS (1 μg/ml), followed by measurement of luciferase activity (F). Luciferase data were normalized to the activity obtained with non-treated cells and represented as fold induction. All data shown are means plus S.D. of three-four experiments.
The Inhibitory Effect of TCM on CCL5 Expression Is Mediated by Tumor-secreted PGE2
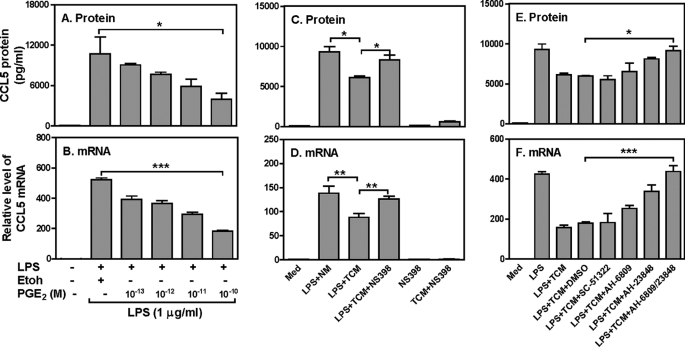
Because the above data showing that PGE2 has similar effect as 4T1 TCM on CCL5 primary transcription, next we wanted to know whether PGE2, like 4T1 TCM, inhibited LPS-induced CCL5 production. Indeed, PGE2 suppressed LPS-induced CCL5 protein (Fig. 4A) and mRNA (Fig, 4B) expression in a dose-dependent manner. Thus, we treated the 4T1 cells with NS398, a specific COX2 inhibitor which can effectively block PGE2 production, collected the culture supernatants, and stimulated RAW cells with the supernatants collected from the NS398-treated 4T1 cells, followed by measuring CCL5 protein (Fig. 4C) and mRNA (Fig. 4D) expression by ELISA and qRT-PCR, respectively. As shown in Fig. 4, C and D, the inhibitory effect of TCM on CCL5 expression was almost completely reversed by blocking tumor PGE2 production, suggesting that PGE2 secreted by mammary gland tumor cells inhibits CCL5 expression in macrophages stimulated with LPS. To confirm the effect of NS398 on PGE2 production, we measured the concentration of PGE2 in 4T1 TCM. 4T1 TCM secreted high amounts of PGE2 and NS398 treatment almost completely abrogated PGE2 secretion from the 4T1 tumor cells (supplemental Fig. S3).
FIGURE 4.
The inhibitory effect of TCM on CCL5 expression is mediated by tumor-secreted PGE2. 1 × 106 RAW cells were stimulated with LPS (1 μg/ml) and various concentrations of PGE2 as indicated for different times to measure CCL5 protein (24 h after treatment) by ELISA (A) and mRNA expression (4 h after treatment) by qRT-PCR (B), respectively. PGE2 was dissolved in ethanol and added to a final concentration as indicated (each contained 0.01% ethanol in final concentration). 1 × 106 RAW cells were cultured with normal media (NM), 400 μl/ml of 4T1 TCM, or 400 μl/ml of TCM collected from 4T1 cells treated with NS398 in the presence or absence of LPS (1 μg/ml) for 4 h. Note that in the fifth and sixth bar from the left, NS398 was added to the macrophage culture, not to the 4T1 culture. CCL5 protein production and mRNA expression were measured by ELISA (C) and qRT-PCR (D), respectively. 1 × 106 RAW cells were pretreated with 10 μm SC51322 (EP1 antagonist), 3 μm AH6809 (EP2 antagonist) and 30 μm AH 23848 (EP4 antagonist) or AH6809 plus Ah23848 for 30 min, then stimulated with LPS (1 μg/ml) and 400 μl 4T1 TCM for different times for detection of CCL5 protein and mRNA expression by ELISA (E) and qRT-PCR (F), respectively. Data shown are means plus S.D. of three-four experiments. qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the sample from the untreated group, which was set as 1. *, p < 0.05 and **, p < 0.01 between two groups as indicated.
To further characterize the molecular mechanism whereby 4T1 TCM inhibits CCL5 production through PGE2, we used various PGE2 receptor antagonists to block PGE2 signaling, followed by testing whether the inhibitory effect of 4T1 TCM was affected. Because there are four prostanoid receptors (EP1-EP4) involved in PGE2 signaling, we blocked these PGE2 receptors individually with SC-51322 (a selective EP1 antagonist), AH-6809 (the EP2 receptor antagonist), and AH-23848 (a selective antagonist for EP4) prior to LPS and 4T1 TCM stimulation. EP1 antagonist had no effect on TCM-mediated CCL5 inhibition whereas EP2 and EP4 antagonists each could partially block the inhibitory effect of TCM on CCL5 protein (Fig. 4E) and mRNA (Fig. 4F) expression. More importantly, blocking both EP2 and EP4 receptors with AH23848 and AH 6809 could completely abolish the inhibitory effect of TCM on CCL5 protein (Fig. 4E) and mRNA (Fig. 4F) expression, further demonstrating that the PGE2 secreted from 4T1 tumor cells mediates the inhibition through binding to the EP2 and EP4 receptors.
The Inhibitory Effect of TCM on CCL5 Expression Is Mediated through cAMP/PKA Signaling Pathway
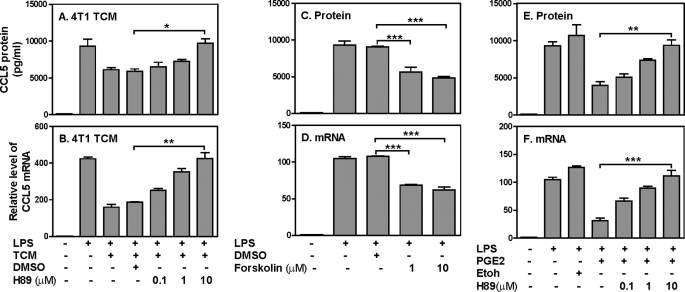
To determine whether the cAMP/PKA signaling pathway is involved in TCM-mediated CCL5 inhibition, we first blocked PKA function by H89 prior to 4T1 TCM and LPS stimulation, followed by measuring CCL5 expression. Blocking PKA by H89 dose-dependently reversed the inhibitory effect of 4T1 TCM on LPS-induced CCL5 protein (Fig. 5A) and mRNA (Fig. 5B) expression, with a complete recovery of CCL5 expression when 10 μm H89 used. In addition, forskolin, a cAMP analog, suppressed LPS-induced CCL5 protein (Fig. 5C) and mRNA (Fig. 5D) expression similar to the 4T1 TCM, further supporting the involvement of cAMP/PKA signaling in 4T1 TCM-mediated CCL5 inhibition. Moreover, PGE2-mediated CCL5 inhibition also signals through PKA because H89 could dose-dependently rescue PGE2-inhibited CCL5 protein (Fig. 5E) and mRNA (Fig. 5F) expression. Taken together, these results indicate that the PGE2 secreted from tumor cells inhibits LPS-induced CCL5 expression through cAMP/PKA signaling pathway.
FIGURE 5.
The inhibitory effect of TCM on CCL5 expression is mediated through cAMP/PKA signaling pathway. 1 × 106 RAW cells were pretreated with various amounts of H89 as indicated for 60 min, then stimulated with LPS (1 μg/ml) and 400 μl 4T1 TCM for different times followed by collection of culture supernatants (24 h after treatment) and total RNA (4 h after treatment) to measure CCL5 protein by ELISA (A) and mRNA expression by qRT-PCR (B), respectively. 1 μm and 10 μm of forskolin were added to LPS-stimulated RAW cells to measure CCL5 protein (24 h after treatment) by ELISA (C) and mRNA expression (4 h after treatment) by qRT-PCR (D), respectively. Same amount of dissolvent, DMSO was used as a negative control. H89 at various concentrations as indicated was added to 1 × 106 RAW cells 60 min prior to LPS (1 μg/ml) and PGE2 (1 × 10−10 m) treatments. CCL5 protein and mRNA expression were measured by ELISA (E) and qRT-PCR (F), respectively. Same amount of ethanol was used as a negative control. Data shown are means plus S.D. of 2–4 independent experiments. qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the sample from the untreated group, which was set as 1. ***, p < 0.001 between two groups as indicated.
Blocking PGE2 Synthesis by NS398 in vivo Partially Recovers CCL5 Production during Endotoxic Shock
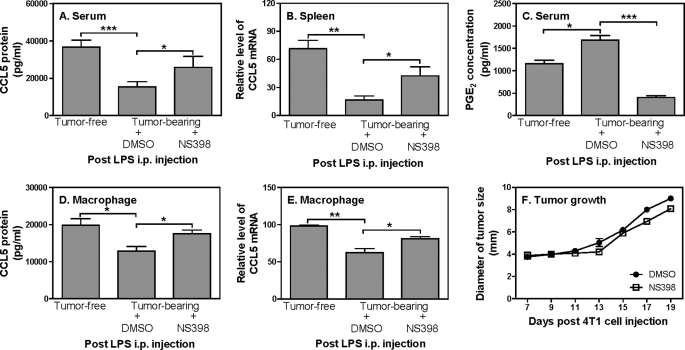
Our results suggest that tumor-secreted PGE2 suppresses LPS-induced CCL5 production in vitro. To verify whether PGE2 also mediates CCL5 inhibition in vivo, we blocked the PGE2 synthesis by NS398 in tumor-bearing mice and then challenged the mice with LPS, followed by measuring CCL5 expression in serum and spleen. Similar as the data shown in Fig. 1B-E, tumor-bearing mice displayed reduced CCL5 protein (Fig. 6A) and mRNA (Fig. 6B) expression compared with tumor-free mice. More importantly, the reduced CCL5 expression was partially recovered after blocking PGE2 synthesis by NS398 (Fig. 6, A and B), suggesting that PGE2 mediates the suppression of CCL5 in tumor-bearing host during infection. Indeed, tumor-bearing mice produced more PGE2 than tumor-free mice and the treatment with NS398 blocked 70% PGE2 production compared with DMSO-treated tumor-bearing mice (Fig. 6C). Because macrophages are a major producer of CCL5 in response to LPS stimulation, we isolated peritoneal macrophages from tumor-free and tumor-bearing mice treated, respectively, with DMSO and NS398, and collected the culture supernatants and total RNA from these cells to measure CCL5 protein and mRNA expression. As shown in Fig. 6, D and E, macrophages isolated from tumor-bearing mice secreted less CCL5 and the levels of CCL5 were enhanced after NS398 treatment compared with the cells isolated from the DMSO treated mice. To evaluate the effects of NS398 on mammary gland tumor growth, we measured the sizes of tumors in DMSO and NS398 treated mice every 3 days after 4T1 cell inoculation. The sizes of tumors in NS398-treated mice were slightly smaller than DMSO-treated mice though the difference had no statistical significance (Fig. 6F). Taken together, these data demonstrate that PGE2 secreted from tumor cells inhibits host CCL5 production in macrophages during infection.
FIGURE 6.
Blockade of PGE2 synthesis by NS398 in vivo partially recovers CCL5 production in tumor-bearing mice during endotoxic shock. A, 1 × 105 4T1 tumor cells were injected s.c. in the abdominal mammary gland in 0.1 ml PBS as described previously in Fig. 1. Seven days later, NS398 (10 mg/kg) and an equal amount of DMSO were given intraperitoneal every 3 days for total four times. 19 days post 4T1 cell injection, blood was drawn by cardiac puncture, and serum was collected for measurement of CCL5 concentration by ELISA. Meanwhile spleens were used to extract total RNA for detection of CCL5 mRNA expression by qRT-PCR (B) and the levels of PGE2 in serum were measured (C). Peritoneal macrophages were collected from tumor-free and tumor-bearing mice treated with DMSO and NS398 after LPS injection and cultured for 24h in vitro, followed by collection of culture supernatants to measure CCL5 protein by ELISA (D) or by extraction of total RNA to test CCL5 mRNA expression by qRT-PCR (E). qRT-PCR data were normalized relative to GAPDH mRNA expression levels in each respective sample and further normalized to the sample from the tumor-free group, which was set as 1. F, tumor growth was measured every 2–3 days after 4T1 cell injection in both DMSO and NS398 injected mice as described previously in Fig. 1. Each group includes four mice and data shown represent one of two experiments with similar results.
DISCUSSION
Previous studies with large numbers of patients at multi-centers or hospitals in the US and Europe reported that tumor patients have a high risk of infection (32, 33). Our study further demonstrates that tumor-bearing mice are more vulnerable to bacterial infection than tumor-free mice (Fig. 1F), and that the reduced CCL5 production during endotoxic shock (Fig. 1, D and E and Fig. 6, D and E) might be responsible for the impaired protective immunity in tumor-bearing mice. Because macrophages are known to play essential roles in infection, inflammation, and bacterial clearance by producing a large variety of mediators, including CCL5 (40, 41), in this study we focused on the effect of mammary gland tumor on CCL5 expression in macrophages. Kinetics of CCL5 mRNA expression are different from LPS and TNF-α treatments in that TNF-α-induced CCL5 mRNA expression peaked at 8 h (supplemental Fig. S4A) and LPS-induced CCL5 peaked at 2 h (supplemental Fig. S4B) after stimulation. We found that tumor-secreted molecules specifically inhibited LPS-induced CCL5 expression and had the opposite effect of TNF-α stimulation in macrophages (Fig. 2, E and F). These differential effects of TCM on LPS- and TNF-α-induced CCL5 expression further support our hypothesis that tumor-bearing host have impaired immunity against pathogen infection. Using normal mammary gland epithelial cells, we further demonstrate that the inhibition of CCL5 expression is restricted to tumor cells because conditioned medium collected from the CommaD cells did not suppress CCL5 expression (Fig. 2, C and D). Consistent with previous reports that breast tumor cells secrete CCL5 (27, 28, 42–44), our data show that 4T1 conditioned medium contained large amounts of CCL5 and this tumor-derived CCL5 did not mediate the inhibition of host CCL5 expression (Fig. 2, G–I). Considering the detrimental role of CCL5 in breast cancer development and its protective role in host defense against infection, it will be interesting to further dissect the differential roles of tumor-derived and host-derived CCL5 in tumorgenesis and host defense.
It has been reported that transcription factors such as interferon regulatory factor-3, STAT1, and NF-κB mediate CCL5 transcription and production (45, 46). Using an Helicobacter pylori infection model, Kudo et al. (47) showed that maximal H. pylori-induced CCL5 gene transcription required the presence of the interferon-stimulated response element (ISRE) and the cyclic AMP-responsive element. Our previous studies further mapped the IFNγ-inducible ISRE in the CCL5 promoter as IRF1-RE (40, 48). Collectively, these data suggest that transcription factors induced by multiple stimuli are responsible for control of CCL5 expression. However, to our surprise, 4T1 conditioned medium did not inhibit LPS-induced CCL5 primary transcript (Fig. 3, A and B) and CCL5 promoter activity (Fig. 3C) but rather increased it, indicating that tumor-mediated CCL5 inhibition is regulated at the post-transcriptional level. We are currently investigating the molecular mechanisms of tumor-mediated post-transcriptional inhibition of CCL5 expression in macrophages.
Significant elevations of COX-2 protein levels were observed in 43% of human invasive breast cancers and 63% of ductal carcinoma in situ (49). Because the effects of 4T1 TCM on CCL5 transcription were similar to PGE2 (Fig. 3A), we reasoned that PGE2 secreted from tumor cells may be involved in the inhibition of CCL5 production. In line with our hypothesis, the inhibitory effect of tumor-secreted molecules on CCL5 expression was reversed by blocking PGE2 synthesis with COX-2 inhibitor (Fig. 4B) and by blocking PGE2 receptor EP2 and EP4 (Fig. 4, E and F). Though we did not use the EP3 antagonist in the blocking experiments, a complete rescue of the CCL5 expression by using EP2 and EP4 antagonists indicates the sole involvement of these two PGE2 receptors in signaling the cells (Fig. 4, E and F). These results are also consistent with a previous study that PGE2 suppressed LPS-stimulated IFN-β and CCL5 expression in J774A.1 cells (50). Our results further demonstrate that 4T1 TCM-mediated CCL5 inhibition is mediated by cAMP/PKA signaling pathway since blockade of PKA by H89 completely rescued TCM-inhibited CCL5 expression (Fig. 5, A and B) and cAMP analog displayed a similar suppression on CCL5 expression as the 4T1 TCM (Fig. 5, C and D). Taken together, our data demonstrate that PGE2 secreted by tumor cells suppresses CCL5 production in LPS-stimulated macrophages through cAMP/PKA signaling pathway.
To confirm the in vitro findings that PGE2 is responsible for tumor-mediated CCL5 inhibition, we treated the tumor-bearing mice with NS398 to block PGE2 synthesis. Though the NS398 treatment significantly inhibited the PGE2 synthesis in tumor-bearing mice to a level even lower than tumor-free mice (Fig. 6C), the levels of CCL5 in tumor-bearing mice were not recovered to the levels of CCL5 in tumor-free mice (Fig. 6, A and B), suggesting that other molecules other than PGE2 may also involve in inhibition of CCL5 expression during endotoxic shock. Because it has been shown by previous studies that PGE2 is involved in tumor growth (51), we wanted to know whether NS398 had any effect on tumor growth in our systems. Our data indicate that blocking PGE2 synthesis by NS398 had little effect on tumor growth (Fig. 6F) but rather increasing CCL5 expression (Fig. 6, A–E), which is also consistent with our previous report (38).
In summary, our data demonstrate that tumor-bearing hosts display impaired CCL5 production during endotoxemia, which may partially explain why cancer patients are vulnerable to infection. The inhibition of CCL5 expression in tumor-bearing hosts is mediated by tumor-secreted PGE2 at the post-transcriptional level through cAMP/PKA signaling pathway. These findings will likely have a significant impact on our understanding of the orchestration of an immune response in breast cancer patients.
Supplementary Material
Acknowledgments
We thank Dr. Daniel F. Hoft and Nicole Sullivan for critical reading of the manuscript.
This work was supported, in whole or in part, by Grant Number K22CA137126 from the NCI of the National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- TCM
- tumor-conditioned medium
- LPS
- lipopolysaccharide.
REFERENCES
- 1. Nicolini A., Carpi A. (2009) Med. Res. Rev. 29, 436–471 [DOI] [PubMed] [Google Scholar]
- 2. Baggiolini M. (1998) Nature 392, 565–568 [DOI] [PubMed] [Google Scholar]
- 3. Luster A. D. (1998) N. Engl. J. Med. 338, 436–445 [DOI] [PubMed] [Google Scholar]
- 4. Rollins B. J. (1997) Blood 90, 909–928 [PubMed] [Google Scholar]
- 5. Schall T. J., Jongstra J., Dyer B. J., Jorgensen J., Clayberger C., Davis M. M., Krensky A. M. (1988) J. Immunol. 141, 1018–1025 [PubMed] [Google Scholar]
- 6. Appay V., Rowland-Jones S. L. (2001) Trends Immunol. 22, 83–87 [DOI] [PubMed] [Google Scholar]
- 7. Levy J. A. (2009) J. Immunol. 182, 3945–3946 [DOI] [PubMed] [Google Scholar]
- 8. Lin P., Buxton J. A., Acheson A., Radziejewski C., Maisonpierre P. C., Yancopoulos G. D., Channon K. M., Hale L. P., Dewhirst M. W., George S. E., Peters K. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8829–8834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Villalta F., Zhang Y., Bibb K. E., Kappes J. C., Lima M. F. (1998) Infect Immun. 66, 4690–4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. VanOtteren G. M., Strieter R. M., Kunkel S. L., Paine R., 3rd, Greenberger M. J., Danforth J. M., Burdick M. D., Standiford T. J. (1995) J. Immunol. 154, 1900–1908 [PubMed] [Google Scholar]
- 11. Cavaillon J. M., Adib-Conquy M., Fitting C., Adrie C., Payen D. (2003) Scand J. Infect Dis. 35, 535–544 [DOI] [PubMed] [Google Scholar]
- 12. Schrum S., Probst P., Fleischer B., Zipfel P. F. (1996) J. Immunol. 157, 3598–3604 [PubMed] [Google Scholar]
- 13. Siveke J. T., Hamann A. (1998) J. Immunol. 160, 550–554 [PubMed] [Google Scholar]
- 14. Makino Y., Cook D. N., Smithies O., Hwang O. Y., Neilson E. G., Turka L. A., Sato H., Wells A. D., Danoff T. M. (2002) Clin Immunol. 102, 302–309 [DOI] [PubMed] [Google Scholar]
- 15. Dorner B. G., Scheffold A., Rolph M. S., Huser M. B., Kaufmann S. H., Radbruch A., Flesch I. E., Kroczek R. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6181–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller F. R., Miller B. E., Heppner G. H. (1983) Invasion Metastasis 33, 22–31 [PubMed] [Google Scholar]
- 17. Pulaski B. A., Ostrand-Rosenberg S. (1998) Cancer Res. 58, 1486–1493 [PubMed] [Google Scholar]
- 18. Mi Z., Guo H., Wai P. Y., Gao C., Wei J., Kuo P. C. (2004) J. Biol. Chem. 279, 46659–46667 [DOI] [PubMed] [Google Scholar]
- 19. Aslakson C. J., Miller F. R. (1992) Cancer Research 52, 1399–1405 [PubMed] [Google Scholar]
- 20. Pulaski B. A., Clements V. K., Pipeling M. R., Ostrand-Rosenberg S. (2000) Cancer Immunol Immunother 49, 34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Christensen C. R., Klingelhöfer J., Tarabykina S., Hulgaard E. F., Kramerov D., Lukanidin E. (1998) Cancer Res. 58, 1238–1244 [PubMed] [Google Scholar]
- 22. Wang H., Mohammad R. M., Werdell J., Shekhar P. V. (1998) Int. J. Mol. Med. 1, 915–923 [DOI] [PubMed] [Google Scholar]
- 23. Morecki S., Yacovlev E., Gelfand Y., Trembovler V., Shohami E., Slavin S. (2000) Cancer Immunol. Immunother. 48, 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morecki S., Yacovlev L., Slavin S. (1998) Int. J. Cancer 75, 894–899 [DOI] [PubMed] [Google Scholar]
- 25. Rakhmilevich A. L., Janssen K., Hao Z., Sondel P. M., Yang N. S. (2000) Cancer Gene Ther. 7, 826–838 [DOI] [PubMed] [Google Scholar]
- 26. Shi X., Cao S., Mitsuhashi M., Xiang Z., Ma X. (2004) J. Immunol. 172, 4111–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Niwa Y., Akamatsu H., Niwa H., Sumi H., Ozaki Y., Abe A. (2001) Clin. Cancer Res. 7, 285–289 [PubMed] [Google Scholar]
- 28. Luboshits G., Shina S., Kaplan O., Engelberg S., Nass D., Lifshitz-Mercer B., Chaitchik S., Keydar I., Ben-Baruch A. (1999) Cancer Res. 59, 4681–4687 [PubMed] [Google Scholar]
- 29. Bièche I., Lerebours F., Tozlu S., Espie M., Marty M., Lidereau R. (2004) Clin. Cancer Res. 10, 6789–6795 [DOI] [PubMed] [Google Scholar]
- 30. Wigler N., Shina S., Kaplan O., Luboshits G., Chaitchik S., Keydar I., Ben-Baruch A. (2002) Isr. Med. Assoc. J. 4, 940–943 [PubMed] [Google Scholar]
- 31. Robinson S. C., Scott K. A., Wilson J. L., Thompson R. G., Proudfoot A. E., Balkwill F. R. (2003) Cancer Res. 63, 8360–8365 [PubMed] [Google Scholar]
- 32. Taccone F. S., Artigas A. A., Sprung C. L., Moreno R., Sakr Y., Vincent J. L. (2009) Crit. Care 13, R15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Williams M. D., Braun L. A., Cooper L. M., Johnston J., Weiss R. V., Qualy R. L., Linde-Zwirble W. (2004) Crit. Care 8, R291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hollmann C. A., Kittrell F. S., Medina D., Butel J. S. (2001) Oncogene 20, 7645–7657 [DOI] [PubMed] [Google Scholar]
- 35. Jayasinghe M. M., Golden J. M., Nair P., O'Donnell C. M., Werner M. T., Kurt R. A. (2008) Breast Cancer Res. Treat 111, 511–521 [DOI] [PubMed] [Google Scholar]
- 36. Rittirsch D., Huber-Lang M. S., Flierl M. A., Ward P. A. (2009) Nat. Protoc. 4, 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu J., Guan X., Tamura T., Ozato K., Ma X. (2004) J. Biol. Chem. 279, 55609–55617 [DOI] [PubMed] [Google Scholar]
- 38. Mitsuhashi M., Liu J., Cao S., Shi X., Ma X. (2004) J. Leukoc. Biol. 76, 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi X., Liu J., Xiang Z., Mitsuhashi M., Wu R. S., Ma X. (2004) Int. J. Cancer 110, 570–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu J., Ma X. (2006) J. Biol. Chem. 281, 19188–19195 [DOI] [PubMed] [Google Scholar]
- 41. Werts C., le Bourhis L., Liu J., Magalhaes J. G., Carneiro L. A., Fritz J. H., Stockinger S., Balloy V., Chignard M., Decker T., Philpott D. J., Ma X., Girardin S. E. (2007) Eur. J. Immunol. 37, 2499–2508 [DOI] [PubMed] [Google Scholar]
- 42. Azenshtein E., Luboshits G., Shina S., Neumark E., Shahbazian D., Weil M., Wigler N., Keydar I., Ben-Baruch A. (2002) Cancer Res. 62, 1093–1102 [PubMed] [Google Scholar]
- 43. Yaal-Hahoshen N., Shina S., Leider-Trejo L., Barnea I., Shabtai E. L., Azenshtein E., Greenberg I., Keydar I., Ben-Baruch A. (2006) Clin. Cancer Res. 12, 4474–4480 [DOI] [PubMed] [Google Scholar]
- 44. Brault M. S., Kurt R. A. (2003) Int. Rev. Immunol. 22, 199–228 [DOI] [PubMed] [Google Scholar]
- 45. Ohmori Y., Schreiber R. D., Hamilton T. A. (1997) J. Biol. Chem. 272, 14899–14907 [DOI] [PubMed] [Google Scholar]
- 46. Lee A. H., Hong J. H., Seo Y. S. (2000) Biochem. J. 350, 131–138 [PMC free article] [PubMed] [Google Scholar]
- 47. Erickson L., Crews G., Pan F., Fisniku O., Jang M. S., Wynn C., Kobayashi M., Jiang H. (2004) Transpl. Immunol. 13, 169–175 [DOI] [PubMed] [Google Scholar]
- 48. Liu J., Guan X., Ma X. (2005) J. Biol. Chem. 280, 24347–24355 [DOI] [PubMed] [Google Scholar]
- 49. Half E., Tang X. M., Gwyn K., Sahin A., Wathen K., Sinicrope F. A. (2002) Cancer Res. 62, 1676–1681 [PubMed] [Google Scholar]
- 50. Xu X. J., Reichner J. S., Mastrofrancesco B., Henry W. L., Jr., Albina J. E. (2008) J. Immunol. 180, 2125–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Howe L. R. (2007) Breast Cancer Res. 9, 210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.