Abstract
The cell fate determination factor Dachshund was cloned as a dominant inhibitor of the hyperactive epidermal growth factor receptor ellipse. The expression of Dachshund is lost in human breast cancer associated with poor prognosis. Breast tumor-initiating cells (TIC) may contribute to tumor progression and therapy resistance. Here, endogenous DACH1 was reduced in breast cancer cell lines with high expression of TIC markers and in patient samples of the basal breast cancer phenotype. Re-expression of DACH1 reduced new tumor formation in serial transplantations in vivo, reduced mammosphere formation, and reduced the proportion of CD44high/CD24low breast tumor cells. Conversely, lentiviral shRNA to DACH1 increased the breast (B)TIC population. Genome-wide expression studies of mammary tumors demonstrated DACH1 repressed a molecular signature associated with stem cells (SOX2, Nanog, and KLF4) and genome-wide ChIP-seq analysis identified DACH1 binding to the promoter of the Nanog, KLF4, and Lin28 genes. KLF4/c-Myc and Oct4/Sox2 antagonized DACH1 repression of BTIC. Mechanistic studies demonstrated DACH1 directly repressed the Nanog and Sox2 promoters via a conserved domain. Endogenous DACH1 regulates BTIC in vitro and in vivo.
Keywords: Breast Cancer, Gene Regulation, Stem Cell, Transcription Regulation, Tumor Suppressor, DACH1
Introduction
Recent studies have suggested the presence of self-renewing stem-like cells within tumors. These tumor-initiating cells (TICs)3 have been described in the hematopoietic system, the breast, colon, brain, and prostate tumors (1–5). Mouse mammary stem cells express specific cell surface markers and show self-renewing properties (6). Only a small number of primary breast cancer cells form secondary tumors. These TICs form mammospheres similar to normal mammary gland stem cells (7). When cultured under specific conditions, these TIC or cancer stem cells can be enriched by fluorescence-activated cell sorting for CD44+/CD24−/low cells (2).
The molecular circuitry controlling embryonic stem cells may also be active in certain tumors. Several key regulators of embryonic stem cell identity (Oct4 and Sox2, EKLF4, Lin28, and Nanog) (8) are expressed in a subset of specific tumors (9, 10). Genes known to regulate features of mammary stem cells include expression of Twist (11). Although self-renewal of primitive hematopoietic stem cells requires p21CIP1 (12), the role of tumor suppressors in regulating TICs, particularly in breast cancer, is poorly understood.
Clinical studies have demonstrated a correlation between poor prognosis breast cancer and reduced expression of the cell fate determination factor DACH1 (13), and loss of DACH1 expression has been observed in prostate and endometrial cancer (14, 15). Several lines of evidence suggest Dachshund may function as a tumor suppressor. Initially cloned as a dominant inhibitor of Ellipse in Drosophila, the mammalian DACH1 gene inhibits breast cancer cellular DNA synthesis and proliferation in cultured cells (13). DACH1 regulates gene expression of target genes in part through interacting with DNA-binding transcription factors (c-Jun, Smads, Six, and ERα) and in part through intrinsic DNA binding (13, 14, 16). Recent studies have demonstrated that DACH1 conveys intrinsic DNA sequence-specific binding properties through elements resembling binding sites for the Forkhead family of proteins (17).
The Drosophila dac gene is a key member of the retinal determination gene network that specifies eye tissue identity. In Drosophila, there is a coordinated system of genes, including dachshund (dac), eyes absent (eya), ey, twin of eyeless (toy), teashirt (tsh), and sin oculis (so). dac is expressed in progenitor cells and neurons of the mushroom body, a brain structure present in most arthropods, and Dac expression can induce ectopic eye formation in Drosophila (18). The mammalian homologue of so is known as Six, and altered expression of the Six family and DACH1 occurs in a variety of human tumors (13, 19–23). The current experiments were conducted to examine a possible role for DACH1 in regulating breast tumor stem cells.
MATERIALS AND METHODS
Mammosphere Formation and FACS Analysis of Stem Cell Surface Markers
Mammosphere formation assays were conducted as described previously (24). Aldefluor and immunostaining of cell surface markers by FACS analysis for breast cancer stem cells was based on prior publications (25). Before labeling, the cells were blocked with normal mouse IgG in 1:100 dilution for 30 min and then incubated with phycoerythrin-labeled mouse anti-human CD24 (1:5) (clone ML5, Pharmingen) and/or phycoerythrin/Cy5-labeled rat anti-human/mouse CD44 (1:200) (clone IM7, BioLegend, San Diego) for 1 h. All experiments were conducted at 4 °C. Cell sorting was performed on a FACSCalibur cell sorter (BD Biosciences). The data were analyzed with FlowJo software (Tree Star, Inc., Ashland, OR).
Cell Culture, Plasmid Construction, Reporter Genes, Expression Vectors, DNA Transfection, and Luciferase Assays
Cell culture, DNA transfection, and luciferase assays using the Sox-2-Luc and Nanog-Luc reporter genes were performed as described previously (26–29). Expression vectors encoding KLF4/c-Myc and Oct4/Sox 2 were from Addgene. The Met-1 cells were cultured in DMEM supplemented with 10% fetal calf serum, 1% penicillin, and 1% streptomycin. The MCF10A and MCF10A-Myc lines were previously described (13). The expression plasmids encoding an N-terminal FLAG peptide linked to DACH1, DACH1 DS (DACH1-Sno/Ski) domain alone (DS), or DACH1 DS domain deleted (ΔDS) were previously described. Lentiviral DACH1 shRNA was from Open BioSystems. Transfection and infection were per standard protocols. GFP-positive cells were selected by FACS. Met-1 cells were plated at a density of 1 × 105 cells in a 24-well plate on the day prior to transfection with Superfect according to the manufacturer's protocol (Qiagen, Valencia, CA). A dose response was determined in each experiment with 50 and 200 ng of expression vector and the promoter reporter plasmids (0.5 μg). Luciferase activity was normalized for transfection efficiency using β-galactosidase reporters as an internal control. The fold effect of expression vector was determined with comparison to the effect of the empty expression vector cassette, and statistical analyses were performed using the Student's t test.
RNA Isolation, RT-PCR, and Quantitative Real Time PCR
Total RNA was isolated from Met-1 cells infected with the DACH1 expression vector system, using TRIzol (30). SYBR Green-based real time PCRs were performed using QuantiTect SYBR Green PCR kit (Qiagen) and Quantitect pre-validated primer assays for mouse and 18 S rRNA as internal control following the manufacturer's recommendations on an ABI Prism 7900HT system (Applied Biosystems Inc., Foster City, CA). Oligonucleotides used for RT-PCR include the following: Nanog, forward CAGAAAAACCAGTGGTTGAAGACTAG and reverse GCAATGGATGCTGGGATACTC; Oct4, forward CTGTAGGGAGGGCTTCGGGCACTT and reverse CTGAGGGCCAGGCAGGAGCACGAG; Sox2, forward GGCAGCTACAGCATGATGCAGGAGC and reverse CTGGTCATGGAGTTGTACTGCAGG; KLF4, forward TGCCAGACCAGATGCAGTCAC and reverse GTAGTGCCTGGTCAGTTCATC; c-Myc, forward TGAGCCCCTAGTGCTGCAT and reverse AGCCCGACTCCGACCTCTT; 18 S rRNA oligonucleotides were used as control (30). The oligonucleotides for chromatin immunoprecipitation (ChIP) were directed to the murine SOX2 as follows: distant site, forward 5′-gcagtgagaggggtggacta-3 and reverse 5′-ctcccctcatctaccccaac-3; proximal site (sox2-binding site), forward 5′-cgcagaaacaatggcacaccac-3′ and reverse 5′-ccgttttcagcaacaggtcacg-3′; Nanog distant site, forward 5′-ggcaaactttgaacttgggatgtggaaata-3′ and reverse 5′-ctcagccgtctaagcaatggaagaagaaat-3′; proximal site (oct4-sox2-binding site), forward 5′-ggatgtctttagatcagaggatgccc-3 and reverse 5′-ccacagaaagagcaagacacca-acc-3′ promoters.
Microarray and Cluster Analysis
DNA-free total RNA isolated from Met-1 cells expressing GFP or DACH1 were used to probe Affymetrix Gene 1.0 arrays (Affymetrix, Santa Clara, CA). RNA quality was determined by gel electrophoresis. Probe synthesis and hybridization were performed as described previously (31). Analysis of the arrays was performed using GeneSpring. Arrays were normalized using robust multiarray analysis, and the p value of 0.05 was applied as a statistical criteria for differentially expressed genes. These genes were then grouped using hierarchical clustering with “complete” agglomeration, and each cluster was further analyzed based upon the known function of the genes contained in the cluster. Expression profiles are displayed using Treeview (32). Classification and clustering for pathway level analysis were performed by using gene sets ASSESS (Analysis of Sample Set Enrichment Scores), available on line (33). ASSESS provides a measure of enrichment of each gene set in each sample. Gene set enrichment was dependent on a concordance of at least two samples within the replicates that was opposite between phenotypes.
Immunohistochemistry, Chromatin Immunoprecipitation, and ChIP-seq Analysis
Immunohistochemical analysis of human breast cancer was conducted using a polyclonal DACH1 antibody (13). Human breast cancer tissue arrays were from Biomax. Chromatin immunoprecipitation assays were conducted as described previously (34, 35) using antibodies directed to the FLAG epitope of the DACH1 protein. ChIP-seq was conducted as described previously (17).
Nude Mice Study
1 × 105 Met-1 cells expressing GFP control or DACH1 were implanted subcutaneously into 4–6-week-old athymic female nude mice purchased from NCI, National Institutes of Health. The tumor growth was measured twice weekly for 7 weeks by using a digital caliper. Tumor weight was measured when mice were sacrificed on day 35 after cells implantation.
RESULTS
DACH1 Expression Is Reduced in Breast Cancer Cell Lines Enriched for Cancer Stem Cells
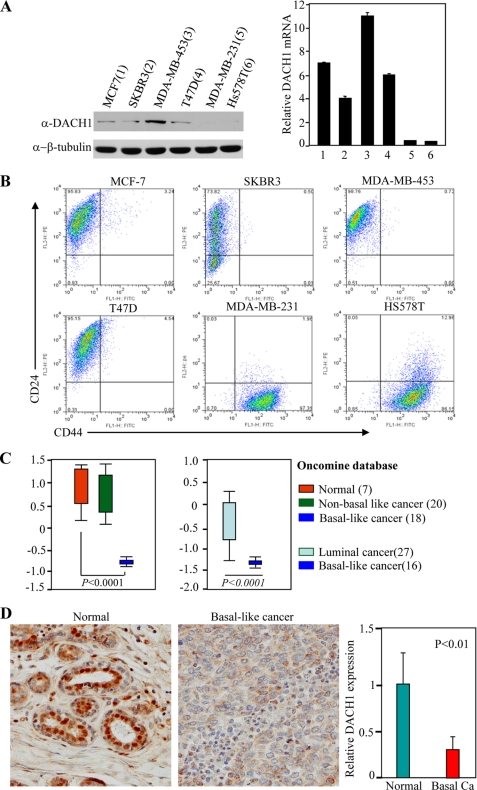
Recent studies have demonstrated the loss of DACH1 expression correlates with poor prognosis in human breast cancer, and DACH1 inhibits MCF7 cell proliferation in tissue culture (13). To characterize further the expression of DACH1 in breast cancer cell types, Western blot analysis was conducted using a previously characterized polyclonal antibody (Fig. 1A) (13). Quantitation of relative abundance from multiple experiments demonstrated a reduction of DACH1 abundance in the MDA-MB231 and HS578T cells. Immunoepitope staining for the breast cancer stem cell markers CD44+/CD24− demonstrated a relative increase in the proportion of CD44+/CD24− cells in the MDA-MB231 and HS578T cells (Fig. 1B). Distinct subtypes of human breast cancer include the basal-like, luminal (A and B), Her2+, and normal breast-like carcinomas with distinct prognostic significance. Basal-like breast carcinomas are of high grade with a distinctive proclivity to metastasize and express genes associated with the maintenance of the stem cell phenotype (36). Comparison between normal human breast epithelial cells and basal-like versus nonbasal-like showed a significant reduction in mRNA expression (Fig. 1C) and in DACH1 abundance (Fig. 1D) in the basal-like tumors.
FIGURE 1.
DACH1 expression in human breast cancer cell lines and breast carcinoma tissues. A, left panel, Western blot for DACH1 abundance of breast cancer cell lines, and β-actin was used as a loading control. Right panel, normalized mRNA expression of DACH1 in breast cancer cell lines. B, CD24/CD44 staining of breast cancer cell lines. C, normalized DACH1 expression from Oncomine databases. D, representative image and semi-quantization of DACH1 expression in normal breast epithelium and triple negative invasive human breast cancer samples using DACH1-specific antibodies. Basal Ca, basal-type cancer.
DACH1 Expression Inhibits the Proportion of Breast Cancer Cells Expressing Cancer Stem Cell Markers in Vivo
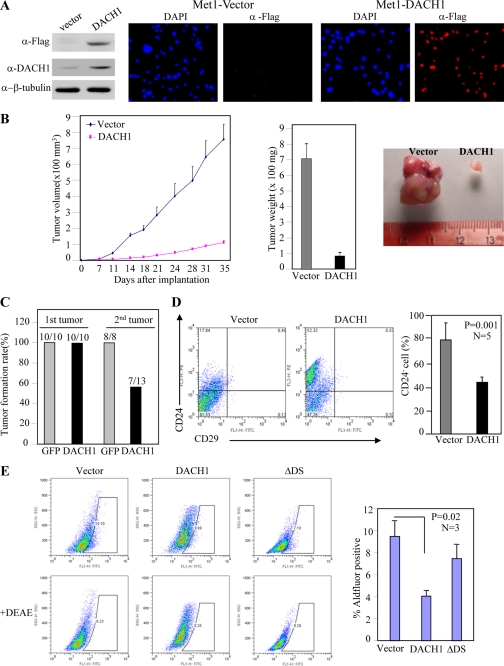
Given the association between low DACH1 expression and increased expression of cancer stem cells, we examined whether DACH1 expression could preferentially inhibit breast cancer stem cells. Met-1 cells were transduced with a DACH1 expression vector resulting in an ∼2-fold increase in DACH1 expression by Western blot analysis (Fig. 2A). Immunohistochemistry demonstrated the presence of the DACH1-tagged FLAG epitope throughout the cell population. The effect of DACH1 on mammary tumor growth in vivo was assessed by implantation in nude mice (Fig. 2B). DACH1 expression reduced the volume of tumors by ∼80%. Tumor weight was reduced by ∼90% (Fig. 2B). Serial transplantation experiments demonstrated an ∼50% reduction in new tumor formation of DACH1 expressing Met-1 breast cancer cells (Fig. 2C).
FIGURE 2.
DACH1 blocks Met-1 tumor growth in vivo and inhibits properties associated with stem cell expansion. (A) Western blot and immunohistochemistry of Met-1 cells transduced with vector or DACH1 expression vector tagged with Flag. (B) Nude mice were injected with equal numbers of Met-1 breast cancer cells co-transduced with either control vector or DACH1 expression plasmid. The tumor volume and tumor weight of Met-1 cells implanted in nude mice. Analysis was conducted of n = 5. Data are mean ± SEM. (C) Serial implantation study of Met-1 cells transduced with either vector or expression plasmid. (D) FACS display of CD24/CD29 double staining of cells isolated from Met-1-GFP or Met-1-DACH1 tumors (Data are mean ± SEM, n = 5, p < 0.001). (E) Aldefluor staining of Met-1 cells transduced either with DACH1 or a mutant of DACH1 which is defective in DNA binding (DACH ΔDS).
DACH1 Inhibits Mammosphere Formation and the CD44+/CD24− Phenotypes
Cancer stem cells can be enriched by sorting for CD24−/low cells (2). To determine whether DACH1 expression regulated the relative proportion of CD24−/low breast tumor cells in vivo, Met-1 cells transduced with either a DACH1 expression vector or a control vector were implanted into nude mice. Tumors were grown for 3 weeks in mice and subsequently analyzed for CD24−/low cells. Induction of DACH1 expression reduced the proportion of CD24−/low cells by ∼50% (Fig. 2D).
As a complementary assay of the BTIC phenotype, Aldefluor staining was conducted as described previously (37). The stem cell marker aldehyde dehydrogenase is thought to regulate stem cell differentiation through metabolism of retinal to retinoic acid. The fluorescent Aldefluor assay measures aldehyde dehydrogenase activity and has been used to isolate cancer stem cells from brain tumors, multiple myeloma, acute myeloid leukemia, and breast cancer. DACH1 expression reduced the proportion of Aldefluor-positive cells by ∼60% (Fig. 2E). Expression of a DNA-binding defective mutant of DACH1 (ΔDS) was defective in reducing Aldefluor staining (Fig. 2E).
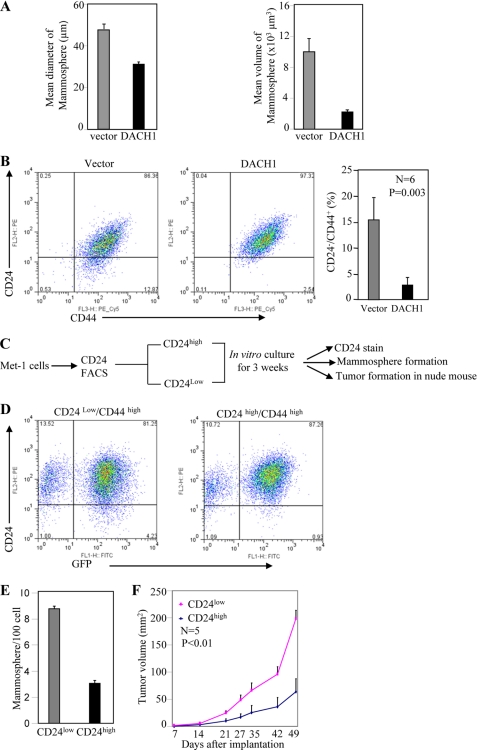
The cancer stem cell hypothesis suggests that many cancers are maintained in a hierarchical organization of cancer “stem cells” or tumor-initiating cells, rapidly dividing amplifying cells (early precursor cells) and differentiated tumor cells (38). Cancer stem cells are thought to contribute to tumor progression and therapy resistance and recurrence (1) and can be enriched by cell sorting for CD44high/CD24−/low cells (2). A small number of primary breast cancer cells, TIC, or cancer stem cells form secondary tumors (8). TICs form nonadherent mammospheres when cultured under specific conditions in the absence of serum. To examine further the role of DACH1 in TIC, mammosphere assays were conducted with the Met-1 mammary tumor cell lines. Induction of DACH1 reduced mammosphere number by >60% in cell lines (Fig. 3A).
FIGURE 3.
DACH1 reduces the proportion of CD24low/CD44high cells. (A) Mammosphere assays of Met-1 cells transduced with vector control or DACH1. (B) FACS based CD24/CD44 double staining of Met-1 cells in vitro expressing DACH1 or control vector. (C) The cellular potential of CD24low/CD44high versus CD24high/CD44high cells was determined. Schematic representation of methods for analysis of Met-1 population. (D) FACS analysis. (E) Mammosphere formation and (F) tumor growth in nude mice studies comparing CD24low/CD44high and CD24high/CD44high cells.
DACH1 expression in Met-1 cells reduced the relative proportion of CD44high/CD24low cells by ∼80% (Fig. 3B, 15% versus 3%, n = 6, p < 0.003). To examine the biological significance of DACH1-mediated inhibition of the CD24 population, Met-1 cells transduced with DACH1 were grown in tissue culture and subjected to FACS analysis for the CD24high versus CD24low populations. Multipotentiality of the CD24high and CD24low populations was determined by their ability to form CD24high and CD24low populations and to form mammospheres as a surrogate measure of stem cell expansion (Fig. 3C). CD24low/CD44+ cells and CD24high/CD44+ cells were separated by FACS analysis and grown in cultures for 3 weeks. Restaining by FACS demonstrated CD24low/CD44high gave rise to both CD24high/CD44high and CD24low/CD44high, whereas CD24high/CD44high gave rise to only the parental CD24high/CD44high population (Fig. 3D). The CD24low and CD24high Met-1 cells were next examined for mammosphere formation. The CD24low cells gave a 4-fold greater yield of mammospheres (Fig. 3E). These studies suggest the CD24low and CD44high cells maintain multipotentiality. To determine the tumor growth characteristics of these two distinct Met-1 cell populations, tumor implantation analysis was conducted. The CD24low/CD44high grew ∼4 times larger tumors than CD24high/CD44high Met-1 cells (Fig. 3F).
Endogenous DACH1 Inhibits the Stem Cell Phenotype
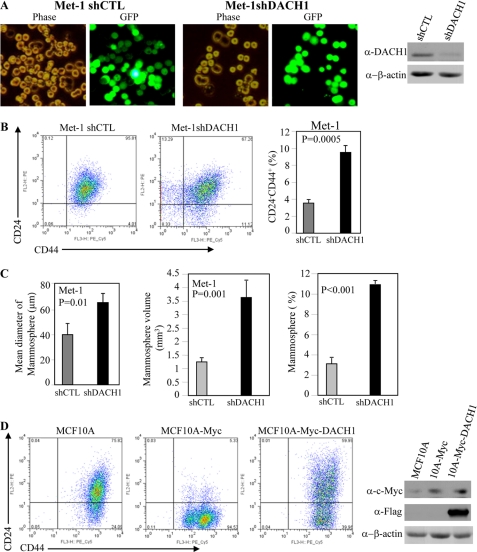
These studies suggested that a modest induction of DACH1 expression was sufficient to inhibit mammosphere formation and the relative proportion of cells with features of breast cancer stem cells. To determine whether endogenous DACH1 functioned to inhibit cancer stem cells, a lentivirus encoding DACH1 shRNA linked via an internal ribosome entry site to GFP was used to transduce Met-1 cells (Fig. 4A). Comparison was made to the control vector. Reduction of DACH1 abundance with DACH1 shRNA in multiplicate experiments increased the proportion of CD44+/CD24− cells ∼2.2-fold (Fig. 4B). The number of mammospheres reflects the relative proportion of progenitor cells, whereas the size of the mammosphere may also be affected in part by the proliferative capacity of the cells. Mammosphere volume was increased 3.5-fold by DACH1 shRNA expression (Fig. 4C). The relative number of mammospheres was increased 350% by DACH1 shRNA (Fig. 4C). c-Myc transduction of the immortal human MCF10A cells induced cells with contact-independent growth properties (13) and increased the proportion of CD44+/CD24− cells (Fig. 4D) from ∼24 to 95%. Transduction of MCF10-c-Myc cells with DACH1 inhibited the proportion of breast cancer stem cells from 95 to ∼40% (Fig. 4D). The relative number of mammospheres was increased 350% by DACH1 shRNA (Fig. 4C). These findings suggest endogenous DACH1 is a key determinant of mammosphere number and therefore of BTIC.
FIGURE 4.
Endogenous DACH1 inhibits the stem cell phenotype. A, knocking down endogenous Dach1 expression by lentivirus shRNA vector to Dach1. B, FACS-based CD24/CD44 double staining and mammosphere assay of Met-1 cells expressing shCTL or shDACH1. C, mammosphere number and size of Met-1 cells expressing shDACH1. Data are mean ± S.E. of >5 spearate experiments. D, FACS display of CD24/CD44 double staining of MCF10A and Myc transformed MCF10A with vector control or DACH1. Western blot of MCF10A-transduced cells. Each cell type also expresses GFP from the vector as a marker.
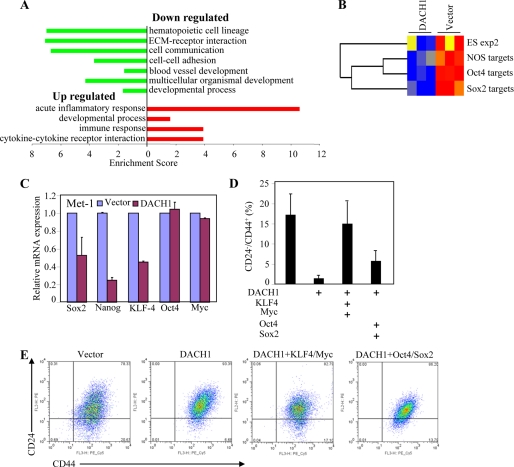
To examine further the mechanisms by which DACH1 inhibited cellular growth and angiogenesis, genome-wide expression studies were conducted of DACH1-transduced cells. Molecular pathway analysis was conducted with DAVID using Gene Ontology and KEGG pathway sets. DACH1 repressed gene expression of signaling pathways governing hematopoietic cell lineage, cellular communication, blood vessel development, and multicellular organismal development. DACH1 induced an acute inflammation response and cytokine-cytokine receptor interaction (Fig. 5A). Several recent studies have suggested the molecular circuitry controlling stem cells may be active in certain tumors. Some of the key regulators of embryonic stem (ES) cell identity, Oct4, Sox2, and Nanog, are expressed in specific tumors (9, 10). An embryonic stem cell-like gene expression signature has been identified in poorly differentiated aggressive human tumors (36). Oct4, Sox2, and Nanog are required for propagation of ES cells in culture. Comparison of Dach1-regulated genes in Met-1 cells to gene sets associated with ES cell identity via gene set enrichment analysis demonstrated Dach1 down-regulates expression of Sox2 and Oct4 gene targets, NOS targets (genes common to Nanog, Oct4, and Sox2), and a gene set overexpressed in human ES cell lines (Fig. 5B).
FIGURE 5.
Pathway analysis. A, pathway analysis of microarray data from Met-1 cells expressing DACH1 or control vector by DAVID using Gene Ontology and KEGG pathway sets. Pathways are graphically rprsented by enrichment score. Red indicates up-regulated and blue indicates down-regulated. B, gene sets enrichment analysis of microarray from Met-1 cells using gene targets of Sox2, Oct4, or NOS (Nanog, Oct4, and Sox2),which are enriched in ES cells (A). C, real time RT-PCR detection of ES cell-related gene expression (data are mean ± S.E., n = 3). D, quantitation of CD24−/CD44+ staining of multiplicate transductions. E, FACS analysis of Met-1 cells transduced with viral vectors encoding KLF/c-Myc or Oct4/Sox2.
Increased expression of the Sox2, Oct4, Nanog, and EKLF4 gene is associated with the stem cell phenotype. We examined whether expression of DACH1 could inhibit expression of genes associated with the cancer stem cell phenotype. Quantitation of the relative expression of the ES cell markers (Sox2, Oct4, Nanog, KLF4 and c-Myc) was conducted using mRNA from the Met-1 tumors expressing DACH1 or control (Fig. 5C). RT-PCR analysis demonstrated a reduction in the abundance of Sox2, Nanog, and KLF4. Each of these genes promotes stem cell expansion.
To determine the functional significance of KLF4 and Sox2 repression, DACH1-transduced Met-1 cells were transfected with expression vectors encoding KLF4 or Sox2. A FACS analysis was conducted to examine the relative proportion of CD24−/CD44+ cells. DACH1 reduced the proportion of CD24−/CD44+ by ∼80%. Re-expression of KLF4/Myc or Sox2/Oct4 partially reversed the phenotype (Fig. 5, D and E).
DACH1 Binds Promoters of Genes Governing Progenitor Cell Expansion in ChIP and ChIP-Seq
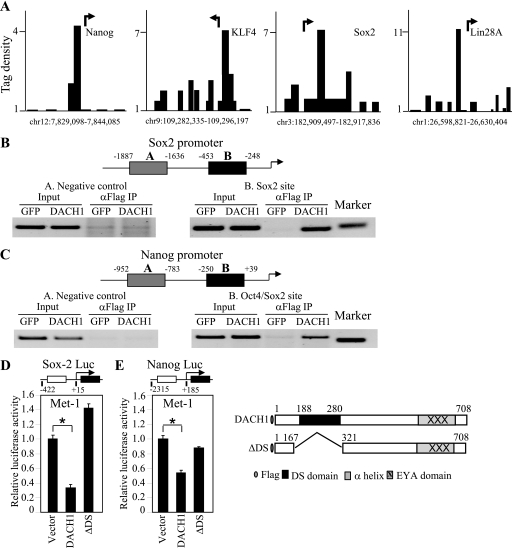
ChIP-Seq analysis was conducted of MDA-MB-231 cells expressing DACH1 to determine whether DACH1 bound the promoters of stem cell regulatory genes. DACH1 occupancy was identified at the Sox2, Nanog, KLF4, and Lin28 promoters (Fig. 6A). Sox2, KLF4, and Lin28 are known to play an important role in the maintenance of stem cell pluripotency (39, 40). To examine further DACH1 physical association with the promoters of the Sox2 and Nanog genes in the context of local chromatin immunoprecipitation, assays were conducted. Comparison was made using Met-1 cells expressing FLAG-tagged DACH1 or control vector. ChIP of the Sox2 promoter was conducted using oligonucleotides directed to either the distal or the proximal promoter. ChIP for DACH1 at the distal promoter failed to identify chromatin-associated DACH1; however, oligonucleotides directed to the proximal promoter, including the Sox2-binding site, demonstrated the recruitment of DACH1 (Fig. 6B). Similarly, the ChIP analysis of the Nanog promoter identified DACH1 recruitment to the proximal but not distal promoter region (Fig. 6C).
FIGURE 6.
DACH1 represses Sox2 and Nanog transcription. (A) DACH1-dependent tag density at selected gene promoters. Arrow indicates the start site and direction of transcription. (B) Chromatin immunopreciptation assays of the Sox2 and (C) Nanog promoter using Met-1 cells expression Flag-tagged DACH1 or control GFP vector. Schematic representation of oligonucleotide probes directed to the promoters. (D) Luciferase reporter gene assays of Sox-2 and (E) Nanog DACH1 wild type or a mutant of the conserved DS domain were co-transfected. The data are mean ± SEM of n = 6 separate transfections (p < 0.001).
As DACH1 had reduced the expression of Sox2 and Nanog, studies were conducted to determine whether the Sox2 and Nanog genes were directly repressed by DACH1. The promoter of the Sox2, the Nanog genes, and Lin28 were directly repressed by DACH1 expression (Fig. 6D). Deletion of the DACH1 DS domain abrogated transcriptional repression by DACH1 (Fig. 6, D and E).
DISCUSSION
This study provides several lines of evidence that DACH1 inhibits breast tumor stem cell expansion. First, DACH1 reduced the expression of the breast cancer stem cell markers (CD44high/CD24low) within the tumors. DACH1 also reduced the relative proportion of CD44high/CD24low Met-1 cells in tissue culture. Second, DACH1 reduced the number and size of mammospheres. Only a small number of primary breast cancer cells known as breast tumor-initiating cells give rise to secondary tumors, and the growth of BTIC as nonadherent mammospheres serves as a useful surrogate of BTIC. Third, shRNA to DACH1 reduced endogenous DACH1 and increased the number of mammospheres and the proportion of CD44+/CD24− cells. Fourth, DACH1 abundance was reduced in cell lines with features of breast cancer stem cells and in the basal phenotype of human breast cancer. Fifth, the expression profile of genes regulated by DACH1, and the genes known to regulate ES cell features showed significant overlap (36). Expression of the key regulators of ES cell function Sox and Nanog was repressed by DACH1 in Met-1 cells, and DACH1 repressed the Sox and Nanog gene promoters. DACH1 associated with the Nanog, Sox2, and KLF4 promoters in ChIP assays or in ChIP-seq analysis. Collectively, this study is consistent with a model in which DACH1 reduces the proportion of breast cancer stem cells.
DACH1 is known to regulate gene transcription indirectly through binding to DNA binding transcription factors (c-jun, Smad, and SIX) (13, 35, 41, 42). Here, the transcriptional repression of the Sox and Nanog genes by DACH1 required a domain that is highly conserved from Drosophila to humans. The Dachshund box N (DS domain) shares significant amino acids with the Ski/Sno family. The DS domain is required for transcriptional repression of a subset of target genes and is required for HDAC1 recruitment by DACH1 in the context of local chromatin using ChIP assays (14, 35) and for binding to DNA (17). The DNA binding domain of DACH1 was required for repression of the BTIC phenotype assessed using Aldefluor staining. DACH1 was recruited in the context of local chromatin to the proximal promoters of the Sox2 and Nanog promoters. Collectively, these studies suggest DACH1 represses expression of Sox2, Nanog, and KLF4. These findings are consistent with the finding that DACH1 expression is reduced in the basal breast cancer phenotype, that the basal phenotype is known to overexpress Sox2, and that the basal phenotype displays features of breast cancer stem cells (9). Sox2 maintains stem cell properties, and Sox2 down-regulation correlates with loss of pluripotency and the induction of differentiation (9). Re-expression of KLF4/c-Myc or Sox2/Oct4 partially reversed the inhibition of the BTIC phenotype. Further studies will be required to distinguish the functional interactions between DACH1 and the gene products governing stem cell phenotypes in breast cancer.
This study demonstrated that DACH1 reduced the proportion of CD44high/CD24low cells. Cancer stem cells can be enriched by sorting for CD44high/CD24low cells and are characterized by their multipotentiality and their ability to self-renew (1). We demonstrated the population of CD44high/CD24low was enriched in their capacity to produce mammospheres. The population of CD44high/CD24low cells was multipotential giving rise to both CD44high/CD24low and CD44high/CD24high populations after serial passage in tissue culture. These studies suggest DACH1 represses the proportion of breast cancer cells with multipotentiality characteristic of cancer stem cells.
These findings are consistent with prior studies suggesting a role for Dac in progenitor cell function. In Drosophila, dac is expressed in progenitor or stem cells that give rise to several distinct organocellular populations, including muscle, neurons, and gonadal germ cells. dac is expressed in neural progenitors (neuroblasts) of the mushroom body, a brain structure present in most arthropods. These neuroblasts divide in a stem cell mode and produce lineages of 10–20 neurons. Dac is thought to play a role in specifying the structural fate of Kenyon cell axons, and mushroom body neuropils are drastically abolished in the pupa of dac null mutants (43). In mammalian cells, DACH1 is expressed in the developing eye, ear, limb, and mammary epithelium. Although Dach1 gene deletion in the mouse is perinatal lethal, expression studies in the murine embryo suggest an important role for Dach1 in cell fate determination. Thus Dac is expressed in embryonic progenitor cells, and expression is lost upon terminal differentiation. DACH1 expression is reduced in tumors (breast, prostate, and uterus) (13, 14) correlating with poor prognosis. DACH1 re-expression in breast cancer cells reduced the proportion of cells with features of cancer stem cells.
In this study, DACH1 reduced cellular invasiveness and reduced the proportion of CD44high/CD24low cells. Analysis of the mechanism by which DACH1 regulates the proportion of BTIC demonstrated the role for a secreted factor. The conditioned medium from DACH1-transduced Met-1 cells recapitulated the effect of DACH1 transduction of Met-1 cells to reduce the proportion of CD44+/CD24−/low cells. In previous studies, we demonstrated the conditioned medium of DACH1-transduced breast cancer cells was sufficient to reduce migration of Met-1 cells (44). It will be of interest in future studies to determine whether similar secreted factors regulate the distinct properties of repressing features of BTIC and inhibiting cellular migration.
Supplementary Material
Acknowledgment
We thank Atenssa L. Cheek for the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA70896, R01CA75503, and R01CA86072 (to R. G. P.) and Grant P30CA56036 (Kimmel Cancer Center Core grant to R. G. P.). This work was also supported by a grant from Dr. Ralph and Marian C. Falk Medical Research Trust (to R. G. P.), Margaret Q. Landenberger Research Foundation (to K. W.), in part by a grant from the Pennsylvania Department of Health (to R. G. P.), and in part by National Natural Science Foundation of China Grant 81072169 (to K. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- TIC
- tumor-initiating cell
- BTIC
- breast tumor-initiating cell
- ES
- embryonic stem.
REFERENCES
- 1. Al-Hajj M. (2007) Curr. Opin. Oncol. 19, 61–64 [DOI] [PubMed] [Google Scholar]
- 2. Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. O'Brien C. A., Pollett A., Gallinger S., Dick J. E. (2007) Nature 445, 106–110 [DOI] [PubMed] [Google Scholar]
- 4. Ricci-Vitiani L., Lombardi D. G., Pilozzi E., Biffoni M., Todaro M., Peschle C., De Maria R. (2007) Nature 445, 111–115 [DOI] [PubMed] [Google Scholar]
- 5. Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 6. Visvader J. E., Lindeman G. J. (2006) Cancer Res. 66, 9798–9801 [DOI] [PubMed] [Google Scholar]
- 7. Dontu G., Abdallah W. M., Foley J. M., Jackson K. W., Clarke M. F., Kawamura M. J., Wicha M. S. (2003) Genes Dev. 17, 1253–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodriguez-Pinilla S. M., Sarrio D., Moreno-Bueno G., Rodriguez-Gil Y., Martinez M. A., Hernandez L., Hardisson D., Reis-Filho J. S., Palacios J. (2007) Mod. Pathol 20, 474–481 [DOI] [PubMed] [Google Scholar]
- 10. Gidekel S., Pizov G., Bergman Y., Pikarsky E. (2003) Cancer Cell 4, 361–370 [DOI] [PubMed] [Google Scholar]
- 11. Mani S. A., Guo W., Liao M. J., Eaton E. N., Ayyanan A., Zhou A. Y., Brooks M., Reinhard F., Zhang C. C., Shipitsin M., Campbell L. L., Polyak K., Brisken C., Yang J., Weinberg R. A. (2008) Cell 133, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., Scadden D. T. (2000) Science 287, 1804–1808 [DOI] [PubMed] [Google Scholar]
- 13. Wu K., Li A., Rao M., Liu M., Dailey V., Yang Y., Di Vizio D., Wang C., Lisanti M. P., Sauter G., Russell R. G., Cvekl A., Pestell R. G. (2006) Mol. Cell. Biol. 26, 7116–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu K., Katiyar S., Witkiewicz A., Li A., McCue P., Song L. N., Tian L., Jin M., Pestell R. G. (2009) Cancer Res. 69, 3347–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nan F., Lü Q., Zhou J., Cheng L., Popov V. M., Wei S., Kong B., Pestell R. G., Lisanti M. P., Jiang J., Wang C. (2009) Cancer Biol. Ther. 8, 1534–1539 [DOI] [PubMed] [Google Scholar]
- 16. Popov V. M., Zhou J., Shirley L. A., Quong J., Yeow W. S., Wright J. A., Wu K., Rui H., Vadlamudi R. K., Jiang J., Kumar R., Wang C., Pestell R. G. (2009) Cancer Res. 69, 5752–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou J., Wang C., Wang Z., Dampier W., Wu K., Casimiro M. C., Chepelev I., Popov V. M., Quong A., Tozeren A., Zhao K., Lisanti M. P., Pestell R. G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6864–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silver S. J., Rebay I. (2005) Development 132, 3–13 [DOI] [PubMed] [Google Scholar]
- 19. Winchester C., Robertson S., MacLeod T., Johnson K., Thomas M. (2000) J. Clin. Pathol. 53, 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coletta R. D., Christensen K., Reichenberger K. J., Lamb J., Micomonaco D., Huang L., Wolf D. M., Müller-Tidow C., Golub T. R., Kawakami K., Ford H. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Micalizzi D. S., Christensen K. L., Jedlicka P., Coletta R. D., Barón A. E., Harrell J. C., Horwitz K. B., Billheimer D., Heichman K. A., Welm A. L., Schiemann W. P., Ford H. L. (2009) J. Clin. Invest. 119, 2678–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laflamme C., Filion C., Bridge J. A., Ladanyi M., Goldring M. B., Labelle Y. (2003) Cancer Res. 63, 449–454 [PubMed] [Google Scholar]
- 23. Yu Y., Khan J., Khanna C., Helman L., Meltzer P. S., Merlino G. (2004) Nat. Med. 10, 175–181 [DOI] [PubMed] [Google Scholar]
- 24. Jiao X., Katiyar S., Willmarth N. E., Liu M., Ma X., Flomenberg N., Lisanti M. P., Pestell R. G. (2010) J. Biol. Chem. 285, 8218–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindsay J., Jiao X., Sakamaki T., Casimiro M. C., Shirley L. A., Tran T., Ju X., Liu M., Li Z., Wang C., Katiyar S., Rao M., Allen K. G., Glazer R. I., Ge C., Stanley P., Lisanti M., Rui H., Pestell R. G. (2008) Clin. Translat. Sci. 1, 107–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shackleton M., Vaillant F., Simpson K. J., Stingl J., Smyth G. K., Asselin-Labat M. L., Wu L., Lindeman G. J., Visvader J. E. (2006) Nature 439, 84–88 [DOI] [PubMed] [Google Scholar]
- 27. Fu M., Rao M., Wang C., Sakamaki T., Wang J., Di Vizio D., Zhang X., Albanese C., Balk S., Chang C., Fan S., Rosen E., Palvimo J. J., Jänne O. A., Muratoglu S., Avantaggiati M. L., Pestell R. G. (2003) Mol. Cell. Biol. 23, 8563–8575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fu M., Wang C., Wang J., Zhang X., Sakamaki T., Yeung Y. G., Chang C., Hopp T., Fuqua S. A., Jaffray E., Hay R. T., Palvimo J. J., Jänne O. A., Pestell R. G. (2002) Mol. Cell. Biol. 22, 3373–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu M., Wang C., Reutens A. T., Wang J., Angeletti R. H., Siconolfi-Baez L., Ogryzko V., Avantaggiati M. L., Pestell R. G. (2000) J. Biol. Chem. 275, 20853–20860 [DOI] [PubMed] [Google Scholar]
- 30. Sakamaki T., Casimiro M. C., Ju X., Quong A. A., Katiyar S., Liu M., Jiao X., Li A., Zhang X., Lu Y., Wang C., Byers S., Nicholson R., Link T., Shemluck M., Yang J., Fricke S. T., Novikoff P. M., Papanikolaou A., Arnold A., Albanese C., Pestell R. (2006) Mol. Cell. Biol. 26, 5449–5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z., Wang C., Jiao X., Katiyar S., Casimiro M. C., Prendergast G. C., Powell M. J., Pestell R. G. (2008) J. Biol. Chem. 283, 7007–7015 [DOI] [PubMed] [Google Scholar]
- 32. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edelman E., Porrello A., Guinney J., Balakumaran B., Bild A., Febbo P. G., Mukherjee S. (2006) Bioinformatics 22, e108–e116 [DOI] [PubMed] [Google Scholar]
- 34. Hulit J., Wang C., Li Z., Albanese C., Rao M., Di Vizio D., Shah S., Byers S. W., Mahmood R., Augenlicht L. H., Russell R., Pestell R. G. (2004) Mol. Cell. Biol. 24, 7598–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu K., Liu M., Li A., Donninger H., Rao M., Jiao X., Lisanti M. P., Cvekl A., Birrer M., Pestell R. G. (2007) Mol. Biol. Cell 18, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ben-Porath I., Thomson M. W., Carey V. J., Ge R., Bell G. W., Regev A., Weinberg R. A. (2008) Nat. Genet. 40, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Charafe-Jauffret E., Ginestier C., Iovino F., Wicinski J., Cervera N., Finetti P., Hur M. H., Diebel M. E., Monville F., Dutcher J., Brown M., Viens P., Xerri L., Bertucci F., Stassi G., Dontu G., Birnbaum D., Wicha M. S. (2009) Cancer Res. 69, 1302–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dalerba P., Cho R. W., Clarke M. F. (2007) Annu. Rev. Med. 58, 267–284 [DOI] [PubMed] [Google Scholar]
- 39. Yamanaka S. (2008) Philos. Trans. R Soc. Lond. B Biol. Sci. 363, 2079–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Viswanathan S. R., Daley G. Q. (2010) Cell 140, 445–449 [DOI] [PubMed] [Google Scholar]
- 41. Li X., Perissi V., Liu F., Rose D. W., Rosenfeld M. G. (2002) Science 297, 1180–1183 [DOI] [PubMed] [Google Scholar]
- 42. Wu K., Yang Y., Wang C., Davoli M. A., D'Amico M., Li A., Cveklova K., Kozmik Z., Lisanti M. P., Russell R. G., Cvekl A., Pestell R. G. (2003) J. Biol. Chem. 278, 51673–51684 [DOI] [PubMed] [Google Scholar]
- 43. Noveen A., Daniel A., Hartenstein V. (2000) Development 127, 3475–3488 [DOI] [PubMed] [Google Scholar]
- 44. Wu K., Katiyar S., Li A., Liu M., Ju X., Popov V. M., Jiao X., Lisanti M. P., Casola A., Pestell R. G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6924–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.