Abstract
Desmosomes and adherens junctions are cadherin-based protein complexes responsible for cell-cell adhesion of epithelial cells. Type 1 cadherins of adherens junctions show specific homophilic adhesion that plays a major role in developmental tissue segregation. The desmosomal cadherins, desmocollin and desmoglein, occur as several different isoforms with overlapping expression in some tissues where different isoforms are located in the same desmosomes. Although adhesive binding of desmosomal cadherins has been investigated in a variety of ways, their interaction in desmosome-forming epithelial cells has not been studied. Here, using extracellular homobifunctional cross-linking, we provide evidence for homophilic and isoform-specific binding between the Dsc2, Dsc3, Dsg2, and Dsg3 isoforms in HaCaT keratinocytes and show that it represents trans interaction. Furthermore, the cross-linked adducts are present in the detergent-insoluble fraction, and electron microscopy shows that extracellular cross-linking probably occurs in desmosomes. We found no evidence for either heterophilic or cis interaction, but neither can be completely excluded by our data. Mutation of amino acid residues Trp-2 and Ala-80 that are important for trans interaction in classical cadherin adhesive binding abolished Dsc2 binding, indicating that these residues are also involved in desmosomal adhesion. These interactions of desmosomal cadherins may be of key importance for their ordered arrangement within desmosomes that we believe is essential for desmosomal adhesive strength and the maintenance of tissue integrity.
Keywords: Adhesion, Cell Adhesion, Cell Junctions, Epithelial Cell, Protein Cross-linking, Cadherin, Desmosome
Introduction
The desmosomal cadherins, desmocollin (Dsc)4 and desmoglein (Dsg), are the adhesion molecules of desmosomes (1–9), intercellular junctions that provide strong adhesion in epithelia and cardiac muscle (10, 11). Adhesion capable of resisting mechanical stress occurs because desmosomes adopt a “hyperadhesive” state not found in other junctions (4, 12, 13). Desmosomes are symmetrical with dense plaques in the cytoplasm of adjacent cells and an intercellular space of about 35 nm wide that shows a zipper-like appearance with a dense midline (14–16), representing an ordered arrangement of the extracellular domains of desmosomal cadherins (16, 17). In this paper we address the molecular mechanism of intercellular binding by desmosomal cadherins.
Dsc and Dsg occur as multiple genetic isoforms; in human tissues there are three Dscs and four Dsgs (18). Dsc2 and Dsg2 are ubiquitous in tissues containing desmosomes. The other isoforms are largely confined to stratified epithelia where they show differentiation-specific expression (18–20). Desmosomes in cells expressing multiple isoforms contain a mixture of those isoforms (20, 21). It is not clear why multiple isoforms of desmosomal cadherins are functionally necessary. Do they have specific adhesive functions or do they carry out specific roles in tissue differentiation and morphogenesis? The evidence is contradictory (2, 5).
Homology modeling indicates that the extracellular (EC) domains of both Dsc and Dsg closely resemble the crystal structure of C-cadherin (12, 22). Adhesion mediated by classical cadherins takes place by strand exchange between the EC1 repeats of cadherin molecules on adjacent cells to form “strand dimers” (22, 23). This involves insertion of the side chain of a tryptophan residue near the extreme N terminus (Trp-2) into a hydrophobic pocket within the EC1 β-barrel (22, 24).
It seems likely that adhesive interaction between desmosomal cadherins involves a similar mechanism. First, the key amino acids involved are conserved in desmosomal cadherins (25). Second, much evidence suggests that desmosomal cadherins interact at their tips or EC1 domains (14–17, 26). Third, anti-adhesion peptides derived from the sequences of the so-called cell adhesion recognition sites in the EC1 domain block adhesion of both classical and desmosomal cadherins (27–32). Fourth, a mis-sense mutation in the human Dsg4 gene of Ala-80, a key residue in the hydrophobic pocket, underlies localized autosomal recessive hypotrichosis (33).
The different classical cadherins show tissue-specific, developmentally regulated expression, and homophilic interaction is believed to regulate tissue segregation during embryogenesis (34, 35). Whereas homophilic adhesion by classical cadherins is believed to be the rule, they can interact heterophilically under some circumstances (36, 37). Chen et al. (38) have suggested how the low affinity interactions between classical cadherins favor homophilic over heterophilic binding.
So-called “cell-based” adhesion assays using cells that cannot form desmosomes or biophysical studies using recombinant EC domains of Dsg and Dsc found evidence for heterophilic or both heterophilic and homophilic binding (31, 38, 39). Biophysical studies of interaction between partial recombinant EC domains of Dsg and Dsc found evidence for both heterophilic and homophilic interaction (40). However, studies with desmosome-forming cells have given results consistent with homophilic binding. Thus anti-adhesion peptides to both Dsc and Dsg were required to block morphogenesis of mammary epithelial cells (30), and a cell type expressing Dsg but not Dsc could form apparently complete desmosomes (41). Homophilic binding is also indicated by atomic force microscopy with tethered recombinant Dsg1 EC domains (42).
Surprisingly, there have been no attempts to determine the mode of desmosomal cadherin binding in desmosome-forming cells. Here, we show that the desmosomal cadherins Dsc2, Dsg2, Dsc3, and Dsg3 in a human keratinocyte cell line interact homophilically and isoform-specifically despite being co-localized at the cell surface and probably present in the same desmosomes. This interaction is dependent on cell-cell adhesion, occurs in trans, and probably involves “strand dimer” formation similar to that of type 1 cadherins.
EXPERIMENTAL PROCEDURES
Plasmids, Mutagenesis
All plasmids were constructed by cutting pEHA (a plasmid containing full-length mouse E-cadherin, a gift from Dr. M. Ozawa) with NotI/EcoRV and ligating with NotI/EcoRV-digested PCR-generated inserts, including full-length mDsg2 (GenBankTM accession number AB072269) and hDsc2a (19) (GenBankTM accession number NM-024422). To detect their expression in HaCaT cells, an HA or FLAG tag sequence was added to the C terminus. The primers used are shown in Table 1. Constructs were then termed pcAGG/mDsg2-HA and pcAGG/mDsg2-FLAG or pcAGG/hDsc2a-HA and pcAGG/hDsc2a-FLAG. W2A and A80I mutants of hDsc2a-HA were generated by PCR-based QuikChange mutagenesis kit (Stratagene) from plasmid pcAGG/hDsc2a-HA. All plasmids were confirmed by DNA sequencing.
TABLE 1.
Primers used to make constructs containing full-length mDsg2 or hDsc2a or for mutagenesis
The enzyme sites NotI or EcoRV are in italics. Start codon and stop codon are boldface. To make pC-mDsg2FLAG, a FLAG sequence (underlined) was incorporated into the antisense primer. Because the original vector pC-E-cadHA contains an in-frame HA tag, no additional HA sequence was added to antisense primers for making pC-mDsg2HA and pC-hDsc2aHA. For mutagenesis, the base pairs being changed are shown in boldface.
| Constructs or mutations | Primers | |
|---|---|---|
| pC-mDsg2HA | Sense | 5′-gacaaGCGGCCGCGGCGGATCGAGGCGATGGCGCGGAGC-3′ |
| Antisense | 5′-gacGATATCGGAGTAAGAATGTTGCATGGTGC-3′ | |
| pC-mDsg2FLAG | Sense | Same as used for construction of pcAGG/mDsg2HA |
| Antisense | 5′-gacaaGATATCTCACTTGTCATCGTCGTCCTTGTAATGGGAGTAAGAATGTTGCATGGTGC-3′ | |
| pC-hDsc2aHA | Sense | 5′-gacaaGCGGCCGCCCTGCCCCGAGCCCTCTCCATGGAGGCAGCCCGCCCCTCCGGC-3′ |
| Antisense | 5′-gacGATATCTCTCTTCATGCATGCTTCTGCTAG-3′ | |
| Mutagenesis | ||
| pC-hDSc2aHAW2A | Sense | 5′-GCGCGCCAAGAGAAGAGCGGCTCCAATTCCTTG-3′ |
| Antisense | 5′-CAAGGAATTGGAGCCGCTCTTCTCTTGGCGCGC-3′ | |
| pC-hDsc2aHAA80I | Sense | 5′-GAGATAATTGCCTTTATAACAACTCCAGATGGG-3′ |
| Antisense | 5′-CCCATCTGGAGTTGTTATAAAGGCAATTATCTC-3′ | |
Cell Culture and Transfection
HaCaT cells, an immortalized, nontumorigenic human keratinocyte line, were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS). They were transfected using Lipofectamine 2000 (Invitrogen). Three days after transfection, cells were cultured in DMEM with 10% FBS containing 0.1 mg/ml G418 for selection. G418-resistant clones were isolated 2 weeks after transfection and screened by immunofluorescence and Western blotting with anti-HA or anti-FLAG antibody. Positive stably transfected HaCaT cell lines were selected with both immunofluorescence and immunoblotting using the anti-tag antibodies. We obtained at least two, and usually three, strongly expressing cell lines for each plasmid used in this study. These positive cells were all tested, and the results were identical.
Chemical Cross-linking
To analyze the intercellular binding of desmosomal cadherins, HaCaT cells were cultured to confluence (unless otherwise stated) and then cross-linked by addition of the membrane-impermeable cross-linker, ethylene glycolbis(sulfosuccinimidylsuccinate) (SEGS) (Pierce) in the presence of 1 mm calcium for the times indicated at room temperature. The reaction was stopped with 50 mm Tris-buffered saline (pH 8.0).
Antibodies, Confocal Immunofluorescence, and Western Blotting and Immunoprecipitation
Monoclonal antibody (mAb) 33-3D against the cytoplasmic domain of Dsg2 and mAb 32-2B against the cytoplasmic domain of Dsg1 and -3 were made in the laboratory (43). mAb P23 against the extracellular domain (EC2–EC4) of hDsg1, polyclonal antibody against the extracellular domain of hDsc2, mAb U100 against the cytoplasmic domain of Dsc1, and mAb U114 against the extracellular domain of hDsc3 were purchased from R&D Systems. Polyclonal anti-HA and mAb anti-FLAG M2 were obtained from Sigma. mAb 12CA5 against HA was a gift from Professor Iain Hagen. Anti-α-catenin was obtained from Sigma and anti-plakoglobin from Chemicon.
For immunofluorescence, confluent cells were fixed with ice-cold methanol/acetone at −20 °C for 10 min. After washing with PBS, primary antibodies were applied. Fluorescence was observed under a Zeiss microscope after incubating cells with FITC- or rhodamine X-labeled secondary antibodies. To visualize the co-localization of different isoforms of desmosomal cadherins or the co-localization of exogenous Dsg2 or Dsc2 or Dsc2 mutants with endogenous desmoplakin, cells were stained with isoform-specific desmosomal antibodies, or anti-tag HA or FLAG antibodies and anti-desmoplakin antibodies (11–5F) (44), or a polyclonal anti-DP antibody from Research Diagnostics and analyzed by Leica SP2 confocal microscope.
Cross-linked adducts were detected by Western blotting. HaCaT cells were cross-linked with SEGS as above and lysed with SDS sample buffer (62.5 mm Tris, 2% SDS, 10% mercaptoethanol, 10% glycerol, and trace of bromphenol blue). Aliquots were loaded onto 2–8% gradient gels. After electrophoresis and transfer to nitrocellulose, membrane proteins were detected with isoform-specific desmosomal cadherin antibodies, anti-E-cadherin, or anti-α-catenin.
To confirm the homophilic and isoform-specific binding of desmosomal cadherins, an immunoprecipitation experiment was used. Cross-linked HaCaT cells were lysed in RIPA buffer (0.4% SDS, 1% sodium deoxycholate, 1% Triton X-100, 2 mm EDTA) (0.5 ml per 75-cm2 flask). Cell lysates were sonicated (Jencons Scientific Ltd., Bedfordshire, UK) three times at level 3, 50% duty for 30 s on ice, and cleared by centrifugation at 10,000 × g for 2 min at room temperature. One ml of pre-cleared supernatant was mixed with 33-3D (1:20), 32-2D (1:10), a polyclonal antibody against Dsc2 (1:25), or U114 (1:2) overnight at 4 °C to form the antibody-antigen complex. Then 80 μl of protein G beads were added to the mixture and incubated at room temperature for 1 h. For 33-3D immunoprecipitation, 50 μl of rabbit anti-mouse IgM (ICN, UK) was added and incubated for a further hour before addition of protein G beads. Then the beads were washed six times with RIPA buffer and twice with 50 mm TBS (pH 8.0) by centrifuging at 100 × g for 1 min and boiled in 100 μl of SDS sample buffer for 10 min. After a centrifugation pulse to pellet the beads, the supernatant was loaded onto 2–8% gel, and electrophoresis and Western blotting were carried out as above.
To investigate trans interaction, a HaCaT cell line expressing HA-tagged mDsg2 was mixed in 1:2 ratio with another expressing FLAG-tagged mDsg2 or vice versa. Mixed cells were cultured for another day until confluence before cross-linking. Immunoprecipitation and Western blotting were carried out as above.
Cell Aggregation Assay
The cell aggregation assay was largely as described previously (45). A single cell suspension was obtained by gently passing trypsin/EDTA-dissociated HaCaT cells through a 40-μm cell strainer twice. More than 95% of cells obtained were single and were >95% viable by trypan blue exclusion. 800 cells were suspended in 100 μl each of media containing different calcium concentrations, added to 2% BSA-coated wells of a 96-well plat, and cultured by shaking at 80 rpm under 5% CO2, 37 °C for 3 h. The extent of cell aggregation was determined by counting the number of the single cells remaining after 3 h.
Electron Microscopy
Cells cultured to confluence on glass coverslips were either untreated or treated with low Ca2+ medium (LCM) for 1 h with or without prior cross-linking with 1 mm SEGS for 10 min. They were then washed with serum-free medium (with or without calcium) and fixed with 2.5% glutaraldehyde in 100 mm phosphate buffer (pH 7.2) followed by 1% osmium tetroxide for 1 h each. Dehydration though an acetone series was followed by treatment with propylene oxide and embedding in TAAB VL resin. The coverslip was removed with tweezers, and thin sections were cut from the block face parallel to the substratum. Sections were stained with 1% uranyl acetate and 0.3% lead citrate before examination on a Tecnai 12 BioTwin electron microscope.
RESULTS
Expression Profile of Desmosomal Cadherins in HaCaT Cells
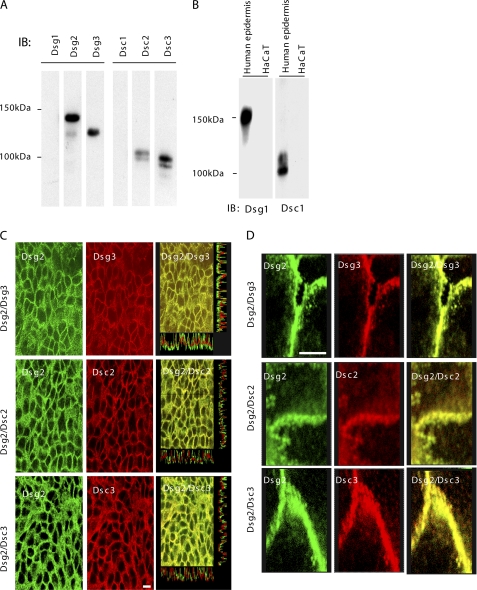
Whole HaCaT cell extracts were probed by immunoblotting with isoform-specific antibodies to determine which desmosomal cadherins were expressed by 24-h confluent cells (Fig. 1A). Dsc2, Dsc3, Dsg2, and Dsg3, but not Dsc1 or Dsg1, were expressed by HaCaT cells under our conditions, and this is consistent with previous findings (46).
FIGURE 1.
HaCaT cells express the Dsc2, Dsc3, Dsg2, and Dsg3 isoforms of desmosomal cadherins. A, extract of confluent HaCaT cells Western-blotted with isoform-specific desmosomal cadherin antibodies. The cells contain Dsg2, Dsg3, Dsc2, and Dsc3 but not Dsc1 and Dsg1. IB, immunoblot. B, positive controls for Dsc1 and Dsg1 antibodies on extract of human epidermis. C, desmosomal cadherins are co-localized on the cell surface. Confluent cells were double-stained with anti-Dsg2 and anti-Dsg3, anti-Dsg2 and anti-Dsc2, or anti-Dsg2 and anti-Dsc3 antibodies and examined by confocal microscopy. The merged images (right) demonstrate that Dsg2 is co-localized with each of the other isoforms. Scale bar, 10 μm. D, images are combined projections of a series of sequential slices and have been enlarged to show the precision of the co-localization. Scale bar, 5 μm.
To determine whether the multiple isoforms in HaCaT cells are co-localized, we carried out immunofluorescence and confocal microscopy. The results show clear co-localization of all isoforms. Dsg2, for example, was co-localized with Dsg3, Dsc2, and Dsc3 (Fig. 1, C and D). On the basis of previous findings (21), this probably indicates that the four isoforms expressed in these cells occur in the same desmosomes.
Desmosomal Cadherins Can Be Cross-linked Extracellularly
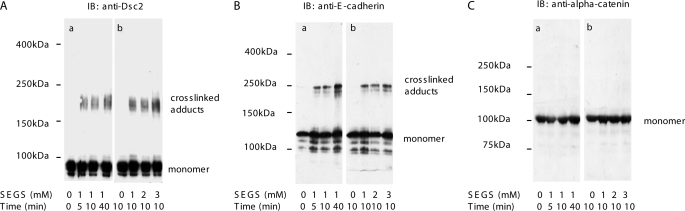
To study the adhesive interactions of desmosomal cadherins, we determined whether they could be cross-linked with the homobifunctional cross-linker SEGS, which has a spacer arm length of 16.1 Å and has been successfully used in the investigation of ligand-receptor interaction of fibronectin (47). SEGS was added to confluent cells at differing concentrations for various periods of time prior to immunoblotting of whole cell lysates for Dsc2 (Fig. 2A, panels a and b). Polypeptide bands at the corresponding to a molecular weight approximately double that of the uncross-linked protein were obtained at all time points following SEGS addition and tested at each concentration. Very similar results were found when the extracts were blotted for E-cadherin (Fig. 2B, panels a and b), resembling those reported previously (48). The cross-linked adducts of both Dsc2 and E-cadherin appeared as multiple but closely similar bands.
FIGURE 2.
Desmosomal cadherins of HaCaT cells can be cross-linked extracellularly with SEGS. A, confluent HaCaT cells were cross-linked with 1 mm SEGS for the times shown (panel a) or for 10 min at the SEGS concentrations shown (panel b), and whole cell extracts were then examined by Western blotting for Dsc2. The Dsc2a monomer had a mass of about 116 kDa. After cross-linking at all SEGS concentrations and all time points, cross-linked adducts of approximately double the size of the monomer were present. B, panels a and b, E-cadherin displayed a similar pattern under the same cross-linking conditions. C, cytoplasmic adherens junction protein α-catenin was not cross-linked under these conditions demonstrating that SEGS acts extracellularly. IB, immunoblot.
Heavier loading of gels sometimes revealed cross-linked adducts of higher molecular weight apparently corresponding to trimers (supplemental Fig. 1A). In some cases, these larger adducts could be enhanced by using higher concentrations of cross-linker (supplemental Fig. 1B). In others, however, such enhancement was not seen (supplemental Fig. 1C). Because these larger adducts were less abundant than the dimers and their appearance less consistent, this paper focuses on the dimers, leaving the others for later investigation.
To confirm that the cross-linking occurred extracellularly, cell extracts were blotted for the cytoplasmic adherens junction protein α-catenin. No cross-linking of α-catenin was observed (Fig. 2C, panels a and b). However, when the membrane-permeable cross-linker ethylene glycolbis(succinimydylsuccinate) was used in place of SEGS, multiple high molecular weight adducts containing α-catenin were formed (data not shown). These results show that the desmosomal cadherin Dsc2 can be extracellularly cross-linked with the membrane-impermeable cross-linker SEGS to form adducts, the most abundant of which approximates in molecular weight to a dimer.
Cross-linking of Desmosomal Cadherins Is Cell-Cell Adhesion-dependent
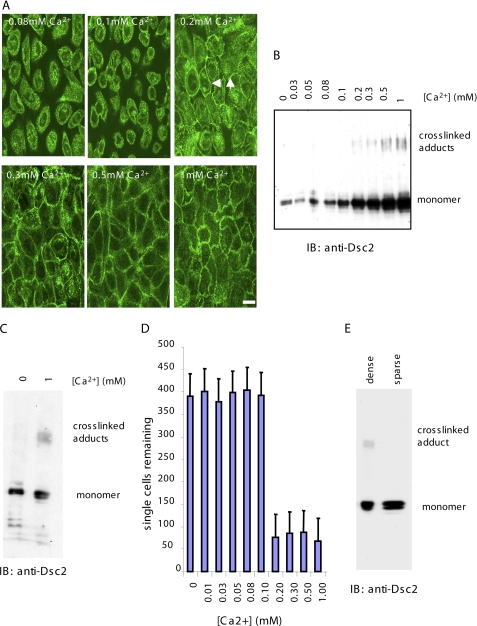
To determine whether the cross-linked adduct of Dsc2 was functionally relevant to cell-cell adhesion, we studied its appearance in response to Ca2+-induced desmosome assembly. Cells were cultured at confluent density in LCM where they formed no intercellular junctions, and the extracellular Ca2+ concentration then was raised to the levels indicated in Fig. 3 for 16 h to allow time for desmosome formation. SEGS (1 mm) was added to parallel cultures for 10 min, and the cells were extracted for analysis by Western blotting with anti-Dsc2 antibody. The lowest extracellular Ca2+ concentration at which desmosome formation was detectable by immunofluorescence was 0.2 mm (Fig. 3A). Similarly, 0.2 mm was the lowest Ca2+ concentration at which cross-linking of Dsc2 was detectable (Fig. 3B). (Similar results were obtained for the other three desmosomal cadherins (supplemental Fig. 2).)
FIGURE 3.
Desmosomal cadherin cross-linking occurs in a cell-cell adhesion-dependent manner. A, HaCaT cells were cultured for 24 h at the extracellular calcium concentrations indicated, and immunofluorescence was carried out for Dsc2. The threshold calcium concentration for desmosome assembly was 0.2 mm as indicated by the presence of Dsc2 staining at the cell periphery (white arrows). Scale bar, 15 μm. B, HaCaT cells were cultured at the same range of extracellular calcium concentrations as in A and cross-linked for 10 min with 1 mm SEGS, and cell extracts were examined by Western blotting for Dsc2. The threshold extracellular calcium concentration for appearance of cross-linked adducts was 0.2 mm, identical to that for desmosome assembly. C, no cross-linking was detected in cells cultured in calcium-free medium. Cells grown in the absence of calcium and in 1 mm calcium were cross-linked with 1 mm SEGS for 10 min. Western blotting for Dsc2 showed cross-linked adducts in the 1 mm extract only. D, dissociated HaCaT cells were aggregated in media containing the calcium concentrations shown, and the number of single cells remaining after 3 h was determined. As for desmosome assembly and cross-linking, the threshold calcium concentration for cells aggregation was 0.2 mm. E, no cross-linking was detectable in cells cultured in standard medium (1.8 mm calcium) at low density (>95% single cells) (sparse). IB, immunoblot.
To confirm that no cross-linking of Dsc2 occurred in LCM, gels were loaded with the cross-linked LCM extract until the amount of Dsc2 monomer was similar to that found in normal culture medium. Still no cross-linked adduct was detectable (Fig. 3C). Because it is possible that significant amounts of desmosomal cadherins may not be present on the surfaces of cell and thus available for cross-linking in LCM, we determined whether Dsc2 was susceptible to trypsin cleavage in cells in LCM. We found that >50% of the Dsc2 present in cells in LCM is susceptible to cleavage by trypsin and therefore exposed on the cell surfaces (data not shown).
To provide a functional demonstration of the Ca2+ dependence of cell-cell adhesion, an aggregation assay was carried out over the same range of Ca2+ concentrations. Aggregation began at an extracellular Ca2+ concentration of 0.2 mm, the same concentrations at which desmosome assembly and cross-linking of Dsc2 were initiated (Fig. 3D).
Because cadherins are not functional in LCM (49, 50), it may be that Ca2+ switching experiments are not sufficient to show that cross-linking of desmosomal cadherins requires cell-cell adhesion; cis interaction may occur, and cross-linking may be obtainable in normal culture medium even in the absence of cell-cell adhesion. To test this, cells were cultured at very low density (1 × 104 cells/cm2 compared with 2 × 105 cells/cm2) such that fewer than 5% of the cells were able to form a contact with a neighbor. SEGS was added to these cells as in the Ca2+ switching experiments and extracts prepared for Western blotting. No cross-linking of Dsc2 was detected, even when gels were loaded until the amount of Dsc2 monomer was comparable with that found in confluent cells (Fig. 3E). Trypsinization of cells at low density followed by Western blotting showed that about 50% of the Dsc2 was at the cell surface under these conditions and therefore available for cross-linking (data not shown). Taken together, these results indicate that formation of the cross-linked Dsc2 adduct was dependent upon cell-cell adhesion and desmosome assembly.
Desmosomal Cadherins Interact Homophilically and Isoform-specifically
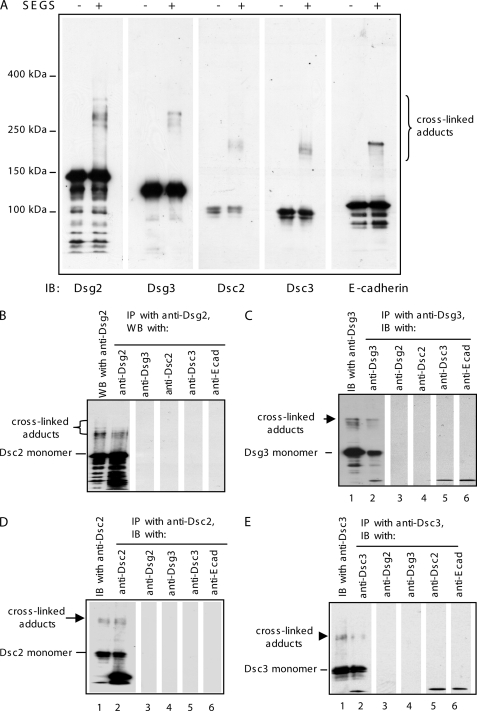
To determine how desmosomal cadherins interact in cell-cell adhesion, cross-linking was carried out with confluent HaCaT cells, and extracts were examined by Western blotting for all four desmosomal cadherins. Cross-linked adducts similar to those obtained for Dsc2 were also obtained for the Dsc3, Dsg2, and Dsg3 (Fig. 4A). In each case, the molecular weight of the cross-linked adduct was consistent with the formation of a dimer. Strikingly, the adducts formed by each molecule were slightly different in mobility from those formed by the others, and by E-cadherin. (Essentially similar results were obtained using another membrane-impermeable cross-linker, bis(sulfosuccinimidyl) suberate, which has a spacer arm length of 11.4 Å, showing that such adduct formation is not a specific property of SEGS (supplemental Fig. 3).) This suggested that the molecules may be interacting in a homophilic and isoform-specific manner; if they were interacting in a nonspecific manner, identical adducts extending over a broader range of mobilities would be expected.
FIGURE 4.
Desmosomal cadherins bind homophilically and isoform-specifically. A, cross-linking produced slightly different adducts for each of the desmosomal cadherins and E-cadherin. Cell extracts from HaCaT cells with (+) or without (−) treatment with 1 mm SEGS for 10 min were blotted with desmosomal cadherin isoform-specific antibodies and E-cadherin. The cross-linked adducts were approximately double the size of the monomers and were slightly different in molecular weight in each case suggesting that they may correspond to homophilic dimers. B–E, cross-linked adducts contain only one desmosomal cadherin. Extracts of cross-linked HaCaT cells (1 mm SEGS for 10 min) were immunoprecipitated (IP) with each of the specific antibodies for the expressed desmosomal cadherins, and then equal amounts of extract were Western-blotted (WB) for one of the desmosomal cadherins and E-cadherin. Lane 1 in each panel shows a Western blot of the cross-linked cell extract with the antibody used for IP. Lanes 2–6 show Western blots of the immunoprecipitated material with the antibodies indicated at the top of the lanes. In each case only the antibody used for IP detected the cross-linked adduct. IB, immunoblot.
To test this hypothesis, confluent HaCaT cells Ca2+ were cross-linked and extracted for immunoprecipitation with antibodies specific for each of the four desmosomal cadherins. Each immunoprecipitate was then Western-blotted with each of the four desmosomal cadherin antibodies. In every case, the cross-linked adduct reacted only with the same antibody that was used for immunoprecipitation (Fig. 4, B–E). Thus, only one desmosomal cadherin was detectable in each cross-linked adduct, indicating that these molecules show adhesion-dependent, isoform-specific, homophilic interaction.
Desmosomes Are Extracellularly Cross-linked by SEGS
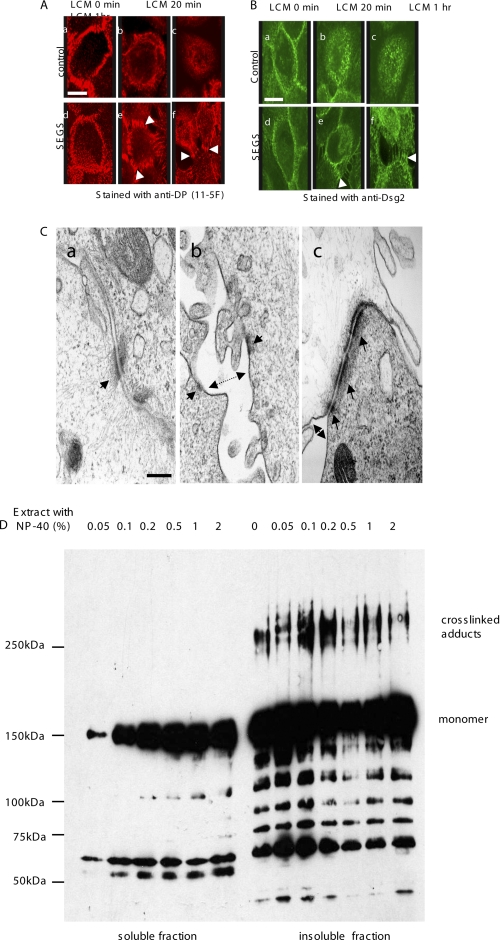
To provide evidence that the cross-linked adducts are significant for intercellular adhesive binding, we sought direct evidence that cross-linking detects trans binding interactions in desmosomes. First, after cross-linking with 1 mm SEGS, HaCaT cells were further treated with LCM. Uncross-linked HaCaT cells in LCM lost cell-cell adhesion, and desmosomal staining for desmoplakin or Dsg2 (Fig 5, A, panels a–c, and B, panels a–c) was largely internalized within 20 min. By contrast, in cross-linked HaCaT cells, although there was some loss of adhesion and internalization of staining, bridge-like connections that were intensely stained for desmosomal components persisted indefinitely between the cells (Fig. 5, A, panels d–f, and B, panels d–f, arrowheads). SEGS treatment for 10 min did not affect cell viability as 100% of the cells excluded trypan blue. The bridge-like desmosome staining suggests that SEGS cross-linked trans interactions in desmosomal adhesion.
FIGURE 5.
Desmosomes are cross-linked by SEGS. A and B, confluent HaCaT were either untreated (control) or treated with 1 mm SEGS for 10 min and exposed to calcium-free medium for 1 h, and then immunofluorescence was carried out for desmoplakin (A) or Dsg2 (B). (standard medium). In untreated cells, the majority of staining was internalized after LCM exposure (A, panels a–c, and B, panels a–c), but after SEGS treatment, a substantial amount of staining remained in intercellular processes at the cell surface (A, panels d–f, and B, panels d–f) (arrowheads). The cross-linking effect was seen in 100% of cells. Scale bars, 10 μm. C, electron microscopy confirms that desmosomes are cross-linked by SEGS. Panel a, standard medium control showing typical desmosomes and apposed plasma membranes. Panel b, after 1 h of LCM treatment, plasma membranes (double arrow) and desmosomal halves (arrows) had separated. Panel c, after cross-linking with 1 mm SEGS for 10 min followed by LCM treatment for 1 h, plasma membranes had separated (double arrow) but desmosomes (arrows) remained intact and became grouped at the cell surface. Scale bar, 200 nm. D, cross-linked adducts are in the Nonidet P-40-insoluble fraction. Confluent cells were cross-linked with 1 mm SEGS for 10 min and extracted with 1% Nonidet P-40 for 20 min, and the soluble and insoluble fractions were loaded onto a gel so that the amount of Dsg2 monomer in each lane was approximately equal and then Western-blotted for Dsg2 with monoclonal antibody 33-3D. Cross-linked adducts appeared in the insoluble fraction only.
To confirm that desmosomes were indeed cross-linked by SEGS, cells were examined by electron microscopy (Fig. 5C). Controls showed desmosomes typical of cultured cells (Fig. 5C, panel a, arrow). Treatment with LCM resulted in characteristic separation of cell membranes (Fig. 5C, panel b, double arrow) and of desmosomal halves (Fig. 5C, panel b, arrows) (51). By contrast, cells treated with 1 mm SEGS before LCM treatment showed persistence and accumulation of desmosomes at the cell surface (Fig. 5C, panel c, arrows) even though the nonjunctional cell membranes were separated by this treatment (Fig. 5C, panel c, double arrow). This indicated that cross-linking by SEGS does indeed involve trans interaction in desmosomes.
To exclude the possibility that the cross-linked products were originating from desmosomal cadherins that were present at the cell surface but not incorporated into desmosomes, we prepared the nonionic detergent (Nonidet P-40)-soluble and -insoluble fractions. Because desmosomes are insoluble in nonionic detergent, the cross-linked products should be represented in the detergent-insoluble fraction. Western blotting of the fractions with antibody to Dsg2 showed that a range of Nonidet P-40 concentrations up to 2% extracted a proportion of the Dsg2 monomer but failed to extract any of the cross-linked adducts (Fig. 5D, soluble), which were exclusively located in the insoluble fraction (Fig. 5D, insoluble), most probably in desmosomes.
SEGS Cross-linked Dimers Result from Trans Interaction
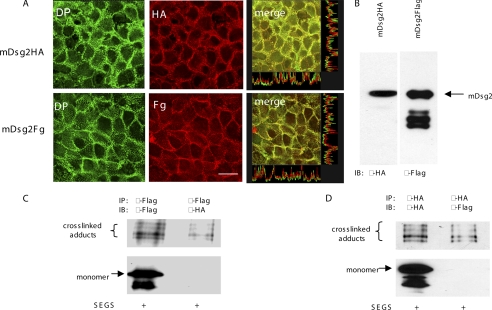
To provide direct evidence for homophilic, isoform-specific trans interaction stable cell lines expressing, respectively, HA-tagged or FLAG-tagged mDsg2 were produced. The cell lines were then plated in mixed cultures, and cross-linking with SEGS was carried out to determine whether dimers contained both tags as could only result from trans interaction. We first showed that the ectopically expressed tagged proteins were expressed in approximately equal amounts (Fig. 6B) and appeared to be incorporated into endogenous desmosomes as revealed by a double staining with anti-tag and anti-desmoplakin antibodies (Fig. 6A). Homophilic trans interaction of mDsg2 was detected by immunoprecipitation with anti-FLAG antibody followed by Western blotting of the immunoprecipitate with anti-HA antibody or vice versa. It was found that cell mixtures that contained twice as many cells of the type to be detected by Western blotting enabled the trans cross-linked adducts to be most readily detected. The anti-FLAG antibody immunoprecipitated both monomer and dimer of mDsg2, whereas the anti-HA antibody detected only the dimer (Fig. 6C), indicating that the cross-linked band contains trans-engaged homophilic dimer. Similarly, the anti-HA antibody immunoprecipitated both monomer and dimer, whereas the anti-FLAG antibody detected only the dimer (Fig. 6D). Thus, in each case the immunoprecipitating antibody detected both monomer and cross-linked adducts, but the other antibody used to blot the immunoprecipitate detected only the adducts, demonstrating unequivocally that the latter contained trans-interacting proteins.
FIGURE 6.
Cross-linked desmosomal cadherins contain trans-interacting molecules. A, mDsg2-HA and mDsg2-FLAG were incorporated into desmosomes. Confluent transfected cells were double-stained with anti-desmoplakin antibody, 11-5F, and anti-HA or anti-FLAG. Co-localization of desmoplakin (DP) with HA-tagged or FLAG-tagged mDsg2 was observed under the confocal microscope. Scale bar, 5 μm. B, whole cell extracts of HaCaT cells with stable expression of pC-mDsg2HA or pC-mDsg2FLAG were Western-blotted (equal loading) and showed approximately equal expression of HA-tagged mDsg2 (mDsg2-HA) or FLAG-tagged mDsg2 (mDsg2-Fg) in the clones selected for the experiment. C and D, detection of trans-interacting cross-linked adducts. Transfected cells were mixed in a ratio of 1:2 of the type to be immunoprecipitated to the type to be Western-blotted after IP and cultured at confluent density. The cultures were cross-linked (1 mm SEGS for 10 min), extracted, and immunoprecipitated with the appropriate anti-tag antibody. The IPs were Western-blotted with the antibody used for IP and with the antibody against the other tag. In each case, the IP antibody recognized both the monomer and the cross-linked adduct, whereas the other antibody recognized only the cross-linked adducts.
Desmosomal and Classical Cadherins Adhere by Similar Mechanisms
Studies of classical cadherins have identified key amino acid residues that are involved in formation of the adhesive interface (52, 53). These include a tryptophan residue in position 2 (Trp-2) and an alanine residue at position 80 (Ala-80). Strand exchange that results in formation of the adhesion or strand dimer involves insertion of the side chain of Trp-2 into a hydrophobic pocket to which Ala-80 contributes. Trp-2 and Ala-80 are conserved in all classical or type I cadherins and desmosomal cadherins. To determine whether a similar adhesion mechanism might apply to both desmosomal and classical cadherins, we mutated Trp-2 and/or Ala-80 in Dsc2, expressed the mutant proteins in HaCaT cells, and studied their interaction by cross-linking.
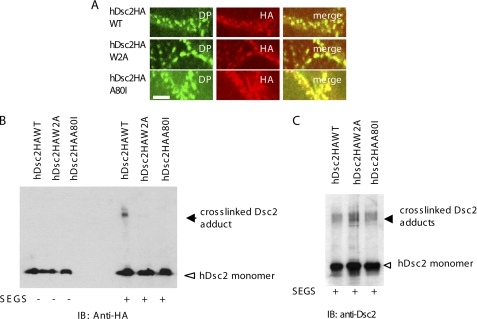
We first showed, by C-terminal HA tagging, that both wild type and mutant exogenous proteins were expressed at the cell surface and co-localized with desmosomes (Fig. 7A), so that any failure to interact could not be attributed to mis-localization. Cross-linking with SEGS in 1 mm Ca2+ showed that cross-linked adducts similar to those found in previous experiments were obtained with the HA-tagged wild type protein but that no such adducts were obtained with any of the mutants (Fig. 7B). Although desmosome formation appeared by immunofluorescence to be normal in cells expressing mutant proteins, we further showed that endogenous Dsc2 could still be cross-linked in these cells (Fig. 7C), indicating that desmosomal adhesion was not disrupted. These results indicate that there is a strong similarity between the mechanisms of adhesion by classical and desmosomal cadherins. They are also consistent with the view that the cross-linked adducts contain material that is trans-interacting through strand dimer formation.
FIGURE 7.
Mutation in either Trp-2 or Ala-80 abolishes cross-linking of hDsc2. A, hDsc2 mutants were incorporated into desmosomes. Confluent cells expressing wild type hDsc2-HA or mutants hDsc2HAW2A or hDsc2HAA80I were double-stained with anti-HA and anti-DP, 11-5F. Co-localization of overexpressed wild type hDsc2 or its mutants with desmoplakin was observed by confocal microscopy. Scale bars, 5 μm. B, Western blot shows that wild type hDsc2 was cross-linked after SEGS treatment but that the mutants, which were expressed in comparable amounts, were not. C, as a control, an identical copy of the blot shown in B was stained with anti-hDsc2 antibody, showing that the endogenous Dsc2 was cross-linked in the presence of both mutants.
DISCUSSION
Our principal finding is that the desmosomal cadherins Dsc2, Dsc3, Dsg2, and Dsg3 bind homophilically and isoform-specifically in HaCaT cells. This binding is functionally important because it depends on desmosome assembly and cell-cell adhesion, occurs in trans, and is abolished by mutation of key residues that are involved in cadherin adhesion. We believe that these interactions probably occur in desmosomes because extracellular cross-linking prevented desmosome disruption by LCM, and the cross-linked adducts were exclusively present in the Nonidet P-40-insoluble cell fraction. We stress that our current findings do not exclude heterophilic or inter-isoform binding, but they may imply that such interactions are less abundant than homophilic, isoform-specific binding.
Research demonstrating heterophilic adhesive binding by desmosomal cadherins has been carried out with either transfected fibroblasts that are incapable of assembling desmosomes (32, 39, 54) or with recombinant protein fragments (40). Such proteins are out of context and thus cannot form the complex cytoplasmic interactions with other desmosomal proteins in the plaque (62). Nevertheless, homophilic binding by desmosomal cadherins has sometimes been reported from studies with transfected L929 cells and recombinant proteins (40, 42, 55, 56). In contrast, work with transfected HT1080 SL-1 fibrosarcoma cells and mammary epithelial cells, both of which form desmosomes, is more consistent with homophilic adhesion (30, 41). The former cells assemble desmosomes with Dsg as the only adhesion molecule, which must therefore bind homophilically, and the latter cell type required anti-adhesion peptides to both Dsg and Dsc to block their adhesive interaction. Our work provides the first direct evidence for homophilic binding between desmosomal cadherins in desmosome-forming epithelial cells.
Furthermore, the binding we have found in HaCaT cells is isoform-specific. Although there is some evidence for nonspecific interaction between different desmosomal cadherin isoforms, for example human Dsg2 and bovine Dsg1, these proteins were again not in desmosomes (54). However, in adhesion-blocking experiments with desmosome-forming mammary epithelial cells, the requirement for isoform-specific anti-adhesion peptides was found (30).
Thus, the overall conclusion from previous and present studies appears to be that in desmosome-forming epithelial cells, in which multiple adhesive interactions are potentially available, homophilic, isoform-specific adhesive binding by desmosomal cadherins may be favored. However, desmosomal cadherins can also participate in heterophilic interaction under some circumstances. In both these respects, desmosomal cadherins appear to resemble type I cadherins (38).
The small proportion of cross-linked desmosomal cadherins in our experiments is similar to other findings using cross-linkers of similar spacer arm length and is consistent with a low efficiency of cross-linking (57). Each cross-linked adduct also consisted of multiple polypeptide bands. This might be expected for Dscs, which have two alternatively spliced variants of different sizes (58), but cannot also be the explanation for Dsgs and E-cadherin. Possible explanations of these multiple bands are as follows. First, SEGS forms covalent bonds with lysine residues of which there are many in the EC domains of desmosomal cadherins. For example, consideration of our homology model for Dsg2 (12) revealed that there are 12 exposed lysine residues in the EC domain. In order for them to be cross-linked by SEGS, they must lie within 16.1 Å of each other, and our analysis showed that by this criterion formation of eight potential intermolecular bonds is possible. The significance of this analysis for this study is that for any desmosomal cadherin the cross-linked dimers are likely to represent several different species with differing cross-linking patterns. These are likely to have different shapes and therefore slightly different electrophoretic mobilities resulting in the appearance of multiple gel bands. A second possibility is that there may be slight degradation of the dimers and/or of the monomers prior to cross-linking.
It is important to consider how the interactions may relate to the organization of desmosomes. We have established previously that different Dsc isoforms can be located in the same individual desmosomes (20, 21), and the same probably is true for Dsgs (59). We detected no patterned arrangement of the different Dsc isoforms in epidermal desmosomes, although the immuno-gold labeling technique may not have sufficient resolution to resolve such a pattern (21). Other ultrastructural data suggest that the extracellular domains of the desmosomal cadherins adopt a highly organized arrangement within the desmoglea (14–16). We have suggested that it is essential for maintaining the hyper-adhesive, calcium-independent state of desmosomes that is important for the stress resistance of tissues (4, 12). The homophilic, isoform-specific binding of desmosomal cadherins may in some way contribute to this ordered arrangement. We speculate that some regular arrangement such as alternating rows of Dscs and Dsgs may be present in mature desmosomes.
We have provided direct evidence for trans adhesive binding, but our data do not exclude the presence of cis dimers. If they are present, our data appear to indicate that cis interaction is also predominantly of a homophilic, isoform-specific nature. We were unable to find evidence for cis interaction in cells in LCM or those prevented from adhering in standard culture medium by plating at low density. This is curious because cis interactions are obviously required to promote the formation of surface-exposed half-desmosomes reported in HaCaT cells maintained in LCM (60, 61). We suggest that such cis interactions are maintained intracellularly through the complex structure of the desmosomal plaque, which contains the cytoplasmic domains of the desmosomal cadherins (62). Different techniques will be required to resolve the issue of whether cis interactions occur between the extracellular domains of desmosomal cadherins. Cis interactions are a feature of the tomographic model of Al-Amoudi et al. (16), but the resolution of that technique is 3.4 nm, or double the spacer arm length of SEGS, so the reported interactions may not be close enough to permit cross-linking.
Molecular modeling studies based on electron tomography of desmosomes have suggested the presence of trimers and tetramers of desmosomal cadherins in addition to dimers (16, 17). These trimers and tetramers are based on the presence of both cis and trans interactions between the desmosomal cadherins. Higher molecular weight adducts were detected in our experiments, but they were less abundant, and their formation was less consistent than that of dimers. Analysis of their composition awaits further investigation. However, our main results are consistent with reports that under standard culture conditions adhesive dimers are the dominant form of classical cadherins (57, 63).
Functional evidence to support EC1-EC1 subdomain interaction between desmosomal cadherins has also been provided through the inhibition of desmosomal adhesion by short sequence-specific peptides derived from the cell adhesion recognition sites of desmosomal cadherins (30, 32). The isoform specificity of such inhibition has been noted above. These peptide sequences are centered on the Ala-80 residue conserved between desmosomal and classical type 1 cadherins. Our finding that mutation of this residue in desmosomal cadherins, together with the conserved Trp-2 residue that is also involved in EC1-EC1 binding, blocks the formation of cross-linked dimers provides further support for this mode of adhesive binding. Similar mechanisms governing the specificity of homophilic adhesion are likely given the striking similarity between desmosomal cadherin adhesion and that of type 1 cadherins. The affinity of a single cadherin-cadherin binding event is very low, and subtle differences in affinity are likely to determine their binding specificity (38). Thus, it appears that homophilic strand exchange to form a strand dimer is energetically favored over strand retention by the monomer or heterophilic binding (38).
A preference for homophilic adhesion between classical cadherins is believed to be functionally important in tissue segregation during development as different cadherins show spatially and temporally different patterns of expression (34, 64). It seems that specific adhesion in desmosomes operates on a smaller scale as the individual desmosomal cadherins can be mixed within junctions that are less than 0.5 μm in diameter (65, 66). Whether specificity of adhesion is important for desmosome structure remains to be seen. However, there is evidence as reviewed above that desmosomal cadherins can contribute to the regulation of cell positioning and epithelial differentiation even though they are restricted to small punctate membrane domains. It is possible that their homophilic, isoform-specific interaction is important for these functions.
Acknowledgments
We thank Stephen High, David Wateridge, Selina McHarg, Michael Richards, and Anthea Scothern for advice and valuable constructive criticism of the manuscript and Amy Rudge and Nicola Aldren for technical assistance.
This work was supported by the Medical Research Council Grants G9630879 and G0700074.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Methods.
- Dsc
- desmocollin
- Dsg
- desmoglein
- SEGS
- ethylene glycolbis(sulfosuccinimidylsuccinate)
- EC
- extracellular
- LCM
- low calcium medium
- IP
- immunoprecipitated.
REFERENCES
- 1. Huber O. (2003) Cell. Mol. Life Sci. 60, 1872–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garrod D. R., Merritt A. J., Nie Z. (2002) Curr. Opin. Cell Biol. 14, 537–545 [DOI] [PubMed] [Google Scholar]
- 3. Garrod D. R., Merritt A. J., Nie Z. (2002) Mol. Membr. Biol. 19, 81–94 [DOI] [PubMed] [Google Scholar]
- 4. Kimura T. E., Merritt A. J., Garrod D. R. (2007) J. Invest. Dermatol. 127, 775–781 [DOI] [PubMed] [Google Scholar]
- 5. Dusek R. L., Godsel L. M., Green K. J. (2007) J. Dermatol. Sci. 45, 7–21 [DOI] [PubMed] [Google Scholar]
- 6. Yin T., Green K. J. (2004) Semin. Cell Dev. Biol. 15, 665–677 [DOI] [PubMed] [Google Scholar]
- 7. Green K. J., Simpson C. L. (2007) J. Invest. Dermatol. 127, 2499–2515 [DOI] [PubMed] [Google Scholar]
- 8. Garrod D., Chidgey M. (2008) Biochim. Biophys. Acta 1778, 572–587 [DOI] [PubMed] [Google Scholar]
- 9. Thomason H. A., Scothern A., McHarg S., Garrod D. R. (2010) Biochem. J. 429, 419–433 [DOI] [PubMed] [Google Scholar]
- 10. Vasioukhin V., Bowers E., Bauer C., Degenstein L., Fuchs E. (2001) Nat. Cell Biol. 3, 1076–1085 [DOI] [PubMed] [Google Scholar]
- 11. Grossmann K. S., Grund C., Huelsken J., Behrend M., Erdmann B., Franke W. W., Birchmeier W. (2004) J. Cell Biol. 167, 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garrod D. R., Berika M. Y., Bardsley W. F., Holmes D., Tabernero L. (2005) J. Cell Sci. 118, 5743–5754 [DOI] [PubMed] [Google Scholar]
- 13. Garrod D. (2010) Dermatol. Res. Pract. 2010;2010:212439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rayns D. G., Simpson F. O., Ledingham J. M. (1969) J. Cell Biol. 42, 322–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Al-Amoudi A., Norlen L. P., Dubochet J. (2004) J. Struct. Biol. 148, 131–135 [DOI] [PubMed] [Google Scholar]
- 16. Al-Amoudi A., Díez D. C., Betts M. J., Frangakis A. S. (2007) Nature 450, 832–837 [DOI] [PubMed] [Google Scholar]
- 17. He W., Cowin P., Stokes D. L. (2003) Science 302, 109–113 [DOI] [PubMed] [Google Scholar]
- 18. Whittock N. V., Bower C. (2003) J. Invest. Dermatol. 120, 523–530 [DOI] [PubMed] [Google Scholar]
- 19. Parker A. E., Wheeler G. N., Arnemann J., Pidsley S. C., Ataliotis P., Thomas C. L., Rees D. A., Magee A. I., Buxton R. S. (1991) J. Biol. Chem. 266, 10438–10445 [PubMed] [Google Scholar]
- 20. Nuber U. A., Schäfer S., Stehr S., Rackwitz H. R., Franke W. W. (1996) Eur. J. Cell Biol. 71, 1–13 [PubMed] [Google Scholar]
- 21. North A. J., Chidgey M. A., Clarke J. P., Bardsley W. G., Garrod D. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7701–7705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boggon T. J., Murray J., Chappuis-Flament S., Wong E., Gumbiner B. M., Shapiro L. (2002) Science 296, 1308–1313 [DOI] [PubMed] [Google Scholar]
- 23. Shapiro L., Fannon A. M., Kwong P. D., Thompson A., Lehmann M. S., Grübel G., Legrand J. F., Als-Nielsen J., Colman D. R., Hendrickson W. A. (1995) Nature 374, 327–337 [DOI] [PubMed] [Google Scholar]
- 24. Häussinger D., Ahrens T., Aberle T., Engel J., Stetefeld J., Grzesiek S. (2004) EMBO J. 23, 1699–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nollet F., Kools P., van Roy F. (2000) J. Mol. Biol. 299, 551–572 [DOI] [PubMed] [Google Scholar]
- 26. Shimizu A., Ishiko A., Ota T., Saito H., Oka H., Tsunoda K., Amagai M., Nishikawa T. (2005) J. Invest. Dermatol. 124, 984–989 [DOI] [PubMed] [Google Scholar]
- 27. Blaschuk O. W., Sullivan R., David S., Pouliot Y. (1990) Dev. Biol. 139, 227–229 [DOI] [PubMed] [Google Scholar]
- 28. Williams E., Williams G., Gour B. J., Blaschuk O. W., Doherty P. (2000) J. Biol. Chem. 275, 4007–4012 [DOI] [PubMed] [Google Scholar]
- 29. Noë V., Willems J., Vandekerckhove J., Roy F. V., Bruyneel E., Mareel M. (1999) J. Cell Sci. 112, 127–135 [DOI] [PubMed] [Google Scholar]
- 30. Runswick S. K., O'Hare M. J., Jones L., Streuli C. H., Garrod D. R. (2001) Nat. Cell Biol. 3, 823–830 [DOI] [PubMed] [Google Scholar]
- 31. Chidgey M., Brakebusch C., Gustafsson E., Cruchley A., Hail C., Kirk S., Merritt A., North A., Tselepis C., Hewitt J., Byrne C., Fassler R., Garrod D. (2001) J. Cell Biol. 155, 821–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tselepis C., Chidgey M., North A., Garrod D. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8064–8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Messenger A. G., Bazzi H., Parslew R., Shapiro L., Christiano A. M. (2005) J. Invest. Dermatol. 125, 1077–1079 [DOI] [PubMed] [Google Scholar]
- 34. Vleminckx K., Kemler R. (1999) BioEssays 21, 211–220 [DOI] [PubMed] [Google Scholar]
- 35. Overduin M., Harvey T. S., Bagby S., Tong K. I., Yau P., Takeichi M., Ikura M. (1995) Science 267, 386–389 [DOI] [PubMed] [Google Scholar]
- 36. Niessen C. M., Gumbiner B. M. (2002) J. Cell Biol. 156, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duguay D., Foty R. A., Steinberg M. S. (2003) Dev. Biol. 253, 309–323 [DOI] [PubMed] [Google Scholar]
- 38. Chen C. P., Posy S., Ben-Shaul A., Shapiro L., Honig B. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8531–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marcozzi C., Burdett I. D., Buxton R. S., Magee A. I. (1998) J. Cell Sci. 111, 495–509 [DOI] [PubMed] [Google Scholar]
- 40. Syed S. E., Trinnaman B., Martin S., Major S., Hutchinson J., Magee A. I. (2002) Biochem. J. 362, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koeser J., Troyanovsky S. M., Grund C., Franke W. W. (2003) Exp. Cell Res. 285, 114–130 [DOI] [PubMed] [Google Scholar]
- 42. Waschke J., Menendez-Castro C., Bruggeman P., Koob R., Amagai M., Gruber H. J., Drenckhahn D., Baumgartner W. (2007) J. Membr. Biol. 216, 83–92 [DOI] [PubMed] [Google Scholar]
- 43. Vilela M. J., Hashimoto T., Nishikawa T., North A. J., Garrod D. (1995) J. Cell Sci. 108, 1743–1750 [DOI] [PubMed] [Google Scholar]
- 44. Parrish E. P., Steart P. V., Garrod D. R., Weller R. O. (1987) J. Pathol. 153, 265–273 [DOI] [PubMed] [Google Scholar]
- 45. Chidgey M. A., Clarke J. P., Garrod D. R. (1996) J. Invest. Dermatol. 106, 689–695 [DOI] [PubMed] [Google Scholar]
- 46. Denning M. F., Guy S. G., Ellerbroek S. M., Norvell S. M., Kowalczyk A. P., Green K. J. (1998) Exp. Cell Res. 239, 50–59 [DOI] [PubMed] [Google Scholar]
- 47. Akimov S. S., Krylov D., Fleischman L. F., Belkin A. M. (2000) J. Cell Biol. 148, 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Troyanovsky S. M., Eshkind L. G., Troyanovsky R. B., Leube R. E., Franke W. W. (1993) Cell 72, 561–574 [DOI] [PubMed] [Google Scholar]
- 49. Nagar B., Overduin M., Ikura M., Rini J. M. (1996) Nature 380, 360–364 [DOI] [PubMed] [Google Scholar]
- 50. Pertz O., Bozic D., Koch A. W., Fauser C., Brancaccio A., Engel J. (1999) EMBO J. 18, 1738–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mattey D. L., Garrod D. R. (1986) J. Cell Sci. 85, 113–124 [DOI] [PubMed] [Google Scholar]
- 52. Tamura K., Shan W. S., Hendrickson W. A., Colman D. R., Shapiro L. (1998) Neuron 20, 1153–1163 [DOI] [PubMed] [Google Scholar]
- 53. Nose A., Tsuji K., Takeichi M. (1990) Cell 61, 147–155 [DOI] [PubMed] [Google Scholar]
- 54. Chitaev N. A., Troyanovsky S. M. (1997) J. Cell Biol. 138, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Amagai M., Kàrpàti S., Klaus-Kovtun V., Udey M. C., Stanley J. R. (1994) J. Invest. Dermatol. 103, 609–615 [DOI] [PubMed] [Google Scholar]
- 56. Waschke J., Bruggeman P., Baumgartner W., Zillikens D., Drenckhahn D. (2005) J. Clin. Invest. 115, 3157–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Troyanovsky R. B., Sokolov E., Troyanovsky S. M. (2003) Mol. Cell. Biol. 23, 7965–7972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Collins J. E., Legan P. K., Kenny T. P., MacGarvie J., Holton J. L., Garrod D. R. (1991) J. Cell Biol. 113, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shimizu H., Masunaga T., Ishiko A., Kikuchi A., Hashimoto T., Nishikawa T. (1995) J. Invest. Dermatol. 105, 153–159 [DOI] [PubMed] [Google Scholar]
- 60. Duden R., Franke W. W. (1988) J. Cell Biol. 107, 1049–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Demlehner M. P., Schäfer S., Grund C., Franke W. W. (1995) J. Cell Biol. 131, 745–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. North A. J., Bardsley W. G., Hyam J., Bornslaeger E. A., Cordingley H. C., Trinnaman B., Hatzfeld M., Green K. J., Magee A. I., Garrod D. R. (1999) J. Cell Sci. 112, 4325–4336 [DOI] [PubMed] [Google Scholar]
- 63. Klingelhöfer J., Laur O. Y., Troyanovsky R. B., Troyanovsky S. M. (2002) Mol. Cell. Biol. 22, 7449–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takeichi M. (1995) Curr. Opin. Cell Biol. 7, 619–627 [DOI] [PubMed] [Google Scholar]
- 65. Mattey D. L., Burdge G., Garrod D. R. (1990) J. Cell Sci. 97, 689–704 [DOI] [PubMed] [Google Scholar]
- 66. Skerrow C. J., Clelland D. G., Skerrow D. (1989) J. Cell Sci. 92, 667–677 [DOI] [PubMed] [Google Scholar]