Abstract
The first intron of FTO contains common single nucleotide polymorphisms associated with body weight and adiposity in humans. In an effort to identify the molecular basis for this association, we discovered that FTO and RPGRIP1L (a ciliary gene located in close proximity to the transcriptional start site of FTO) are regulated by isoforms P200 and P110 of the transcription factor, CUX1. This regulation occurs via a single AATAAATA regulatory site (conserved in the mouse) within the FTO intronic region associated with adiposity in humans. Single nucleotide polymorphism rs8050136 (located in this regulatory site) affects binding affinities of P200 and P110. Promoter-probe analysis revealed that binding of P200 to this site represses FTO, whereas binding of P110 increases transcriptional activity from the FTO as well as RPGRIP1L minimal promoters. Reduced expression of Fto or Rpgrip1l affects leptin receptor isoform b trafficking and leptin signaling in N41 mouse hypothalamic or N2a neuroblastoma cells in vitro. Leptin receptor clusters in the vicinity of the cilium of arcuate hypothalamic neurons in C57BL/6J mice treated with leptin, but not in fasted mice, suggesting a potentially important role of the cilium in leptin signaling that is, in part, regulated by FTO and RPGRIP1L. Decreased Fto/Rpgrip1l expression in the arcuate hypothalamus coincides with decreased nuclear enzymatic activity of a protease (cathepsin L) that has been shown to cleave full-length CUX1 (P200) to P110. P200 disrupts (whereas P110 promotes) leptin receptor isoform b clustering in the vicinity of the cilium in vitro. Clustering of the receptor coincides with increased leptin signaling as reflected in protein levels of phosphorylated Stat3 (p-Stat3). Association of the FTO locus with adiposity in humans may reflect functional consequences of A/C alleles at rs8050136. The obesity-risk (A) allele shows reduced affinity for the FTO and RPGRIP1L transcriptional activator P110, leading to the following: 1) decreased FTO and RPGRIP1L mRNA levels; 2) reduced LEPR trafficking to the cilium; and, as a consequence, 3) a diminished cellular response to leptin.
Keywords: Mouse, Neuron, Obesity, Protease Inhibitor, Receptor Recycling, CUX1, FTO, RPGRIP1L, Cilium, Leptin
Introduction
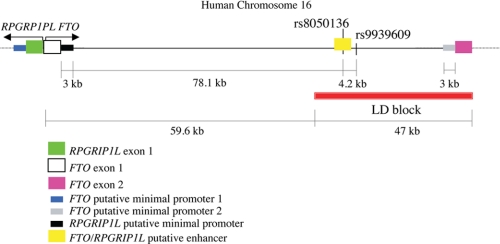
Multiple genome-wide association studies have identified common single nucleotide polymorphisms (SNPs)3 within an ∼47-kb interval in the first intron of FTO (“Fat Mass and Obesity-associated gene”; 16q12.2) by virtue of association with adiposity as reflected in body mass index (Fig. 1) (1–4). FTO is a nuclear protein homologous to the AlkB family of deoxygenases that appears to function as a DNA or RNA demethylase (5–7). Retinitis pigmentosa GTPase regulator-interacting protein-1 like (RPGRIP1L), also known as “FTM,” may also account for some or all of the association, as it lies in close proximity 5′ of and in the opposite orientation as FTO. RPGRIP1L is a component of the basal body of the cilium (8, 9) and might participate in what is now appreciated as an important pathway in human energy homeostasis by virtue of the obesity phenotype in the Bardet-Biedl and Alström syndromes (10).
FIGURE 1.
Genomic organization of the human FTO/RPGRIP1L interval on chromosome 16. Red block denotes region of linkage disequilibrium (LD) that includes SNPs associated with increased body mass index (1, 2). Figure not drawn to scale.
Cilia are conserved from early eukaryotes, such as Chlamydomonas, to humans. Unlike motile cilia, such as those present on the epithelial lung lining responsible for moving mucus, primary cilia are nonmotile microtubular structures found as a single unit on most epithelial and stromal cells throughout the body of mammals (11). Primary cilia, typically ∼5 μm in length, consist of nine doublet microtubules that rise from the centriole after migration and attach to the cell membrane (12). Primary cilia are ubiquitous in the brain (including the hippocampus and olfactory bulb), as well as the hypothalamus (13). In contrast to the emerging role of ciliary function in brain development, as reflected by studies in transgenic mice lacking cilia (14, 15) as well as human “ciliopathies” such as Joubert syndrome characterized by malformations of the cerebellum (16), the roles of cilia in the adult neuron are poorly understood. The somatostatin receptor SSTR3, present in cilia of most adult mammalian neurons, may modulate cAMP signaling within cilia and mediate aspects of object recognition memory (17). The melanin-concentrating hormone receptor 1 (MCHR1), which recognizes as a ligand the orexigenic melanin-concentrating hormone (MCH), localizes to neuronal primary cilia and fails to do so in mice lacking BBS2 or BBS4 implicated in the genetically heterogeneous disorder Bardet-Biedl that includes an obese phenotype (18). Adult mice lacking cilia in pro-opiomelanocortin-expressing neurons responsible for leptin-mediated anorexigenic signals in the hypothalamus display increased food intake leading to obesity (19). Thus, RPGRIP1L, which is expressed in the hypothalamus (20), could also account for some or all of the strong association signal for obesity coming from Chr16 (52.3–52.4 Mb), the interval in which FTO and RPGRIP1L are located.
Our previous studies suggest that FTO and/or RPGRIP1L act in an anorexigenic pathway in the hypothalamus, as expression of these genes decreases upon fasting or cooling of mice (20). In this study, we confirm that FTO and RPGRIP1L mRNA levels decline specifically in the ventromedial and arcuate hypothalamic nuclei of fasted mice and that expression levels of these transcripts are restored by systemic leptin administration. In vitro and in vivo experiments suggest that FTO and RPGRIP1L are involved in leptin receptor trafficking as well as leptin signaling, processes that may account for their apparent influences on adiposity.
Earlier, we found that the transcription factor cut-like homeobox 1 (CUX1), also known as “CUTL1,” binds DNA at SNP rs8050136 within the region of linkage disequilibrium in the first intron of FTO associated with adiposity phenotypes, and it regulates the expression of both FTO and RPGRIP1L in vitro (Fig. 1) (20). Originally characterized as the CCAAT-displacement transcriptional repressor, CUX1 is also processed to isoforms that act as transcriptional activators. Both of these regulatory functions have been implicated in cell cycle progression and cell motility (21). We examined the role of CUX1 as a transcriptional regulator of FTO/RPGRIP1L by performing electrophoretic mobility shift assays (EMSA) and promoter-analysis experiments. Based upon these studies, we propose that isoforms of CUX1 (P200 and P110) display allelic preferential binding at rs8050136 and act as a transcriptional repressor of FTO (P200) or transcriptional activator of FTO and RPGRIP1L (P110). In addition, in vivo and in vitro data presented here suggest that LEPR trafficking to the cilium is important for leptin signaling and that FTO and/or RPGRIP1L modulate LEPR trafficking and thereby leptin signaling. Based upon these data, we propose a molecular mechanism of FTO and RPGRIP1L regulation by CUX1 that mediates control of food intake in both rodents and humans and partly or entirely accounts for the genetic association signal in intron 1 of FTO with adiposity.
EXPERIMENTAL PROCEDURES
Mouse Strains
Lepob (B6.V-Lepob/J) and wild-type (+/+; C57BL/6J) male mice were ordered from The Jackson Laboratory.
Diet and Dietary Treatment
Mice were fed breeder chow (9% kcal from fat; Picolab 5058; Purina Mills) and sacrificed at 4–5 weeks of age. Fasted mice were food-deprived for 42 h, starting at 6 p.m. At 12-h intervals until sacrifice, they received intraperitoneal injections of a saline solution of leptin (purchased from A. F. Parlow, NIDDK, National Institutes of Health), at 1 μg/g of total body weight, or they were co-injected with leptin and with cathepsin L inhibitor I in corn oil (Calbiochem) at 15 μg/g of total body weight. Control mice were injected with saline and/or corn oil. For the thermal challenge experiments, mice were placed singly without bedding in a 4 °C cold room for 4 h. Room temperature was constant at 23 °C on a 12-h light/12-h dark cycle (lights were turned off at 7 p.m.), and mice had ad libitum access to food and water. All protocols were approved by the Columbia University Institutional Animal Care and Use Committee and were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Tissue Culture
Murine neuroblastoma N2a (ATCC ccl-131) and embryonic mouse hypothalamic N41 (mHypoE-N41; Cedarlane Laboratories Ltd., Burlington, NC) cells were grown in Dulbecco's modified Eagle's medium (Invitrogen), 10% (v/v) fetal bovine serum (Invitrogen) in a humidified atmosphere at 37 °C and 5% CO2. 2 days after transfection with p200- or p110-overexpressing vectors, N41 or N2a embryo hypothalamic cells were treated with 5 μg/ml blasticidin (Invitrogen) prior to total RNA extraction. For the phosphorylated Stat3 assay or immunofluorescence studies, subconfluent N41 or N2a cells were made quiescent 36 h after transfection, by incubation in DMEM with 0.5% BSA overnight. Cells were then treated with leptin for 6 h prior to protein extraction.
Primary Neuronal Culture
The hypothalami of ∼40, 3–4 week-old C57BL/6J male and female mice were dissected as described elsewhere (22). The arcuate nucleus was micropunched and added directly to the papain solution provided by the papain dissociation system (Worthington). Cells were dissociated according to the manufacturer's instructions, plated in DMEM/F-12 + GlutaMAXTM-I (supplemented with 20% fetal bovine serum; Invitrogen) on poly-d-lysine/laminin 2-well culture slides (BD Biosciences) for 7 days. Cells were treated with leptin (1 μg/ml) every 24 h for 2 days and/or cathepsin L inhibitor I (10 μm).
Immunohistochemistry and Cellular Immunofluorescence
Four-week-old C57BL/6J male mice were perfused with 4% paraformaldehyde. Tissues were excised, dehydrated in 30% sucrose, frozen, and placed on slides as 10-μm medial coronal hypothalamic sections. Cultured cell slides were fixed with ice-cold methanol/acetone (1:1). Primary antibodies against LEPR (2 μg/ml for immunohistochemistry, 0.5 μg/ml for immunofluorescence; Mouse; catalog no. AF497, R & D Systems, Minneapolis, MN), adenylyl cyclase III (AcIII; 1:500; mouse; catalog no. sc-588, Santa Cruz Biotechnology, Santa Cruz, CA), pericentrin (1:500; rabbit; catalog no. ab4448, Abcam, Cambridge, MA), and GFP (1:500; mouse; catalog no. 11814460001, Roche Applied Science) were used in conjunction with Alexa Fluor® 488, 555, and 647 secondary antibodies (Invitrogen). ProLong® Gold antifade reagent with either DAPI (Invitrogen) or DRAQ5 (Axxora, San Diego) was used for mounting and nuclear staining.
Isolation of Total RNA and Nuclear Protein Extracts
All mice were sacrificed between 2 and 4 p.m. Within 2 min of decapitation, brains were submerged in O.C.T. compound (Sakura Finetek, Torrance, CA) and frozen in an isopentane/dry ice bath. Blocks were brought to −6 °C and sectioned into 0.3-mm-thick slices in a Microm HM 525 cryotome (Thermo Fisher Scientific, Waltham, MA). ARH, VMH, PVN, and DMH were isolated by multiple micro-punches (23) using the coordinates −0.7 to −2.4 (24). Total RNA was extracted and DNase-treated using the RNAqueous® micro kit according to the manufacturer's instructions. Nuclear protein extracts were isolated in the presence of Halt protease and phosphatase inhibitor mixture (Pierce) using the NE-PER® nuclear and cytoplasmic extraction reagents (Pierce) according to the manufacturer's instructions. Total protein levels were measured using the Bradford assay (Pierce).
N41 or N2a cells were plated to 90% confluency and transfected with 20 ng of plasmid DNA (96-well tissue culture dish; BD Biosciences) or 2 μg of plasmid DNA (T25 tissue culture dish; BD Biosciences) using Lipofectamine 2000TM (Invitrogen), and cells were subsequently grown for 48 h. All cultured cells were washed with PBS, scraped off the surface of the Petri dish, and collected by centrifugation for 5 min at ∼500 × g. Total RNA extraction, DNase treatment, isolation of nuclear protein extracts, and total protein level measurement were performed as above.
cDNA Synthesis
cDNA synthesis from 1.0 μg of total RNA was performed at 50 °C for 50 min utilizing the transcriptor first strand cDNA synthesis kit (Roche Applied Science) with oligo(dT)20 primers.
Quantitative PCR
Quantitative PCR was performed as described elsewhere (20). The geometric mean of Gapdh and Actb expression levels was used as loading controls. Cux1-specific primers are described elsewhere (supplemental Table 1).
Antibodies
Anti-CDP (p200 and p110) (1:500; mouse; catalog no. sc-6327X; Santa Cruz Biotechnology) was used for Western blotting and supershifting. Phospho-Stat3 (Tyr-705) antibody was purchased from Cell Signaling Technology (1:1000; catalog no. 9131, Danvers, MA) and utilized for Western blotting. β-Tubulin (1:2000; catalog no. 05-661; Millipore, Bedford, MA) and nucleolin (0.5 μg/ml; catalog no. ab50279; Abcam) were used as internal controls.
Plasmid Construction
The full-length human CUX1 (P200) cDNA overexpression construct was purchased from Open Biosystems (MHS1010-97228334; Huntsville, AL). The P110 cDNA was obtained by digesting the above construct with HindIII/NotI. The fragment was N-filled and cloned into pCMV-Sport6 (Open Biosystems) that had previously been digested with SalI/NotI, N-filled as above, and treated with calf intestinal alkaline phosphatase (New England Biosystems, Ipswich, MA). The full-length mouse Cux1 (p200) cDNA was cloned by OriGene in pCMV6 (Rockville, MD). Mouse p110 was excised with ScaI/NotI, N-filled, and cloned in pENTRTM 4 (Invitrogen) at the N-filled EcoRI site. p110 was subsequently cloned downstream of the CMV promoter in the pLenti 6.3/V5-DEST Gateway® vector (Invitrogen) utilizing the Gateway® LR ClonaseTM II enzyme mix (Invitrogen). The mouse leptin receptor isoform b (Lepr-b) enhanced GFP translational fusion was amplified by PCR using as template a plasmid kindly provided by C. Bjorbaek (Harvard Medical School, Boston). The consensus ribosomal binding sequence (CACC) was introduced via PCR (for primer sequence see supplemental Table 1) upstream of the Lepr-b cDNA to improve expression efficiency and facilitate directional subcloning into pENTRTM/D-TOPO (Invitrogen) as a blunt-ended DNA fragment. The directionally cloned blunt-PCR product was then used as template for entry downstream of the CMV promoter into pLenti6.3/V5-DEST (Invitrogen). The FTO and RPGRIP1L minimal promoters (Fig. 1) were amplified utilizing genomic DNA from skin-derived human primary fibroblasts homozygous for the A or C allele of rs8050136, using primers described elsewhere (supplemental Table 1). The resulting fragments were cloned into pGL3 (Promega Corp., Madison, WI), at the respective restriction sites. The site containing rs8050136 was obtained by PCR from human DNA homozygous for A or C (supplemental Table 1) and cloned upstream of or the absence of the FTO or RPGRIP1L minimal promoters. pRL (Promega Corp., Madison, WI) was also used as a transfection efficiency control.
Dual-Luciferase Assays
Assays were performed with the Dual-Luciferase reporter system (Promega Corp., Madison, WI). All reagents were prepared as described by the manufacturer. N2a cells were transfected in a 96-well Petri dish with 20 ng of each plasmid (2 ng of Enh(C):FTO1p, Enh(C):FTO2p, and Enh(C):RPGRIP1Lp were used) at 50% confluency using the Lipofectamine 2000TM reagent 48 h before lysis in passive lysis buffer supplied by the manufacturer. Measurement of luciferase activity was conducted in a BD Monolight 3096 microplate luminometer (BD Biosciences). 100 μl of the firefly luciferase reagent (LARII) was added to the test sample, followed by addition of 100 μl of the Renilla luciferase reagent and firefly quenching (Stop & Glo). A 10-s equilibration time was allowed, and measurement of luminescence with a 10-s integration time was performed. The data are represented as the ratio of firefly to Renilla luciferase activity (Fluc/Rluc).
EMSA
N2a cells at 75% confluency in a T25 flask were transfected with 2 μg of overexpression construct using Lipofectamine 2000TM reagent. Cells were collected, and nuclear protein extracts were isolated and quantified as described earlier. Oligonucleotides with the human CUX1-binding sequence containing the A, C, or mutated alleles as well as the mouse Cux1-binding sequence containing the native mouse (A) or human (C) alleles were synthesized by Invitrogen (supplemental Table 1). Double-stranded oligonucleotides were obtained by priming at 95 °C for 5 min in TE buffer containing 50 nm NaCl and cooled gradually for 2 h. Each lane of a 4–20% TBE gel (Invitrogen) was loaded with 20 ng of double-stranded oligonucleotide and/or 300 ng of total protein. Supershifting was achieved by loading 3 μg of antibody. DNA-protein binding and DNA staining with SYBR Green were performed using the nonradioactive EMSA kit (Invitrogen) according to the manufacturer's instructions. Visualization of the DNA was performed in a Kodak Gel Logic 212 Imaging System using a SYBR Green filter. Band intensities were measured using ImageJ 1.36B (National Institutes of Health).
Cathepsin L Activity Assay
Cathepsin L activity was measured in 50 μg of total nuclear extracts of the arcuate nucleus using the cathepsin L activity assay kit (Abcam) according to the manufacturer's instructions. Fluorescence intensity was measured in a white flat-bottom Costar 96-well plate (catalog no. 3693; Corning, Lowell, MA) using an AnalystTM AD 96-384 plate reader (LJL Biosystems/Molecular Devices; Sunnyvale, CA) with a 360-nm excitation filter and 520-nm emission filter. Interval between flashes and delay after flash were set at 1000 and 10 ms, respectively. Lamp was set at flash. Background readings were subtracted from sample values.
Fto and Rpgrip1l Knockdown
siRNA transfection has been described elsewhere (20). siRNA was co-transfected with 0.5 μg/ml of the Lepr-b-overexpressing vector. For Fto or Rpgrip1l, a combination of three siRNA or scrambled species was used (supplemental Table 1).
Statistical Analysis
Data are expressed as means ± S.D. Statistical analysis was performed using Student's t test (StatView 5.0, SAS Institute Inc.). Levels of statistical significance were set at two-tailed pα <0.05. In all figures, error bars are S.D.
RESULTS
Expression Analysis of Fto and Rpgrip1l in Mouse Hypothalamic Nuclei
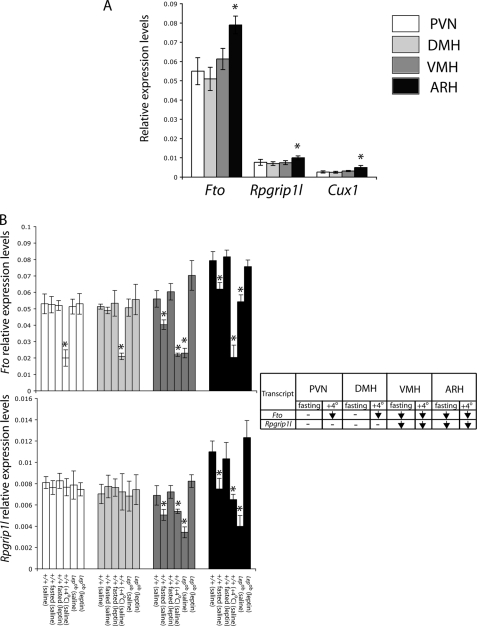
We have previously reported that in whole hypothalami of mice exposed to 4 °C or fasted, there is a decrease of Fto and Rpgrip1l expression in Lepob versus +/+ mice (20). In this study, we used RT-PCR to examine Fto and Rpgrip1l transcript levels in isolated hypothalamic PVN, DMH, VMH, and ARH nuclei of lean and Lepob mice. Fto expression was ∼7-fold higher than Rpgrip1l expression in all nuclei (p < 0.001) (Fig. 2A). In the ARH, Fto and Rpgrip1l expression levels were, respectively, ∼30% (p < 0.001) and ∼25% (p < 0.001) higher than in any other nuclei tested (p < 0.001). In Lepob mice versus +/+ controls, Fto and Rpgrip1l expression was reduced in the VMH (∼55 and ∼25%, respectively; p < 0.001) and ARH (∼30 and ∼55%, respectively; p < 0.001) (Fig. 2B). In contrast, Fto and Rpgrip1l transcript levels in the PVN and DMH were indistinguishable between +/+ lean and Lepob mice (p > 0.1), suggesting that Fto and Rpgrip1l may play specific roles regarding energy homeostasis in neurons of the VMH and ARH.
FIGURE 2.
Fto/Rpgrip1l hypothalamic expression. A, Fto, Rpgrip1l, and Cux1 transcript levels, assessed by RT PCR, in the PVN, DMH, VMH, and arcuate hypothalamic nuclei of lean (+/+) C57BL/6J mice. *, ARH versus PVN, DMH, or VMH. B, assessment of Fto and Rpgrip1l mRNA levels in the PVN, DMH, VMH, and ARH of +/+ C57BL/6J mice compared with fasted +/+ mice, Lepob, as well as mice exposed to 4 °C. Mice were either administered leptin (fasted +/+) or saline (+/+, fasted +/+, 4 °C +/+, Lepob) intraperitoneally. Error bars represent one S.D. Asterisk indicates statistical significance (p < 0.05). Each column represents the mean of measurements from eight mice.
In the ARH, food restriction and cold exposure increase expression of the orexigens NPY/AGRP and suppress the anorexigen pro-opiomelanocortin (25). Fto and Rpgrip1l expression were measured in hypothalamic nuclei of +/+ fasted mice and mice exposed to 4 °C for 4 h (summarized in Table in Fig. 2B). Fto and Rpgrip1l expression levels were decreased in the VMH (∼25 and ∼30% respectively; p < 0.001) and ARH (∼25 and ∼40%, respectively; p < 0.001) of fasted +/+ mice compared with fed +/+ mice, and expression was restored in fasted mice by peripheral administration of leptin. Moreover, Fto and Rpgrip1l expression levels in the VMH and ARH of Lepob mice were restored by peripheral administration of leptin. These results suggest that FTO and RPGRIP1L may act as tonic anorexigens, declining in response to fasting in neurons of the VMH and ARH that mediate leptin signals from the peripheral circulation. In mice exposed to 4 °C, Fto expression decreased in the PVN (∼60%; p < 0.001), DMH (∼60%; p < 0.001), VMH (∼60%; p < 0.001), and ARH (∼75%; p < 0.001) nuclei, whereas decreased Rpgrip1l expression was restricted to the VMH (∼20%; p < 0.001) and ARH (∼40%; p < 0.001) in these animals. The effect of ambient cooling on Fto expression in the PVN may implicate PVN neurons that receive inputs from the ARH and regulate the pituitary-thyroid axis in response to thermal challenge (26).
CUX1 Influences FTO and RPGRIP1L Expression
We have previously identified and confirmed by chromatin immunoprecipitation (ChIP) (20) a CUX1-binding site that includes rs8050136, an SNP associated with adiposity within the first intron of FTO (2).
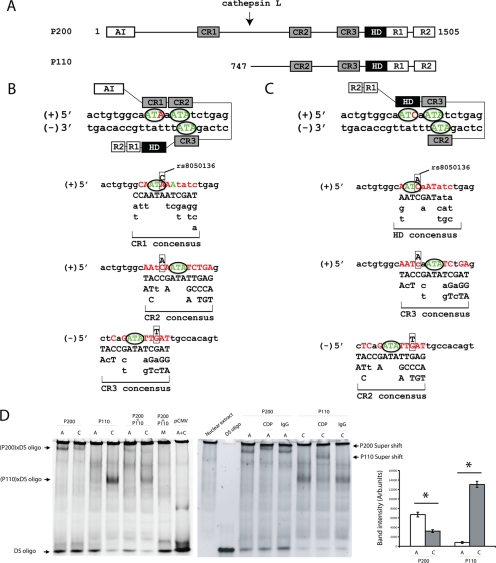
As noted, CUX1 is a transcription factor implicated primarily in the control of cell-autonomous functions, such as cell cycle progression and mobility (21). CUX1 interacts with DNA via three cut-like domains (CR1–3) and a homeodomain (HD) (Fig. 3A). The full-length CUX1 (P200; 1505 amino acids) is proteolytically processed by a nuclear isoform of cathepsin L, generating a C-terminal peptide of 110 kDa that lacks CR1 (27, 28). Based on CR1, CR2, CR3, and HD consensus recognition sequences derived from previous DNA binding studies (27, 29), the rs8050136 A (obesity risk) and C (protective) alleles were predicted to favor binding of P200 CR1 (Fig. 3B) and P110 HD (Fig. 3C) domains, respectively. Electrophoretic mobility shift assays confirmed higher binding affinity of the P200 protein for the A (over C) allele, whereas P110 bound with greater affinity to the C allele (Fig. 3D). Alteration of the ATA-repeat recognition sequence in vitro to GGG (Fig. 3, A and B; sequence in green) abolished binding of both P200 and P110 (Fig. 3D), pointing to the critical role of these repeats in P200 and P110 DNA binding. To assess whether P110 competed with P200 for binding to the A and/or C allele, we tested allelic binding by combining nuclear extracts from N2a cells overexpressing P200 with nuclear extracts from N2a cells overexpressing P110. The mixture exhibited much lower binding affinity for both alleles (Fig. 3D), suggesting that higher molecular weight P200/P110 multimers may have formed which we were unable to resolve by electrophoresis.
FIGURE 3.
A, CUX1 is composed of an N-terminal autoinhibitory domain (AI), DNA-interacting cut-like repeats (CR) 1–3, and cut HD, as well as two repressor domains (R1 and R2) that do not interact with DNA. Cathepsin L cleaves P200 (at a site between CR1 and CR2) to P110 (21). Modeling of P200 (B) and P110 (C) binding affinities for the A (obesity risk) or C alleles of rs8050136. Consensus recognition sequences for CR1–3 and HD were determined from previous reports of in vitro binding experiments (27, 29). CR2 and CR3 domains recognize a degenerate sequence suggesting flexibility in DNA interaction, whereas CR1 or HD determines binding specificity. DNA consensus binding site for each DNA-binding domain (except HD domain) consists of an obligatory ATA sequence (in green and circled). Thus, the rs8050136 site has either three ATA sequences, including the A obesity-risk allele (two in the top and one in the reverse strand), that align with the predicted DNA binding consensus for P200, including CR1, CR2, and CR3, or two ATA and one ATC core sequence, including the rs8050136 C protective allele that aligns with the predicted DNA binding consensus for P110, including HD, CR2, and CR3. Boxed bases show the position of rs8050136 (A/C). D, nonradioactive EMSA using SYBR Green for DNA staining, Left panel, N2a cellular extracts enriched with human P200 or P110 mixed with double-stranded (DS) oligonucleotides carrying the A or C alleles. A double-stranded oligonucleotide in which the three predicted ATA recognition sequences were replaced with GGG (“M”) was also used as a control (described in supplemental Table 1). Band intensities were measured using Image J 1.36B (National Institutes of Health). Right panel, EM supershift assay using an antibody that recognizes the HD domain present in P200 and P110. Mouse IgG was used as a negative control. *, statistically significant (p = 0.001), comparing band intensity between 1st and 2nd and 3rd and 4th gel lanes (gel on left).
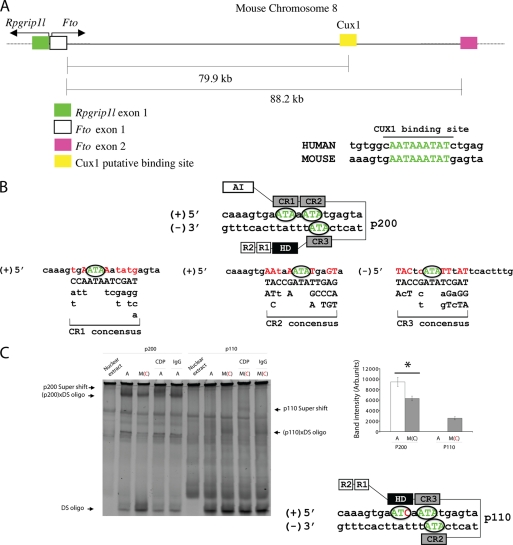
In an effort to establish the presence of a regulatory mechanism similar to human that controls Fto/Rpgrip1l expression in the mouse, we identified a Cux1-binding site in mouse Fto intron 1 in a remarkably similar location to that present in human (Fig. 4A). The mouse carries the A allele at the position equivalent to rs8050136 in humans and is predicted to bind p200 (Fig. 4B). As in humans, we confirmed by EMSA that mouse p200 protein preferentially bound A, whereas mouse p110 preferentially bound the double-stranded oligonucleotide with the mouse sequence in which the human C allele had been introduced (Fig. 4C). These data suggest that, in the mouse, Cux1 may regulate Fto/Rpgrip1l expression as its homolog does in humans.
FIGURE 4.
Sequence preference of p200 (Fto transcriptional repressor) and p110 (Fto and Rpgrip1l transcriptional activator) in the mouse. A, genomic organization of the mouse Fto/Rprgrip1l interval and Cux1-binding site on chromosome 8. B, modeling of p200 binding affinity for the mouse binding site. C, EMSA using N2a cellular extracts enriched with mouse p200 or p110 mixed with double-stranded (DS) oligonucleotides carrying the A or human C [M(C)] alleles. EM supershift assay was performed using an antibody that recognizes the HD domain present in p200 and p110. Mouse IgG was used as a negative control. *, statistically significant (p = 0.003), comparing major band intensity between 2nd and 3rd gel lanes.
The CUX1 P200 isoform has been reported to function exclusively as a transcriptional repressor, whereas P110 has been found predominantly to activate transcription of genes associated with cell cycle progression (30). Based on the distances between rs8050136 and the first exon of RPGRIP1L (∼78-kb 5′) and FTO exon 2 (∼28-kb 3′; Fig. 1), we hypothesized that CUX1 may bind to a single enhancer that regulates the expression of both FTO and RPGRIP1L. We cloned a 3-kb fragment, present in the first intron of FTO (Fig. 1), containing the CUX1-binding site upstream of a putative minimal promoter for FTO (3-kb fragment 5′ of exon 1), an alternative putative minimal promoter for FTO (3-kb fragment 5′ of exon 2), and a putative minimal promoter for RPGRIP1L (3-kb fragment 3′ of exon 1), and assayed promoter activity of those sequences in N2a cells. Overexpression of P200 or P110 had no effect on FTO or RPGRIP1L minimal promoter activity in the absence of the putative enhancer (3-kb fragment including rs8050136); the same was true in the presence of the putative enhancer and absence of either FTO or RPGRIP1L minimal promoters (Fig. 5A). P200 repressed the activity of FTO minimal promoters (by ∼60%, p < 0.001) in the presence of the putative enhancer sequence (carrying the A obesity-risk allele of rs8050136). Overexpression of P200 failed to repress the FTO minimal promoter in the presence of the putative enhancer carrying the C (protective) allele (Fig. 5A), presumably because of weaker P200 binding to the enhancer with the C allele (Fig. 3D). Moreover, FTO minimal promoter activity was enhanced upon P110 overexpression in the presence of the putative enhancer carrying the A allele of rs8050136, and even more so in the presence of the putative enhancer carrying the C allele of rs8050136, to which P110 binds with higher affinity than the A allele in vitro (Fig. 3D). These data suggest the following: 1) The 3-kb fragment that includes rs8050136 acts as an enhancer of FTO expression. 2) P200 acts as a repressor of FTO expression at the A (obesity risk) allele of rs8050136. 3) P110 acts as an enhancer of FTO expression at the A (obesity risk) and more so at the C (protective) allele of rs8050136.
FIGURE 5.
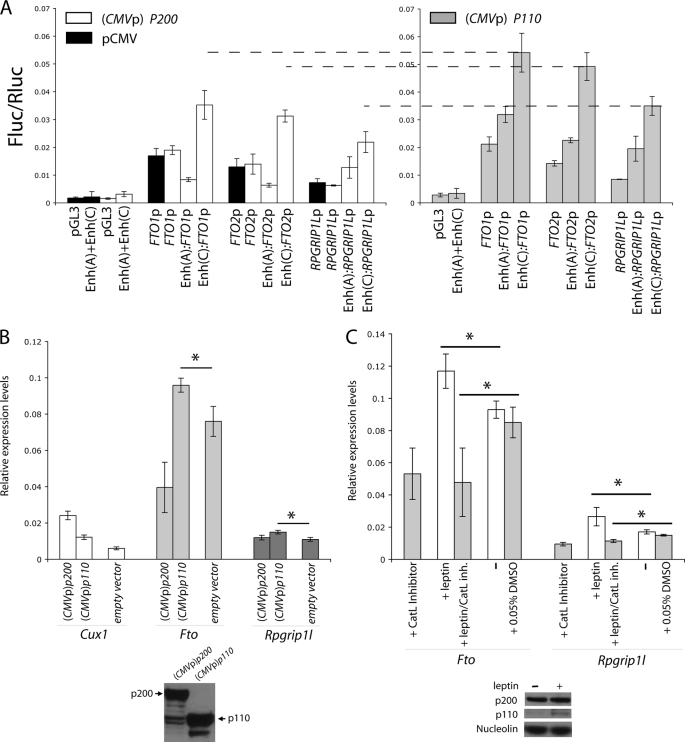
A, luciferase assay used to measure putative minimal FTO (FTO1p and FTO2p) and RPGRIP1L (RPGRIP1Lp) promoter activity upon human P200 ((pCMV) P200) or P110 ((pCMV) P110) overexpression or transfection with empty pCMV and in the presence or absence of the putative enhancer carrying the CUX1-binding A (Enh(A)) or C (Enh(C)) alleles. To control for background, extracts from cells transfected with empty pGL3 or pGL3 carrying the putative enhancer (Enh(A)+Enh(C)) in the absence of FTO1p, FTO2p, and RPGRIP1Lp were also assayed for luciferase activity. Transfection with 20 ng of Enh(C):FTO1p, Enh(C):FTO2p, or Enh(C):RPGRIP1Lp pGL3-based plasmids resulted in off-scale fluorescence intensity; only 2 ng of the above plasmids was used in this experiment. B, expression analysis in N41 cells overexpressing p200 or p110. C, expression analysis in primary neuronal cultures treated with leptin and/or cathepsin L inhibitor I. Each bar represents n = 3. Experiments were repeated twice. *, statistically significant. Used only for comparisons of data within close range (p values <0.01). Error bars represent S.D.
Overexpression of P110 also increased activity of the RPGRIP1L minimal promoter in the presence of the enhancer containing the C allele of rs8050136, more so than in the presence of the enhancer carrying the A allele of rs8050136. Overexpression of P200 failed to repress RPGRIP1L minimal promoter activity in the presence of the putative enhancer carrying either the A or C allele. These data suggest the following: 1) P200 does not repress RPGRIP1L expression; and 2) P110, which preferentially binds the C allele of rs8050136, enhances RPGRIP1L minimal promoter activity in the presence of C (protective allele) more so than in the presence of the A (obesity risk) allele. In short, P200 appears to act exclusively as a repressor of FTO, whereas P110 functions as an activator of FTO and RPGRIP1L. In agreement with these inferences, in vitro transfection of the N41 (or N2a; data not shown) hypothalamic cell line with a vector containing the p200 cDNA led to an ∼4-fold increase in p200 expression and an ∼50% decrease (p < 0.001) in Fto mRNA levels, whereas Rpgrip1L expression levels remained unchanged (Fig. 5B). An ∼2-fold increase in p110 expression, achieved by transfection of N41 (or N2a; data not shown) cells with CMVp-p110, led to an ∼25% increase (p < 0.02) in Fto and Rpgrip1L expression (Fig. 5B).
Cathepsin L Controls FTO and RPGRIP1L Expression
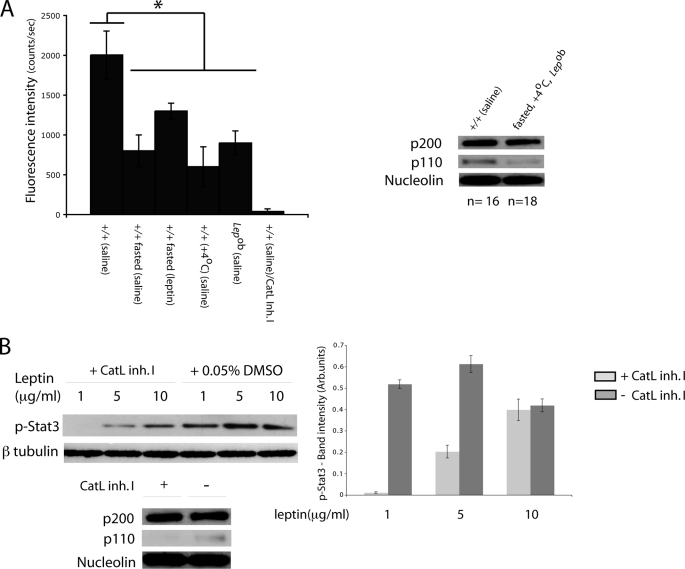
Cathepsin L is a ubiquitous cysteine protease located in vertebrate lysosomes (31). In vitro studies suggest that translational initiation at an alternative internal site leads to the production of a shorter cathepsin L protein species devoid of an endoplasmic reticulum signal peptide that localizes to the nucleus and cleaves CUX1 (P200) to P110 (28). In addition, increased P110 protein in tumor cell lines correlates with increased nuclear cathepsin L (32). The identification of P200 as a transcriptional repressor of FTO and of P110 as an activator of FTO and RPGRIP1L suggested that proteolytic cleavage of CUX1 might modulate FTO and RPGRIP1L expression in response to metabolic circumstance. In mouse ARH primary neurons, we found that Fto and Rpgrip1L expression declined by ∼60 and ∼40%, respectively (p < 0.001), after treatment with a cathepsin L inhibitor (10 μm) in vitro (Fig. 5C). Fto and Rpgrip1L mRNA levels increased by ∼30 and ∼40%, respectively (p < 0.02), after leptin (1 μg/ml) was added to the medium for 24 h but not in cells treated with leptin plus the cathepsin L inhibitor (Fig. 5C). p110 protein levels increased in leptin-treated neurons (Fig. 5C), suggesting that p110 may be responsible for increased Fto and Rpgrip1l expression in response to leptin. Moreover, we found that cathepsin L enzymatic activity declined by ∼60% in nuclear extracts of the ARH obtained from fed Lepob or +/+ mice that were fasted or exposed to 4 °C (Fig. 6A). Leptin administration to fasted mice partly restored cathepsin L activity. The decrease of p110 protein in pooled ARH nuclear extracts of Lepob, fasted and cooled +/+ mice, compared with +/+ controls (Fig. 6A) was proportional to the decline in cathepsin L activity measured for each treatment, suggesting that cathepsin L modulates p110 levels in neuronal nuclei of the mouse hypothalamus. On the other hand, p200 levels appeared unchanged in Lepob, fasted or cooled mice. These results suggest that leptin regulates cathepsin L activity that, in turn, controls FTO and RPGRIP1L expression by modulating CUX1 protein processing to P110. Considered in the context of the promoter assays, we conclude that declining cathepsin L activity levels (as during fasting) result in decreased P110 protein levels leading to decreased FTO expression. The decreased P110 protein levels would also lead to decreased RPGRIP1L expression. The net result would be decreased FTO and RPGRIP1L expression seen in the ARH of Lepob, fasted or cooled +/+ mice.
FIGURE 6.
A, nuclear cathepsin L activity in the arcuate nucleus of lean (+/+), fasted +/+, and mice exposed to 4 °C and Lepob mice injected with saline intraperitoneally or fasted +/+ mice administered leptin intraperitoneally. p110 protein was measured in pooled nuclear extracts of fasted +/+, Lepob, and +/+ mice and mice exposed to 4 °C by Western blotting. *, statistically significant (p < 0.01).+/+ saline, significantly higher that +/+ fasted (saline); +/+ fasted (leptin), +/+ (4 °C) (saline), or Lepob (saline). B, Western blot showing p-Stat3 levels in whole protein extracts from primary neuronal cultures treated with cathepsin L inhibitor I (Cat. Inh. I) or DMSO. We failed to detect p110 species in nuclear fractions from neuronal cultures treated with cathepsin L inhibitor I. Experiments were repeated twice. Each error bar represents one standard deviation. p200, p110, nucleolin, p-Stat3, and β-tubulin-specific bands were variably exposed to film to achieve optimal quantitation range. Therefore, no comparisons should be made between different protein species.
Cathepsin L, Cux1, Fto, and Rpgrip1l Control Lepr-b Trafficking to the Cilium
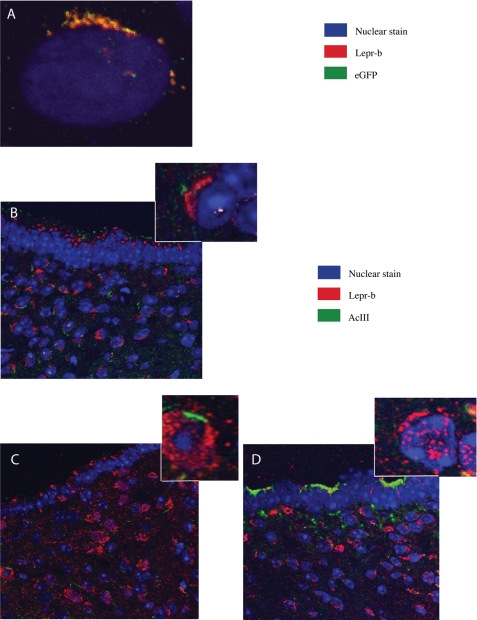
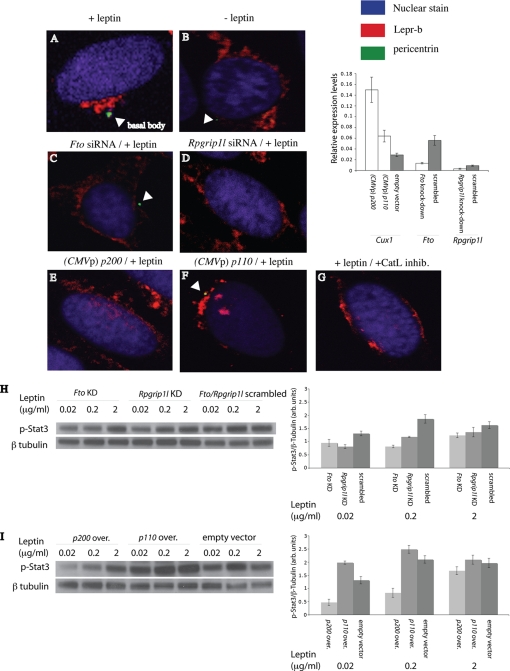
Restoration of Fto and Rpgrip1l expression levels in fasted mice by leptin administration suggested that Fto and/or Rpgrip1l may participate in the leptin signaling pathway. Bardet-Biedl syndrome, which includes obesity as a prominent phenotype, results from mutations in genes that are structural components of the primary cilium (10). BBS1 and BBS2 have been implicated in LEPR trafficking (33). The fact that RPGRIP1L is a ciliary protein suggested a possible role of RPGRIP1L in LEPR trafficking. In support of this inference, in vitro treatment of leptin-supplemented ARH primary neurons with a cathepsin L inhibitor resulted in decreased p110 protein levels, as well as decreased phosphorylated Stat3 (p-Stat3) protein levels (Fig. 6B), suggesting that cathepsin L may modulate LEPR signaling activity through P110 and RPGRIP1L. Moreover, we tested the specificity of a LEPR antibody in the mouse neuronal hypothalamic N41 cell line overexpressing the mouse b isoform of Lepr (Lepr-b) translationally fused to enhanced GFP (Fig. 7A). We then assessed the cellular location of Lepr in the ARH of fasted mice and of mice administered peripherally with leptin or leptin plus cathepsin L inhibitor. Lepr did not display a discrete localization pattern in the ARH of fasted mice (Fig. 7C), whereas Lepr localized exclusively in the vicinity of the cilium in ARH neurons from fasted mice administered leptin (Fig. 7B). The majority of the ARH neurons in fasted mice administered leptin plus cathepsin L inhibitor appeared to have truncated or no cilia, and Lepr was only partially “polarized” toward those cilia (Fig. 7D). Similarly, quiescent N41 (or N2a; data not shown) cells transfected with a Lepr-b overexpression vector displayed distribution of Lepr-b toward the cilium in the presence of leptin (Fig. 8A) but not in the absence of leptin or in the presence of leptin plus the cathepsin L inhibitor (Fig. 8, B and G).
FIGURE 7.
A, N41 cells transfected with the Lepr-b::eGFP overexpression vector and treated with leptin. B, immunohistochemistry showing the arcuate hypothalamic region adjacent to the third ventricle from mice administered leptin peripherally. C, fasted mice; D, mice administered leptin and cathepsin L inhibitor I peripherally. Cilia are stained with an Adenylyl cyclase III (AcIII)-specific antibody.
FIGURE 8.
Immunofluorescence of N41 cells co-transfected with the Lepr-b::eGFP overexpression vector. A, treated with 200 ng(/ml) of leptin for 6 h after becoming quiescent; B, no leptin treatment after becoming quiescent; C, co-transfected with Fto siRNA, grown for 48 h, and treated with leptin for 6 h after becoming quiescent; D, co-transfected with Rpgrip1l, grown for 48 h, and treated with leptin for 6 h after becoming quiescent; E, co-transfected with p200 overexpressing vector, grown for 48 h, and treated with leptin for 6 h after becoming quiescent; F, co-transfected with p110-overexpressing vector, grown for 48 h, and treated with leptin for 6 h after becoming quiescent; and G, treated with 200 ng(/ml) of leptin and cathepsin L inhibitor I (CatL inhib.) (10 μm) for 6 h after becoming quiescent. In vitro assessment of leptin receptor activity was by measurement of p-Stat3 levels in N41 cells co-transfected with the Lepr-b::eGFP overexpression vector. H, Fto-specific or Rpgrip1l-specific siRNA, grown for 48 h, and treated with leptin prior to becoming quiescent; I, p200 or p110 overexpressing (over.) vector, grown for 48 h, and treated with leptin prior to becoming quiescent. Quiescent N41 cells treated with Fto-specific scrambled siRNA displayed near identical p-Stat3 levels to N41 cells treated with Rpgrip1l-specific siRNA. p-Stat3 and β-tubulin-specific bands were variably exposed to film to achieve optimal quantitation range. Therefore, no comparisons should be made between different protein species.
We examined whether RPGRIP1L and/or FTO are involved in trafficking of LEPR to the cilium by knocking down Rpgrip1l or Fto expression in quiescent N41 mouse hypothalamic cells overexpressing Lepr-b. Cells treated with either the Rpgrip1l- or Fto-specific siRNA failed to localize Lepr-b toward the base of the cilium upon leptin stimulation (200 ng/ml for 6 h) (Fig. 8, C and D). ∼70% of N41 cells treated with the Rpgrip1l siRNA did not have detectable ciliary basal bodies. Similarly, ∼70% of N41 cells (overexpressing Lepr-b) transfected with the p200 overexpression vector did not have detectable ciliary basal bodies and failed to cluster Lepr-b (Fig. 8E) in response to leptin stimulation. In contrast, transfection of the N41 mouse hypothalamic line overexpressing Lepr-b with the p110 overexpression vector did not affect Lepr-b trafficking to the vicinity of the cilium (Fig. 8F). These results were reproduced in N2a cells (data not shown). Finally, knockdown of Fto or Rpgrip1l, or overexpression of p200, in quiescent N41 (or N2a; data not shown) cells overexpressing Lepr-b resulted in diminished responses to leptin, as measured by p-Stat3 protein levels in the presence of leptin (Fig. 8, H and I). In contrast, p110 overexpression in N41 cells overexpressing Lepr-b led to enhanced leptin signaling (Fig. 8I). These data implicate P200 and P110 in control of LEPRB trafficking and leptin signaling by regulation of FTO and RPGRIP1L expression.
DISCUSSION
Evidence presented here suggests that SNP rs8050136 in intron 1 of FTO, which lies within a binding site for the transcription factor CUX1, plays a role in the control of FTO and RPGRIP1L expression and implies that functional consequences of the allelic variation at rs8050136 could be, in part or exclusively, responsible for the association of this region of linkage disequilibrium with adiposity in humans. We propose that preferential affinity of P110 for the C (protective) rather than the A (obesity-risk allele) allele at rs8050136 may result in altered leptin sensitivity that could result in increased food intake in individuals carrying the rs8050136 A allele. Supporting this inference, a number of association studies have suggested that the primary cause of the association of adiposity with FTO SNPs (1) is due to increased energy intake (34–39). In a recent study, mice lacking Fto systemically were lean due to increased energy expenditure (40), suggesting that FTO hypermorphic alleles are the cause of increased adiposity in humans. But, the mice lacking Fto also had higher postnatal mortality rates, decreased physical activity, and hyperphagia, implying that reduced adipose tissue mass may be due to developmental abnormalities related to Fto. Mice homozygous for a dominant-negative Fto mutation that does not cause increased perinatal mortality or stunting display reduced fat mass as well as increased energy expenditure unaccompanied by changes in physical activity or food intake (41). In humans, FTO expression was positively correlated in fibroblasts (72) and uncorrelated in fat (73, 74) with the FTO susceptibility allele. Meyre et al. (42) identified several FTO heterozygous apparent loss-of-function mutations at equal frequency both in lean and obese humans, suggesting that the FTO protein itself may not account for the association of the SNPs in intron 1 with adiposity. Tung et al. (43) recently reported that a 2.5-fold increase or 40% decrease of Fto expression, via AAV-mediated transfer of an Fto overexpression cassette or an Fto-specific shRNA, in the ARH of rats, led to a 14% decrease and 16% increase in energy intake, respectively. Fto overexpression was accompanied by a 4-fold increase in levels of Stat3 mRNA in the ARH, implying that Fto overexpression is compatible with neuronal leptin hypersensitivity and a decrease in energy intake, a finding contradicting the implication of previous FTO inactivation studies that Fto overexpression leads to obesity. In our study, leptin reversed the decreases in Fto and Rpgrip1l expression that occur with fasting; and reducing Fto or Rpgrip1l mRNA levels with siRNA led to diminished leptin signaling in N41 cells. These results are consistent with the formulation that decreased FTO and RPGRIP1L expression in the ARH may lead to decreased leptin sensitivity, and thus increased food intake.
Similar to mice, humans homozygous for an inactivating mutation of FTO display growth retardation and severe malformations, including microcephaly, lissencephaly, hydrocephalus, cardiac, and facial malformations (44). Interestingly, targeted disruption of Cux1 in the mouse also causes generalized somatic growth retardation, consistent with our data indicating that CUX1 modulates FTO expression (45–47). Moreover, skin fibroblasts from patients without functional FTO displayed decreased proliferative ability (44). We have also observed that Fto knockdown or p200 overexpression resulted in decreased proliferation of N41 or N2a cells (data not shown). Based on these reports, it is conceivable that CUX1 P200 may also modulate somatic growth through FTO during development. If FTO/CUX1 play a role in adipocyte development/proliferation, extreme hypomorphs or overexpressors could obscure somatic consequences of primary hypothalamic effects on energy intake.
The in vivo and in vitro demonstrations of leptin receptor localization in the vicinity of the cilium of leptin-stimulated ARH neurons, as well as changes in leptin receptor localization upon fasting with respect to the cilium in vivo, suggest that the cilium may participate in leptin signaling. In support of this inference, mice deleted for adenylyl cyclase III, localized inside hypothalamic primary cilia (13), exhibit obesity that is apparently caused by hyperphagia due to reduced responsiveness to leptin manifested as a lack of weight loss in response to exogenously administrated leptin (48). Moreover, mice lacking ciliary structural genes mutated in individuals with Bardet-Biedl syndrome (9) have diminished leptin-induced Stat3 phosphorylation in the hypothalamus (33).
Knocking down BBS1 or BBS2 in human retinal pigment epithelial cells disrupts Lepr-b trafficking to the post-Golgi network, resulting in partitioning of Lepr-b molecules in large vesicle-like compartments (33). In retinal cells, RPGRIP1L acts as a scaffolding component of a multiprotein complex that includes the retinitis pigmentosa GTPase regulator, implicated in retinitis pigmentosa, and CEP290 (centrosomal protein 290 kDa) (49, 50). Indirect evidence that RPGRIP1L may be part of a protein complex that includes Lepr-b includes the observations that CEP290 is part of a ciliary basal body-associated protein complex that includes BBS1 (51) and that Lepr-b binds BBS1 in vitro (33). In our study, siRNA-mediated reduction of Rpgrip1l mRNA levels in N41 hypothalamic cells reduced p-Stat3 levels and abolished leptin receptor segregation to the base of the cilium. It has been proposed that leptin receptor clustering facilitates leptin signaling (52, 53). GTP-bound Arl6, a small GTPase encoded by Bardet-Biedl-associated BBS3 gene, recruits the Bardet-Biedl syndrome complex (“BBsome”) to the cell membrane resulting in clustering of the transmembrane protein SSTR3 before its targeting to the cilium (54). Thus, it is conceivable that LEPR may be clustered and targeted in a similar fashion by elements of the ciliary complex.
Hypofunctional mutations of RPGRIP1L cause the Joubert and Meckel syndromes, characterized by abnormalities that include cerebellar ataxia and cystic renal dysplasia (55) but not obesity. An emerging role of CUX1 in cell-cell interactions has been inferred from altered ciliary morphology and polycystic kidney disease in mice overexpressing the CUX1 homeobox domain, or cpk mice segregating for a Cux1 mutation resulting in ectopic Cux1 expression in renal tissue, supporting the notion that CUX1 may participate in ciliary assembly/function by regulating RPGRIP1L (56, 57). Polycystic kidney disease is also common in patients with Bardet-Biedl syndrome (58).
The presence of a region in mouse Fto intron 1 that interacts with both p200 and p110 supports the finding that Fto and Rpgrip1l expression in the mouse is controlled by Cux1 isoforms in a similar fashion to human, p200 (acting as a repressor of Fto) and p110 (acting as an activator of Fto and Rpgrip1l). Preliminary data suggest that SNP rs36839403, located 7 bp upstream of the Cux1-binding site in the mouse, affects binding affinity of the CCAAT enhancer-binding protein (transcriptional regulator displaced by Cux1) (59), which acts as a transcriptional activator of Fto and Rpgrip1l in vitro (data not shown). Moreover, a comparable CCAAT enhancer-binding protein-binding site is conserved in humans. These findings imply the possibility that rs36839403 affects Fto/Rpgrip1l expression and predisposes to increased adiposity in mice, as rs8050136 does in humans.
The fact that the proposed FTO/RPGRIP1L regulatory site at rs8050136 is distant from their respective transcriptional start sites suggests that the rs8050136 site operates as an enhancer. Using a GAL4 reporter system, it has been reported that CUX1 is transcriptionally active at a large distance from a transcriptional start site (60).
A vector expressing CUX1 cut-like repeat (CR) 1 and CR2 (present in P200) alone has been shown to bind preferentially two CRAT motifs in either orientation and at variable distance, whereas a vector expressing CUX1 without CR1 (i.e. P110) binds stably to the ATCRAT (R = A or G) sequence (61). According to our current model of the rs8050136 site, ATA repeats are crucial for binding of CR1–3 (two repeats in top DNA strand and one in antisense DNA strand), and the CUX1 HD prefers ATC. In support of this model, substitution of either the ATA repeats or ATC abolishes binding of P200 or P110 at rs8050136 in our gel-shift assay. As predicted, P200 (including CR1) prefers ATA (obesity-risk allele), whereas P110 (excluding CR1) prefers ATC (protective allele) of rs8050136.
More p200 than p110 protein was consistently seen in ARH nuclear extracts, cultured ARH primary neurons, or cell lines deployed in this study. Considering that CR1–CR2, as well as an N-terminal autoinhibitory domain present in P200 but absent in P110, reduces CUX1 DNA binding affinity and transcriptional activity (62), it is conceivable that the presence of P110 at relatively lower levels can successfully compete against P200 protein bound to DNA at a lower affinity and more so in individuals with the protective (C) allele of rs8050136, given that P110 has higher affinity than P200 for the protective allele.
We have also shown that in the fed state, increased hypothalamic cathepsin L activity (which is responsive to ambient leptin) coincides with increased Fto/Rpgrip1l expression and increased p110 protein levels. During fasting, a decline in circulating leptin reduced cathepsin L enzymatic activity, decreased p110 levels, and decreased Fto/Rpgrip1l expression. RPGRIP1L may control food intake by participating in an anchoring complex that facilitates leptin receptor clustering, thus modulating leptin sensitivity at the cellular level. How FTO might participate in LEPRB clustering is unknown; its demethylase activity may be required to control expression of downstream genes required for clustering LEPRB in the vicinity of the cilium.
Presumably, reduced ambient leptin accounts for decreased Fto and Rpgrip1l expression in the ARH of Lepob mice. Even though we did not further investigate the cooling-induced decrease in Fto and Rpgrip1l expression in the hypothalamus, it is conceivable that, at least in the ARH, Fto and Rpgrip1l expression is regulated similarly to the fasted state, as circulating leptin levels also decrease in rats exposed to cold (∼30%) (63).
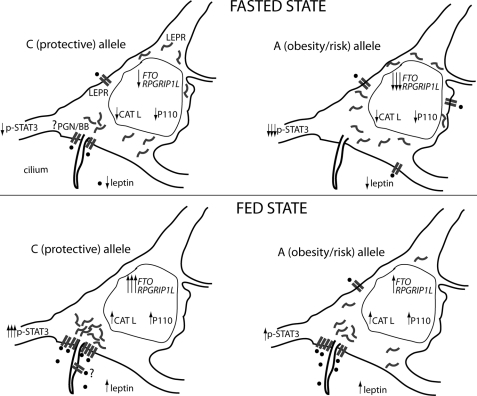
In the model proposed here (Fig. 9), reduced circulating leptin (as a result of weight loss or acute calorie restriction) results in decreased cathepsin L activity in the ARH cell nucleus. This leads to reduced enzymatic processing of CUX1 P200, resulting in decreased CUX1 P110 protein levels, which in turn results in decreased RPGRIP1L and FTO expression. Low RPGRIP1L and FTO expression levels would reduce leptin signaling (as seen in N41 cells treated with Fto or Rpgrip1l siRNA that led to decreased p-Stat3 levels) increasing the drive to eat (64). CUX1 P110, which we show here, enhances leptin signaling in the mouse and has reduced affinity for the putative enhancer of RPGRIP1L or FTO in individuals segregating for the rs8050136 obesity-risk (A) allele, leading to relative down-regulation of FTO and RPGRIP1L. Because P110 has higher affinity for the protective (C) allele, individuals with the C allele would have higher FTO and RPGRIP1L expression levels and thus be relatively more leptin-sensitive than individuals with the obesity risk (A) allele. We are proposing that FTO, RPGRIP1L, and CUX1 facilitate the leptin response in arcuate neurons; P200 acts as a break of leptin congregation in the vicinity of the neuronal cilium in the absence of leptin, and P110 is made (via cathepsin L) by the cell as an initial response to STAT3 activation by leptin, leading to increased LEPR recruited at the cilium, resulting in increased leptin sensitivity, which, in turn, leads to a further increase in p-STAT3. The predicted positive feedback loop in STAT3 activation by leptin may constitute a mechanism through which catabolic signals remain activated in the basal state (65).
FIGURE 9.
Schematic representing the proposed model of FTO/RPGRIP1L rs8050136 allele-specific transcriptional regulation in response to feeding or fasting. Increased circulating leptin upon feeding leads to increased nuclear cathepsin L (CATL) activity in arcuate neurons expressing LEPR, increased P110 levels upon P200 cleavage by CATL, and in turn increased FTO/RPGRIP1L expression. By an unknown mechanism, FTO/RPGRIP1L facilitates LEPR clustering close to the base of the cilium at a site that may be the post-Golgi network (33, 70, 71) and/or the ciliary basal body (33). LEPR trafficking to the basal body may be facilitated by RPGRIP1L that localizes to the basal body protein complex (BBsome) and by FTO that may control expression of gene(s) implicated in LEPR trafficking. LEPR may multicluster at the membrane in the region of the cilium, thus enhancing leptin signaling. In some photoreceptor connecting cilia, RPGRIP1L localizes to the basal body as well as inside the cilium (9). Although we did not see LEPR inside the cilium, it is possible that the experimental conditions employed in this study limited our ability to visualize a small number of LEPR molecules transported into the cilium by RPGRIP1L. Fasting (low-leptin ambient condition) leads to decreased nuclear cathepsin L enzymatic activity in LEPR-positive arcuate neurons, decreased P110 levels, and decreased FTO/RPGRIP1L expression, resulting in dispersal of LEPR throughout the cell. Individuals with rs8050136 A (obesity risk), as opposed to individuals with rs8050136 C (protective) allele, display lower p110 binding, and thus lower FTO/RPGRIP1L expression levels causing decreased clustering of LEPR in close proximity to the cilium, resulting in less efficient leptin signaling. Abbreviations: PGN, post-Golgi network; BB, basal body.
In many cells, ciliary formation is coordinated with the cell cycle so that cilia start to form during G1, are most abundant through G0, and disassemble during mitosis or before S-phase (66). The cilium itself is involved in transmitting mitotic signals, thereby, promoting cell cycle progression (67, 68). Thus, it is not surprising that CUX1, involved in cell proliferation, can modify ciliary assembly. However, it is not clear how dynamic ciliary assembly might occur in nonproliferating cells such as mature neurons. Our study suggests a dynamic role of the primary cilium in LEPR-expressing neurons of the ARH that may be manifested by assembly/disassembly of the cilium in response to extracellular stimuli. In Caenorhabditis elegans, specialized neuronal cilia in the olfactory epithelium require sensory signaling to maintain their architecture (69), suggesting that extracellular stimuli, such as leptin, may regulate the assembly of neuronal cilia of the ARH.
In conclusion, our data suggest that FTO and RPGRIP1L participate in the control of food intake by modulating leptin sensitivity in the ARH, and that, in humans, either or both may account for the association of alleles of rs8050136 with relative adiposity.
Supplementary Material
Acknowledgments
We thank David Barth Penn for technical assistance and Stuart G. Fischer and Yiying Zhang for informative discussions.
This work was supported in part by National Institutes of Health Grants RO1 DK52431-15, P30 DK26687-30, and P30 DK63608-08, an American Diabetes Association-mentored fellowship award, a grant from the Russell Berrie Foundation, and a gift from the Lasky Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- SNP
- single nucleotide polymorphism
- ARH
- arcuate hypothalamic
- PVN
- paraventricular
- DMH
- dorsomedial
- VMH
- ventromedial
- Lepr-b
- leptin receptor isoform b
- HD
- homeodomain.
REFERENCES
- 1. Frayling T. M., Timpson N. J., Weedon M. N., Zeggini E., Freathy R. M., Lindgren C. M., Perry J. R., Elliott K. S., Lango H., Rayner N. W., Shields B., Harries L. W., Barrett J. C., Ellard S., Groves C. J., Knight B., Patch A. M., Ness A. R., Ebrahim S., Lawlor D. A., Ring S. M., Ben-Shlomo Y., Jarvelin M. R., Sovio U., Bennett A. J., Melzer D., Ferrucci L., Loos R. J., Barroso I., Wareham N. J., Karpe F., Owen K. R., Cardon L. R., Walker M., Hitman G. A., Palmer C. N., Doney A. S., Morris A. D., Smith G. D., Hattersley A. T., McCarthy M. I. (2007) Science 316, 889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scuteri A., Sanna S., Chen W. M., Uda M., Albai G., Strait J., Najjar S., Nagaraja R., Orrú M., Usala G., Dei M., Lai S., Maschio A., Busonero F., Mulas A., Ehret G. B., Fink A. A., Weder A. B., Cooper R. S., Galan P., Chakravarti A., Schlessinger D., Cao A., Lakatta E., Abecasis G. R. (2007) PLoS Genet. 3, e115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meyre D., Delplanque J., Chèvre J. C., Lecoeur C., Lobbens S., Gallina S., Durand E., Vatin V., Degraeve F., Proença C., Gaget S., Körner A., Kovacs P., Kiess W., Tichet J., Marre M., Hartikainen A. L., Horber F., Potoczna N., Hercberg S., Levy-Marchal C., Pattou F., Heude B., Tauber M., McCarthy M. I., Blakemore A. I., Montpetit A., Polychronakos C., Weill J., Coin L. J., Asher J., Elliott P., Järvelin M. R., Visvikis-Siest S., Balkau B., Sladek R., Balding D., Walley A., Dina C., Froguel P. (2009) Nat. Genet. 41, 157–159 [DOI] [PubMed] [Google Scholar]
- 4. Thorleifsson G., Walters G. B., Gudbjartsson D. F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., Jonsdottir T., Olafsdottir E. J., Olafsdottir G. H., Jonsson T., Jonsson F., Borch-Johnsen K., Hansen T., Andersen G., Jorgensen T., Lauritzen T., Aben K. K., Verbeek A. L., Roeleveld N., Kampman E., Yanek L. R., Becker L. C., Tryggvadottir L., Rafnar T., Becker D. M., Gulcher J., Kiemeney L. A., Pedersen O., Kong A., Thorsteinsdottir U., Stefansson K. (2009) Nat. Genet. 41, 18–24 [DOI] [PubMed] [Google Scholar]
- 5. Gerken T., Girard C. A., Tung Y. C., Webby C. J., Saudek V., Hewitson K. S., Yeo G. S., McDonough M. A., Cunliffe S., McNeill L. A., Galvanovskis J., Rorsman P., Robins P., Prieur X., Coll A. P., Ma M., Jovanovic Z., Farooqi I. S., Sedgwick B., Barroso I., Lindahl T., Ponting C. P., Ashcroft F. M., O'Rahilly S., Schofield C. J. (2007) Science 318, 1469–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia G., Yang C. G., Yang S., Jian X., Yi C., Zhou Z., He C. (2008) FEBS Lett. 582, 3313–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han Z., Niu T., Chang J., Lei X., Zhao M., Wang Q., Cheng W., Wang J., Feng Y., Chai J. (2010) Nature 464, 1205–1209 [DOI] [PubMed] [Google Scholar]
- 8. Vierkotten J., Dildrop R., Peters T., Wang B., Rüther U. (2007) Development 134, 2569–2577 [DOI] [PubMed] [Google Scholar]
- 9. Khanna H., Davis E. E., Murga-Zamalloa C. A., Estrada-Cuzcano A., Lopez I., den Hollander A. I., Zonneveld M. N., Othman M. I., Waseem N., Chakarova C. F., Maubaret C., Diaz-Font A., MacDonald I., Muzny D. M., Wheeler D. A., Morgan M., Lewis L. R., Logan C. V., Tan P. L., Beer M. A., Inglehearn C. F., Lewis R. A., Jacobson S. G., Bergmann C., Beales P. L., Attié-Bitach T., Johnson C. A., Otto E. A., Bhattacharya S. S., Hildebrandt F., Gibbs R. A., Koenekoop R. K., Swaroop A., Katsanis N. (2009) Nat. Genet. 41, 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baker K., Beales P. L. (2009) Am. J. Med. Genet. C. Semin. Med. Genet. 151C, 281–295 [DOI] [PubMed] [Google Scholar]
- 11. Wheatley D. N., Wang A. M., Strugnell G. E. (1996) Cell Biol. Int. 20, 73–81 [DOI] [PubMed] [Google Scholar]
- 12. Marshall W. F. (2008) Curr. Top. Dev. Biol. 85, 1–22 [DOI] [PubMed] [Google Scholar]
- 13. Bishop G. A., Berbari N. F., Lewis J., Mykytyn K. (2007) J. Comp. Neurol. 505, 562–571 [DOI] [PubMed] [Google Scholar]
- 14. Gorivodsky M., Mukhopadhyay M., Wilsch-Braeuninger M., Phillips M., Teufel A., Kim C., Malik N., Huttner W., Westphal H. (2009) Dev. Biol. 325, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Breunig J. J., Sarkisian M. R., Arellano J. I., Morozov Y. M., Ayoub A. E., Sojitra S., Wang B., Flavell R. A., Rakic P., Town T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13127–13132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cantagrel V., Silhavy J. L., Bielas S. L., Swistun D., Marsh S. E., Bertrand J. Y., Audollent S., Attié-Bitach T., Holden K. R., Dobyns W. B., Traver D., Al-Gazali L., Ali B. R., Lindner T. H., Caspary T., Otto E. A., Hildebrandt F., Glass I. A., Logan C. V., Johnson C. A., Bennett C., Brancati F., Valente E. M., Woods C. G., Gleeson J. G. (2008) Am. J. Hum. Genet. 83, 170–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Einstein E. B., Patterson C. A., Hon B. J., Regan K. A., Reddi J., Melnikoff D. E., Mateer M. J., Schulz S., Johnson B. N., Tallent M. K. (2010) J. Neurosci. 30, 4306–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berbari N. F., Lewis J. S., Bishop G. A., Askwith C. C., Mykytyn K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4242–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Davenport J. R., Watts A. J., Roper V. C., Croyle M. J., van Groen T., Wyss J. M., Nagy T. R., Kesterson R. A., Yoder B. K. (2007) Curr. Biol. 17, 1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stratigopoulos G., Padilla S. L., LeDuc C. A., Watson E., Hattersley A. T., McCarthy M. I., Zeltser L. M., Chung W. K., Leibel R. L. (2008) Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sansregret L., Nepveu A. (2008) Gene 412, 84–94 [DOI] [PubMed] [Google Scholar]
- 22. Kamberi I. A., Kobayashi Y. (1970) J. Neurochem. 17, 261–268 [DOI] [PubMed] [Google Scholar]
- 23. Palkovits M. (1973) Brain Res. 59, 449–450 [DOI] [PubMed] [Google Scholar]
- 24. Slotnick B. M., Leonard C. M. (eds) (1975) A Stereotaxic Atlas of the Albino Mouse Forebrain, United States Department of Health, Education, and Welfare, Public Health Service, Alcohol, Drug Abuse, Rockville, MD [Google Scholar]
- 25. Overton J. M., Williams T. D. (2004) Physiol. Behav. 81, 749–754 [DOI] [PubMed] [Google Scholar]
- 26. Herwig A., Ross A. W., Nilaweera K. N., Morgan P. J., Barrett P. (2008) Obes. Facts 1, 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moon N. S., Premdas P., Truscott M., Leduy L., Bérubé G., Nepveu A. (2001) Mol. Cell. Biol. 21, 6332–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goulet B., Baruch A., Moon N. S., Poirier M., Sansregret L. L., Erickson A., Bogyo M., Nepveu A. (2004) Mol. Cell 14, 207–219 [DOI] [PubMed] [Google Scholar]
- 29. Aufiero B., Neufeld E. J., Orkin S. H. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7757–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harada R., Vadnais C., Sansregret L., Leduy L., Bérubé G., Robert F., Nepveu A. (2008) Nucleic Acids Res. 36, 189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collette J., Bocock J. P., Ahn K., Chapman R. L., Godbold G., Yeyeodu S., Erickson A. H. (2004) Int. Rev. Cytol. 241, 1–51 [DOI] [PubMed] [Google Scholar]
- 32. Goulet B., Sansregret L., Leduy L., Bogyo M., Weber E., Chauhan S. S., Nepveu A. (2007) Mol. Cancer Res. 5, 899–907 [DOI] [PubMed] [Google Scholar]
- 33. Seo S., Guo D. F., Bugge K., Morgan D. A., Rahmouni K., Sheffield V. C. (2009) Hum. Mol. Genet. 18, 1323–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Speakman J. R., Rance K. A., Johnstone A. M. (2008) Obesity 16, 1961–1965 [DOI] [PubMed] [Google Scholar]
- 35. Cecil J. E., Tavendale R., Watt P., Hetherington M. M., Palmer C. N. (2008) N. Engl. J. Med. 359, 2558–2566 [DOI] [PubMed] [Google Scholar]
- 36. Wardle J., Carnell S., Haworth C. M., Farooqi I. S., O'Rahilly S., Plomin R. (2008) J. Clin. Endocrinol. Metab. 93, 3640–3643 [DOI] [PubMed] [Google Scholar]
- 37. Haupt A., Thamer C., Staiger H., Tschritter O., Kirchhoff K., Machicao F., Häring H. U., Stefan N., Fritsche A. (2009) Exp. Clin. Endocrinol. Diabetes 117, 194–197 [DOI] [PubMed] [Google Scholar]
- 38. Goossens G. H., Petersen L., Blaak E. E., Hul G., Arner P., Astrup A., Froguel P., Patel K., Pedersen O., Polak J., Oppert J. M., Martinez J. A., Sorensen T. I., Saris W. H. (2009) Int. J. Obes. 33, 669–679 [DOI] [PubMed] [Google Scholar]
- 39. Wardle J., Llewellyn C., Sanderson S., Plomin R. (2009) Int. J. Obes. 33, 42–45 [DOI] [PubMed] [Google Scholar]
- 40. Fischer J., Koch L., Emmerling C., Vierkotten J., Peters T., Brüning J. C., Rüther U. (2009) Nature 458, 894–898 [DOI] [PubMed] [Google Scholar]
- 41. Church C., Lee S., Bagg E. A., McTaggart J. S., Deacon R., Gerken T., Lee A., Moir L., Mecinović J., Quwailid M. M., Schofield C. J., Ashcroft F. M., Cox R. D. (2009) PLoS Genet. 5, e1000599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyre D., Proulx K., Kawagoe-Takaki H., Vatin V., Gutiérrez-Aguilar R., Lyon D., Ma M., Choquet H., Horber F., Van Hul W., Van Gaal L., Balkau B., Visvikis-Siest S., Pattou F., Farooqi I. S., Saudek V., O'Rahilly S., Froguel P., Sedgwick B., Yeo G. S. (2010) Diabetes 59, 311–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tung Y. C., Ayuso E., Shan X., Bosch F., O'Rahilly S., Coll A. P., Yeo G. S. (2010) PLoS One 5, e8771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boissel S., Reish O., Proulx K., Kawagoe-Takaki H., Sedgwick B., Yeo G. S., Meyre D., Golzio C., Molinari F., Kadhom N., Etchevers H. C., Saudek V., Farooqi I. S., Froguel P., Lindahl T., O'Rahilly S., Munnich A., Colleaux L. (2009) Am. J. Hum. Genet. 85, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ellis T., Gambardella L., Horcher M., Tschanz S., Capol J., Bertram P., Jochum W., Barrandon Y., Busslinger M. (2001) Genes Dev. 15, 2307–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luong M. X., van der Meijden C. M., Xing D., Hesselton R., Monuki E. S., Jones S. N., Lian J. B., Stein J. L., Stein G. S., Neufeld E. J., van Wijnen A. J. (2002) Mol. Cell. Biol. 22, 1424–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sinclair A. M., Lee J. A., Goldstein A., Xing D., Liu S., Ju R., Tucker P. W., Neufeld E. J., Scheuermann R. H. (2001) Blood 98, 3658–3667 [DOI] [PubMed] [Google Scholar]
- 48. Wang Z., Li V., Chan G. C., Phan T., Nudelman A. S., Xia Z., Storm D. R. (2009) PLoS One 4, e6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao Y., Hong D. H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A., Li T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3965–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S. K., Cheng H., Scott A., Hurd R. E., Sayer J. A., Otto E. A., Attanasio M., O'Toole J. F., Jin G., Shou C., Hildebrandt F., Williams D. S., Heckenlively J. R., Swaroop A. (2006) Hum. Mol. Genet. 15, 1847–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim J., Krishnaswami S. R., Gleeson J. G. (2008) Hum. Mol. Genet. 17, 3796–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zabeau L., Defeau D., Van der Heyden J., Iserentant H., Vandekerckhove J., Tavernier J. (2004) Mol. Endocrinol. 18, 150–161 [DOI] [PubMed] [Google Scholar]
- 53. Peelman F., Iserentant H., De Smet A. S., Vandekerckhove J., Zabeau L., Tavernier J. (2006) J. Biol. Chem. 281, 15496–15504 [DOI] [PubMed] [Google Scholar]
- 54. Jin H., White S. R., Shida T., Schulz S., Aguiar M., Gygi S. P., Bazan J. F., Nachury M. V. (2010) Cell 141, 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Devuyst O., Arnould V. J. (2008) Nephrol. Dial. Transplant. 23, 1500–1503 [DOI] [PubMed] [Google Scholar]
- 56. Cadieux C., Harada R., Paquet M., Côté O., Trudel M., Nepveu A., Bouchard M. (2008) J. Biol. Chem. 283, 13817–13824 [DOI] [PubMed] [Google Scholar]
- 57. Alcalay N. I., Sharma M., Vassmer D., Chapman B., Paul B., Zhou J., Brantley J. G., Wallace D. P., Maser R. L., Vanden Heuvel G. B. (2008) Am. J. Physiol. Renal Physiol. 295, F1725–F1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zaghloul N. A., Katsanis N. (2009) J. Clin. Invest. 119, 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Khanna-Gupta A., Zibello T., Sun H., Gaines P., Berliner N. (2003) Blood 101, 3460–3468 [DOI] [PubMed] [Google Scholar]
- 60. Mailly F., Bérubé G., Harada R., Mao P. L., Phillips S., Nepveu A. (1996) Mol. Cell. Biol. 16, 5346–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moon N. S., Bérubé G., Nepveu A. (2000) J. Biol. Chem. 275, 31325–31334 [DOI] [PubMed] [Google Scholar]
- 62. Truscott M., Raynal L., Wang Y., Bérubé G., Leduy L., Nepveu A. (2004) J. Biol. Chem. 279, 49787–49794 [DOI] [PubMed] [Google Scholar]
- 63. Hardie L. J., Rayner D. V., Holmes S., Trayhurn P. (1996) Biochem. Biophys. Res. Commun. 223, 660–665 [DOI] [PubMed] [Google Scholar]
- 64. Villanueva E. C., Myers M. G., Jr. (2008) Int. J. Obes. 32, Suppl. 7, s8–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schwartz M. W., Woods S. C., Seeley R. J., Barsh G. S., Baskin D. G., Leibel R. L. (2003) Diabetes 52, 232–238 [DOI] [PubMed] [Google Scholar]
- 66. Santos N., Reiter J. F. (2008) Dev. Dyn. 237, 1972–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kenney A. M., Rowitch D. H. (2000) Mol. Cell. Biol. 20, 9055–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., Chuang P. T., Reiter J. F. (2008) Nat. Cell Biol. 10, 70–76 [DOI] [PubMed] [Google Scholar]
- 69. Mukhopadhyay S., Lu Y., Shaham S., Sengupta P. (2008) Dev. Cell 14, 762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Diano S., Kalra S. P., Horvath T. L. (1998) J. Neuroendocrinol. 10, 647–650 [DOI] [PubMed] [Google Scholar]
- 71. Belouzard S., Delcroix D., Rouillé Y. (2004) J. Biol. Chem. 279, 28499–28508 [DOI] [PubMed] [Google Scholar]
- 72. Berulava T., Horsthemke B. (2010) Eur. J. Hum. Genet. 9, 1054–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Klöting N., Schleinitz D., Ruschke K., Berndt J., Fasshauer M., Tönjes A., Schön M. R., Kovacs P., Stumvoll M., Blüher M. (2008) Diabetologia 14, 641–647 [DOI] [PubMed] [Google Scholar]
- 74. Grunnet L. G., Nilsson E., Ling C., Hansen T., Pedersen O., Groop L., Vaag A., Poulsen P. (2009) Diabetes 10, 2402–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.