Abstract
Keratin polypeptide 8 (K8) associates noncovalently with its partners K18 and/or K19 to form the intermediate filament cytoskeleton of hepatocytes and other simple-type epithelial cells. Human K8, K18, and K19 variants predispose to liver disease, whereas site-specific keratin phosphorylation confers hepatoprotection. Because stress-induced protein phosphorylation regulates sumoylation, we hypothesized that keratins are sumoylated in an injury-dependent manner and that keratin sumoylation is an important regulatory modification. We demonstrate that K8/K18/K19, epidermal keratins, and vimentin are sumoylated in vitro. Upon transfection, K8, K18, and K19 are modified by poly-SUMO-2/3 chains on Lys-285/Lys-364 (K8), Lys-207/Lys-372 (K18), and Lys-208 (K19). Sumoylation affects filament organization and stimulus-induced keratin solubility and is partially inhibited upon mutation of one of three known K8 phosphorylation sites. Extensive sumoylation occurs in cells transfected with individual K8, K18, or K19 but is limited upon heterodimerization (K8/K18 or K8/K19) in the absence of stress. In contrast, keratin sumoylation is significantly augmented in cells and tissues during apoptosis, oxidative stress, and phosphatase inhibition. Poly-SUMO-2/3 conjugates are present in chronically injured but not normal, human, and mouse livers along with polyubiquitinated and large insoluble keratin-containing complexes. Notably, common human K8 liver disease-associated variants trigger keratin hypersumoylation with consequent diminished solubility. In contrast, modest sumoylation of wild type K8 promotes solubility. Hence, conformational changes induced by keratin natural mutations and extensive tissue injury result in K8/K18/K19 hypersumoylation, which retains keratins in an insoluble compartment, thereby limiting their cytoprotective function.
Keywords: Intermediate Filaments, Liver Injury, Oxidative Stress, Post-translational Modification, Sumoylation
Introduction
Sumoylation is an important regulatory modification for an ever-increasing number of proteins implicated in various human diseases, including cancer, neurodegeneration, and cardiovascular disease (1–3). This reversible process of addition and removal of small ubiquitin-like modifier (SUMO)2 polypeptides (SUMO-1, -2, or -3) targets protein lysine residues and affects protein localization, interactions with binding partners, and degradation (4). Sumoylation typically modifies lysine residues located within a classical SUMO consensus motif ΨKX(D/E), where Ψ is a hydrophobic amino acid and X is any residue. In addition, some proteins harbor a phosphorylation-dependent SUMO motif, in which there is an adjacent proline-directed phosphorylation site next to the SUMO consensus site (ΨKXEXXSP) (5). Whereas SUMO-2 and 3 are nearly identical, each shares ∼50% sequence homology with SUMO-1, with which they have both common and distinct substrates (6–9).
Although most substrates identified to date are nuclear proteins, SUMO also regulates proteins found in other cellular compartments (4). Sumoylation of IFB-1, a cytoplasmic intermediate filament (IF) protein in Caenorhabditis elegans, regulates filament assembly via sequestration and maintenance of a cytoplasmic pool of unpolymerized IFB-1 (10). Additionally, defective sumoylation of the human nuclear IF lamin A may potentially be involved in the pathophysiology behind dilated cardiomyopathy associated with lamin A mutations (11). Sumoylation of mammalian cytoplasmic IFs, many of which are implicated in human disease (12), has not been investigated to date. Given the strong association of IF mutations with human disease (12–14), a better understanding of the role of sumoylation in the function of the various IFs is needed.
IF proteins comprise 73 unique human gene products grouped into six major types: types I–IV, which are cytoplasmic and include the epithelial and hair keratins, myocyte desmin, and neurofilaments, among others; type V, which are the nuclear lamins; and type VI, which are found in the fiber cells of the lens (14, 15). Keratins are the IFs of epithelial cells and exist as obligate heteropolymers of relatively acidic (type I) and relatively basic (type II) keratins. Mutations in the genes KRT8, KRT18, and KRT19, which encode K8, K18, and K19, respectively, predispose their carriers to acute (16) and chronic (17) liver disease. In the liver, K8/K18 and K8/(K18/K19) heteropolymers are found in hepatocytes and ductal cells, respectively. Keratin post-translational modifications, such as phosphorylation, are closely linked to keratin cytoprotective function during liver injury (18). For example, mice that overexpress human K8 S74A, which cannot be phosphorylated by stress-activated kinases at Ser-74, have a marked predisposition to Fas-mediated liver injury relative to WT mice (19). Notably, overexpression of the human liver disease-associated K8 variant G62C in mice leads to a marked decrease in K8 Ser-74 phosphorylation and a similar injury phenotype to the K8 S74A mutant (19). Therefore, understanding keratin post-translational modifications is critical because it may provide a mechanistic link between clinically relevant mutations and their disease manifestations.
The types of post-translational modifications that keratins may undergo are largely impacted by their structural characteristics. The tertiary structure of keratins (and other IFs) is comprised of a central coil-coil α-helical rod domain flanked by the non-α-helical head and tail domains, whereas their quaternary filamentous structure involves the association of multiple tetramers composed of two keratin heterodimers and is facilitated via interactions between the rod domains (15, 20). Although the head and tail domains contain motifs for stress-induced post-translational modifications, including phosphorylation and glycosylation (18), there is limited experimental evidence that post-translational modifications occur within the keratin rod domain, as exemplified by caspase-mediated cleavage of type I keratins and other IFs during apoptosis (21–24).
We hypothesized that sumoylation may also be an important regulatory modification for keratins because sumoylation of lamin A occurs on a rod domain lysine residue that, along with its associated SUMO consensus motif, is conserved across selected type I IFs (including K18 and K19; supplemental Fig. S1). We tested this hypothesis in the context of mouse and human chronic liver injury, cell culture models, and in vitro and demonstrated that K8, K18, and K19 undergo oxidative and other stress-induced sumoylation. We also identified sumoylation sites on K8, K18, and K19 within their rod domains and showed that phosphorylation plays a regulatory role in keratin sumoylation. Sumoylation of WT K8 leads to increased solubility, whereas human mutations in K8 that predispose their carriers to liver disease are hypersumoylated and exhibit decreased solubility.
EXPERIMENTAL PROCEDURES
Antibodies
The antibodies we used included: rat anti-mouse Troma I (K8) and Troma III (K19) (Developmental Studies Hybridoma Bank) and rabbit anti mouse/human 4668 (K18); mouse monoclonal antibodies TS1 (K8), DC10 (K18) (Labvision, Fremont, CA), and 4.62 (K19) (Sigma-Aldrich) to human keratins; rabbit polyclonal antibodies to mouse/human SUMO-1 and SUMO-2/3 (Abcam); mouse monoclonal antibodies against K5, K14, and vimentin (Labvision); anti-ubiquitin antibody (Santa Cruz Biotechnology); and Alexa 488- and Alexa 594-conjugated goat anti-rabbit antibodies (Invitrogen).
Animal and Human Liver Experiments
Animal and human tissue use was approved by the corresponding oversight committees at the University of Michigan. To determine the effect of 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC)-induced liver injury (after a 90-day DDC treatment) on sumoylation, the livers of FVB/N female mice were used as described (25). To determine the effect of the phosphatase inhibitor microcystin LR (MLR) on sumoylation, human K18-overexpressing mice (26) were injected with 30 μg/kg of MLR intraperitoneally followed by harvesting of the livers after 195 min. The human liver samples have been described previously (17) and were used under an approved human subjects protocol.
Isolation and Treatment of Primary Mouse Hepatocytes
Male mice overexpressing human keratin 18 (26) were anesthetized with 50 mg/kg Nembutal (Ovation Pharmaceuticals, Inc.). The liver was first perfused with 30 ml of perfusion solution I (Hanks' balanced salt solution containing 0.5 mm EGTA, 5.5 mm glucose, and 1% penicillin-streptomycin) through the portal vein with a flow rate of 7 ml/min, followed by perfusion with 25 ml of perfusion solution II (Hanks' balanced salt solution containing 1.5 mm CaCl2, 5.5 mm glucose, 1% penicillin-streptomycin, and ∼2000 units of collagenase IV (Worthington)) at the same flow rate. The perfused liver was placed in a sterile Petri dish containing William's medium E supplemented with 1% penicillin-streptomycin and subjected to mechanical breakdown. The cell suspension was filtered through a 70-μm cell strainer and pelleted by centrifugation (500 rpm, 2 min, 4 °C). The cell pellet was washed two times, and the cells were cultured in William's medium E supplemented with 10% FBS and 1% penicillin-streptomycin. All of the solutions were prewarmed to 37 °C before use, and the cells were plated at a density of 0.5 million/ml onto collagen I-coated plates (BD BioCoat). The cells were allowed to attach for 8 h (37 °C, 5.0% CO2) before the addition of anisomycin (10 μg/ml, 6 or 24 h), H2O2 (1 mm, 45 min), or okadaic acid (OA; 1 μm, 45 min).
In Vitro Sumoylation Assay
An in vitro sumoylation kit (Biomol) was used to determine the ability of K8, K18, K5, K14, and vimentin to be modified by SUMO-1, SUMO-2, and SUMO-3 in the presence of ATP. Bacterially expressed purified human keratins and vimentin (27) were used at 200 nm in the assay, and the reaction was performed as recommended by manufacturer. Analysis of keratin solubility was performed by separation of the soluble and insoluble fractions of the sumoylation reaction by centrifugation at 45,000 rpm (1 h) and subsequent analysis of the supernatant and pellet contents by immunoblotting.
Site-directed Mutagenesis
The human K8 and K18 cDNA in vector pcDNA3.1 and K19 cDNA in vector pMRB101 were mutated to generate single-point lysine to arginine mutations using the QuikChange site-directed mutagenesis kit (Stratagene). The phospho-mutant keratin constructs were generated as described previously (28). The WT and mutant keratin constructs were confirmed by DNA sequencing.
Cell Cultures and Transfection
For the biochemical studies, BHK-21 (baby hamster kidney) cells were used because of their robust keratin expression after transfection. We chose NIH3T3 (mouse fibroblast) cells for immunofluorescence experiments because, upon transfection, they form more normal-appearing keratin filaments as compared with BHK-21 cells. BHK-21, NIH3T3, and HT29 (human colon) cells were obtained from American Type Culture Collection and cultured as recommended by the supplier. Lipofectamine 2000 (for BHK-21 transfections) or Lipofectamine LTX (for NIH3T3 transfections) (Invitrogen) were used. Cell transfections used for immunoblots were carried out in six-well plates using 3–4 μg of total DNA/well and for immunostaining were performed in four-chamber cell culture slides (BD Biosciences) using 0.5 μg of total DNA/well. Biochemical and immunofluorescence staining analyses were performed 18–22 h post-transfection.
Immunofluorescence Staining and Confocal Imaging
Keratin-transfected NIH3T3 cells were either left untreated or incubated with 1 mm H2O2 for 45 min followed by methanol fixation (10 min, −20 °C). After fixation, the cells were-air dried, and nonspecific binding was blocked by incubation in blocking buffer (PBS with 2.5% (w/v) BSA and 2% normal goat serum). Primary antibodies were diluted 1:100 (4.62), 1:150 (SUMO-1 and SUMO-2/3), or 1:200 (TS1 and DC10) in blocking buffer and incubated with the cells for 1 h at room temperature, followed by three 5-min washes in PBS and a 30-min incubation (in the dark) with secondary Alexa-conjugated antibodies (1:1000 dilution). HT29 cells were handled essentially the same as NIH3T3 cells except that some of the cells were treated with 1 μm OA for 45 min prior to immunostaining. Following secondary antibody incubation, the cells were rinsed three times for 5 min in PBS, mounted with Prolong Gold containing DAPI (Invitrogen), viewed using an Olympus FluoView 500 confocal microscope with a 60× oil immersion (1.4 NA) objective, and magnified 2.5 times with FluoView software (version 5.0). DAPI, Alexa Fluor 488, and Alexa Fluor 594 were excited with a 405-nm laser diode, a 488-nm argon laser, and a 543-nm HeNe green laser, respectively, and sequential scans were used to maximize signal separation.
High Salt Extraction of Keratins, Immunoprecipitation, and Immunoblot analysis
To obtain enriched keratin fractions from liver tissues and cultured cells, we used high salt extraction (HSE), as described previously (29), with the addition of 20 mm N-ethylmaleimide to the lysis and homogenization buffers to inhibit desumoylation during extraction. In some cases, the initial lysates in Triton X-100-containing buffer, representing the soluble fraction, were analyzed alongside the HSEs to determine keratin solubility. Anisomycin treatment (10 μg/ml) of HT29 cells was carried out for 24 h, at which point the caspase-generated K18 fragment was observed. HSE samples were resolved on 4–20% gradient SDS-PAGE gels that were subsequently stained with GelCode Blue reagent (Thermo Scientific). For immunoblot analysis, the gels were transferred onto polyvinylidene difluoride membranes, which were subsequently blocked and incubated with the designated antibodies. Antibody signals were detected using ECL Plus reagents for SUMO blots or ECL reagents for all other blots (PerkinElmer Life Sciences). For immunoprecipitation experiments, HT29 cells were grown to 80% confluency and were collected untreated or after treatment for 45 min with H2O2 (1 mm) or okadaic acid (1 μm). The cells were scraped in prewarmed PBS-EDTA and centrifuged, and the lysates were prepared using 1% Empigen BB/PBS-EDTA as the lysis buffer. Rabbit polyclonal antibody targeting the C terminus of SUMO-2 (Abgent) was conjugated to protein G Dynabeads (Invitrogen), which were then incubated with the HT29 cell lysates.
Data Analysis
The pixel intensities of the scanned immunoblot images were estimated using Adobe Photoshop CS2 version 9.0 and used to obtain the relative intensity values shown in Figs. 2 and 5.
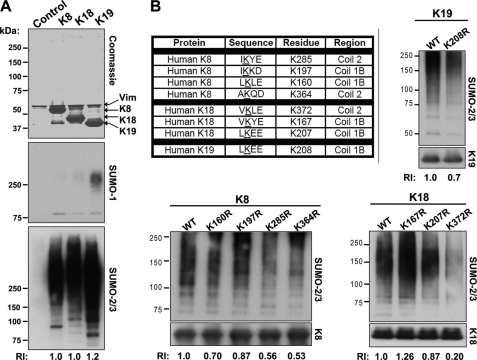
FIGURE 2.
Sumoylation of K8, K18, and K19 on rod domain lysine residues. A, HSEs were prepared from BHK-21 cells transiently transfected with empty vector (control) or the cDNA of human K8, K18, or K19 and then analyzed by Coomassie staining (loading control), which shows the presence of endogenously expressed vimentin (Vim; ∼57 kDa) in all samples, and only the specific keratins K8 (∼54 kDa), K18 (∼46 kDa), or K19 (∼40 kDa) in samples from keratin-transfected cells. Two additional gels identical to the one shown by the Coomassie stain were analyzed by blotting with antibodies to SUMO-1 or SUMO-2/3. No detectable immunoreactivity was observed below 75 kDa (not shown). The relative intensity (RI) of the SUMO-2/3 signal, compared with K8, is included for K18 and K19. B, in silico identification of consensus sumoylation sites (ΨKX(E/D)) on human K8, K18, and K19 using the SUMOplotTM program, ordered from highest to lowest probability scores, and experimental validation of the predicted sites. An identical analysis such as the one shown in A was carried out using HSEs of BHK-21 cells expressing WT K8, K18, and K19 proteins individually, or their corresponding predicted SUMO target lysine-to-arginine mutants. Equal loading was estimated initially based on Coomassie-stained gels of the HSEs (not shown) and subsequently confirmed by blotting for K8, K18, and K19. Western blotting for the presence of SUMO-2/3 conjugates shows differences between the WT proteins and some of their mutants, which are quantified and expressed as relative intensity values (normalized to WT controls).
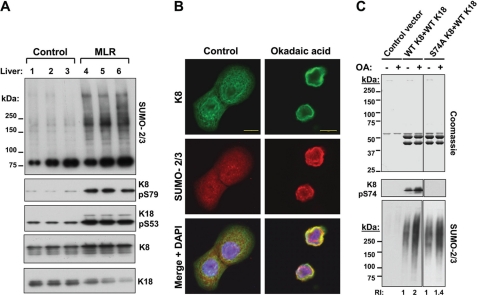
FIGURE 5.
Keratin sumoylation in association with phosphorylation in vivo and in vitro. A, immunoblot analysis of liver HSEs of untreated (control) or MLR-treated mice analyzed for SUMO-2/3, phosphorylated K8 (Ser(P)-79), and K18 (Ser(P)-53), which serve as functional readouts of the effect of MLR, and total K8 and K18 (loading controls). B, immunofluorescence analysis demonstrating the localization of K8/K18 heteropolymers (green) and SUMO-2/3 (red) in untreated (control) and OA-treated HT29 cells. Scale bars, 10 μm. C, Coomassie stain (loading control) and immunoblot analysis of the HSEs of untreated (−) and OA-treated (+) BHK-21 cells transfected with control vector. WT human K8/K18, or S74A K8/WT K18. The relative intensity (RI) of the SUMO-2/3 signal increases after OA treatment 2-fold in WT K8-transfected cells and 1.4-fold in S74A-transfected cells, relative to the corresponding untreated control (representative of three separate transfection experiments). Note that human K8 Ser-74 corresponds to mouse K8 Ser-79.
RESULTS
In Vitro Sumoylation of K8, K18, K5, K14, and Vimentin
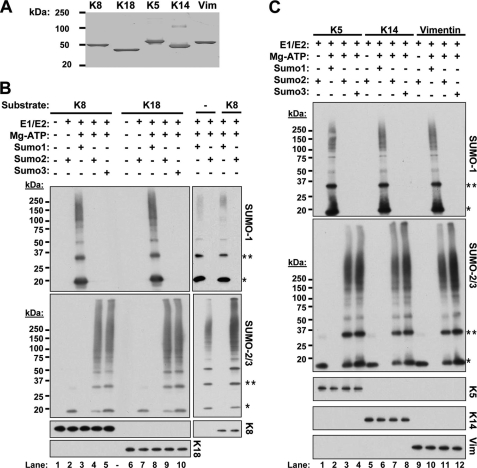
SUMO proteins are added enzymatically to target lysines by SUMO E1 (ATP-dependent) and E2 ligases (4). This process can be recapitulated in vitro using purified substrates. We used five different recombinantly expressed purified human cytoplasmic IFs (K8, K18, K5, K14, and vimentin) as substrates (Fig. 1A) to examine their possible sumoylation. As shown in Fig. 1B, both K8 and K18 were modified by polymeric chains of all three SUMO isoforms in an ATP-dependent manner. However, the extent of modification by SUMO-2/3 appeared to be greater than by SUMO-1, based upon the relative signal intensities of the unconjugated to conjugated SUMO protein (Fig. 1B). To determine whether the effect seen with K8 and K18 extends to other cytoplasmic IFs, we also investigated the in vitro sumoylation of the epidermal keratins K5 and K14 and the type III mesenchymal IF, vimentin. The results shown in Fig. 1C indicate that these IFs, like K8 and K18, are also sumoylated preferentially by SUMO-2/3 over SUMO-1, and in the case of K14 and vimentin, modification by SUMO-3 may occur more extensively than that by SUMO-2.
FIGURE 1.
In vitro sumoylation of keratins and vimentin. A, Coomassie stain of purified recombinant human K8, K18, K5, K14, and vimentin (Vim) resolved on a 4–20% gradient SDS-PAGE gel. B and C, purified keratins, vimentin, or no substrate (−) were incubated with E1/E2 SUMO ligases, ATP, and either SUMO-1, SUMO-2, or SUMO-3. The resulting reaction products were analyzed by immunoblotting for the ATP-dependent formation of SUMO-1 and SUMO-2/3-containing intermediate filament conjugates. The asterisks denote SUMO monomers (*) or dimers (**). Keratin and vimentin blots serve as loading controls.
Sumoylation of Human K8, K18, and K19 on Rod Domain Lysines within SUMO Consensus Sites
Sumoylation of keratins was examined further by transient transfection of human K8, K18, and K19 into BHK-21 cells and subsequent biochemical analysis of the cellular HSE fractions. HSEs contain primarily the highly insoluble intermediate filaments because most other cellular proteins are eliminated during the extraction procedure, which involves detergent solubilization followed by a high salt buffer extraction of the detergent-insoluble fractions (29). The HSEs of the empty vector control- and keratin-transfected cells were initially resolved on a gel, which was stained by Coomassie Blue, confirming the expression of each keratin (Fig. 2A, top panel). Immunoblot analysis using antibodies against SUMO-1 and SUMO-2/3 showed the presence of high molecular weight (HMW) complexes in the keratin-containing HSEs but not those of control transfections (Fig. 2A, middle and bottom panels), which contained vimentin (confirmed by immunoblotting; not shown). In agreement with the in vitro sumoylation analysis (Fig. 1), there was preferential modification by SUMO-2/3 over SUMO-1 (Fig. 2A).
Analysis of potential SUMO consensus motif sites using the SUMOplotTM program revealed four such sites in K8, three in K18, and one in K19 (Fig. 2B). Of note, all potential sumoylation sites are located within the rod domains of each protein, specifically in coils 1B and 2. Site-directed mutagenesis was performed to convert each of the predicted lysine residues to a nonsumoylatable arginine, and the mutant proteins were transiently expressed in BHK-21 cells. The HSEs of the WT and mutant keratin-expressing cells were analyzed biochemically for the presence of SUMO-2/3-containing complexes. Mutation of residues Lys-285 and Lys-364 in K8; Lys-372 and Lys-207 in K18; and Lys-208 in K19 to arginines decreased sumoylation in comparison with the respective WT proteins when equal protein amounts were analyzed (Fig. 2B). Therefore, K8, K18, and K19 are sumoylated on several lysine residues located within the coiled-coil α-helical rod domain. In addition, most of the Lys to Arg mutants displayed abnormal filament networks (supplemental Fig. S2), but the severity of filament disorganization was more apparent in those mutants that significantly interfere with sumoylation (K8 Lys-285/364, K18 Lys-372, and K19 Lys-208) based on the biochemical evidence in Fig. 2B.
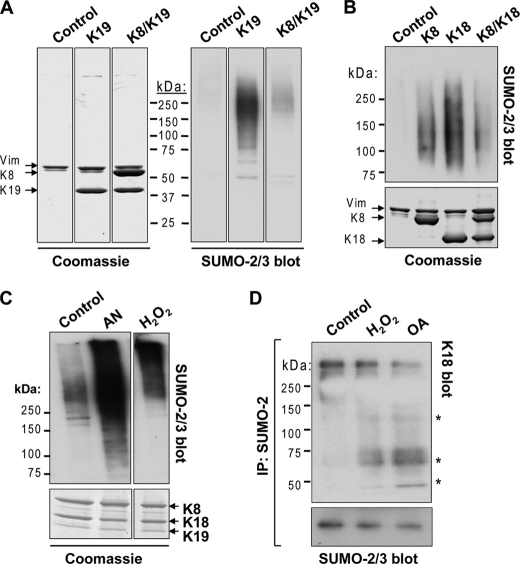
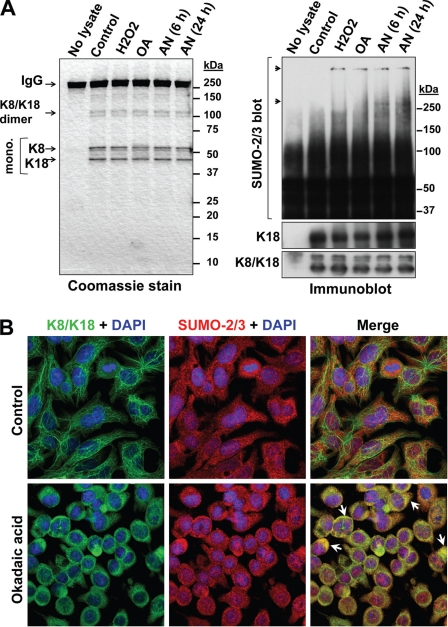
Keratin Sumoylation Is Low under Basal Conditions and Significantly Induced during Apoptosis, Oxidative Stress, and Phosphatase Inhibition
The location of the keratin sumoylation sites within the rod domain suggests that the involved lysines are unlikely to be readily accessible under basal conditions. To test this hypothesis, we examined the extent of sumoylation in HSEs of BHK-21 cells that were transfected with the individual keratins or cotransfected with a combination of K8/K18 or K8/K19. Significant sumoylation occurred when keratins K8, K18, and K19 were individually transfected in comparison with double K8/K18, K8/K19 transfections (Fig. 3, A and B). Similarly, HSEs of untreated human colon carcinoma HT29 cells, which endogenously express all three keratins, showed minimal SUMO-2/3 modification relative to that seen after treatment with either anisomycin to induce apoptosis or with H2O2 to induce oxidative stress (Fig. 3C), suggesting that cellular stress causes increased sumoylation following reorganization of keratin filaments, which occurs under these contexts (21, 30). In addition, SUMO-2 immunoprecipitation from HT29 cell lysates yielded K18-containing complexes after H2O2 treatment, but not in untreated cells (Fig. 3D). We also tested the effect of the phosphatase inhibitor OA, because keratin hyperphosphorylation, which occurs during liver injury (19, 31) and increases filament solubility (32), may affect sumoylation. Similar to the peroxide treatment, SUMO-2 immunoprecipitate of OA-treated HT29 cells contained K18 (Fig. 3D).
FIGURE 3.
Keratin sumoylation under basal conditions and during apoptosis, oxidative stress, and phosphatase inhibition. A, HSEs of BHK-21 cells transiently transfected with empty vector (control), K19, or K8/K19 were resolved by SDS-PAGE and analyzed by Coomassie staining (loading control) or immunoblotting with anti-SUMO-2/3 antibody. Note that control-transfected cell HSEs contain only vimentin (Vim). Separation by black lines indicates nonconsecutive lanes of the same membrane (all lanes were exposed equally). B, identical analysis as in A, except the analysis was done on individually expressed K8 and K18 or the K8/K18 pair. C, SUMO-2/3 immunoblot analysis of HSEs from control, anisomycin (AN)-treated, or H2O2-treated HT29 cells, which express K8, K18, and K19 endogenously, and the corresponding Coomassie stain of the gel showing equal loading. D, detection of keratin-containing complexes (asterisks) from SUMO-2 immunoprecipitates (IP) of HT29 cells treated with H2O2 and OA but not in control cells. Shown are immunoblots for K18 and SUMO-2/3 (loading control).
Colocalization of Keratins with SUMO-2/3 during Oxidative Stress
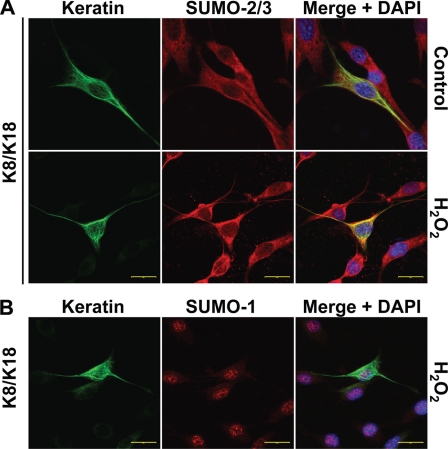
After expressing K8 in combination with either K18 or K19 in NIH3T3 cells, we used immunofluorescence staining to examine SUMO-keratin interactions in the context of oxidative stress. As shown in Fig. 4A, K8/K18 and SUMO-2/3 proteins manifest enhanced colocalization upon induction of oxidative stress using H2O2 as compared with transfected cells in the absence of peroxide treatment. Similar results were obtained with K8/K19-transfected cells (supplemental Fig. S3). We also checked for potential colocalization of keratins with SUMO-1, but its expression was primarily nuclear and did not appear to colocalize with keratin filaments (Fig. 4B), consistent with the biochemical analysis shown in Fig. 2A.
FIGURE 4.
Keratins colocalize with SUMO-2/3 during oxidative stress. NIH3T3 cells were transfected with human K8/K18 and were either left untreated (control) or treated with 1 mm H2O2 for 45 min prior to immunofluorescence staining. Primary mouse antibody recognizing K18 (DC10) was used to localize K8/K18 heteropolymers, respectively, along with rabbit anti-SUMO-1 or SUMO-2/3 antibodies. Secondary goat anti-mouse Alexa Fluor 488 (green) and goat anti-rabbit Alexa Fluor 594 (red) antibodies were used to visualize the localization of keratins and SUMO by confocal microscopy. K8/SUMO coimmunostaining (not shown) demonstrated analogous results. Shown are representative images from the single channels as well as a merged image with DAPI counterstain showing the cell nuclei. Scale bars, 20 μm.
Sumoylation Is Associated with Keratin Phosphorylation in Vivo and in Vitro
Because both keratin phosphorylation (18) and sumoylation (Fig. 3, C and D) are increased upon stress, and SUMO modification of several proteins is known to be phosphorylation-dependent (5), we investigated whether this is also the case for keratins. Intraperitoneal treatment of mice that express human K18 with MLR leads to liver K8 and K18 hyperphosphorylation, as determined using phospho-specific antibodies to mouse K8 pS79 and human transgene K18 Ser(P)-53 (Fig. 5A). SUMO-2/3 immunoblot analysis of the HSE fraction of control and MLR-treated mouse livers shows increased SUMO-2/3-modified HMW complexes in livers of MLR-treated mice relative to control livers (Fig. 5A). Similarly, treatment of HT29 cells with OA leads to colocalization of the keratin and SUMO-2/3 immunofluorescence signals (Fig. 5B). We then asked whether previously identified phosphorylation sites within the head and tail domains of K8 (18) are important for its sumoylation. For this, we cotransfected human WT K18 with WT, S24A, S74A, or S432A human K8 and tested the extent of sumoylation in the HSE fraction of the transfected BHK-21 cells. Although there was no significant difference between WT, S24A, and S432A K8-transfected cells (not shown), a relative decrease in sumoylation was observed in cells transfected with S74A K8 (Fig. 5C; human K8 Ser-74 is homologous to mouse K8 Ser-79). However, because the effect was not dramatic, we hypothesize that there are other critical phosphorylated keratin residues or that sumoylation of keratins is dependent upon the phosphorylation of other proteins.
Keratins Are Sumoylated in Primary Mouse Hepatocytes and in Human Hepatoma HepG2 Cells upon Oxidative Stress, Apoptosis, and/or Phosphatase Inhibition
To validate our findings (with the transfected BHK-21 cells and the human colonic HT29 cells) in cells from hepatic origin, we analyzed keratin sumoylation in isolated hepatocytes from human K18-overexpressing mice. Keratins were immunoprecipitated using an antibody against human K18 from control and hydrogen peroxide-, okadaic acid- and anisomycin-treated cells and initially analyzed by nonreducing SDS-PAGE and Coomassie staining of the gel to confirm the presence of keratins. As shown in Fig. 6A, K8/K18 monomers and dimers are present in all immunoprecipitates in equal amounts, which was also confirmed by immunoblot using antibodies against K18 and K8/K18. SUMO-2/3 immunoblot analysis of the immunoprecipitates (under reducing conditions) revealed that keratins were sumoylated only in the presence of oxidative and apoptotic stresses and phosphatase inhibition but not under basal conditions. Keratin sumoylation results in HMW conjugates (Fig. 6A, right panel). There was no modification by SUMO-1 (not shown). Furthermore, we observed significant keratin-SUMO-2/3 colocalization in okadaic acid-treated but not control HepG2 cells (Fig. 6B). Similar results were obtained using anisomycin treatment (not shown). These data provide direct evidence that keratins are targets for sumoylation in liver cells during stress.
FIGURE 6.
Keratins are sumoylated in primary mouse hepatocytes and in human hepatoma HepG2 cells under oxidative stress, apoptosis, and/or phosphatase inhibition. A, primary hepatocytes from human K18-overexpressing mice were isolated and cultured as described under “Experimental Procedures.” K8/K18 were immunoprecipitated using mouse monoclonal DC10 antibody (anti-human K18), and the immunoprecipitates were resolved, under nonreducing conditions, on a SDS-PAGE gel, which was stained by Coomassie to verify the presence of keratins. Both keratin monomers (mono.) and likely dimers are visible. Immunoblots of the immunoprecipitates verify the presence of keratins, in equal amounts across treatment groups, as detected by rabbit antibodies to human K18 and K8/K18. Immunoblot for SUMO-2/3 (under reducing conditions) shows sumoylated keratins (arrows) in the presence of H2O2, OA, and anisomycin (AN) for 6 or 24 h, but not in control hepatocytes. B, untreated (control) and OA-treated HepG2 cells were stained for K18 (DC10 antibody) and SUMO-2/3 followed by Alexa 488 (green) and Alexa 594 (red)-conjugated secondary antibodies and imaged by confocal microscopy. The arrows point to significant colocalization (yellow signal) of keratins and SUMO-2/3 in the presence of OA.
Increased Sumoylation of Insoluble Liver Proteins during Chronic Mouse Liver Injury
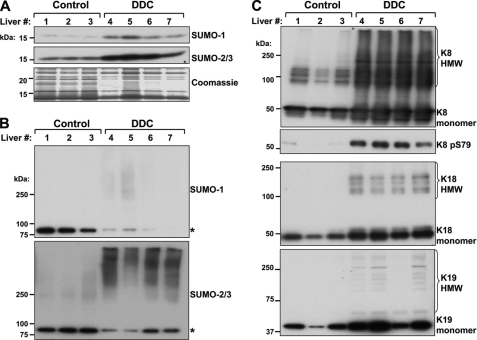
Prolonged feeding of mice with a diet containing the porphyrinogenic compound DDC is a commonly used liver injury model because it reflects some of the hallmarks of chronic human liver injury, such as oxidative stress, matrix accumulation, and inflammation (25, 33). Notably, chronic treatment of mice with DDC resulted in a significant induction in the hepatic expression of SUMO-1 and SUMO-2/3 isoforms, as assessed by immunoblotting (Fig. 7A). Mouse liver HSEs were analyzed biochemically for the presence of SUMO and keratins. Keratin IFs, of which K8, K18, and K19 are most abundantly expressed in liver with a stoichiometry of K8 > K18 ≫ K19, are known to be extensively modified by phosphorylation during liver injury (31). As shown in Fig. 7B, there was a significant increase in HMW complexes reactive with SUMO-2/3 antibody in liver fractions obtained from DDC-treated mice relative to untreated control mice. In contrast, the signal of a single SUMO-1 and SUMO-2/3 reactive band that migrated between 75 and 100 kDa, which may represent sumoylated lamin A (11) or RanGAP1, which forms a stable 90-kDa SUMO complex (34), decreased after DDC treatment (Fig. 7B). Similar results were obtained with SUMO-1, although the signal intensity for the HMW SUMO-1-containing complexes was significantly less (Fig. 7B). Immunoblot analysis of K8, K18, and K19 showed an increase in all three proteins, along with increased K8 phosphorylation at Ser-79 (Fig. 7C), consistent with the known fate of these proteins during liver injury (18, 35). After DDC, there was a strong expression of keratin-containing HMW complexes (Fig. 7C, top panel), which also paralleled the SUMO-2/3 results (Fig. 7B, bottom panel).
FIGURE 7.
Increased SUMO expression and SUMO-2/3 modification of insoluble mouse liver proteins during chronic liver injury. A, total cell lysates were prepared from separate livers of three untreated (1–3) and four DDC-treated (4–7) FVB/N mice. The detergent-soluble fractions were analyzed by SDS-PAGE followed by Coomassie staining (loading control) or immunoblot for the presence of monomeric SUMO-1 and SUMO-2/3 isoforms (∼15 kDa). B, the corresponding HSEs of the liver lysates in A were analyzed by immunoblotting using antibodies to SUMO-1 and SUMO-2/3. The asterisks denote SUMO complexes that likely include lamin A and/or RanGAP1. C, the liver HSEs from control and DDC-treated mice were analyzed for the expression of K8, K18, and K19 (as loading controls). Note that there is keratin induction during liver injury in addition to K8 phosphorylation at Ser79. The monomeric proteins are distinguished from the HMW keratin-containing complexes, which are primarily observed after DDC treatment, similar to the SUMO-2/3-containing HMW complexes (B).
Increased Sumoylation of Insoluble Liver Proteins during Chronic Human Liver Injury
We also tested human chronic liver disease patient samples using HSEs prepared from either a nondiseased human liver or the livers of patients with alcoholic cirrhosis or viral hepatitis. There was strong SUMO-2/3 reactivity in all of the liver disease samples that was absent from the control liver (Fig. 8A). This effect was not due to differences in protein loading, as demonstrated by the levels of K8, K18, and K19 (Fig. 8A, three bottom panels). In agreement with the findings in mouse liver injury (Fig. 7), K8-containing HMW complexes were only detected in the diseased human samples, where the signals partially overlapped with SUMO-2/3, particularly within large complexes visualized at the top of the gel. Because the ubiquitin/proteasome system has been shown to be an important component of the SUMO-2/3 cycle (36, 37), we also analyzed the extent of ubiquitination. Similar to SUMO-2/3, ubiquitin-containing HMW complexes were abundantly present in five of the six patient samples (supplemental Fig. S4).
FIGURE 8.
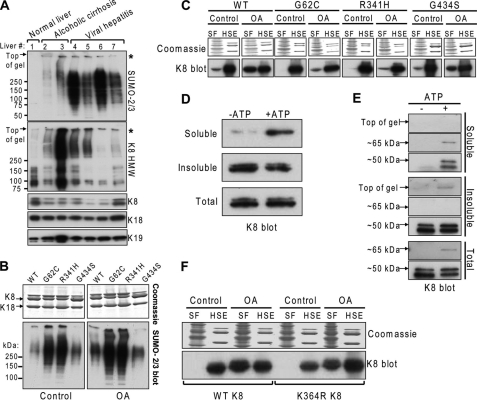
The extent of sumoylation determines keratin solubility in human liver disease and disease-associated K8 mutants. A, SUMO-2/3-modified proteins were preferentially detected in diseased human liver HSEs (2–7) as compared with normal liver (1). The asterisks denote insoluble HMW K8-containing complexes that also contain SUMO-2/3. Immunoblots of monomeric K8 (lower exposure of the K8 HMW blot), K18, and K19 are loading controls. B, Coomassie stain (loading control) and SUMO-2/3 immunoblot of HSEs of untreated and OA-treated BHK-21 cells expressing WT K18 and either WT K8, or mutants of K8 that predispose to human liver disease. C, determination of relative K8 solubility by comparing the soluble fraction (SF) and the insoluble HSE of the same cells shown in B. Coomassie-stained gels are loading controls. Each K8 variant was run on a separate gel. Separation by black boxes between control and OA groups denotes nonconsecutive lanes of the same gel or equally exposed membrane. D, human recombinant K8 was incubated with SUMO-2, E1, and E2 SUMO ligases in the absence (−ATP) or presence (+ATP) of Mg-ATP for 4 h at 37 °C, after which the reaction was separated into an insoluble and soluble fraction by high speed centrifugation. The presence of total amount of K8 (before centrifugation) and in the two fractions was determined by immunoblotting. E, detection of hypersumoylated (Top of gel), monosumoylated (∼65 kDa), and unmodified (∼50 kDa) K8 in the soluble, insoluble, and total fractions from in vitro sumoylation reactions that were done in the absence (−) or presence (+) of ATP for 16 h. Immunoblot for K8 in the total reaction is shown as a loading control. F, comparison of the relative solubility of WT K8 and sumoylation-deficient K364R K8 after treatment with OA.
Hypersumoylation of K8 Variants That Predispose to End Stage Liver Disease
Several mutations in K8, including G62C, R341H, and G434S, predispose their carriers to acute and chronic liver disease, based on animal and human studies (16, 17, 38). The most common amino acid-altering K8 mutations in white and black individuals are R341H and G434S, respectively. The K8 G62C mutation is also a common mutation that, when overexpressed in mice, results in marked predisposition to liver injury (19). Among the potential mechanisms whereby these clinically relevant mutations diminish the cytoprotective function of K8 are interference with stimulus-induced keratin filament reorganization and phosphorylation by stress-activated kinases, as well as structural protein destabilization (19). Therefore, we investigated whether these three K8 human variants exhibited altered sumoylation. Immunoblot analysis of HSEs from WT and mutant K8-transfected cells, all of which were cotransfected with WT K18, showed that K8 G62C and R341H are hypersumoylated under basal conditions, to a level markedly exceeding that of WT K8 under both basal and OA-treated conditions (Fig. 8B). In contrast, the K8 G434S mutant behaved similarly to WT K8. The presence of insoluble complexes at the top of the gel was a common observation between the patient samples and the mutant K8-transfected cells (Fig. 8, compare A with B) and suggested that sumoylation may alter K8 solubility. To investigate that, we analyzed the presence of K8 in the soluble fraction relative to the insoluble HSE fraction under basal and OA treatment conditions. The solubility of the hypersumoylated mutants of K8, G62C and R341H, was significantly less in comparison with WT and G434S K8 (Fig. 8C) when comparing the distribution of K8 between the two fractions.
To determine how sumoylation affects the solubility of WT K8, we analyzed K8 distribution between the soluble and insoluble fractions of in vitro sumoylation reactions. Under sumoylation-permissive reaction conditions (+ATP), the amount of soluble unmodified K8 is increased, whereas insoluble K8 is accordingly decreased (Fig. 8D). Furthermore, longer sumoylation reaction times (16 h; +ATP) allowed us to discriminate between the soluble monosumoylated (∼65 kDa) and the insoluble hypersumoylated (top of gel) forms of K8 (Fig. 8E). In addition, the sumoylation-deficient K8 K364R mutant is less soluble upon OA treatment relative to WT K8 (Fig. 8F). Collectively, these data point to a keratin solubility-promoting role for sumoylation, which is aberrant in the context of disease-associated keratin mutations and human liver injury, whereby hypersumoylation retains keratins in an insoluble compartment.
DISCUSSION
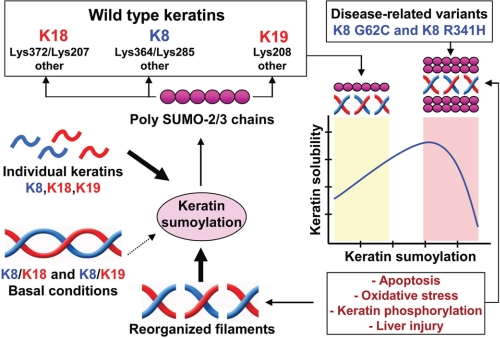
Utilizing multiple experimental systems, including cell-free, cultured cells, and diseased livers, we demonstrate that K8, K18, and K19 undergo sumoylation in a stress- and injury-dependent manner (Fig. 9). In a cell-free system, K8 and K18 are modified by SUMO-2/3 and, to a lesser extent, by SUMO-1 in an ATP-dependent manner. Keratin sumoylation occurs on rod domain lysine residues nested within classical SUMO motif sequences (ΨKX(D/E)), and thus it is not observed in the context of intact keratin heteropolymers but occurs extensively under conditions of oxidative stress and phosphatase inhibition (Fig. 9), as demonstrated biochemically and by immunofluorescence staining. At the tissue level, we show that the insoluble liver protein pool, consisting primarily of keratins, is preferentially modified by polymeric SUMO-2/3 chains during mouse and human chronic liver injury. Furthermore, the poly-SUMO-2/3 conjugates are observed in association with polyubiquitinated and large, insoluble K8-containing complexes that fail to migrate into an SDS-PAGE gel. Notably, some liver disease-associated variants of human K8 (R341H and G62C) exhibit dramatically elevated basal and phosphorylation-induced sumoylation along with diminished solubility, whereas the modest, stimulus-induced sumoylation observed in WT K8 promotes solubility (Fig. 9). Therefore, sumoylation of keratin IFs represents a novel post-translational modification that regulates their solubility and carries potential implications to liver injury as well as extrahepatic pathologies where sumoylation of other keratin intermediate filaments (e.g. K5 and K14) and vimentin may occur in a similar tissue-specific, injury-dependent fashion.
FIGURE 9.
Dual role for sumoylation in regulating the solubility of wild type and disease-associated keratin mutants. K8, K18, and K19 are modified by polymeric SUMO-2/3 chains on multiple rod domain lysine residues. Wild type keratin SUMO modification is highest in the context of individually expressed keratins or upon stress- and phosphorylation-induced filament reorganization. Under basal conditions where type I/II keratin heteropolymers predominate, precluding access to SUMO target lysines, wild type keratin sumoylation is minimal. In contrast, variants of human K8 that predispose individuals to acute and chronic liver diseases, including K8 G62C and K8 R341H, are hypersumoylated under basal conditions, likely in response to post-translational modifications or structurally induced changes in filament organization. Sumoylation regulates keratin solubility, such that low level sumoylation promotes solubility, whereas hypersumoylation, which is seen during liver injury, renders keratins more insoluble.
Sumoylation as a Common Regulatory Modification of Intermediate Filaments
It was recently shown that human lamin A Lys-201 is modified by SUMO-2 within the consensus motif MKEE, and this modification was found to be important for proper lamin A perinuclear localization (11). Mutation of lamin A Lys-201 to an arginine resulted in abnormal lamin A puncta surrounding the nucleus. This aberrant localization of K201R lamin A was similar to sumoylation-deficient E203G and E203K lamin A, which are naturally occurring mutations found in familial dilated cardiomyopathy. Furthermore, cells expressing lamin A devoid of the ability to be sumoylated were found to be more prone to cell death, although the mechanism behind this is not known (11). Because the Lys-207 and Lys-208 residues in the type I keratins K18 and K19, respectively, correspond to the Lys-201 residue in lamin A (supplemental Fig. S1) and because both are targets for sumoylation, the evidence thus far implicates this conserved lysine residue as an important site for sumoylation across IFs.
Regulation of Intermediate Filament Solubility by Sumoylation
Sumoylation regulates the availability of the unpolymerized cytoplasmic pool of the invertebrate intermediate filament IFB-1, aiding in its assembly into epidermal attachment structures (10). Although IFB-1 lacks a specific human ortholog, it is functionally associated with hemidesmosome-like attachment structures and provides epidermal mechanical integrity (39) similar to keratins.
The solubility of mammalian keratin IFs varies, such that epidermal keratins exhibit very low solubility (<1%), whereas ∼5% of K8/K18 in colonic HT29 cells exist in a soluble form (32). The exchange between the soluble and filamentous keratin pools is highly dynamic, as demonstrated by quantitative analyses of the motile properties of live cell tonofibrils, which are composed of bundles of keratin IFs, and squiggles, which are short keratin fibrils (40). Aside from phosphorylation, additional factors that regulate the dynamics of this process are not known, although our findings herein indicate the potential involvement of SUMO.
The availability of soluble keratins in the context of stress-induced filament rearrangement is critical for their cell protective effects, because the interaction of keratins with other proteins is largely dependent upon it. For example, an increase in keratin phosphorylation generates a soluble K8/K18 fraction that binds 14-3-3 adaptor proteins in the context of cell stress (18). Mutation of K18 at the Ser-33 site, which regulates 14-3-3 binding, prevents movement of 14-3-3 from the nucleus and plays a modulating role in mitotic progression during liver regeneration (41). In addition, K18 and 14-3-3 interaction is important in regulating the G2/M checkpoint in Xenopus (42). Furthermore, K17, an IF that is rapidly induced in wounded stratified epithelia, regulates cell growth by binding 14-3-3, leading to 14-3-3 translocation from the nucleus to the cytoplasm and concomitant mammalian target of rapamycin activation (43).
Alterations in the cellular responses that govern the availability of soluble IFs, such as sumoylation, may affect incorporation of IFs into pathologic inclusions. In sumoylation-deficient worms, there was an aberrant localization of IFB-1 such that long and circular ectopic filamentous structures were seen in addition to cytoplasmic inclusions appearing as either large “nucleation sites” or inclusions that were scattered throughout the cytoplasm (10). The formation of cytoplasmic aggregates in this model is relevant, because IFs are major components of intracellular inclusions seen in several human diseases, including the K8/K18-containing Mallory-Denk bodies that are primarily seen in hepatocytes of patients with steatohepatitis and hepatocellular carcinoma, among other liver diseases (33, 38). Examination of the livers of DDC-fed mice, which form human Mallory-Denk body-like inclusions (25, 38) suggested that there is no significant localization of SUMO within Mallory-Denk bodies.3 In light of the study by Kaminsky et al. (10) and our current results demonstrating that modest sumoylation increases K8 solubility (Fig. 9), SUMO conjugation may function to prevent IF-containing inclusion formation, although antibody epitope masking is also possible.
The Significance of Selective Keratin Modification by SUMO-2/3 Polymers
The addition of SUMO to various substrates in the form of monomers and polymers has been documented (44). Although the exact functional significance of each type of modification is poorly defined, there is evidence for a structural role of polymeric SUMO-2/3 chains in kinetochore function during cell division (45, 46) as well as a role for sumoylation in the ubiquitin-proteasome system (36, 37). Unlike SUMO-2 and -3, which form polymeric chains in vivo via internal sumoylation sites, SUMO-1, by lacking such a sites, is not known to polymerize in vivo, but it does so in vitro (47). However, in vivo SUMO-1 can be linked to the end of SUMO-2/3 chains, resulting in a mixed SUMO chain (37, 48). It is likely that the detection of high molecular weight SUMO-1-containing complexes (Figs. 2A and 7B) is reflective of SUMO-1-induced SUMO-2/3 chain termination. Our in vitro sumoylation assay that showed SUMO-1 chain formation with IFs (Fig. 1B) is similar to at least one previous report (47). Additionally, the existence of mixed SUMO/ubiquitin chains has also been reported. For example, phosphorylation-induced sumoylation of the promyelocytic leukemia protein results in a polymeric SUMO chain that, via the action of the E3 ubiquitin ligase activity of RING finger protein 4, is conjugated to polyubiquitin chains, ultimately leading to 26 S proteasome-mediated degradation of promyelocytic leukaemia protein (49). As further evidence for the interplay between SUMO-2/3 and the ubiquitin-proteasome system, a recent quantitative proteomics study found that ubiquitin was enriched selectively with SUMO-2 over SUMO-1 conjugates, and this effect was significantly enhanced upon pharmacologic inhibition of the proteasome (36). Furthermore, K8 and K18 were among 847 proteins that were enriched with the SUMO-2 fraction in a proteasome inhibitor-sensitive manner (36). Therefore, the phosphorylation-associated sumoylation and ubiquitination of keratins reported herein may implicate sumoylation as a functional link between keratins and the ubiquitin-proteasome system.
Cross-talk between Sumoylation and Other Keratin Post-translational Modifications
Phosphorylation has been reported to have substrate-dependent effects on sumoylation. For example, phosphorylation is a negative sumoylation regulator in the case of the polyglutamine repeat protein ataxin-1 (50) and a positive regulator for the microtubule-stabilizing protein Tau (51). In our study we observed a direct correlation between keratin phosphorylation and sumoylation, but abrogating the ability of keratins to be phosphorylated on previously identified serine residues in the head and tail domains did not prevent sumoylation, with the exception of a moderate effect of the Ser-74 residue. Additionally, search of the keratin primary structure did not reveal any phosphorylation-dependent SUMO motifs in K8, K18, or K19. Therefore, the exact mechanism whereby keratin phosphorylation associates with sumoylation remains to be defined, and it may involve other hitherto uncharacterized phosphorylation sites, potentially ones located within the linker and coiled regions of the rod domains, which have been identified via large scale proteomic studies (52, 53). Other post-translational modifications, such as keratin glycosylation (54) may also play a role.
Studying the context-dependent mechanism of IF post-translational modifications is an effort likely to lead to the identification of potential novel pharmacologic strategies for the treatment of IF-related pathologies and one that would require decoding of complex post-translational modification cross-talk. By identifying keratins as targets for solubility-altering sumoylation during liver injury and cellular stress, our study aids in that effort while also providing new physiological contexts for sumoylation.
Supplementary Material
Acknowledgments
We thank Drs. Matthew Pratt-Hyatt for technical assistance with site-directed mutagenesis and Koksun Looi for help with the mouse MLR treatment.
This work was supported by National Institutes of Health Grant DK52951 (to M. B. O.) and by University of Michigan Postdoctoral Translational Scholars Program Award UL1RR024986 (to N. T. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
N. T. Snider and M. B. Omary, unpublished data.
- SUMO
- small ubiquitin-like modifier
- DDC
- 3,5-diethoxycarbonyl-1,4-dihydrocollidine
- HMW
- high molecular weight
- HSE
- high salt extract
- IF
- intermediate filament
- K
- keratin
- MLR
- microcystin LR
- OA
- okadaic acid.
REFERENCES
- 1. Sarge K. D., Park-Sarge O. K. (2009) Trends Biochem. Sci. 34, 200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bergink S., Jentsch S. (2009) Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
- 3. Mukherjee S., Thomas M., Dadgar N., Lieberman A. P., Iniguez-Lluhi J. A. (2009) J. Biol. Chem. 284, 21296–21306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 5. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., Lamond A. I. (2006) Mol. Cell Proteomics 5, 2298–2310 [DOI] [PubMed] [Google Scholar]
- 7. Mahajan R., Delphin C., Guan T., Gerace L., Melchior F. (1997) Cell 88, 97–107 [DOI] [PubMed] [Google Scholar]
- 8. Lapenta V., Chiurazzi P., van der Spek P., Pizzuti A., Hanaoka F., Brahe C. (1997) Genomics 40, 362–366 [DOI] [PubMed] [Google Scholar]
- 9. Saitoh H., Hinchey J. (2000) J. Biol. Chem. 275, 6252–6258 [DOI] [PubMed] [Google Scholar]
- 10. Kaminsky R., Denison C., Bening-Abu-Shach U., Chisholm A. D., Gygi S. P., Broday L. (2009) Dev. Cell 17, 724–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y. Q., Sarge K. D. (2008) J. Cell Biol. 182, 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Omary M. B. (2009) J. Clin. Invest. 119, 1756–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuchs E., Cleveland D. W. (1998) Science 279, 514–519 [DOI] [PubMed] [Google Scholar]
- 14. Eriksson J. E., Dechat T., Grin B., Helfand B., Mendez M., Pallari H. M., Goldman R. D. (2009) J. Clin. Invest. 119, 1763–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herrmann H., Strelkov S. V., Burkhard P., Aebi U. (2009) J. Clin. Invest. 119, 1772–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strnad P., Zhou Q., Hanada S., Lazzeroni L. C., Zhong B. H., So P., Davern T. J., Lee W. M., Omary M. B. (2010) Gastroenterology 139, 828–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ku N. O., Lim J. K., Krams S. M., Esquivel C. O., Keeffe E. B., Wright T. L., Parry D. A., Omary M. B. (2005) Gastroenterology 129, 885–893 [DOI] [PubMed] [Google Scholar]
- 18. Omary M. B., Ku N. O., Tao G. Z., Toivola D. M., Liao J. (2006) Trends Biochem. Sci. 31, 383–394 [DOI] [PubMed] [Google Scholar]
- 19. Ku N. O., Omary M. B. (2006) J. Cell Biol. 174, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S., Coulombe P. A. (2007) Genes Dev. 21, 1581–1597 [DOI] [PubMed] [Google Scholar]
- 21. Ku N. O., Liao J., Omary M. B. (1997) J. Biol. Chem. 272, 33197–33203 [DOI] [PubMed] [Google Scholar]
- 22. Caulín C., Salvesen G. S., Oshima R. G. (1997) J. Cell Biol. 138, 1379–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ku N. O., Omary M. B. (2001) J. Biol. Chem. 276, 26792–26798 [DOI] [PubMed] [Google Scholar]
- 24. Marceau N., Schutte B., Gilbert S., Loranger A., Henfling M. E., Broers J. L., Mathew J., Ramaekers F. C. (2007) Exp. Cell Res. 313, 2265–2281 [DOI] [PubMed] [Google Scholar]
- 25. Hanada S., Snider N. T., Brunt E. M., Hollenberg P. F., Omary M. B. (2010) Gastroenterology 138, 1607–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abe M., Oshima R. G. (1990) J. Cell Biol. 111, 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herrmann H., Kreplak L., Aebi U. (2004) Methods Cell Biol. 78, 3–24 [DOI] [PubMed] [Google Scholar]
- 28. Ku N. O., Michie S. A., Soetikno R. M., Resurreccion E. Z., Broome R. L., Omary M. B. (1998) J. Cell Biol. 143, 2023–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ku N. O., Toivola D. M., Zhou Q., Tao G. Z., Zhong B., Omary M. B. (2004) Methods Cell Biol. 78, 489–517 [DOI] [PubMed] [Google Scholar]
- 30. Ku N. O., Gish R., Wright T. L., Omary M. B. (2001) N. Engl. J. Med. 344, 1580–1587 [DOI] [PubMed] [Google Scholar]
- 31. Toivola D. M., Ku N. O., Resurreccion E. Z., Nelson D. R., Wright T. L., Omary M. B. (2004) Hepatology 40, 459–466 [DOI] [PubMed] [Google Scholar]
- 32. Omary M. B., Ku N. O., Liao J., Price D. (1998) Subcell. Biochem. 31, 105–140 [PubMed] [Google Scholar]
- 33. Zatloukal K., French S. W., Stumptner C., Strnad P., Harada M., Toivola D. M., Cadrin M., Omary M. B. (2007) Exp. Cell Res. 313, 2033–2049 [DOI] [PubMed] [Google Scholar]
- 34. Matunis M. J., Coutavas E., Blobel G. (1996) J. Cell Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toivola D. M., Strnad P., Habtezion A., Omary M. B. (2010) Trends Cell Biol. 20, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schimmel J., Larsen K. M., Matic I., van Hagen M., Cox J., Mann M., Andersen J. S., Vertegaal A. C. (2008) Mol. Cell Proteomics 7, 2107–2122 [DOI] [PubMed] [Google Scholar]
- 37. Geoffroy M. C., Hay R. T. (2009) Nat. Rev. Mol. Cell Biol. 10, 564–568 [DOI] [PubMed] [Google Scholar]
- 38. Omary M. B., Ku N. O., Strnad P., Hanada S. (2009) J. Clin. Invest. 119, 1794–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carberry K., Wiesenfahrt T., Windoffer R., Bossinger O., Leube R. E. (2009) Cell Motil. Cytoskeleton 66, 852–864 [DOI] [PubMed] [Google Scholar]
- 40. Yoon K. H., Yoon M., Moir R. D., Khuon S., Flitney F. W., Goldman R. D. (2001) J. Cell Biol. 153, 503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ku N. O., Michie S., Resurreccion E. Z., Broome R. L., Omary M. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4373–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Margolis S. S., Perry J. A., Forester C. M., Nutt L. K., Guo Y., Jardim M. J., Thomenius M. J., Freel C. D., Darbandi R., Ahn J. H., Arroyo J. D., Wang X. F., Shenolikar S., Nairn A. C., Dunphy W. G., Hahn W. C., Virshup D. M., Kornbluth S. (2006) Cell 127, 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim S., Wong P., Coulombe P. A. (2006) Nature 441, 362–365 [DOI] [PubMed] [Google Scholar]
- 44. Tatham M. H., Jaffray E., Vaughan O. A., Desterro J. M., Botting C. H., Naismith J. H., Hay R. T. (2001) J. Biol. Chem. 276, 35368–35374 [DOI] [PubMed] [Google Scholar]
- 45. Zhang X. D., Goeres J., Zhang H., Yen T. J., Porter A. C., Matunis M. J. (2008) Mol Cell 29, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mukhopadhyay D., Arnaoutov A., Dasso M. (2010) J. Cell Biol. 188, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang M., Hsu C. T., Ting C. Y., Liu L. F., Hwang J. (2006) J. Biol. Chem. 281, 8264–8274 [DOI] [PubMed] [Google Scholar]
- 48. Ulrich H. D. (2008) Mol Cell 32, 301–305 [DOI] [PubMed] [Google Scholar]
- 49. Lallemand-Breitenbach V., Zhu J., Puvion F., Koken M., Honoré N., Doubeikovsky A., Duprez E., Pandolfi P. P., Puvion E., Freemont P., de Thé H. (2001) J. Exp. Med. 193, 1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riley B. E., Zoghbi H. Y., Orr H. T. (2005) J. Biol. Chem. 280, 21942–21948 [DOI] [PubMed] [Google Scholar]
- 51. Dorval V., Fraser P. E. (2006) J. Biol. Chem. 281, 9919–9924 [DOI] [PubMed] [Google Scholar]
- 52. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Körner R., Greff Z., Kéri G., Stemmann O., Mann M. (2008) Mol. Cell 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 54. Ku N. O., Toivola D. M., Strnad P., Omary M. B. (2010) Nat. Cell Biol. 12, 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.