Abstract
Plasma triglyceride (TG) concentration is reemerging as an important cardiovascular disease risk factor. More complete understanding of the genes and variants that modulate plasma TG should enable development of markers for risk prediction, diagnosis, prognosis, and response to therapies and might help specify new directions for therapeutic interventions. Recent genome-wide association studies (GWAS) have identified both known and novel loci associated with plasma TG concentration. However, genetic variation at these loci explains only ∼10% of overall TG variation within the population. As the GWAS approach may be reaching its limit for discovering genetic determinants of TG, alternative genetic strategies, such as rare variant sequencing studies and evaluation of animal models, may provide complementary information to flesh out knowledge of clinically and biologically important pathways in TG metabolism. Herein, we review genes recently implicated in TG metabolism and describe how some of these genes likely modulate plasma TG concentration. We also discuss lessons regarding plasma TG metabolism learned from various genomic and genetic experimental approaches. Treatment of patients with moderate to severe hypertriglyceridemia with existing therapies is often challenging; thus, gene products and pathways found in recent genetic research studies provide hope for development of more effective clinical strategies.
Keywords: hypertriglyceridemia, hypotriglyceridemia, dyslipidemias, genetic variation, genomics, lipoproteins, resequencing
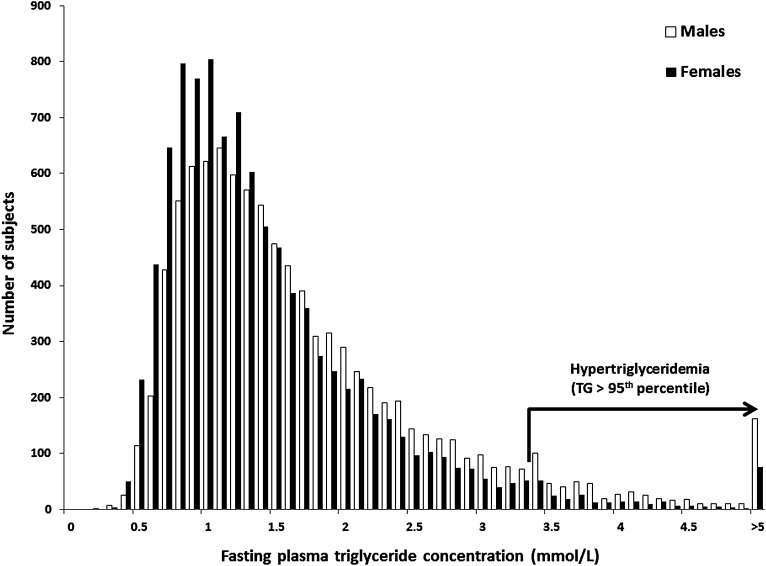
Plasma triglyceride (TG) concentration is a complex polygenic trait that follows a rightward-skewed distribution in the population (Fig. 1). As a clinical measurement, it integrates multiple TG-rich lipoprotein (TRL) species that circulate in plasma, predominantly intestinally synthesized chylomicrons (CMs) in the postprandial state and hepatically synthesized very low density lipoproteins (VLDL) in the fasted state. Epidemiological evidence indicates that plasma TG concentration is a strong independent risk factor for cardiovascular disease (CVD), suggesting that prolonged residence of plasma TRLs, especially in the postprandial state, may contribute to CVD susceptibility (1–7). Together with environmental influences, common and rare variants in multiple genes may collectively determine a patient's plasma TG concentration. Identifying genes and genetic variants associated with plasma TG concentration will enrich our understanding of biochemical pathways involved in TRL metabolism, enabling identification of subjects with increased susceptibility to disordered metabolism, and development of therapeutic interventions to improve plasma TG concentration and ameliorate CVD risk.
Fig. 1.
Frequency distribution of fasting plasma triglyceride (TG) concentrations. White bars represent male subjects, and black bars represent female subjects. Subjects with plasma TG concentration >3.37 mmol·l−1 are in the 95th percentile, considered the threshold for hypertriglyceridemia (HTG). To convert mmol·l−1 to mg·dl−1, multiply by 88.6. The maximum plasma TG concentration in this sample was 45 mmol·l−1. Data were obtained from the Canadian Heart Health Survey, a cross-sectional population-based study including >26,000 participants of multiple ancestries and ages from 18 to 74 years old, from metropolitan, urban, and rural areas of Canada (170).
Our understanding of the genetic architecture, that is, the integrated contribution of all genetic variants to interindividual variations of plasma TG concentrations, has increased substantially following reports of genome-wide association studies (GWAS) in the literature. Briefly, GWAS test for associations between common genetic variants with frequencies >1% (called single nucleotide polymorphisms [SNPs]) and either quantitative or discrete traits (8). Common variants used in GWAS are chosen to “tag” additional variants (tagSNP) by virtue of blocks of linkage disequilibrium (LD), which cover large regions of the genome and often contain multiple genomic elements including variants, genes, and regulatory elements. GWAS cannot impute causation to a particular gene or variant underlying an association, although they can implicate new genomic regions for further study. Herein, we define a GWAS locus as a genomic region marked by a common variant statistically associated with TG. Methodological features of GWAS will not be discussed here, as comprehensive reviews of study design, execution, and interpretation can be found elsewhere (9–11).
GWAS have identified signals associated with regions containing both classically established genes and previously unknown genomic regions as determinants of plasma TG concentration. However, complementary experimental approaches, including rare variant sequencing, study of animal models, linkage and family-based studies, and functional cellular and biochemical experiments, will be required to more fully elucidate the genetic architecture of plasma TG concentration. Herein, we review recent advances in genetics of plasma TG, summarizing recent GWAS findings supplemented with results from complementary lines of genetic investigation.
TRIGLYCERIDE-ASSOCIATED LOCI IDENTIFIED FROM GENOME-WIDE ASSOCIATION STUDIES
Genome-wide association studies of population-based TG concentration
Because fasting plasma TG is a stable biochemical analyte, GWAS can combine findings from multiple epidemiologic studies to identify associated genetic loci (12–18). These earlier findings have been confirmed and expanded by the recent definitive study from the Global Lipids Genetics Consortium (GLGC) (19), which reported a meta-analysis of >100,000 subjects encompassing multiple ethnic groups and multiple lipid and CVD phenotypes. The GLGC analysis has provided the most comprehensive list of TG-associated loci to date, identifying 32 loci harboring common variants that contribute to variations in plasma TG concentrations (Table 1). Many genes at GWAS-identified loci were familiar from previous studies of TG metabolism, increasing confidence in the potential biological relevance of novel loci (Table 1).
TABLE 1.
GLGC-identified loci harboring common genetic variants associated with fasting plasma TG concentration
| Locus | CHR | SNP | Associated traitsa | Risk allele | Allele frequency | TG effect (mg/dl) | P | eQTLb | New GWAS-identified locic |
|---|---|---|---|---|---|---|---|---|---|
| APOA5 | 11 | rs964184 | TC, LDL, HDL | G | 0.13 | 16.95 | 7.0 × 10−240 | N | N |
| GCKR | 2 | rs1260326 | TC | T | 0.41 | 8.76 | 6.0 × 10−133 | Y | Y |
| LPL | 8 | rs12678919 | HDL | A | 0.88 | 13.64 | 2.0 × 10−115 | N | N |
| MLXIPL | 7 | rs7811265 | HDL | A | 0.81 | 7.91 | 9.0 × 10−59 | Y | N |
| TRIB1 | 8 | rs2954029 | TC, LDL, HDL | A | 0.53 | 5.64 | 3.0 × 10−55 | N | Y |
| APOB | 2 | rs1042034 | TC, LDL, HDL | T | 0.78 | 5.99 | 1.0 × 10−45 | N | Y |
| ANGPTL3 | 1 | rs2131925 | TC, LDL | T | 0.68 | 4.94 | 9.0 × 10−43 | Y | N |
| APOE | 19 | rs439401 | TC, LDL, HDL | C | 0.64 | 5.50 | 1.0 × 10−30 | Y | N |
| CILP2 | 19 | rs10401969 | TC, LDL | T | 0.93 | 7.83 | 2.0 × 10−29 | N | Y |
| FADS1-2-3 | 11 | rs174546 | TC, LDL, HDL | T | 0.34 | 3.82 | 5.0 × 10−24 | Y | Y |
| PLTP | 20 | rs4810479 | HDL | T | 0.24 | 3.32 | 5.0 × 10−18 | N | Y |
| HLA | 6 | rs2247056 | TC, LDL | C | 0.75 | 2.99 | 2.0 × 10−15 | N | Y |
| NAT2 | 8 | rs1495743 | TC | G | 0.22 | 2.97 | 4.0 × 10−14 | N | Y |
| GALNT2 | 1 | rs1321257 | HDL | G | 0.39 | 2.76 | 2.0 × 10−14 | N | Y |
| LIPC | 15 | rs261342 | TC, LDL, HDL | G | 0.22 | 2.99 | 2.0 × 10−13 | N | N |
| CETP | 16 | rs7205804 | TC, LDL, HDL | G | 0.55 | 2.88 | 1.0 × 10−12 | N | Y |
| JMJD1C | 10 | rs10761731 | A | 0.57 | 2.38 | 3.0 × 10−12 | N | Y | |
| TIMD4 | 5 | rs1553318 | TC, LDL | C | 0.64 | 2.63 | 4.0 × 10−12 | N | Y |
| KLHL8 | 4 | rs442177 | T | 0.59 | 2.25 | 9.0 × 10−12 | N | Y | |
| FRMD5 | 15 | rs2929282 | T | 0.05 | 5.13 | 2.0 × 10−11 | Y | Y | |
| MAP3K1 | 5 | rs9686661 | T | 0.20 | 2.57 | 1.0 × 10−10 | N | Y | |
| COBLL1 | 2 | rs10195252 | HDL | T | 0.60 | 2.01 | 2.0 × 10−10 | Y | Y |
| LRP1 | 12 | rs11613352 | HDL | C | 0.77 | 2.70 | 4.0 × 10−10 | N | N |
| TYW1B | 7 | rs13238203 | C | 0.96 | 7.91 | 1.0 × 10−9 | N | Y | |
| PINX1 | 8 | rs11776767 | C | 0.37 | 2.01 | 1.0 × 10−8 | N | Y | |
| ZNF664 | 12 | rs12310367 | HDL | A | 0.66 | 2.42 | 1.0 × 10−8 | N | Y |
| CAPN3 | 15 | rs2412710 | A | 0.02 | 7.00 | 2.0 × 10−8 | N | Y | |
| CYP26A1 | 10 | rs2068888 | G | 0.54 | 2.28 | 2.0 × 10−8 | N | Y | |
| IRS1 | 2 | rs2943645 | HDL | T | 0.63 | 1.89 | 2.0 × 10−8 | Y | Y |
| CTF1 | 16 | rs11649653 | C | 0.60 | 2.13 | 3.0 × 10−8 | Y | Y | |
| MSL2L1 | 3 | rs645040 | T | 0.78 | 2.22 | 3.0 × 10−8 | N | Y | |
| PLA2G6 | 22 | rs5756931 | T | 0.60 | 1.54 | 4.0 × 10−8 | N | Y |
eQTL, expression quantitative trait locus; CHR, chromosome; GWAS, genome-wide association study; HDL, high-density lipoprotein cholesterol; LDL, low-density lipoprotein cholesterol; SNP, single nucleotide polymorphism; TC, total cholesterol; TG, triglyceride.
Other traits are associated with each locus, but not necessarily at the same SNP, at genome-wide significance levels.
eQTLs are SNPs that are associated with ≥1 transcript within 500-kb associated at P < 0.001 in liver and omental or subcutaneous fat.
“Y” indicates locus was first implicated in TG metabolism identified by GWAS; “N” indicates locus was identified by other means prior to GWAS.
The effect sizes of lead SNPs, i.e., those SNPs most strongly associated from a specific locus, in recently identified TG-associated GWAS-identified loci are small (Table 1), typically 1/5 to 1/10 the effect size per allele of loci identified earlier such as APOA5 or LPL. However, while small in effect, the associated loci are statistically robust and replicable, given the unprecedented statistical power derived from GLGC. The lead GWAS-discovered SNPs at each of 32 TG -associated loci cumulatively explained 9.6% of the total variation in plasma TG concentrations, corresponding to 25–30% of the total genetic contribution to TG variability (19). The 21 loci newly discovered by GLGC, most of which contain novel TG-associated genes, adds only ∼2.2% to the total variation that was accounted for in previous GWAS meta-analyses (17). The remaining sources of “missing variability” are the subject of intense speculation (20).
GLGC also provided in vitro and in vivo evidence supporting the involvement of specific genes in plasma TG metabolism (19). First, each locus was analyzed as a putative expression quantitative trait locus (eQTL) by testing for the association of common variants with mRNA transcript abundance in hepatic or adipose tissue of nearby genes. Common variants at 9/32 TG-associated loci were associated with transcript abundance at one or more genes (Table 1). Identification of an eQTL at a locus implicates promoter or regulatory elements in the modulation of plasma TG and provides strong evidence favoring involvement of genes whose expression is altered. Second, functional analysis of the HDL-cholesterol (HDL-C) and TG-associated GALNT2 locus in mouse models demonstrated that Galnt2 overexpression decreased HDL-C concentration, while RNA interference-mediated knockdown of Galnt2 increased HDL-C concentration. These results provide strong evidence for a causal association between GALNT2 and plasma lipids.
Pleiotropy or simply correlation.
One lesson learned from GLGC GWAS was that pleiotropy, referring to the involvement of a single gene or genetic variant in multiple biological phenotypes, is a relatively common feature of the genetic determinants of plasma lipids. For instance, 21 of 32 TG-associated loci were associated with at least one additional lipid trait at the genome-wide significance threshold for association, according to a P-value < 5 × 10−8 (Table 1): e.g., 6 loci were associated with total or LDL-cholesterol (LDL-C) only; 8 loci were associated with HDL-C only; and 7 loci were associated with total cholesterol, LDL-C, and HDL-C; while 11 loci had specific associations with TG only. Astonishingly, virtually every TG-associated locus is associated with another lipid phenotype, when using a looser significance threshold of P-value < 0.05. However, some of these findings may represent false-positive associations rather than true biological effects. Only GCKR was exclusively associated with plasma TG, although this locus is pleiotropic for nonlipid traits involved in carbohydrate metabolism.
The complexity of lipoprotein metabolism means that genes and biochemical pathways can be involved in metabolism of several lipoprotein classes. Thus, it is not surprising that 15/32 TG-associated loci in GLGC studies are also associated with HDL-C at genome-wide significance levels (Table 2) , without further adjustment for related lipid phenotypes (19). Among jointly associated loci, one-third were associated with TG and HDL-C with similar strengths of association. Most loci, however, seemed preferentially associated with one phenotype, notably, APOA5 and MLXIPL with TG, and CETP and LIPC with HDL-C, with P values differing by 233, 49, 368, and 83 orders of magnitude, respectively. These findings cannot distinguish between a mechanistic model in which genomic variations primarily modulate one lipoprotein variable, with subsequent secondary effects on the others, or in which a common single mechanism primarily affects multiple lipoprotein species concurrently. Postgenomic biochemical interactions among circulating TRLs and HDL may ultimately underlie such associations. Interestingly, the directions of genetic associations with TG and HDL-C were opposite at most loci, except at LIPC and APOE. Furthermore, common variants at LIPC and APOE are also eQTLs, suggesting that single regulatory element loci at these loci may control multiple gene products.
TABLE 2.
GLGC meta-analysis of genetic loci associated with concentrations of both plasma TG and HDL-C
| Locus | Lead traita | Role in lipoprotein metabolism | TG P | HDL P | SNPb |
|---|---|---|---|---|---|
| APOA5 | TG | Activator of lipoprotein lipase | 7 × 10−240 | 5 × 10−47 | rs964184 |
| LPL | TG | Hydrolysis of TG-rich lipoproteins | 2 × 10−115 | 9 × 10−98 | rs12678919 |
| MLXIPL | TG | Activation of glycolytic and lipogenic enzymes | 9 × 10−59 | 1 × 10−9 | TG rs7811265HDL rs17145738 |
| TRIB1 | TG | Unknown | 3 × 10−55 | 6 × 10−19 | TG rs2954029HDL rs10808546 |
| APOB | TG | Backbone of atherogenic lipoproteins | 1 × 10−45 | 1 × 10−30 | rs1042037 |
| APOE | TG | TG-rich lipoprotein receptor ligand for the LDLR and LDLR-related protein (LRP1) | 1 × 10−30 | 4 × 10−21 | TG rs439401HDL rs4420638 |
| FADS | TG | Fatty acid desaturation | 5 × 10−24 | 2 × 10−22 | TG rs174546HDL rs174601 |
| PLTP | HDL-C | Transfer phospholipid between TG-rich lipoprotein and HDL | 5 × 10−18 | 2 × 10−22 | HDL rs6065906TG rs4810479 |
| GALNT2 | HDL-C | O-linked glycosylation of proteins | 2 × 10−14 | 4 × 10−21 | HDL rs4846914TG rs1321257 |
| LIPC | HDL-C | TG lipase | 2 × 10−13 | 3 × 10−96 | HDL rs1532085TG rs261342 |
| CETP | HDL-C | Exchanges cholesteryl ester for TG between HDL and TG-rich lipoproteins | 1 × 10−12 | 7 × 10−380 | HDL rs3764261TG rs7205804 |
| COBLL1 | TG | Unknown | 2 × 10−10 | 3 × 10−10 | TG rs10195252HDL rs12382675 |
| LRP1 | TG | TG-rich lipoprotein receptor via apoE | 4 × 10−10 | 2 × 10−8 | TG rs11613352HDL rs3741414 |
| IRS1 | HDL-C | Involved in insulin signaling | 2 × 10−8 | 2 × 10−9 | HDL rs2972146TG rs2943645 |
| ZNF664 | HDL-C | Unknown | 1 × 10−8 | 3 × 10−10 | HDL rs4765127TG rs12310367 |
HDL-C, high-density lipoprotein cholesterol; LDLR, low-density lipoprotein receptor; SNP, single nucleotide polymorphism; TG, triglyceride.
“Lead trait” is the strongest associated trait.
SNP column is the strongest trait-associated SNP for TG and HDL-C; a single SNP is the strongest associated variant for both traits.
Given the limitations of the GWAS approach, the mechanism(s) that underlies joint associations between TG and HDL-C cannot be simply explained. Furthermore, the strength and effective size of significantly associated loci only minimally reflect the potential biologic importance of the true causal gene; they only suggest that genetic variation in a genomic region appears to modulate the trait of interest. Therefore, loci jointly associated with >1 trait may prove to have biological effects that are quite different from what has been suggested by GWAS P values. Ultimately, biochemical experiments are needed to explain the (dys)function underlying GWAS-discovered signals.
Studies in multiethnic populations.
A general limitation of GWAS is that they have been conducted predominantly with European subjects to date, although the extended patterns of LD in European populations have facilitated GWAS identification of TG-associated loci. A few replication studies have confirmed some TG associations in small multiethnic cohorts (21–23), although this has begun to change with GLGC studies in which 22/32 TG associations were replicated based on concordant direction of effect in European (n = 7,000), East Asian (n = 15,000), South Asian (n = 9,700), and African (n = 8,000) populations (19). However, substantial GWAS have still not been conducted with multiethnic populations. Differences in allele frequencies and more complex patterns of LD among populations of varying ancestry could also affect, either positively or negatively, the statistical power necessary to discover novel TG-associated loci (24), providing additional opportunities to identify loci and variants specific to ethnic groups.
The focus on European subjects also fails to identify functional variants that are restricted to populations of non-European ancestry. For example, the APOA5 variant causing a glycine-to-cysteine substitution at position 185 (G185C), which causes a 23% reduction in LPL activity, is found in nearly 27% of Taiwanese subjects with high TG compared with only 4.2% of controls (25). This important variant is completely absent from European subjects (26), illustrating how some common or rare private variants are not identifiable in studies restricted to Europeans. Thus, GWAS and resequencing studies of multiethnic populations will help to develop a more complete understanding of the genetic determinants of plasma TG.
Genome-wide association study of hypertriglyceridemia
Do the same genes and variants that modulate plasma TG concentration in GWAS performed in the general population also contribute to pathological TG levels in patients ascertained through specialty clinics? Hypertriglyceridemia (HTG) is defined as a fasting plasma TG concentration >95th percentile (Fig. 1). Clinical and biochemical features in HTG subjects include eruptive, tuberous or palmar crease xanthomas, lipemia retinalis, hepatosplenomegaly, and pancreatitis (27, 28). HTG is thought to result from the accumulation of genetic determinants of plasma TG associated with defective TRL lipolysis (29) combined with environmental and metabolic stressors such as alcohol consumption, medications, renal disease, nonalcoholic fatty-liver disease, pregnancy, obesity, metabolic syndrome, and type 2 diabetes (T2D) (27), which culminate in a clinical phenotype. Together, HTG and accompanying comorbidities also significantly increase CVD risk (30). However, while both common and rare genetic determinants of HTG have been identified, the majority of genetic susceptibility in people with an HTG diagnosis remains unattributed (31). The study of HTG patients could improve understanding of the mechanisms by which genetic burden contributes to elevated TG.
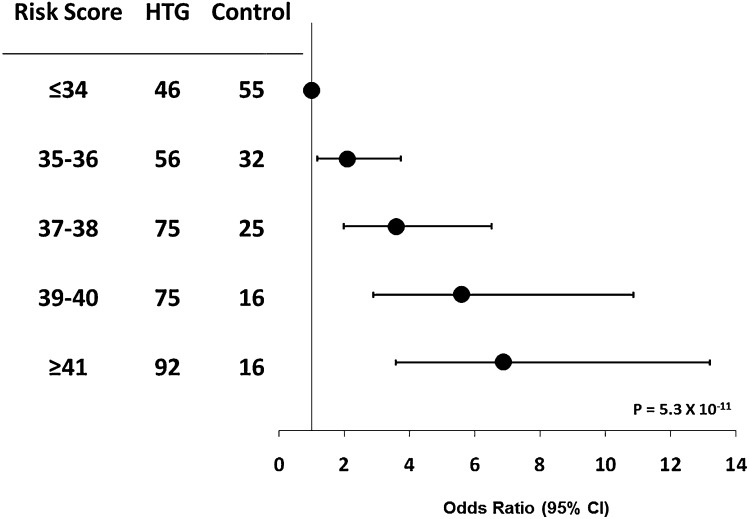
Only one GWAS study evaluating subjects at the extremes of lipid phenotypes has been conducted to date. Using a case-control design comparing ∼500 HTG patients with ∼1,200 normotriglyceridemic population-based controls (32), we showed genome-wide significant associations among the APOA5, GCKR, LPL, and APOB loci, and replicated the MLXIPL, TRIB1, ANGPTL3, and NCAN loci, with trends toward significance as determined by P values near significance at the FADS1-FADS2-FADS3 (FADS1-2-3) cluster, XKR6/PINX1, and PLTP. Thus, loci that determine plasma TG concentration in the general population also underlie HTG susceptibility in patients. Furthermore, we demonstrated that 29/32 TG-raising alleles associated with TG in the GLGC analyses were overrepresented in HTG patients compared with controls, leading to an increased genetic burden of risk (Fig. 2) (19). However, replication of all 32 newly identified TG-associated loci will be required in a larger HTG cohort, because larger sample sizes will more likely detect variants with clinically relevant effects. Although our study identified and replicated several TG-associated loci in HTG pathophysiology, it was not exhaustive and does not exclude additional loci that potentially modulate plasma TG concentration or determine HTG risk. A complete list of HTG-associated loci is shown in Table 3.
Fig. 2.
An increased genetic burden of triglyceride (TG)-raising alleles is characteristic of hypertriglyceridemia (HTG). Unweighted risk scores were constructed from the sum of TG-raising alleles at 32 TG-associated loci (19), generating a minimum possible score of 0 and a maximum possible score of 64; actual risk scores ranged between 26 and 47 alleles in HTG patients and 27 and 45 alleles in controls. Risk scores were compared among risk score bins in 344 HTG patients and 144 controls by using Fisher's exact test; the P value is derived from the Cochrane-Armitage test for trend. These results were generated based on the HTG subject cohort reported in the GLGC (19) but without adjustment for population-based effect estimates or clinical covariates. CI, confidence interval.
TABLE 3.
TG-associated genes identified by various methodologies involved in HTG
| Locus | Identified by GWAS of HTG | Replication of population-based TG-associated loci | HTG-causing rare variants | Mouse models of HTG | Family based and linkage studies |
|---|---|---|---|---|---|
| APOA5 | X | X | X | ||
| GCKR | X | X | |||
| LPL | X | X | X | ||
| APOB | X | X | |||
| APOE | X | X | X | ||
| ANGPTL3 | X | ||||
| MLXIPL | X | ||||
| TRIB1 | X | ||||
| NCAN | X | ||||
| GALNT2 | X | ||||
| LIPC | X | ||||
| APOC2 | X | X | |||
| GPIHBP1 | X | X | |||
| LMF1 | X | X | |||
| USF1 | X |
GWAS, genome-wide association study; HTG, hypertriglyceridemia; TG, triglyceride; X, the locus was identified.
We next searched for rare variations in genes under GWAS peaks by resequencing the protein coding exons of APOA5, GCKR, LPL, and APOB (32). We demonstrated a significant accumulation of rare variants in HTG patients, defined by a minor allele frequency of <1% in controls. Several variants found in HTG patients were either truncation mutations or were predicted to have deleterious effects in silico or were previously demonstrated in vitro to be dysfunctional, particularly in LPL, a long-established functional candidate for TG metabolism. In aggregate, 154 rare missense and nonsense variants were seen in 28.1% of 438 HTG patients compared with 53 rare variants found in 15.3% of 327 controls. A more stringent analysis restricted to variants found exclusively in HTG patients or controls confirmed the mutation skew: 47 rare variants were found in 10.3% of HTG patients, while only 9 rare variants were found in 2.8% of controls. Carrying rare variants thus decreased the unattributed variation in the diagnosis of HTG. Further studies, especially functional characterization, will help refine the estimate of variation attributable to rare variants in HTG patients.
An emerging mosaic genetic model of hypertriglyceridemia.
The overall HTG phenotype is heterogeneous due to distinct differences in populations of TRLs (33), classically defined using the Fredrickson HLP phenotype system based on lipoprotein fractions quantified by ultracentrifugation and electrophoresis (28). However, genetic studies suggest that HLP phenotypes are essentially indistinguishable at the level of common SNP genotypes (34). We suggest that genetic susceptibility requires a minimal burden (a “quorum”) of common HTG risk alleles, with rare loss-of-function variants that also predispose to HTG (Table 3). This minimal number of common variants is associated with susceptibility to develop moderate HTG (HLP type 4). The “stacking” of additional HTG risk alleles on top of the quorum leads to severe HTG (HLP type 5). The presence of the APOE ϵ2/ϵ2 genotype in the background of the quorum of HTG risk alleles is associated with dysbetalipoproteinemia (HLP type 3). The presence of LDL-associated alleles drives the phenotype toward HLP type 2B. Thus, HLP phenotypes are similar at the level of HTG risk alleles, and differences between them may arise from a variable combination of genetic determinants and presence of environmental factors such as nutritional stress, obesity, hormonal disturbances, or additional metabolic stress such as diabetes. So far, these findings have not been replicated, and future studies will require collaborations in order to generate HTG sample sizes sufficient to determine the validity of this hypothesis.
Clinical hypotriglyceridemia.
Studies of patients with very low plasma TG concentrations, severe hypotriglyceridemia, using GWAS criteria are less straightforward because such individuals are usually healthy, and their conditions are not ascertained through medical clinics. Monogenic autosomal recessive disorders characterized by very low plasma TG concentration include abetalipoproteinemia (ABL), homozygous hypobetalipoproteinemia (HHBL), and familial combined hypolipidemia (FCH) (35) caused by rare homozygous loss-of-function mutations in MTP, APOB and ANGPTL3, respectively. Patients with ABL and HHBL have virtually complete absence of apolipoprotein B (apoB)-containing lipoproteins, including both TRLs and LDL, so it is difficult to attribute the clinical subphenotypes to low TG alone.
ApoB is the backbone of atherogenic lipoproteins. Two different apoB isoforms, namely apoB-48 and apoB-100, are synthesized by enterocytes and hepatocytes, respectively, and result from transcription of a message that has been edited by the product of the APOBEC1 gene (36). TRL synthesis is initiated by cotranslation of apoB concurrent with the lipid loading action of the endoplasmic reticulum chaperone microsomal TG transfer protein (MTP) (37), producing CMs from dietary lipids and VLDL from endogenously synthesized TG. Secreted TRLs reach systemic circulation where they are acted upon by lipoprotein lipase (LPL) to distribute fatty acids to metabolically active tissues.
The multisystem clinical phenotypes for ABL and HHBL include retinopathy, disturbed bone metabolism, neuropathy, myopathy, and coagulopathy and stem from deficiency in fat-soluble vitamins. Atherosclerosis risk in ABL and HHBL patients is low, probably due to reduced LDL-C rather than to reduced plasma TG levels. Additional features of ABL and HHBL, and also heterozygous hypobetalipoproteinemia, is hepatosteatosis, suggesting that the pathogenic accumulation of TG within hepatocytes is related to a deficiency of circulating TRLs. The biochemical phenotype of FCH resembles that of HHBL (35), although mutations causing FCH likely do not interfere with TRL assembly and export, because hepatosteatosis and other clinical phenotypes characteristic of ABL and HHBL are not observed.
New treatments directed against these molecular targets, for example, pharmacological inhibition of MTP in the case of lomitapide (38) and RNA interference of APOB in the case of mipomersen (39), should lower both LDL-C and TG levels, with a potential increased risk of hepatosteatosis. Molecular inhibition of ANGPTL3 would be predicted to reduce circulating lipoproteins, potentially without off-target effects including hepatosteatosis, given the absence of additional pathologies in FCH patients with loss-of-function mutations in ANGPTL3 (35).
Functional variants in classical triglyceride-associated genes
Lipoprotein lipase.
The gene encoding LPL is located on chromosome 8, and its product has long been recognized as the fulcrum for hydrolysis of plasma TRLs (40). LPL is highly synthesized in tissues requiring free fatty acids for energy metabolism, predominantly heart, adipose tissue, and skeletal muscle (41). It is secreted into the vasculature supplying these tissues, where it binds to glycosylphosphatidylinositol-anchored HDL-binding protein 1 (GPIHBP1) on endothelial cell surfaces (42). LPL hydrolyzes TRLs to release free fatty acids, which may be used for TG resynthesis in adipose tissue or β-oxidation in muscle. LPL is under strong dietary and hormonal regulation (43), and its activity depends upon additional cofactors, most notably apoC-II which is absolutely required for hydrolysis (44). Murine models lacking Lpl have markedly increased plasma TG concentration, with VLDL accumulation after birth and chylomicronemia after suckling (45). TRL accumulation is lethal within 18 h of onset of feeding. Conversely, overexpression of Lpl in mice confers a protective phenotype, improving clearance of both CM and VLDL and reducing plasma TG concentration nearly 75% (46).
The lead TG-associated LPL variant (rs12678919) is an intergenic SNP located downstream of LPL, the only gene within a large block of LD spanning the GWAS-described locus. The minor allele frequencies range from 9% to 14%, lowest in African and highest in European and Japanese populations. In population-based GWAS (19), this variant increases plasma TG concentration by 0.16 mmol·l−1 (13.95 mg·dl−1). A second HTG-associated LPL variant (rs7016880) downstream of LPL is strongly associated with the population-based associated SNP (r2 = 0.86) conferring nearly 3-fold increase in risk of HTG (32). Some tagSNPs associated with the LPL locus have further been associated with plasma TG in multiple ethnicities, with allele frequencies differing by up to 10% in various populations (21, 22). The association of lead LPL-associated variants with plasma TG concentration indicates that common variants at this locus affect plasma TG; however, the precise functional variants have not been identified.
Several LPL variants have systematically been identified as determinants of plasma TG concentration and CVD risk (47). Notably, functional coding sequence changes include aspartic acid-to-asparagine substitution at position 9 (D9N), which decreases both LPL activity and mass, resulting in a 25% increase in plasma TG concentration among carriers (48); the asparagine-to-serine substitution at position 291 (N291S), which decreases LPL activity and promotes monomerization (49, 50); and the premature stop codon in place of serine at position 447 (S447X) (51), which increases LPL activity nearly 2-fold and improves TRL clearance (52). The LPL D9N variant is significantly overrepresented in patients with HTG, while the N291S variant is not (53). Conversely, the LPL S447X variant has been repeatedly associated with lower TG and higher HDL-C concentrations (54) and is less prevalent in HTG patients (34, 55). Interestingly, the S447X variant is now the basis of a gene therapy strategy, alipogene tiparvovec, for treatment of severe chylomicronemia (56).
Rare loss-of-function LPL variants are also associated with HTG. Both homozygous and compound heterozygous loss-of-function LPL mutations cause childhood-onset HTG, with prevalence of ∼1 case in 1,000,000 population (27, 28). Numerous functional variants reduce LPL activity when in the homozygous state, preventing hydrolysis and resulting in TRL accumulation (57–61). Interestingly, homozygous APOC2 mutations also cause HTG (31, 62). The APOC2 mutations were first definitively shown to cause HTG after they were characterized at the amino acid level (40). Rare heterozygous LPL mutations also accumulate in patients with late-onset HTG (32) and were observed in ∼6.4% of these patients (53). Both common and rare LPL variants are thus strongly associated with both population-based plasma TG concentration and diagnosis of HTG.
Apolipoprotein A-V.
The APOA5 gene was bioinformatically identified within the APOA5-APOA4-APOC3-APOA1 gene cluster on chromosome 11 (63, 64). It encodes the liver-expressed apoA-V, which enhances LPL activity (65–67). About 80% of apoA-V associates with CMs, VLDL, and HDL at very low concentrations (68), while ∼20% is retained in an intrahepatic pool associated with lipid droplets (69). Plasma apoA-V likely affects the distribution of apoC-III molecules on VLDL and thus promotes lipolysis (70); however, the role of intracellular apoA-V is less clear. In mice, overexpression of the human APOA5 gene markedly decreases plasma TG concentration, whereas mice lacking the Apoa5 gene become severely hypertriglyceridemic (63, 71). APOA5 is regulated by several transcription factors that are involved in plasma lipoprotein and glucose homeostasis, suggesting it responds to multiple environmental cues (72); however, such responses may differ between mice and humans. Peroxisome proliferator-activator receptor α (PPARα) agonists, such as fibrates, that induce APOA5 expression in humans do not equivalently induce expression of Apoa5 or transgenic human APOA5 in mice, the result of a nonfunctional PPARα response element in the mouse Apoa5 promoter sequence (73). Regardless, the APOA5 gene is clearly a crucial determinant of TRL metabolism in both mice and humans, although its regulation and molecular function are incompletely understood.
The lead TG-associated APOA5 variant (rs964184) is an intergenic SNP located downstream of the gene cluster containing APOA5. Its minor allele frequency ranges from 12% to 33%, lowest in European and highest in Japanese individuals. In GWAS of population-based samples (19), each APOA5 risk allele increases plasma TG by 0.19 mmol·l−1 (16.95 mg/dl). Interestingly, the same variant was also associated with HTG: the minor allele frequency was 33% in HTG patients of European descent compared with 14% in controls, increasing the HTG odds ratio by 3.3-fold (32). The APOA5 variants are also associated with plasma TG in multiple ethnicities (21, 22). As with LPL, the precise functional variant(s) that leads to the GWAS-determined association by virtue of LD, and the nature of their dysfunction, are not known.
Several common functional APOA5 variants have been identified (74). However, because these variants are largely not genotyped by the International HapMap Consortium, it is impossible to determine whether they underlie the associations with the APOA5 lead SNP (rs964184). For instance, a coding sequence variant causing a serine-to-tryptophan substitution at amino acid residue 19 (S19W) in the signal peptide of apoA-V (75) impairs protein translocation and secretion (76). Several promoter variants have also been identified (63, 75), including −3A>G in the APOA5 Kozak sequence and −1131T>C. Functional analyses in vitro suggest that it is primarily the -3A>G variant that decreases APOA5 expression, whereas –1131T>C has minimal effect on expression (76, 77). Interestingly, the −1131T>C variant is located on a haplotype distinct from S19W and is in strong LD with functional APOC3 variants (75, 78), suggesting that could underlie the APOC3 association with plasma TG (78, 79). Both the S19W and the −1131T>C variants are also strong predictors of HTG (34, 55, 80) and increased CVD risk (81, 82). The APOA5 S19W carriers are >6 times more likely to be diagnosed with severe HTG than controls (80). Rare loss-of-function APOA5 variants are also associated with HTG (32, 53, 83). Functional analysis of such variants has generally revealed loss of LPL activity, particularly with C-terminal truncation mutations that interfere with apoA-V ability to interact with lipid and lipoproteins (61). Finally, 41.8% of patients with severe HTG were carriers for either the APOA5 S19W mutation or at least one of the above-mentioned rare APOA5, LPL, or APOC2 variants compared with only 8.9% of controls who carried these variants (53).
Apolipoprotein C-III.
The APOC3 gene is also part of the APOA5-APOA4-APOC3-APOA1 gene cluster on chromosome 11. ApoC-III inhibits LPL-mediated TG hydrolysis by opposing the effect of apoC-II, and the apoC-II/apoC-III ratio determines net activation or inhibition of LPL (84, 85). Intracellular apoC-III also promotes assembly and secretion of VLDL particles (86). Both putative functions of apoC-III are consistent with in vivo studies showing HTG in mice overexpressing APOC3 (87) and hypotriglyceridemia and enhanced postprandial TG clearance in APOC3-deficient mice (88). In humans, linkage studies have implicated APOC3 as a candidate gene for familial combined hyperlipidemia (FCHL); however, specific common variants have not been associated with disease (89).
APOC3 is not strictly speaking a GWAS-identified locus. The APOA5-APOA4-APOC3-APOA1 locus produces the strongest association signal in GWAS of plasma TG concentration, although there is ambiguity regarding those causal genes and variants that underlie this signal. Strong yet complex patterns of LD encompass APOA5 and APOC3; therefore, common functional APOC3 variants may also contribute to the GWAS-identified signal (75, 78). Regardless, APOC3 is a crucial determinant of plasma TG concentration. Two functional variants, namely -482C>T and -455T>C, in the APOC3 regulatory sequences have been shown to attenuate APOC3 insulin responsiveness in vitro (90) and to increase fasting plasma TG concentration by ∼2-fold in Asian Indian men (91). These APOC3 variants are further associated with increased risk of metabolic syndrome in multiethnic studies (92, 93).
Because APOC3 inhibits TRL lipolysis, rare APOC3 variants increase lipolysis and reduce plasma TG. For example, GWAS of fasting and postprandial TG in 806 Old Order Amish subjects implicated APOC3, and resequencing identified a heterozygous nonsense mutation at residue 19 (R19X) in 2.8% of subjects; plasma apoC-III concentrations were reduced 50% compared with that in noncarriers (94). This variant was associated with a favorable plasma lipoprotein profile: decreased plasma TG and LDL and increased plasma HDL-C. Subclinical atherosclerosis, as measured by electron beam computed tomography, was also significantly reduced in carriers. Another rare APOC3 variant, the alanine-to-threonine missense mutation at residue 23 (A23T), was identified in three Yucatan Indian subjects with apoC-III deficiency (95). Functional characterization of A23T in vitro demonstrated attenuated VLDL assembly and secretion from hepatocytes, with TG accumulation within the microsomal lumen that was independent of MTP activity (96). Perhaps pharmacologic inhibition of APOC3 could improve elevated plasma TG phenotypes, but additional mechanistic evidence is required to definitively establish the benefit of inhibiting APOC3 expression or function, given the complexity of phenotypes and mechanisms of the genes and gene products at this locus.
Angiopoietin-like 3 protein.
Angiopoietin-like 3 protein (ANGPTL3) was initially identified as a regulator of lipid metabolism in mice (97, 98). A mutant hypolipidemia phenotype was mapped to this locus, revealing that disruption of Angptl3 decreased plasma TG concentrations in addition to improving other lipid parameters including total cholesterol and nonesterified fatty acids, while ANGPTL3 overexpression significantly worsened the lipoprotein profile (98). ANGPTL3 appears to reversibly inhibit LPL catalytic activity (99, 100), which is inhibited in turn by LPL-stabilizing proteins such as GPIHBP1 (101). ANGPTL3 is abundant in plasma (102), which may reflect its low affinity and reversible LPL binding, and larger concentrations may be required to effectively inhibit LPL activity (100). ANGPTL3 expression is regulated by the promoter liver X receptor (LXR) response element, increasing plasma concentration in response to LXR agonists (103).
The lead SNP identifying ANGTPL3 as the TG-associated locus (rs2131925) is an intronic SNP in the DOCK7 gene, corresponding to a location upstream of ANGPTL3. The frequency of this allele varies among different populations, ranging from 88% in Japanese to 25% in African individuals; thus, in addition to variable LD patterns, variable allele frequencies might explain nonreplication of ANGPTL3 associations in multiethnic studies (21, 22). Interestingly, the ANGPTL3 GWAS-described signal is an eQTL for ANGPTL3 expression (17, 19), and the risk allele associated with increased expression is also associated with increased plasma TG concentration. This suggests that common ANGPTL3 variants likely lie within regulatory elements; whether they affect multiple genes in TRL metabolism remains to be determined.
Population-based resequencing in the Dallas Heart Study (n = 3,551 subjects) has revealed several heterozygous nonsynonymous sequence variants that compromised protein synthesis or secretion of ANGPTL3, in turn decreasing plasma TG concentration (104). In ANGPTL3, 15 nonsynonymous variants were identified in subjects at the lower 25th percentile of population-based TG concentration versus 5 variants identified in subjects at the upper 25th percentile (104). Furthermore, ANGPTL3 belongs to a family of seven angiopoietin-like proteins involved in angiogenesis and lipoprotein metabolism, including ANGTPL4, which also carries nonsynonymous sequence variants with synthesis or secretion defects that modulate plasma concentrations of TG and HDL (104, 105). Similar extreme sampling within this cohort revealed 13 nonsynonymous variants in subjects with low TG versus 2 variants in subjects with higher TG concentrations (105). Such variants likely extend to subjects with very low circulating plasma TG in other populations, such as in subjects with FCH (35).
Novel TG-associated loci
Identification and replication of established genes in lipoprotein metabolism, serving as “positive controls” for the GWAS approach, have supported GWAS methodology as the tools with which to identify novel TG-associated loci of likely biological relevance. However, unlike classically established loci, the precise genes and variants responsible for novel GWAS peaks are unclear. The genes described below are likely biological candidates underlying their respective TG-associated signals, either because of prior implication in lipoprotein metabolism or because they are the sole gene at the GWAS-identified locus. Future studies using larger samples, fine mapping, deep resequencing, and bioinformatic analyses will be required to elucidate the function of novel genes in TG metabolism.
Glucokinase regulatory protein.
The GCKR locus, encoding the glucokinase (GCK) regulatory protein (GCKR), is the strongest completely novel locus implicated in TG metabolism (106). It has been robustly replicated in GWAS of plasma TG concentration (19) and HTG (32) and implicated in HTG by an excess of rare variants in patients (32). GCKR is expressed in the liver and acts as an allosteric regulator of GCK, allowing it to be rapidly mobilized in response to increased cellular glucose concentrations (107). A functional GCKR variant, a proline-to-leucine substitution at position 446 (P446L) that is commonly genotyped using high-density microarrays, may be the basis of this association (108). Studies in vitro suggest that allosteric regulation of GCK is attenuated by this GCKR variant, disrupting its sensitivity to physiological concentrations of fructose-6-phosphate (109). It mediates a dampened response to GCK inhibition in response to endogenous agonists, indirectly promoting GCK activity (109), which might increase glycolytic flux, increasing cellular glucose uptake, de novo TG synthesis and inhibition fatty acid oxidation.
Interestingly, GCKR is also associated with diverse traits that could modulate CVD risk and link carbohydrate with lipoprotein metabolism. The same risk allele for increased plasma TG concentration is associated with improved glycemia indices, including reduced plasma glucose, improved insulin sensitivity, and decreased T2D risk (108, 110–113). The functional GCKR variant is associated with other phenotypes, such as plasma C-reactive protein (CRP) and serum uric acid (108, 114). How the functional GCKR variant mediates these other interactions is unknown.
Carbohydrate response element binding protein.
The MLXIPL locus encodes the carbohydrate response element (CRE) binding protein (CHREBP). The lead TG-associated SNP (rs7811265) is located downstream of MLXIPL, in a block of LD encompassing several genes; however, MLXIPL is a strong candidate gene. The lead SNP is associated with MLXIPL transcript abundance in subcutaneous fat, although with an atypical U-shaped direction of effect across genotypes, and will require replication and functional evaluation (19). In any event, the minor allele is robustly associated with lower TG concentration (19) and protection from HTG (32). CHREBP is a basic helix-loop-helix transcription factor expressed in lipogenic tissues, responsible for the coordinate activation of glycolytic enzymes such as GCK and lipogenic enzymes such as fatty acid synthase, required to convert dietary carbohydrate to TG (115).
CHREBP activates target genes in TG synthesis by binding to a CRE in the promoter (116, 117). The phenotype of Mlxipl knockout mice is consistent with the protective effect of the minor allele, as seen in humans, showing delaying activation of CHREBP and dampened cellular glycolytic and lipogenic programs (118). These mice have decreased hepatic TG content, decreased plasma free fatty acids, and suppression of both glycolysis and lipogenesis. The association between CHREBP and TRL metabolism should invigorate interest in metabolic pathways downstream from MLXIPL, as responsive genes may suggest potential targets for intervention to reduce plasma TG concentration.
Drosophila Tribbles homolog 1.
The lead TG-associated SNP (rs2954029) is located downstream of the gene, in a block of LD spanning only the TRIB1 locus, suggesting it is probably the gene underlying the GWAS association signal. TRIB1 is expressed in most tissues but is most abundant in skeletal muscle, thyroid, pancreas, blood leukocytes, and bone marrow (119). TRIB1 modulates mitogen-activated protein kinase activity in vascular smooth muscle cell proliferation and also chemotaxis (120). Trib1 knockout mice have impaired macrophage activation of signal transduction pathways involved in toll-like receptor activation in response to lipopolysaccharides, but lipoprotein phenotypes were not investigated in model systems (121).
Apolipoprotein B.
The APOB gene is another familiar player in lipoprotein metabolism. Prior to GWAS, variation in APOB was most often associated with LDL-C (122). Only recently has APOB been implicated as a TG-associated locus. GWAS have identified two signals at APOB: 1) a missense variant (rs1042034) causing a serine-to-isoleucine substitution at position 4338 (S4338I) was associated with population-based TG levels (19); and 2) a variant ∼120 kb upstream of APOB (rs4635554) was associated with increased HTG risk (32). Neither variant has been functionally implicated in the modulation of plasma TG; however, we anticipate that they are merely tagSNPs with sufficient power to detect associations at this locus. Additional direct evidence is provided by the observation that rare variants in APOB accumulate in HTG patients (32), suggesting that by an unknown mechanism, mutations are capable of increasing plasma TG concentration in addition to causing disorders such as familial defective apoB-100, where LDL-C is increased but TRLs are typically not affected (123), and hypobetalipoproteinemia, where TRLs and LDL-C are decreased (124).
Phospholipid transfer protein.
The phospholipid transfer protein (PLTP) locus is considered to be involved primarily in HDL metabolism but is similarly associated with plasma TG concentration (Table 2). The lead SNP in PLTP (rs4810479) is located upstream of the gene; however, it was not among eQTLs identified by the GLGC (19), suggesting that the underlying functional variant likely affects protein function. PLTP is secreted into the plasma by multiple tissues, where it shuttles phospholipids from TRLs to HDL and may remodel HDL producing pre-β HDL particles (125). Its expression is modulated by bound farnesoid X receptor (FXR) and LXR (126). Several common variants carried by PLTP are associated with decreased PLTP expression, decreased activity, increased number of small HDL molecules, and a protective cardiovascular effect, including the lead SNP identified by GLGC; however, measures of TRL metabolism were not investigated in depth (127, 128). In mice, overexpression of Pltp modestly increased HDL-C, while Pltp knockout mice had reduced HDL-C, phospholipids, and apoA-I (129, 130). Other studies in mice have suggested that Pltp contributes to TG-rich particle assembly by increasing VLDL secretion (131, 132). The emergence of PLTP as a TG-associated GWAS-discovered locus solidifies the rationale to better understand its role in lipoprotein metabolism.
Fatty acid desaturases.
The FADS1-2-3 gene cluster is associated with plasma TG and inversely with plasma HDL-C concentrations (Table 2) (17, 19). These three genes each encode fatty acid desaturases responsible for the metabolism of polyunsaturated fatty acids involved in cell signaling (133); however, little is known about the role of desaturases in TG or HDL metabolism. Interestingly, the lead TG-associated SNP (rs174546) is found within the 3′ untranslated region of FADS1, and the lead HDL-C-associated SNP (rs174601) is within an intron of FADS2. However, both SNPs are located within a large LD block and are in strong LD with each other (r2 = 0.86). The FADS1-associated SNP is also an eQTL in liver and omental fat (19). Common variants at this locus have also been associated with serum phospholipid concentrations including arachidonic acid (134, 135), desaturase activity (increased arachidonic-to-linoleic acid ratio), increased CRP concentration and CVD risk (136), phospholipid subfractions including sphingomyelin species (137), and increased plasma glucose and T2D risk (112). These findings suggest a proinflammatory response mediated by increased desaturase activity, although the mechanistic link between these pathways and lipoprotein metabolism is unclear. Similarly, it is uncertain whether one or all of these genes are involved in TRL metabolism.
GENETIC DETERMINANTS OF TRIGLYCERIDE FOUND USING OTHER APPROACHES
Important genes are not always identified by GWAS. There are several possible reasons for this: 1) a locus encodes a crucial protein, but there is no structural or functionally relevant genetic variation in humans; 2) only very rare functional variants exist at a locus, and these have no consistent LD relationship with the common genetic variation that is genotyped on microarrays; 3) common functional genetic variation at a locus affects TG metabolism but is not included on the genotyping microarray nor is it in strong LD with surrogate markers on microarrays; 4) common functional variation has too small an effect size to be significantly associated; 5) GWAS studies that evaluate an additive mode of inheritance may miss associations with variants that act through simple dominant or recessive models, or nonadditive gene-gene or gene-environment interactions, or non-Mendelian mechanisms including inheritance of mitochondrial DNA; or 6) common variation is present at a biologically important locus, detectable and included on genotyping microarrays, but it has no functional consequence. Thus, non-GWAS methodologies such as linkage and family based studies or analysis of human homologs of animal models may help with gene discovery. In the following sections we describe TG-associated genes identified using such complementary methodologies.
Linkage and family studies
Upstream transcription factor 1.
Linkage analysis is a classical genetic approach to statistically implicate chromosomal loci (and ultimately genes) that cosegregate with disease phenotypes which show vertical transmission in multigenerational families or kindreds. The approach is most powerful for disorders in which a single gene completely explains the phenotype.
Among familial HTG disorders in which linkage analysis has been attempted is FCHL. FCHL is a relatively common familial dyslipidemia defined by increased plasma concentrations of total and LDL-C, TG, and apoB and by decreased plasma concentrations of HDL-C (89). The mechanism underlying FCHL has been considered the overproduction of apoB-containing lipoproteins together with delayed clearance of TRLs. Although it does not demonstrate pure autosomal dominant inheritance in most affected families, it is nonetheless strongly heritable; the phenotypic variability in family members has sometimes been attributed to variable penetrance or to gene-diet or gene-hormone interactions. Classical genetic linkage approaches have been applied in specific FCHL families (89). Notably, linkage mapping was used to identify the 1q21-q23 locus as associated with the FCHL-associated locus reported in multiple independent cohorts (138–140), shown to be mediated by upstream transcription factor 1 (USF1) (141).
USF1 encodes a transcription factor responsible for the regulation of many proteins involved in glucose and lipid metabolism, including apoA-V and apoC-III and metabolic enzymes such as fatty acid synthase and GCK (142, 143). Recently, the risk allele of a strongly associated intronic variant in USF1 was shown to attenuate the insulin induction of USF1, thus preventing activation of responsive genes (144). Coding sequence variants in USF1 are rare, suggesting that this intronic variant is the functional variant underlying dyslipidemia. The potential functional variant is found in a highly conserved nucleotide in a putative transcriptional regulatory element, which may alter expression of USF1 target genes including APOE (145).
Human homologs of mouse models of hypertriglyceridemia
Combined lipase deficiency (lipase maturation factor 1).
Combined lipase deficiency (cld) was first described in a spontaneous mouse mutant that was deficient in both LPL and hepatic lipase (HL) activity (146). Mice carrying autosomal recessive mutations in Cld develop postpartum chylomicronemia and death from ischemia and cyanosis from increased blood viscosity after nursing (146, 147). Using linkage analysis, the cld phenotype was mapped to murine chromosome 17, and further fine mapping identified the Tmem112 gene, renamed lipase maturation factor 1 (Lmf1), as the cause (147). Subsequent studies showed that Lmf1 is coexpressed in tissues that express LPL or HL; its gene product localizes to the membrane of endoplasmic reticulum, stimulating maturation of both LPL and HL (147, 148).
The cld mutation is a premature truncation of Lmf1 affecting a conserved domain of uncertain function (DUF1222) that cripples maturation of the target lipases, apparently independent of its subcellular localization and without requiring any direct interaction (148). Naturally occurring murine mutations in Lmf1 have also been demonstrated to compromise Lmf1 function in vitro (149). For instance, the Lmf1 glycine-to-glutamic acid missense mutation at position 181 (G181E), located next to a transmembrane domain, has severely compromised function (149).
The HTG phenotype in cld-carrying mice suggested that the human ortholog LMF1 on chromosome 16 was a candidate gene for TG metabolism (Table 3). In one study, 11 patients with chylomicronemia and defective lipase activity were resequenced for mutations in LMF1. A tyrosine-to-stop codon (Y439X) mutation in LMF1 exon 9 was identified in an HTG patient with recurrent pancreatitis resulting from a plasma TG concentration of ∼30 mmol·l−1, caused by a 93% reduction in post-heparin plasma activity of LPL (147). In a second study, another homozygous mutation was identified in a patient with recurrent pancreatitis and a plasma TG concentration of ∼27 mmol·l−1 (150). The tryptophan-to-stop codon (W464X) mutation was found to have reduced LPL and HL activities by 76% and 27%, respectively (150). Thus, LMF1 appears to be a bona fide gene that regulates TG metabolism, as indicated by phenotypic and functional consequences of rare large-effect mutations in humans and mice, while its common variants interrogated using GWAS failed to produce an association signal.
Homologous protein sequences that share significant sequences or even protein domain structures may point toward genes with similar function or common interactions in TG metabolism. Thus, evaluation of protein families may help identify additional genes involved in lipoprotein metabolism. For instance, lipase maturation factor 2 (LMF2) is a potential TG metabolism candidate gene (147). Lmf2 is homologous to Lmf1, with 42% protein sequence identity, including the conserved C-terminal DUF1222 (147). Both Lmf1 and Lmf2 are conserved across vertebrates, although Lmf2 cannot complement or rescue Lmf1-deficient cells, suggesting it has a physiologically relevant and nonredundant role (147).
Glycosylphosphatidylinositol-anchored HDL-binding protein 1 deficiency.
Gpihbp1 was identified through a large scale murine cDNA library screen (151). Gpihbp1 knockout mice have chylomicronemia (152). Gpihbp1 deficiency results in progressive onset of HTG from decreased ability of LPL to efficiently metabolize CMs (152). Gpihbp1 is expressed primarily in capillaries of heart, adipose tissue, and skeletal muscle, where it localizes to the luminal surface of endothelial cells (151, 152). Gpihbp1 probably directs fat toward energy sinks, such as the heart and adipose tissue (153). Gpihbp1 is regulated by PPARγ, with increased expression during fasting and return to baseline after refeeding (154). Gpihbp1 contains an N-terminal acidic domain and a lymphocyte antigen 6 (Ly6) domain. The 25-residue acidic domain appears to bind for heparin binding substrates, including LPL, apoA-V and CMs (155). The ∼80-residue Ly6 domain is required for N-linked glycosylation, forming a three-fingered structural motif (156), essential for translocation to the endothelial cell surface (157, 158).
Defining the role of the human ortholog GPIHBP1 in TG metabolism was inspired by its role in murine TG metabolism. For instance, the coding region of GPIHBP1 from 60 HTG patients without known disease-causing mutations was sequenced to find potential GPIHBP1 mutations (159). In a patient whose plasma TG was ∼38 mmol·l−1, a homozygous glutamine-to-proline substitution at amino acid residue 115 (Q115P) was found; this mutation was within the Ly6 domain of GPIHBP1. Follow-up in vitro analyses demonstrated that the GPIHBP1 mutant was properly shuttled to the endothelial cell surface but was deficient in binding both LPL and CMs. Additional GPIHBP1 mutations were found in three Swedish siblings whose plasma TG concentrations were between 18 and 27 mmol·l−1: each sibling was a compound heterozygote for a cysteine-to-serine mutation at amino acid residue 65 (C65S) and a cysteine-to-glycine mutation at amino acid residue 68 (C68G) (160). Another patient was homozygous for a cysteine-to-tyrosine mutation also at residue 65 (C65Y) (161). These mutations all replace cysteine residues required for disulfide bonding in the GPIHBP1 Ly6 domain. Functional analyses in vitro showed that each mutant protein could reach the cell surface but that each was defective in LPL and CM binding (160, 161). Finally, screening of 160 patients with severe HTG identified a homozygous glycine-to-arginine mutation at GPIHBP1 residue 56 (G56R) in two siblings, localized to a linker domain between the N-terminal acidic domain and the Ly6 domain (162). This mutation cosegregated with the clinical and biochemical phenotype (162): the proband had an average lifetime plasma TG concentration of ∼40 mmol·l−1, with recurrent pancreatitis beginning at 22 years of age, while her older brother had a similar medical history and lipid profile. Furthermore, several heterozygote relatives had less severe HTG, while relatives who were homozygous for the wild-type gene had normal lipid profiles. Analysis of this variant in vitro demonstrated that GPIHBP1-56R reached the cell surface and bound all ligands normally (163), suggesting that some other function of GPIHBP1 was affected. Cumulatively, evidence from mouse models and rare human mutations supports a role for GPIHBP1 in TG metabolism (Table 3).
FUTURE DIRECTIONS
The legacy of GWAS will be gauged by the number of genetic loci that help to elucidate and functionally annotate disease-associated metabolic pathways (Fig. 3). However, the greatly expanded number of TG-associated genes may challenge our ability to rapidly study their functions. Taking full advantage of the new opportunities that have arisen through GWAS discoveries will require analogous technological advances allowing fine mapping of association peaks, deep resequencing for functional variants, and finally high-throughput, robust functional validation at all stages: in vitro, in vivo, and ultimately in clinical trials. In the interim, TG-associated loci may prove useful for personalized health management strategies.
Fig. 3.
Sequence of experimental approaches required to achieve clinical benefit from current TG-associated loci.
Genetic risk prediction of HTG
Early identification of subjects at risk for developing HTG could prompt early lifestyle modification or evidence-based pharmacological intervention to reduce risk of clinical end points. We have shown that there is a significant difference in mean score of risk alleles in HTG patients compared with those in population-based controls (19) (Fig. 2). TG risk scoring could help identify subjects at risk for developing HTG and who might benefit from early intervention. However, while few HTG patients have very low risk allele scores and few normal individuals have very high risk allele scores, there is substantial overlap toward the centers of the distributions of risk allele scores. Thus, the risk allele scores tend to discriminate primarily between normal and HTG for those with extreme values. Emerging methodologies such as whole-genome sequencing, including known common functional variants and rare deleterious or protective alleles, may help produce more discriminating TG risk scores.
Genetic risk prediction of coronary artery disease
The GLGC studied coronary artery disease (CAD) risk in ∼25,000 cases and ∼66,000 controls (19). Six TG-associated loci from GLGC studies were associated with CAD, including APOA5, LPL, TRIB1, APOE, CILP2, NAT2, and IRS1, compared with 11 LDL-C-associated loci and 8 HDL-associated loci. Interestingly, only two TG-associated loci that were independent of TG or LDL-C were associated with CAD, specifically LPL and IRS1, both of which were also HDL-C-associated loci. IRS1 is similarly associated with increased risk of T2D, insulin resistance, and hyperinsulinemia (164). Thus, it has been hypothesized that some mechanisms which increase plasma TG concentration may also directly increase CAD risk. In support of this observation, a recent Mendelian randomization experiment with the APOA5 functional variant −1131T>C has suggested a causal relationship between increased plasma TG concentration and CAD (81).
Genetic variables effectively determine a subject's “genetic burden” for developing an unfavorable TRL profile; this factor may improve existing risk prediction algorithms for CAD, such as the Framingham risk score (165). Genetic risk scores could be composed of risk alleles exclusively for a single trait such as plasma TG or risk alleles from multiple independent traits, such as a general lipoprotein risk score. Cumulative risk scores constructed as a weighted sum of allelic effect estimates have been associated with increased CVD risk, significantly improving risk stratification of patients by 6.1% overall and by 26% for subjects in the intermediate risk category (166). Increased lipoprotein risk scores are associated with increasing plasma lipids, including plasma TG (17, 21, 167). The modest improvement of risk prediction provided by genetics, while biologically important, may be due to small effect sizes. Many additional genetic variables, or improved composite genetic risk scores, might improve the performance of risk prediction algorithms (168).
Pharmacogenomics
Fibrates and niacin are the primary pharmacotherapies for HTG. Interindividual variations in plasma TG response have been reported to differ according to genotype of common APOA5 SNPs (169) and to the presence of rare loss-of-function variants of candidate genes in TG metabolism (53). A complete genetic profile related to TG metabolism and possibly plasma TG responsiveness will include common SNP genotypes and eventually rare variants and genomic copy number variation, from GWAS of clinical trials of TG-lowering strategies. As genetic information is included as covariates in clinical trials, an individual's genetic score might predict both biochemical TG and CVD outcome response to drugs, diet, exercise, or other interventions.
CONCLUSION
Plasma TG concentration appears to be an independent predictor of CVD risk. A combination of genetic approaches, including study of genes that encode proteins identified from classical biochemistry, genes identified through GWAS of normal and HTG samples, and genes identified by studies of families and animal models, together accounts for a relatively large proportion of variation in plasma TG. Clinical HTG has a complex genetic basis that includes a burden of both common variants, such that each has a relatively small effect on TG levels, and rare variants, such that each has a relatively large effect on TG levels. A more complete understanding of the genes and variants that modulate plasma TG will enable development of markers for risk prediction, diagnosis, prognosis, and response to therapies and might help specify new directions for therapeutic interventions.
Footnotes
Abbreviations:
- ABL
- abetalipoproteinemia
- ANGPTL3
- angiopoietin-like 3 protein
- CAD
- coronary artery disease
- CHREBP
- carbohydrate response element binding protein
- Cld
- combined lipase deficiency
- CM
- chylomicrons
- CRE
- carbohydrate response element
- CRP
- C-reactive protein
- CVD
- cardiovascular disease
- eQTL
- expression quantitative trait locus
- FCH
- familial combined hypolipidemia
- FCHL
- familial combined hyperlipidemia
- GCK
- glucokinase
- GCKR
- glucokinase regulatory protein
- GLGC
- Global Lipids Genetics Consortium
- GPIHBP1
- glycosylphosphatidylinositol-anchored HDL-binding protein 1
- GWAS
- genome-wide association studies
- HHBL
- homozygous hypobetalipoproteinemia
- HL
- hepatic lipase
- HLP
- hyperlipoproteinemia
- HTG
- hypertriglyceridemia
- LD
- linkage disequilibrium
- Lmf1
- lipase maturation factor 1
- LXR
- liver X receptor
- Ly6
- lymphocyte antigen 6
- MTP
- microsomal TG transfer protein
- PLTP
- phospholipid transfer protein
- PPARα
- peroxisome proliferator-activator receptor α
- SNP
- single nucleotide polymorphism
- TG
- triglyceride
- TRL
- TG-rich lipoprotein
- USF1
- upstream transcription factor 1
This work was supported by a Canadian Institutes of Health Research (CIHR) Banting and Best Canada Graduate Scholarship Doctoral Research Award (CTJ); by operating grants from CIHR (MOP-13430, MOP-79523, CTP-79853) and the Heart and Stroke Foundation of Ontario (NA-6059, T-6018, PRG-4854) (RAH), and by a Pfizer Jean Davignon Distinguished Cardiovascular and Metabolic Research Award and Genome Canada through the Ontario Genomics Institute. C. T. J. is a University of Western Ontario MD/PhD student and a CIHR fellow in vascular research. R. A. H. is the Jacob J. Wolfe Distinguished Medical Research Chair, University of Western Ontario, the Edith Schulich Vinet Canada Research Chair in Human Genetics (Tier I), and the Martha G. Blackburn Chair in Cardiovascular Research.
REFERENCES
- 1.Cullen P. 2000. Evidence that triglycerides are an independent coronary heart disease risk factor. Am. J. Cardiol. 86: 943–949. [DOI] [PubMed] [Google Scholar]
- 2.Morrison A., Hokanson J. E. 2009. The independent relationship between triglycerides and coronary heart disease. Vasc. Health Risk Manag. 5: 89–95. [PMC free article] [PubMed] [Google Scholar]
- 3.Labreuche J., Touboul P. J., Amarenco P. 2009. Plasma triglyceride levels and risk of stroke and carotid atherosclerosis: a systematic review of the epidemiological studies. Atherosclerosis. 203: 331–345. [DOI] [PubMed] [Google Scholar]
- 4.Hokanson J. E., Austin M. A. 1996. Plasma triglyceride level is a risk factor for cardiovascular disease independent of high-density lipoprotein cholesterol level: a meta-analysis of population-based prospective studies. J. Cardiovasc. Risk. 3: 213–219. [PubMed] [Google Scholar]
- 5.Bansal S., Buring J. E., Rifai N., Mora S., Sacks F. M., Ridker P. M. 2007. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. JAMA. 298: 309–316. [DOI] [PubMed] [Google Scholar]
- 6.Nordestgaard B. G., Benn M., Schnohr P., Tybjaerg-Hansen A. 2007. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 298: 299–308. [DOI] [PubMed] [Google Scholar]
- 7.Freiberg J. J., Tybjaerg-Hansen A., Jensen J. S., Nordestgaard B. G. 2008. Nonfasting triglycerides and risk of ischemic stroke in the general population. JAMA. 300: 2142–2152. [DOI] [PubMed] [Google Scholar]
- 8.Altshuler D., Daly M. J., Lander E. S. 2008. Genetic mapping in human disease. Science 322: 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attia J., Ioannidis J. P., Thakkinstian A., McEvoy M., Scott R. J., Minelli C., Thompson J., Infante-Rivard C., Guyatt G. 2009. How to use an article about genetic association: A: Background concepts. JAMA. 301: 74–81. [DOI] [PubMed] [Google Scholar]
- 10.Attia J., Ioannidis J. P., Thakkinstian A., McEvoy M., Scott R. J., Minelli C., Thompson J., Infante-Rivard C., Guyatt G. 2009. How to use an article about genetic association: B: Are the results of the study valid? JAMA. 301: 191–197. [DOI] [PubMed] [Google Scholar]
- 11.Attia J., Ioannidis J. P., Thakkinstian A., McEvoy M., Scott R. J., Minelli C., Thompson J., Infante-Rivard C., Guyatt G. 2009. How to use an article about genetic association: C: What are the results and will they help me in caring for my patients? JAMA. 301: 304–308. [DOI] [PubMed] [Google Scholar]
- 12.Wallace C., Newhouse S. J., Braund P., Zhang F., Tobin M., Falchi M., Ahmadi K., Dobson R. J., Marcano A. C., Hajat C., et al. 2008. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 82: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N. P., Rieder M. J., Cooper G. M., Roos C., Voight B. F., Havulinna A. S., et al. 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40: 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aulchenko Y. S., Ripatti S., Lindqvist I., Boomsma D., Heid I. M., Pramstaller P. P., Penninx B. W., Janssens A. C., Wilson J. F., Spector T., et al. 2009. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 41: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabatti C., Service S. K., Hartikainen A. L., Pouta A., Ripatti S., Brodsky J., Jones C. G., Zaitlen N. A., Varilo T., Kaakinen M., et al. 2009. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 41: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., et al. 2009. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooner J. S., Chambers J. C., Aguilar-Salinas C. A., Hinds D. A., Hyde C. L., Warnes G. R., Gomez Perez F. J., Frazer K. A., Elliott P., Scott J., et al. 2008. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat. Genet. 40: 149–151. [DOI] [PubMed] [Google Scholar]
- 19.Teslovich T. M. 2010. Biological, clinical, and population relevance of 95 loci mapped for serum lipid concentrations. Nature. 466: 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manolio T. A. 2010. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 363: 166–176. [DOI] [PubMed] [Google Scholar]
- 21.Lanktree M. B., Anand S. S., Yusuf S., Hegele R. A. 2009. Replication of genetic associations with plasma lipoprotein traits in a multiethnic sample. J. Lipid Res. 50: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keebler M. E., Sanders C. L., Surti A., Guiducci C., Burtt N. P., Kathiresan S. 2009. Association of blood lipids with common DNA sequence variants at 19 genetic loci in the multiethnic United States National Health and Nutrition Examination Survey III. Circ. Cardiovasc. Genet. 2: 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keebler M. E., Deo R. C., Surti A., Konieczkowski D., Guiducci C., Burtt N., Buxbaum S. G., Sarpong D. F., Steffes M. W., Wilson J. G., et al. 2010. Fine-mapping in African Americans of eight recently discovered genetic loci for plasma lipids: the Jackson Heart Study. Circ. Cardiovasc. Genet. 3: 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teo Y. Y., Small K. S., Kwiatkowski D. P. 2010. Methodological challenges of genome-wide association analysis in Africa. Nat. Rev. Genet. 11: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao J. T., Wen H. C., Chien K. L., Hsu H. C., Lin S. W. 2003. A novel genetic variant in the apolipoprotein A5 gene is associated with hypertriglyceridemia. Hum. Mol. Genet. 12: 2533–2539. [DOI] [PubMed] [Google Scholar]
- 26.Hubacek J. A., Adamkova V., Ceska R., Poledne R., Horinek A., Vrablik M. 2004. New variants in the apolipoprotein AV gene in individuals with extreme triglyceride levels. Physiol. Res. 53: 225–228. [PubMed] [Google Scholar]
- 27.Yuan G., Al-Shali K. Z., Hegele R. A. 2007. Hypertriglyceridemia: its etiology, effects and treatment. CMAJ. 176: 1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegele R. A., Pollex R. L. 2009. Hypertriglyceridemia: phenomics and genomics. Mol. Cell. Biochem. 326: 35–43. [DOI] [PubMed] [Google Scholar]
- 29.Plomin R., Haworth C. M., Davis O. S. 2009. Common disorders are quantitative traits. Nat. Rev. Genet. 10: 872–878. [DOI] [PubMed] [Google Scholar]
- 30.Kannel W. B., Vasan R. S. 2009. Triglycerides as vascular risk factors: new epidemiologic insights. Curr. Opin. Cardiol. 24: 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegele R. A. 2009. Plasma lipoproteins: genetic influences and clinical implications. Nat. Rev. Genet. 10: 109–121. [DOI] [PubMed] [Google Scholar]
- 32.Johansen C. T. 2010. Excess of rare variats in genes identified by genome-wide association study of patients with hypertriglyceridemia. Nat. Genet. 42: 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sniderman A., Couture P., de Graaf J. 2010. Diagnosis and treatment of apolipoprotein B dyslipoproteinemias. Nat. Rev. Endocrinol. 6: 335–346. [DOI] [PubMed] [Google Scholar]
- 34.Hegele R. A., Ban M. R., Hsueh N., Kennedy B. A., Cao H., Zou G. Y., Anand S., Yusuf S., Huff M. W., Wang J. 2009. A polygenic basis for four classical Fredrickson hyperlipoproteinemia phenotypes that are characterized by hypertriglyceridemia. Hum. Mol. Genet. 18: 4189–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musunuru K., Pirruccello J. P., Do R., Peloso M. S., Guiducci C., Sougnez C., Garimella K. V., Fisher S., Abreu J., Barry A. J., et al. 2010. Exome Sequencing, Mutations in ANGPTL3, and Familial Combined Hypolipidemia. N. Engl. J. Med. Oct 13, 2010 [Epub ahead of print] ; doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anant S., Davidson N. O. 2001. Molecular mechanisms of apolipoprotein B mRNA editing. Curr. Opin. Lipidol. 12: 159–165. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Z. G., Liu Y., Hussain M. M., Atkinson D., McKnight C. J. 2008. Reconstituting initial events during the assembly of apolipoprotein B-containing lipoproteins in a cell-free system. J. Mol. Biol. 383: 1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samaha F. F., McKenney J., Bloedon L. T., Sasiela W. J., Rader D. J. 2008. Inhibition of microsomal triglyceride transfer protein alone or with ezetimibe in patients with moderate hypercholesterolemia. Nat. Clin. Pract. Cardiovasc. Med. 5: 497–505. [DOI] [PubMed] [Google Scholar]
- 39.Raal F. J., Santos R. D., Blom D. J., Marais A. D., Charng M. J., Cromwell W. C., Lachmann R. H., Gaudet D., Tan J. L., Chasan-Taber S., et al. 2010. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 375: 998–1006. [DOI] [PubMed] [Google Scholar]
- 40.Havel R. J. 2010. Triglyceride-rich lipoproteins and plasma lipid transport. Arterioscler. Thromb. Vasc. Biol. 30: 9–19. [DOI] [PubMed] [Google Scholar]
- 41.Kirchgessner T. G., LeBoeuf R. C., Langner C. A., Zollman S., Chang C. H., Taylor B. A., Schotz M. C., Gordon J. I., Lusis A. J. 1989. Genetic and developmental regulation of the lipoprotein lipase gene: loci both distal and proximal to the lipoprotein lipase structural gene control enzyme expression. J. Biol. Chem. 264: 1473–1482. [PubMed] [Google Scholar]
- 42.Weinstein M. M., Yin L., Beigneux A. P., Davies B. S., Gin P., Estrada K., Melford K., Bishop J. R., Esko J. D., Dallinga-Thie G. M., et al. 2008. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J. Biol. Chem. 283: 34511–34518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Eckel R. H. 2009. Lipoprotein lipase: from gene to obesity. Am. J. Physiol. Endocrinol. Metab. 297: E271–E288. [DOI] [PubMed] [Google Scholar]
- 44.LaRosa J. C., Levy R. I., Herbert P., Lux S. E., Fredrickson D. S. 1970. A specific apoprotein activator for lipoprotein lipase. Biochem. Biophys. Res. Commun. 41: 57–62. [DOI] [PubMed] [Google Scholar]
- 45.Weinstock P. H., Bisgaier C. L., Aalto-Setala K., Radner H., Ramakrishnan R., Levak-Frank S., Essenburg A. D., Zechner R., Breslow J. L. 1995. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J. Clin. Invest. 96: 2555–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimada M., Shimano H., Gotoda T., Yamamoto K., Kawamura M., Inaba T., Yazaki Y., Yamada N. 1993. Overexpression of human lipoprotein lipase in transgenic mice. Resistance to diet-induced hypertriglyceridemia and hypercholesterolemia. J. Biol. Chem. 268: 17924–17929. [PubMed] [Google Scholar]
- 47.Fisher R. M., Humphries S. E., Talmud P. J. 1997. Common variation in the lipoprotein lipase gene: effects on plasma lipids and risk of atherosclerosis. Atherosclerosis. 135: 145–159. [DOI] [PubMed] [Google Scholar]
- 48.Mailly F., Tugrul Y., Reymer P. W., Bruin T., Seed M., Groenemeyer B. F., Asplund-Carlson A., Vallance D., Winder A. F., Miller G. J., et al. 1995. A common variant in the gene for lipoprotein lipase (Asp9→Asn). Functional implications and prevalence in normal and hyperlipidemic subjects. Arterioscler. Thromb. Vasc. Biol. 15: 468–478. [DOI] [PubMed] [Google Scholar]
- 49.Reymer P. W., Gagne E., Groenemeyer B. E., Zhang H., Forsyth I., Jansen H., Seidell J. C., Kromhout D., Lie K. E., Kastelein J., et al. 1995. A lipoprotein lipase mutation (Asn291Ser) is associated with reduced HDL cholesterol levels in premature atherosclerosis. Nat. Genet. 10: 28–34. [DOI] [PubMed] [Google Scholar]
- 50.Fisher R. M., Mailly F., Peacock R. E., Hamsten A., Seed M., Yudkin J. S., Beisiegel U., Feussner G., Miller G., Humphries S. E., et al. 1995. Interaction of the lipoprotein lipase asparagine 291→serine mutation with body mass index determines elevated plasma triacylglycerol concentrations: a study in hyperlipidemic subjects, myocardial infarction survivors, and healthy adults. J. Lipid Res. 36: 2104–2112. [PubMed] [Google Scholar]
- 51.Hata A., Robertson M., Emi M., Lalouel J. M. 1990. Direct detection and automated sequencing of individual alleles after electrophoretic strand separation: identification of a common nonsense mutation in exon 9 of the human lipoprotein lipase gene. Nucleic Acids Res. 18: 5407–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ross C. J., Liu G., Kuivenhoven J. A., Twisk J., Rip J., van Dop W., Excoffon K. J., Lewis S. M., Kastelein J. J., Hayden M. R. 2005. Complete rescue of lipoprotein lipase-deficient mice by somatic gene transfer of the naturally occurring LPLS447X beneficial mutation. Arterioscler. Thromb. Vasc. Biol. 25: 2143–2150. [DOI] [PubMed] [Google Scholar]
- 53.Wang J., Cao H., Ban M. R., Kennedy B. A., Zhu S., Anand S., Yusuf S., Pollex R. L., Hegele R. A. 2007. Resequencing genomic DNA of patients with severe hypertriglyceridemia (MIM 144650). Arterioscler. Thromb. Vasc. Biol. 27: 2450–2455. [DOI] [PubMed] [Google Scholar]
- 54.Rip J., Nierman M. C., Ross C. J., Jukema J. W., Hayden M. R., Kastelein J. J., Stroes E. S., Kuivenhoven J. A. 2006. Lipoprotein lipase S447X: a naturally occurring gain-of-function mutation. Arterioscler. Thromb. Vasc. Biol. 26: 1236–1245. [DOI] [PubMed] [Google Scholar]
- 55.Wang J., Ban M. R., Zou G. Y., Cao H., Lin T., Kennedy B. A., Anand S., Yusuf S., Huff M. W., Pollex R. L., et al. 2008. Polygenic determinants of severe hypertriglyceridemia. Hum. Mol. Genet. 17: 2894–2899. [DOI] [PubMed] [Google Scholar]
- 56.Stroes E. S., Nierman M. C., Meulenberg J. J., Franssen R., Twisk J., Henny C. P.., Maas M. M., Zwinderman A. H., Ross C., Aronica E., et al. 2008. Intramuscular administration of AAV1-lipoprotein lipase S447X lowers triglycerides in lipoprotein lipase-deficient patients. Arterioscler. Thromb. Vasc. Biol. 28: 2303–2304. [DOI] [PubMed] [Google Scholar]
- 57.Ishimura-Oka K., Faustinella F., Kihara S., Smith L. C., Oka K., Chan L. 1992. A missense mutation (Trp86—-Arg) in exon 3 of the lipoprotein lipase gene: a cause of familial chylomicronemia. Am. J. Hum. Genet. 50: 1275–1280. [PMC free article] [PubMed] [Google Scholar]
- 58.Henderson H. E., Ma Y., Hassan M. F., Monsalve M. V., Marais A. D., Winkler F., Gubernator K., Peterson J., Brunzell J. D., Hayden M. R. 1991. Amino acid substitution (Ile194—-Thr) in exon 5 of the lipoprotein lipase gene causes lipoprotein lipase deficiency in three unrelated probands. Support for a multicentric origin. J. Clin. Invest. 87: 2005–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma Y., Henderson H. E., Murthy V., Roederer G., Monsalve M. V., Clarke L. A., Normand T., Julien P., Gagne C., Lambert M., et al. 1991. A mutation in the human lipoprotein lipase gene as the most common cause of familial chylomicronemia in French Canadians. N. Engl. J. Med. 324: 1761–1766. [DOI] [PubMed] [Google Scholar]
- 60.Emi M., Wilson D. E., Iverius P. H., Wu L., Hata A., Hegele R., Williams R. R., Lalouel J. M. 1990. Missense mutation (Gly—-Glu188) of human lipoprotein lipase imparting functional deficiency. J. Biol. Chem. 265: 5910–5916. [PubMed] [Google Scholar]
- 61.Dorfmeister B., Zeng W. W., Dichlberger A., Nilsson S. K., Schaap F. G., Hubacek J. A., Merkel M., Cooper J. A., Lookene A., Putt W., et al. 2008. Effects of six APOA5 variants, identified in patients with severe hypertriglyceridemia, on in vitro lipoprotein lipase activity and receptor binding. Arterioscler. Thromb. Vasc. Biol. 28: 1866–1871. [DOI] [PubMed] [Google Scholar]
- 62.Connelly P. W., Maguire G. F., Little J. A. 1987. Apolipoprotein CIISt. Michael. Familial apolipoprotein CII deficiency associated with premature vascular disease. J. Clin. Invest. 80: 1597–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pennacchio L. A., Olivier M., Hubacek J. A., Cohen J. C., Cox D. R., Fruchart J. C., Krauss R. M., Rubin E. M. 2001. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294: 169–173. [DOI] [PubMed] [Google Scholar]
- 64.van der Vliet H. N., Sammels M. G., Leegwater A. C., Levels J. H., Reitsma P. H., Boers W., Chamuleau R. A. 2001. Apolipoprotein A-V: a novel apolipoprotein associated with an early phase of liver regeneration. J. Biol. Chem. 276: 44512–44520. [DOI] [PubMed] [Google Scholar]
- 65.Merkel M., Loeffler B., Kluger M., Fabig N., Geppert G., Pennacchio L. A., Laatsch A., Heeren J. 2005. Apolipoprotein AV accelerates plasma hydrolysis of triglyceride-rich lipoproteins by interaction with proteoglycan-bound lipoprotein lipase. J. Biol. Chem. 280: 21553–21560. [DOI] [PubMed] [Google Scholar]
- 66.Schaap F. G., Rensen P. C., Voshol P. J., Vrins C., van der Vliet H. N., Chamuleau R. A., Havekes L. M., Groen A. K., van Dijk K. W. 2004. ApoAV reduces plasma triglycerides by inhibiting very low density lipoprotein-triglyceride (VLDL-TG) production and stimulating lipoprotein lipase-mediated VLDL-TG hydrolysis. J. Biol. Chem. 279: 27941–27947. [DOI] [PubMed] [Google Scholar]
- 67.Grosskopf I., Baroukh N., Lee S. J., Kamari Y., Harats D., Rubin E. M., Pennacchio L. A., Cooper A. D. 2005. Apolipoprotein A-V deficiency results in marked hypertriglyceridemia attributable to decreased lipolysis of triglyceride-rich lipoproteins and removal of their remnants. Arterioscler. Thromb. Vasc. Biol. 25: 2573–2579. [DOI] [PubMed] [Google Scholar]
- 68.O'Brien P. J., Alborn W. E., Sloan J. H., Ulmer M., Boodhoo A., Knierman M. D., Schultze A. E., Konrad R. J. 2005. The novel apolipoprotein A5 is present in human serum, is associated with VLDL, HDL, and chylomicrons, and circulates at very low concentrations compared with other apolipoproteins. Clin. Chem. 51: 351–359. [DOI] [PubMed] [Google Scholar]
- 69.Shu X., Nelbach L., Ryan R. O., Forte T. M. 2010. Apolipoprotein A-V associates with intrahepatic lipid droplets and influences triglyceride accumulation. Biochim. Biophys. Acta. 1801: 605–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qu S., Perdomo G., Su D., D'Souza F. M., Shachter N. S., Dong H. H. 2007. Effects of apoA-V on HDL and VLDL metabolism in APOC3 transgenic mice. J. Lipid Res. 48: 1476–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Vliet H. N., Schaap F. G., Levels J. H., Ottenhoff R., Looije N., Wesseling J. G., Groen A. K., Chamuleau R. A. 2002. Adenoviral overexpression of apolipoprotein A-V reduces serum levels of triglycerides and cholesterol in mice. Biochem. Biophys. Res. Commun. 295: 1156–1159. [DOI] [PubMed] [Google Scholar]
- 72.Jakel H., Nowak M., Helleboid-Chapman A., Fruchart-Najib J., Fruchart J. C. 2006. Is apolipoprotein A5 a novel regulator of triglyceride-rich lipoproteins? Ann. Med. 38: 2–10. [DOI] [PubMed] [Google Scholar]
- 73.Prieur X., Lesnik P., Moreau M., Rodriguez J. C., Doucet C., Chapman M. J., Huby T. 2009. Differential regulation of the human versus the mouse apolipoprotein AV gene by PPARalpha. Implications for the study of pharmaceutical modifiers of hypertriglyceridemia in mice. Biochim. Biophys. Acta. 1791: 764–771. [DOI] [PubMed] [Google Scholar]
- 74.Hubacek J. A. 2005. Apolipoprotein A5 and triglyceridemia. Focus on the effects of the common variants. Clin. Chem. Lab. Med. 43: 897–902. [DOI] [PubMed] [Google Scholar]
- 75.Pennacchio L. A., Olivier M., Hubacek J. A., Krauss R. M., Rubin E. M., Cohen J. C. 2002. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum. Mol. Genet. 11: 3031–3038. [DOI] [PubMed] [Google Scholar]
- 76.Talmud P. J., Palmen J., Putt W., Lins L., Humphries S. E. 2005. Determination of the functionality of common APOA5 polymorphisms. J. Biol. Chem. 280: 28215–28220. [DOI] [PubMed] [Google Scholar]
- 77.Palmen J., Smith A. J., Dorfmeister B., Putt W., Humphries S. E., Talmud P. J. 2008. The functional interaction on in vitro gene expression of APOA5 SNPs, defining haplotype APOA52, and their paradoxical association with plasma triglyceride but not plasma apoAV levels. Biochim. Biophys. Acta. 1782: 447–452. [DOI] [PubMed] [Google Scholar]
- 78.Talmud P. J., Hawe E., Martin S., Olivier M., Miller G. J., Rubin E. M., Pennacchio L. A., Humphries S. E. 2002. Relative contribution of variation within the APOC3/A4/A5 gene cluster in determining plasma triglycerides. Hum. Mol. Genet. 11: 3039–3046. [DOI] [PubMed] [Google Scholar]
- 79.Olivier M., Wang X., Cole R., Gau B., Kim J., Rubin E. M., Pennacchio L. A. 2004. Haplotype analysis of the apolipoprotein gene cluster on human chromosome 11. Genomics. 83: 912–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang J., Ban M. R., Kennedy B. A., Anand S., Yusuf S., Huff M. W., Pollex R. L., Hegele R. A. 2008. APOA5 genetic variants are markers for classic hyperlipoproteinemia phenotypes and hypertriglyceridemia. Nat. Clin. Pract. Cardiovasc. Med. 5: 730–737. [DOI] [PubMed] [Google Scholar]
- 81.Sarwar N., Sandhu M. S., Ricketts S. L., Butterworth A. S., Di Angelantonio E., Boekholdt S. M., Ouwehand W., Watkins H., Samani N. J., Saleheen D., et al. 2010. Triglyceride-mediated pathways and coronary disease: collaborative analysis of 101 studies. Lancet. 375: 1634–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dallongeville J., Cottel D., Wagner A., Ducimetiere P., Ruidavets J. B., Arveiler D., Bingham A., Ferrieres J., Amouyel P., Meirhaeghe A. 2008. The APOA5 Trp19 allele is associated with metabolic syndrome via its association with plasma triglycerides. BMC Med. Genet. 9: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Talmud P. J. 2007. Rare APOA5 mutations–clinical consequences, metabolic and functional effects: an ENID review. Atherosclerosis. 194: 287–292. [DOI] [PubMed] [Google Scholar]
- 84.Shachter N. S. 2001. Apolipoproteins C–I and C–III as important modulators of lipoprotein metabolism. Curr. Opin. Lipidol. 12: 297–304. [DOI] [PubMed] [Google Scholar]
- 85.van Dijk K. W., Rensen P. C., Voshol P. J., Havekes L. M. 2004. The role and mode of action of apolipoproteins CIII and AV: synergistic actors in triglyceride metabolism? Curr. Opin. Lipidol. 15: 239–246. [DOI] [PubMed] [Google Scholar]
- 86.Sundaram M., Zhong S., Khalil M. B., Links P. H., Zhao Y., Iqbal J., Hussain M. M., Parks R. J., Wang Y., Yao Z. 2010. Expression of apolipoprotein C–III in McA-RH7777 cells enhances VLDL assembly and secretion under lipid-rich conditions. J. Lipid Res. 51: 150–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ito Y., Azrolan N., O'Connell A., Walsh A., Breslow J. L. 1990. Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science 249: 790–793. [DOI] [PubMed] [Google Scholar]
- 88.Maeda N., Li H., Lee D., Oliver P., Quarfordt S. H., Osada J. 1994. Targeted disruption of the apolipoprotein C–III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. J. Biol. Chem. 269: 23610–23616. [PubMed] [Google Scholar]
- 89.Naukkarinen J., Ehnholm C., Peltonen L. 2006. Genetics of familial combined hyperlipidemia. Curr. Opin. Lipidol. 17: 285–290. [DOI] [PubMed] [Google Scholar]
- 90.Li W. W., Dammerman M. M., Smith J. D., Metzger S., Breslow J. L., Leff T. 1995. Common genetic variation in the promoter of the human apo CIII gene abolishes regulation by insulin and may contribute to hypertriglyceridemia. J. Clin. Invest. 96: 2601–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petersen K. F., Dufour S., Hariri A., Nelson-Williams C., Foo J. N., Zhang X. M., Dziura J., Lifton R. P., Shulman G. I. 2010. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N. Engl. J. Med. 362: 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pollex R. L., Ban M. R., Young T. K., Bjerregaard P., Anand S. S., Yusuf S., Zinman B., Harris S. B., Hanley A. J., Connelly P. W., et al. 2007. Association between the -455T>C promoter polymorphism of the APOC3 gene and the metabolic syndrome in a multi-ethnic sample. BMC Med. Genet. 8: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller M., Rhyne J., Chen H., Beach V., Ericson R., Luthra K., Dwivedi M., Misra A. 2007. APOC3 promoter polymorphisms C-482T and T-455C are associated with the metabolic syndrome. Arch. Med. Res. 38: 444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pollin T. I., Damcott C. M., Shen H., Ott S. H., Shelton J., Horenstein R. B., Post W., McLenithan J. C., Bielak L. F., Peyser P. A., et al. 2008. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science 322: 1702–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu H., Labeur C., Xu C. F., Ferrell R., Lins L., Brasseur R., Rosseneu M., Weiss K. M., Humphries S. E., Talmud P. J. 2000. Characterization of the lipid-binding properties and lipoprotein lipase inhibition of a novel apolipoprotein C–III variant Ala23Thr. J. Lipid Res. 41: 1760–1771. [PubMed] [Google Scholar]
- 96.Sundaram M., Zhong S., Bou Khalil M., Zhou H., Jiang Z. G., Zhao Y., Iqbal J., Hussain M. M., Figeys D., Wang Y., et al. 2010. Functional analysis of the missense APOC3 mutation Ala23Thr associated with human hypotriglyceridemia. J. Lipid Res. 51: 1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Conklin D., Gilbertson D., Taft D. W., Maurer M. F., Whitmore T. E., Smith D. L., Walker K. M., Chen L. H., Wattler S., Nehls M., et al. 1999. Identification of a mammalian angiopoietin-related protein expressed specifically in liver. Genomics. 62: 477–482. [DOI] [PubMed] [Google Scholar]
- 98.Koishi R., Ando Y., Ono M., Shimamura M., Yasumo H., Fujiwara T., Horikoshi H., Furukawa H. 2002. Angptl3 regulates lipid metabolism in mice. Nat. Genet. 30: 151–157. [DOI] [PubMed] [Google Scholar]
- 99.Shimizugawa T., Ono M., Shimamura M., Yoshida K., Ando Y., Koishi R., Ueda K., Inaba T., Minekura H., Kohama T., et al. 2002. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J. Biol. Chem. 277: 33742–33748. [DOI] [PubMed] [Google Scholar]
- 100.Shan L., Yu X. C., Liu Z., Hu Y., Sturgis L. T., Miranda M. L., Liu Q. 2009. The angiopoietin-like proteins ANGPTL3 and ANGPTL4 inhibit lipoprotein lipase activity through distinct mechanisms. J. Biol. Chem. 284: 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sonnenburg W. K., Yu D., Lee E. C., Xiong W., Gololobov G., Key B., Gay J., Wilganowski N., Hu Y., Zhao S., et al. 2009. GPIHBP1 stabilizes lipoprotein lipase and prevents its inhibition by angiopoietin-like 3 and angiopoietin-like 4. J. Lipid Res. 50: 2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robciuc M. R., Tahvanainen E., Jauhiainen M., Ehnholm C. 2010. Quantitation of serum angiopoietin-like proteins 3 and 4 in a finnish population sample. J. Lipid Res. 51: 824–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Inaba T., Matsuda M., Shimamura M., Takei N., Terasaka N., Ando Y., Yasumo H., Koishi R., Makishima M., Shimomura I. 2003. Angiopoietin-like protein 3 mediates hypertriglyceridemia induced by the liver X receptor. J. Biol. Chem. 278: 21344–21351. [DOI] [PubMed] [Google Scholar]
- 104.Romeo S., Yin W., Kozlitina J., Pennacchio L. A., Boerwinkle E., Hobbs H. H., Cohen J. C. 2009. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J. Clin. Invest. 119: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Romeo S., Pennacchio L. A., Fu Y., Boerwinkle E., Tybjaerg-Hansen A., Hobbs H. H., Cohen J. C. 2007. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat. Genet. 39: 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Saxena R., Voight B. F., Lyssenko V., Burtt N. P., de Bakker P. I., Chen H., Roix J. J., Kathiresan S., Hirschhorn J. N., Daly M. J., et al. 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336. [DOI] [PubMed] [Google Scholar]
- 107.Agius L. 2008. Glucokinase and molecular aspects of liver glycogen metabolism. Biochem. J. 414: 1–18. [DOI] [PubMed] [Google Scholar]
- 108.Orho-Melander M., Melander O., Guiducci C., Perez-Martinez P., Corella D., Roos C., Tewhey R., Rieder M. J., Hall J., Abecasis G., et al. 2008. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 57: 3112–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Beer N. L., Tribble N. D., McCulloch L. J., Roos C., Johnson P. R., Orho-Melander M., Gloyn A. L. 2009. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 18: 4081–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vaxillaire M., Cavalcanti-Proenca C., Dechaume A., Tichet J., Marre M., Balkau B., Froguel P. 2008. The common P446L polymorphism in GCKR inversely modulates fasting glucose and triglyceride levels and reduces type 2 diabetes risk in the DESIR prospective general French population. Diabetes. 57: 2253–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Saxena R., Hivert M. F., Langenberg C., Tanaka T., Pankow J. S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A. U., et al. 2010. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 42: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A. U., Wheeler E., Glazer N. L., Bouatia-Naji N., Gloyn A. L., et al. 2010. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sparso T., Andersen G., Nielsen T., Burgdorf K. S., Gjesing A. P., Nielsen A. L., Albrechtsen A., Rasmussen S. S., Jorgensen T., Borch-Johnsen K., et al. 2008. The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia. 51: 70–75. [DOI] [PubMed] [Google Scholar]
- 114.Kolz M., Johnson T., Sanna S., Teumer A., Vitart V., Perola M., Mangino M., Albrecht E., Wallace C., Farrall M., et al. 2009. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 5: e1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iizuka K., Horikawa Y. 2008. ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr. J. 55: 617–624. [DOI] [PubMed] [Google Scholar]
- 116.Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J., Shillinglaw W., Arnot D., Uyeda K. 2001. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA. 98: 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ishii S., Iizuka K., Miller B. C., Uyeda K. 2004. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc. Natl. Acad. Sci. USA. 101: 15597–15602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Iizuka K., Bruick R. K., Liang G., Horton J. D., Uyeda K. 2004. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci. USA. 101: 7281–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kiss-Toth E., Bagstaff S. M., Sung H. Y., Jozsa V., Dempsey C., Caunt J. C., Oxley K. M., Wyllie D. H., Polgar T., Harte M., O'Neill L A., Qwarnstrom E. E., Dower S. K. 2004. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J. Biol. Chem. 279: 42703–42708. [DOI] [PubMed] [Google Scholar]
- 120.Sung H. Y., Guan H., Czibula A., King A. R., Eder K., Heath E., Suvarna S. K., Dower S. K., Wilson A. G., Francis S. E., et al. 2007. Human tribbles-1 controls proliferation and chemotaxis of smooth muscle cells via MAPK signaling pathways. J. Biol. Chem. 282: 18379–18387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yamamoto M., Uematsu S., Okamoto T., Matsuura Y., Sato S., Kumar H., Satoh T., Saitoh T., Takeda K., Ishii K. J., et al. 2007. Enhanced TLR-mediated NF-IL6 dependent gene expression by Trib1 deficiency. J. Exp. Med. 204: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Benn M. 2009. Apolipoprotein B levels, APOB alleles, and risk of ischemic cardiovascular disease in the general population, a review. Atherosclerosis. 206: 17–30. [DOI] [PubMed] [Google Scholar]
- 123.Varret M., Abifadel M., Rabes J. P., Boileau C. 2008. Genetic heterogeneity of autosomal dominant hypercholesterolemia. Clin. Genet. 73: 1–13. [DOI] [PubMed] [Google Scholar]
- 124.Zhong S., Magnolo A. L., Sundaram M., Zhou H., Yao E. F., Leo E. D., Loria P., Wang S., Bamji-Mirza M., Wang L., et al. 2010. Nonsynonymous mutations within APOB in human familial hypobetalipoproteinemia - evidence for feedback inhibition of lipogenesis and post-endoplasmic reticulum degradation of apolipoprotein B. J. Biol. Chem. 285: 6453–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Masson D., Jiang X. C., Lagrost L., Tall A. R. 2009. The role of plasma lipid transfer proteins in lipoprotein metabolism and atherogenesis. J. Lipid Res. 50: S201–S206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mak P. A., Kast-Woelbern H. R., Anisfeld A. M., Edwards P. A. 2002. Identification of PLTP as an LXR target gene and apoE as an FXR target gene reveals overlapping targets for the two nuclear receptors. J. Lipid Res. 43: 2037–2041. [DOI] [PubMed] [Google Scholar]
- 127.Vergeer M., Boekholdt S. M., Sandhu M. S., Ricketts S. L., Wareham N. J., Brown M. J., de Faire U., Leander K., Gigante B., Kavousi M., et al. 2010. Genetic variation at the phospholipid transfer protein locus affects its activity and high-density lipoprotein size and is a novel marker of cardiovascular disease susceptibility. Circulation. 122: 470–477. [DOI] [PubMed] [Google Scholar]
- 128.Jarvik G. P., Rajagopalan R., Rosenthal E. A., Wolfbauer G., McKinstry L., Vaze A., Brunzell J., Motulsky A. G., Nickerson D. A., Heagerty P. J., et al. 2010. Genetic and nongenetic sources of variation in phospholipid transfer protein activity. J. Lipid Res. 51: 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jiang X., Francone O. L., Bruce C., Milne R., Mar J., Walsh A., Breslow J. L., Tall A. R. 1996. Increased prebeta-high density lipoprotein, apolipoprotein AI, and phospholipid in mice expressing the human phospholipid transfer protein and human apolipoprotein AI transgenes. J. Clin. Invest. 98: 2373–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jiang X. C., Bruce C., Mar J., Lin M., Ji Y., Francone O. L., Tall A. R. 1999. Targeted mutation of plasma phospholipid transfer protein gene markedly reduces high-density lipoprotein levels. J. Clin. Invest. 103: 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jiang X. C., Li Z., Liu R., Yang X. P., Pan M., Lagrost L., Fisher E. A., Williams K. J. 2005. Phospholipid transfer protein deficiency impairs apolipoprotein-B secretion from hepatocytes by stimulating a proteolytic pathway through a relative deficiency of vitamin E and an increase in intracellular oxidants. J. Biol. Chem. 280: 18336–18340. [DOI] [PubMed] [Google Scholar]
- 132.Jiang X. C., Qin S., Qiao C., Kawano K., Lin M., Skold A., Xiao X., Tall A. R. 2001. Apolipoprotein B secretion and atherosclerosis are decreased in mice with phospholipid-transfer protein deficiency. Nat. Med. 7: 847–852. [DOI] [PubMed] [Google Scholar]
- 133.Glaser C., Heinrich J., Koletzko B. 2010. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism. 59: 993–999. [DOI] [PubMed] [Google Scholar]
- 134.Schaeffer L., Gohlke H., Muller M., Heid I. M., Palmer L. J., Kompauer I., Demmelmair H., Illig T., Koletzko B., Heinrich J. 2006. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum. Mol. Genet. 15: 1745–1756. [DOI] [PubMed] [Google Scholar]
- 135.Tanaka T., Shen J., Abecasis G. R., Kisialiou A., Ordovas J. M., Guralnik J. M., Singleton A., Bandinelli S., Cherubini A., Arnett D., et al. 2009. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5: e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martinelli N., Girelli D., Malerba G., Guarini P., Illig T., Trabetti E., Sandri M., Friso S., Pizzolo F., Schaeffer L., et al. 2008. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am. J. Clin. Nutr. 88: 941–949. [DOI] [PubMed] [Google Scholar]
- 137.Hicks A. A., Pramstaller P. P., Johansson A., Vitart V., Rudan I., Ugocsai P., Aulchenko Y., Franklin C. S., Liebisch G., Erdmann J., et al. 2009. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 5: e1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coon H., Myers R. H., Borecki I. B., Arnett D. K., Hunt S. C., Province M. A., Djousse L., Leppert M. F. 2000. Replication of linkage of familial combined hyperlipidemia to chromosome 1q with additional heterogeneous effect of apolipoprotein A-I/C-III/A-IV locus. The NHLBI Family Heart Study. Arterioscler. Thromb. Vasc. Biol. 20: 2275–2280. [DOI] [PubMed] [Google Scholar]
- 139.Huertas-Vazquez A., del Rincon J. P., Canizales-Quinteros S., Riba L., Vega-Hernandez G., Ramirez-Jimenez S., Auron-Gomez M., Gomez-Perez F. J., Aguilar-Salinas C. A., Tusie-Luna M. T. 2004. Contribution of chromosome 1q21-q23 to familial combined hyperlipidemia in Mexican families. Ann. Hum. Genet. 68: 419–427. [DOI] [PubMed] [Google Scholar]
- 140.Pei W., Baron H., Muller-Myhsok B., Knoblauch H., Al-Yahyaee S. A., Hui R., Wu X., Liu L., Busjahn A., Luft F. C., et al. 2000. Support for linkage of familial combined hyperlipidemia to chromosome 1q21-q23 in Chinese and German families. Clin. Genet. 57: 29–34. [DOI] [PubMed] [Google Scholar]
- 141.Pajukanta P., Lilja H. E., Sinsheimer J. S., Cantor R. M., Lusis A. J., Gentile M., Duan X. J., Soro-Paavonen A., Naukkarinen J., Saarela J., et al. 2004. Familial combined hyperlipidemia is associated with upstream transcription factor 1 (USF1). Nat. Genet. 36: 371–376. [DOI] [PubMed] [Google Scholar]
- 142.Casado M., Vallet V. S., Kahn A., Vaulont S. 1999. Essential role in vivo of upstream stimulatory factors for a normal dietary response of the fatty acid synthase gene in the liver. J. Biol. Chem. 274: 2009–2013. [DOI] [PubMed] [Google Scholar]
- 143.Vallet V. S., Casado M., Henrion A. A., Bucchini D., Raymondjean M., Kahn A., Vaulont S. 1998. Differential roles of upstream stimulatory factors 1 and 2 in the transcriptional response of liver genes to glucose. J. Biol. Chem. 273: 20175–20179. [DOI] [PubMed] [Google Scholar]
- 144.Naukkarinen J., Nilsson E., Koistinen H. A., Soderlund S., Lyssenko V., Vaag A., Poulsen P., Groop L., Taskinen M. R., Peltonen L. 2009. Functional variant disrupts insulin induction of USF1: mechanism for USF1-associated dyslipidemias. Circ. Cardiovasc. Genet. 2: 522–529. [DOI] [PubMed] [Google Scholar]
- 145.Naukkarinen J., Gentile M., Soro-Paavonen A., Saarela J., Koistinen H. A., Pajukanta P., Taskinen M. R., Peltonen L. 2005. USF1 and dyslipidemias: converging evidence for a functional intronic variant. Hum. Mol. Genet. 14: 2595–2605. [DOI] [PubMed] [Google Scholar]
- 146.Paterniti J. R., Jr., Brown W. V., Ginsberg H. N., Artzt K. 1983. Combined lipase deficiency (cld): a lethal mutation on chromosome 17 of the mouse. Science 221: 167–169. [DOI] [PubMed] [Google Scholar]
- 147.Peterfy M., Ben-Zeev O., Mao H. Z., Weissglas-Volkov D., Aouizerat B. E., Pullinger C. R., Frost P. H., Kane J. P., Malloy M. J., Reue K., et al. 2007. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat. Genet. 39: 1483–1487. [DOI] [PubMed] [Google Scholar]
- 148.Doolittle M. H., Neher S. B., Ben-Zeev O., Ling-Liao J., Gallagher C. M., Hosseini M., Yin F., Wong H., Walter P., Peterfy M. 2009. Lipase maturation factor LMF1, membrane topology and interaction with lipase proteins in the endoplasmic reticulum. J. Biol. Chem. 284: 33623–33633. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 149.Yin F., Doolittle M. H., Peterfy M. 2009. A quantitative assay measuring the function of lipase maturation factor 1. J. Lipid Res. 50: 2265–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cefalu A. B., Noto D., Arpi M. L., Yin F., Spina R., Hilden H., Barbagallo C. M., Carroccio A., Tarugi P., Squatrito S., et al. 2009. Novel LMF1 nonsense mutation in a patient with severe hypertriglyceridemia. J. Clin. Endocrinol. Metab. 94: 4584–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ioka R. X., Kang M. J., Kamiyama S., Kim D. H., Magoori K., Kamataki A., Ito Y., Takei Y. A., Sasaki M., Suzuki T., et al. 2003. Expression cloning and characterization of a novel glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein, GPI-HBP1. J. Biol. Chem. 278: 7344–7349. [DOI] [PubMed] [Google Scholar]
- 152.Beigneux A. P., Davies B. S., Gin P., Weinstein M. M., Farber E., Qiao X., Peale F., Bunting S., Walzem R. L., Wong J. S., et al. 2007. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Beigneux A. P., Davies B. S., Bensadoun A., Fong L. G., Young S. G. 2009. GPIHBP1, a GPI-anchored protein required for the lipolytic processing of triglyceride-rich lipoproteins. J. Lipid Res. 50(Suppl): S57–S62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Davies B. S., Waki H., Beigneux A. P., Farber E., Weinstein M. M., Wilpitz D. C., Tai L. J., Evans R. M., Fong L. G., Tontonoz P., et al. 2008. The expression of GPIHBP1, an endothelial cell binding site for lipoprotein lipase and chylomicrons, is induced by peroxisome proliferator-activated receptor-gamma. Mol. Endocrinol. 22: 2496–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gin P., Yin L., Davies B. S., Weinstein M. M., Ryan R. O., Bensadoun A., Fong L. G., Young S. G., Beigneux A. P. 2008. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J. Biol. Chem. 283: 29554–29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dallinga-Thie G. M., Franssen R., Mooij H. L., Visser M. E., Hassing H. C., Peelman F., Kastelein J. J., Peterfy M., Nieuwdorp M. 2010. The metabolism of triglyceride-rich lipoproteins revisited: New players, new insight. Atherosclerosis. 211: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Beigneux A. P., Vergnes L., Qiao X., Quatela S., Davis R., Watkins S. M., Coleman R. A., Walzem R. L., Philips M., Reue K., et al. 2006. Agpat6–a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J. Lipid Res. 47: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Beigneux A. P., Gin P., Davies B. S., Weinstein M. M., Bensadoun A., Ryan R. O., Fong L. G., Young S. G. 2008. Glycosylation of Asn-76 in mouse GPIHBP1 is critical for its appearance on the cell surface and the binding of chylomicrons and lipoprotein lipase. J. Lipid Res. 49: 1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Beigneux A. P., Franssen R., Bensadoun A., Gin P., Melford K., Peter J., Walzem R. L., Weinstein M. M., Davies B. S., Kuivenhoven J. A., et al. 2009. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler. Thromb. Vasc. Biol. 29: 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Olivecrona G., Ehrenborg E., Semb H., Makoveichuk E., Lindberg A., Hayden M. R., Gin P., Davies B. S., Weinstein M. M., Fong L. G., et al. 2010. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J. Lipid Res. 51: 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Franssen R., Young S. G., Peelman F., Hertecant J., Sierts J. A., Schimmel A. W., Bensadoun A., Kastelein J. J., Fong L., Dallinga-Thie G. M., et al. 2010. Chylomicronemia with Low Postheparin Lipoprotein Lipase Levels in the Setting of GPIHBP1 Defects. Circ. Cardiovasc. Genet. 3: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang J., Hegele R. A. 2007. Homozygous missense mutation (G56R) in glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPI-HBP1) in two siblings with fasting chylomicronemia (MIM 144650). Lipids Health Dis. 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gin P., Beigneux A. P., Davies B., Young M. F., Ryan R. O., Bensadoun A., Fong L. G., Young S. G. 2007. Normal binding of lipoprotein lipase, chylomicrons, and apo-AV to GPIHBP1 containing a G56R amino acid substitution. Biochim. Biophys. Acta. 1771: 1464–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rung J., Cauchi S., Albrechtsen A., Shen L., Rocheleau G., Cavalcanti-Proenca C., Bacot F., Balkau B., Belisle A., Borch-Johnsen K., et al. 2009. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nat. Genet. 41: 1110–1115. [DOI] [PubMed] [Google Scholar]
- 165.Wilson P. W., D'Agostino R. B., Levy D., Belanger A. M., Silbershatz H., Kannel W. B. 1998. Prediction of coronary heart disease using risk factor categories. Circulation. 97: 1837–1847. [DOI] [PubMed] [Google Scholar]
- 166.Kathiresan S., Melander O., Anevski D., Guiducci C., Burtt N. P., Roos C., Hirschhorn J. N., Berglund G., Hedblad B., Groop L., et al. 2008. Polymorphisms associated with cholesterol and risk of cardiovascular events. N. Engl. J. Med. 358: 1240–1249. [DOI] [PubMed] [Google Scholar]
- 167.Talmud P. J., Drenos F., Shah S., Shah T., Palmen J., Verzilli C., Gaunt T. R., Pallas J., Lovering R., Li K., et al. 2009. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am. J. Hum. Genet. 85: 628–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kraft P., Hunter D. J. 2009. Genetic risk prediction–are we there yet? N. Engl. J. Med. 360: 1701–1703. [DOI] [PubMed] [Google Scholar]
- 169.Lai C. Q., Arnett D. K., Corella D., Straka R. J., Tsai M. Y., Peacock J. M., Adiconis X., Parnell L. D., Hixson J. E., Province M. A., et al. 2007. Fenofibrate effect on triglyceride and postprandial response of apolipoprotein A5 variants: the GOLDN study. Arterioscler. Thromb. Vasc. Biol. 27: 1417–1425. [DOI] [PubMed] [Google Scholar]
- 170.MacLean D. R., Petrasovits A., Nargundkar M., Connelly P. W., MacLeod E., Edwards A., Hessel P. 1992. Canadian heart health surveys: a profile of cardiovascular risk. Survey methods and data analysis. Canadian Heart Health Surveys Research Group. CMAJ. 146: 1969–1974. [PMC free article] [PubMed] [Google Scholar]