Abstract
ELOVL2 is a member of the mammalian microsomal ELOVL fatty acid enzyme family, involved in the elongation of very long-chain fatty acids including PUFAs required for various cellular functions in mammals. Here, we used ELOVL2-ablated (Elovl2−/−) mice to show that the PUFAs with 24–30 carbon atoms of the ω-6 family in testis are indispensable for normal sperm formation and fertility in male mice. The lack of Elovl2 was associated with a complete arrest of spermatogenesis, with seminiferous tubules displaying only spermatogonia and primary spermatocytes without further germinal cells. Furthermore, based on acyl-CoA profiling, heterozygous Elovl2+/− male mice exhibited haploinsufficiency, with reduced levels of C28:5 and C30:5n-6 PUFAs, which gave rise to impaired formation and function of haploid spermatides. These new insights reveal a novel mechanism involving ELOVL2-derived PUFAs in mammals and previously unrecognized roles for C28 and C30 n-6 PUFAs in male fertility. In accordance with the function suggested for ELOVL2, the Elovl2−/− mice show distorted levels of serum C20 and C22 PUFAs from both the n-3 and the n-6 series. However, dietary supplementation with C22:6n-3 could not restore male fertility to Elovl2+/− mice, suggesting that the changes in n-6 fatty acid composition seen in the testis of the Elovl2+/− mice, cannot be compensated by increased C22:6n-3 content.
Keywords: fatty acid elongase, omega-6, spermatogenesis
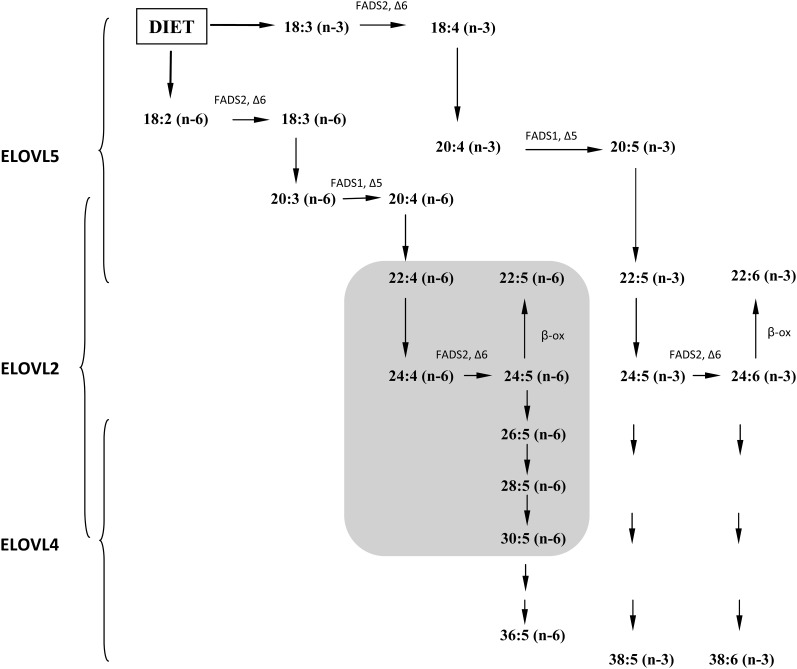
Essential fatty acids, i.e., linoleic (C18:2n-6) and α-linolenic acid (C18:3n-3), which are prerequisites for normal growth, development, and function in mammals, cannot be synthesized de novo and, as such, have to be derived from diet (1). Subsequently, the C18:2n-6 and C18:3n-3 fatty acids can be converted into other PUFAs through a series of elongation and desaturation steps performed by distinct enzymes residing in the endoplasmic reticulum (2). Desaturation steps are performed by the Δ5- and Δ6-desaturases, while chain elongation is controlled by the elongation of very-long-chain fatty acid (ELOVL) enzymes, ELOVL2, ELOVL4, and ELOVL5. ELOVL4 is believed to be involved in the elongation of PUFAs in the retina, brain, skin, and testis (3). Using cultured cells that overexpress this protein, it has been shown that ELOVL4 is involved in the synthesis of saturated C28 and C30 very-long-chain FA (VLCFA), as well as polyunsaturated C28–C38 VLC-PUFAs such as the C34:6n-3 and C36:6n-3 fatty acids (4). ELOVL5 predominantly elongates C18 PUFAs up to 22 carbons in length, while ELOVL2 has been shown to be involved in the elongation of C20 and C22 PUFA in order to produce C24:4n-6 and C24:5n-3 PUFAs (5–7). These latter two are also recognized as precursors for docosapentaenoic acid (C22:5n-6) and docosahexaenoic acid (DHA) (C22:6n-3) synthesis, respectively, via the so-called Sprecher pathway, which includes one step of β oxidation/chain shortening (8).
To assess the in vivo function of ELOVL2, we generated Elovl2+/− and Elovl2−/− mice by homologous recombination and present evidence that ELOVL2 is essential for the formation of C24:5n-6 to C30:5n-6 PUFAs in testis and that these fatty acids are indispensable for normal spermatogenesis and fertility.
EXPERIMENTAL PROCEDURES
Elovl2 gene targeting
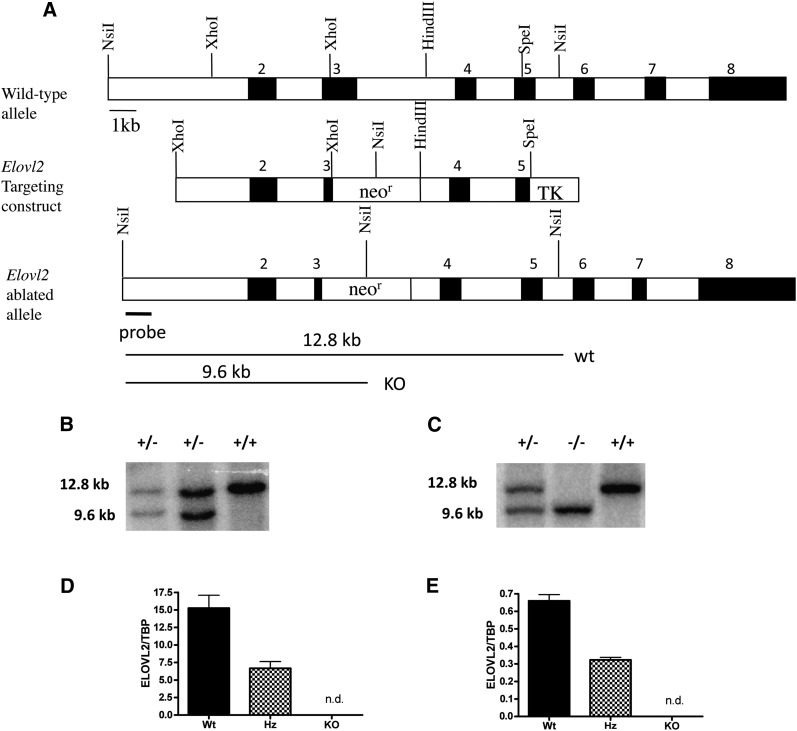
Genomic liver DNA from the 129/Sv mouse strain was used to isolate the Elovl2 gene. A 4.8 kb fragment excised with XhoI and a 3.3 kb fragment excised with SpeI and HindIII was used for the targeting construct (Fig. 1A). A 2.4 kb neomycin resistance (neor) gene fragment was inserted into the construct using SalI, and a thymidine kinase (TK) cassette was inserted using a SpeI site at the 3′ end of the Elovl2 targeting construct. The targeting construct was linearized with NruI and electroporated into R1 embryonic stem (ES) cells derived from male 129/Sv agouti mice (9). ES cells were cultured and subjected to positive/negative selection using G418 and fialuridine, (FIUA) respectively, and were further analyzed by Southern blotting (see below). The Elovl2-ablated ES cells were injected into C57BL/6J blastocysts, which were implanted into foster mothers (F1, CBA x C57/BL6) according to standard procedures. Male offspring, being the most chimeric, were bred with C57BL/6J females to generate offspring heterozygous for the mutation.
Fig. 1.
Generation of Elovl2-ablated mice. A: Elovl2 gene targeting construct. The neomycin resistance (neor) gene was used to replace a major portion of exon 3 in the ELOVL2 gene. The mutated Elovl2 allele differs from that of the wild-type by the addition of an internal NsiI site in the neor gene. Black boxes represent exons and are numbered (2–8) above. TK, thymidine kinase. Genomic DNA from embryonic stem cells (B) and tail DNA (C) was digested with NsiI and analyzed with Southern blotting for the mutated Elovl2 allele. Digestion yielded the 12.8 kb wild-type band and the 9.6 kb targeted band detected by the probe indicated in panel A. Elovl2 expression analysis using real-time PCR is shown in liver (D) and testis (E). Results are means ± SEM of 7 wild-type (Wt) heterozygous (Hz) mice and 2 knockout (KO) animals. *, P < 0.05; **, P < 0.01.
Southern blot analysis
A 1.5 kb fragment at the 5′ end of the targeting construct was used as a probe to confirm Elovl2 disruption in ES cell clones and mouse tail DNA. The probe was labeled with α-dCTP (Amersham Bioscience), using Ready-To-Go DNA labeling beads (GE Healthcare) for >45 min at 37°C. Probes were purified with a Sephadex G-50 DNA-grade nick column (GE Healthcare) and subsequently denatured. In addition, the neor fragment was also used as a probe to confirm a specific targeting event. Approximately 10 μg of ES cell DNA or mouse tail DNA was digested with NsiI and separated using 1% agarose gel electrophoresis. DNA was denatured in a solution containing 0.2 M NaOH, 0.6 M NaCl for 30 min and in a solution of 0.5 M Tris (pH 7.5), 1.5 M NaCl for 30 min. Digested DNA was transferred to Hybond-XL membrane (Amersham Bioscience) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight and cross-linked to the membrane using a UV Stratalinker 1800 unit (Stratagene). Membranes were prehybridized in a solution of 50% formamide, 5× SSC, 5× Denhart's solution, 50 mM sodium phosphate, 0.5% SDS, and 1.5 mg of degraded herring sperm DNA (Sigma Aldrich) at 42°C. After membranes were prehybridized, they were transferred to a similar solution containing the denatured probe. Hybridization was carried out at 42°C overnight. Membranes were washed in a mixture of 2× SSC, 0.2% SDS at room temperature for 15 min and in 0.1× SSC, 0.2% SDS at 43°C for 15 min and analyzed by phosphorimaging using an FLA-3000 reader (Fuji).
Animals
All animals were housed at 22°C and maintained on a 12 h light:12 h dark cycle. Animals were fed ad libitum with Rat and Mouse standard chow (no.1; BeeKay Feeds, B and K Universal, Stockholm, Sweden) and had free access to water. All studies were carried out with permission from the Animal Ethics Board in Stockholm, Sweden.
Dietary supplementation
Male and female Elovl2+/− mice were fed a DHA (C22:6n-3)-enriched diet (19.5% of total) obtained from Polaris (Quimper, France), for 3 months ad libitum. For the fatty acid composition of the diet see Table 3.
TABLE 3.
Fatty acid composition of DHA-enriched diet
| Fatty acid | % of fatty acid in DHA diet |
|---|---|
| C12:0 | 0 |
| C14:0 | 0 |
| C16:0 | 4.44 |
| C18:0 | 2.09 |
| C18:1n-9 | 30.53 |
| C18:2n-6 | 34.69 |
| C18:3n-3 | 3.45 |
| C20:5n-3 | 5.27 |
| C22:6n-3 | 19.53 |
| satfa | 6.53 |
| mufa | 30.53 |
| n-6 | 34.69 |
| n-3 | 28.25 |
Quantitative RT-PCR
RNA was isolated with Ultraspec RNA isolation solution (Biosite, San Diego, CA), and total RNA was isolated according to the manufacturer's procedure. For real-time PCR, 500 ng of total RNA was reverse transcribed using random hexamer primers, deoxynucleoside triphosphates (dNTPs), multiscript, and RNase inhibitor (Applied Biosystems, Foster City, CA). Complementary DNA samples were diluted 1:10, and 2 µl aliquots of sample cDNA were mixed with SYBR Green JumpStart Taq ReadyMix (Sigma Aldrich), prevalidated primers, and diethyl pyrocarbonate DEPC, Sigma)-treated water and measured in duplicate for each sample. Expression analysis was performed with an ABI Prism 7000 sequence detection system (Applied Biosystems). Data were normalized to TFIIb or TBP.
MRI measurement
In vivo MRI using an EchoMRI-100TM machine (Echo Medical Systems, Houston, TX) was performed in order to measure body fat and lean contents.
Lipid profiling
Triglyceride and fatty acid assays were performed as previously described (10). To measure total fatty acid methyl ester (FAME) molecular species, lipid amounts corresponding to an equivalent of 3 mg of tissue were extracted in the presence of glyceryl triheptadecanoate (0.5 μg) as an internal standard. Lipid extract was transmethylated with 1 ml of BF3 in methanol (1/20, v/v) for 150 min at 100°C and evaporated to dryness, and the FAME species were extracted with hexane-water, 1:1. The organic phase was evaporated to dryness and dissolved in 50 μl of ethyl acetate. One microliter of FAME was analyzed by gas-liquid chromatography using a 5890 Hewlett Packard system with Restek Famewax fused silica capillary columns (30 m × 0.32 mm ID; 0.25 mm film thickness). Oven temperature changes were programmed from 110°C to 220°C at a rate of 2°C per min, and the carrier gas was helium (0.5 bar). Injector and detector temperatures were 225°C and 245°C, respectively.
HPLC Analysis of acyl-etheno-CoA derivatives
Twenty milligram portions of material were frozen in liquid nitrogen and subsequently extracted for quantitative analysis of fluorescent acyl-etheno-CoA derivatives by HPLC. Analysis of acyl-CoA was performed using an Agilent 1100 LC system (LUNA 150 × 3.2 mm C18[2] column; Phenomenex); methodology and gradient conditions have been described previously (11–13).
Histology
At necropsy, testis and epididymis tissues were fixed in 10% neutral buffered formalin, sectioned, stained with hematoxylin and eosin, and examined by light microscopy for histological evaluation.
RESULTS
ELOVL2 expression is critical for normal male fertility
Real-time quantitative PCR revealed a reduction of almost exactly 50% of Elovl2 mRNA transcript in the liver and testis of the Elovl2+/− mice (Fig. 1D, E), indicating an Elovl2 haploinsufficiency in these mice. No transcript was found in the Elovl2−/− mice. Breeding of Elovl2+/− mice for further production of Elovl2−/− mice displayed severe fertility problems, as shown by reduced production of offspring.
Elovl2+/− female and Elovl2+/+ male mice gave rise to offspring of both the Elovl2+/+ and the Elovl2+/− genotypes in a normal ratio, while 90% of all Elovl2+/− males tested (36/40) were shown to be infertile during their reproductive period (1 year), and only 2 of the fertile males gave rise to more than one litter. In addition, heterozygous breeding did not produce offspring in a normal 1:2:1 Mendelian ratio. Rather, the ratio between Elovl2+/+ and Elovl2+/− genotypes was close to 1:1, and of about 300 offspring, only 3 Elovl2−/− mice (2 males and 1 female) were born. As no indications of embryonic lethality were observed, this implies a deficiency of spermatogenesis in Elovl2+/− mice. The two male Elovl2−/− mice were both sterile, while the female mouse was able to produce offspring together with wild-type males.
No differences were seen in body weight, fat and lean contents, or liver weight among the wild-type, Elovl2+/−, and Elovl2−/− male mice fed on a standard chow diet (Table 1). However, the two male Elovl2−/− mice exhibited marked hypogonadism, with testis weight reduced by 60% compared with age-matched controls.
TABLE 1.
Phenotypic comparison of wild-type, Elovl2, and Elovl2 male mice
| Parameter | Elovl2+/+ | Elovl2+/− | Elovl2+/+ | Elovl2−/− |
|---|---|---|---|---|
| Sex | Male | Male | Male | Male |
| Age (weeks) | 14 | 14 | 22/52 | 22/52 |
| Number of mice | 7 | 6 | 2 | 2 |
| Body weight (g) | 30.7 ± 0.8 | 30.4 ± 0.7 | 40.6 ± 0.9 | 42.5 ± 6.1 |
| Fat content (g) | 4.4 ± 0.8 | 4.4 ± 0.4 | 11.7 ± 1.0 | 12.2 ± 4.1 |
| Lean content (g) | 23.6 ± 0.4 | 23.1 ± 0.5 | 25.4 ± 0.1 | 25.4 ± 1.6 |
| Liver weight (g) | 1.5 ± 0.1 | 1.4 ± 0.1 | NA | NA |
| Testis weight (g) | 0.22 ± 0.01 | 0.23 ± 0.01 | 0.25 ± 0.01 | 0.10 ± 0.02 |
Characteristics of mice at 14 weeks of age (Elovl2+/+ and Elovl2+/−) or at 22 and 52 weeks of age (Elovl2+/+ and Elovl2−/−, respectively) fed on standard chow diet. Results shown are means ± SEM. NA, not analyzed
ELOVL2 is essential for normal spermatogenesis
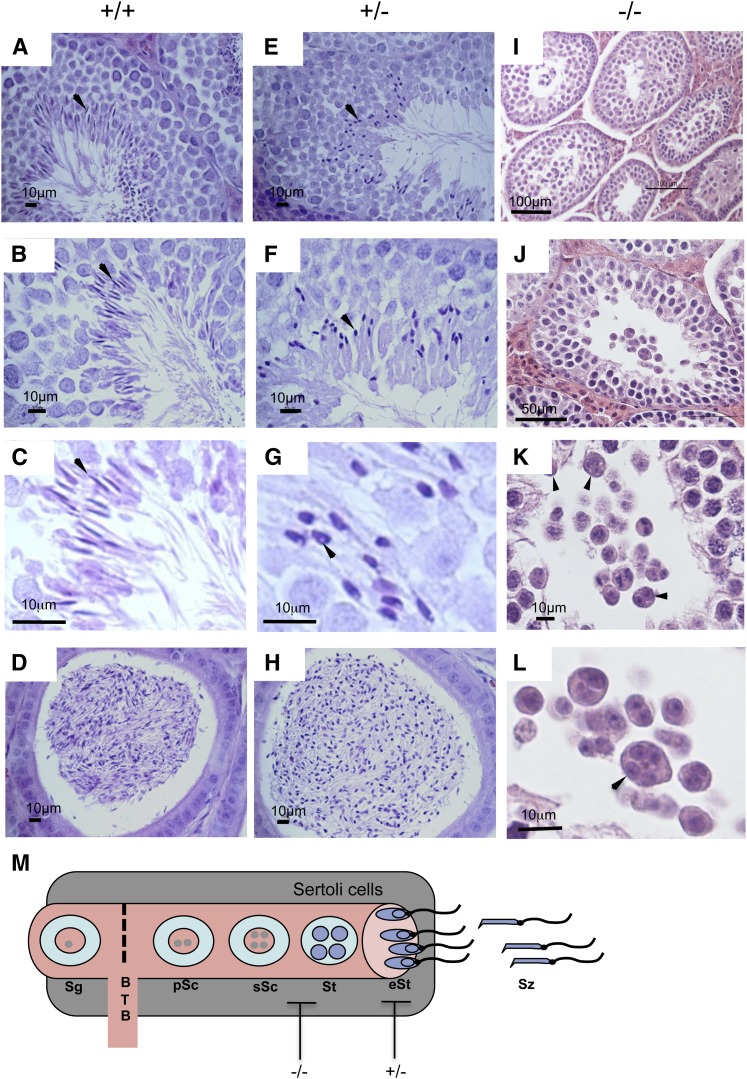
Histological analyses of wild-type testis showed all stages of spermatogenesis (Fig. 2AndashC), and mature spermatozoa were elongated and displayed a marked hook-shaped head (Fig. 2C, arrow). In contrast, seminiferous tubules of the Elovl2−/− mice displayed spermatogonia with numerous mitoses and primary spermatocytes but lacked further germinal cells (Fig. 2IndashL). The primary spermatocytes degenerated and formed multinucleate giant cells that desquamated in the lumen of the seminiferous tubule with an incomplete formation of the four haploid spermatides (Fig. 2K, L, arrows). These changes were observed in every section of seminiferous tubules of both testicles and were consistent with bilateral testicular hypoplasia and arrested spermatogenesis; however, both the Sertoli cells and the Leydig cells appeared normal in the interstitium.
Fig. 2.
Histological analysis of testes in wild-type, Elovl2+/−, and Elovl2−/− mice. Mature spermatozoa are shown with elongated heads in seminiferous tubules (A–C, arrows) and epididymes (D) of wild-type mice. Elovl2+/− testis are seen with numerous spermatozoa displaying rounded, condensed heads (E–G, arrows), and the condensed nucleus frequently displays a small rounded vacuole about 1–2 µm (G, arrowhead). Epididymes of wild-type (D) and Elovl2+/− (H) mice display spermatozoa with rounded condensed heads compared with those of wild-type epididymis. Elovl2−/− mice displayed only spermatogonia and primary spermatocytes (I, J) with multinucleate giant cells in the lumen (K, L, arrows). (M) Spermatogenesis within the seminiferous tubules and blockage by Elovl2 ablation in Elovl2−/− and Elovl2+/− mice. Sg, spermatogonia; pSc, primary spermatocytes; sSc, secondary spermatocytes; BTB, blood testis barrier; St, spermatides; eSt, elongated spermatides; Sz, spermatozoa.
Testis of the Elovl2+/− male mice showed normal seminiferous tubules displaying normal Sertoli cells, and the Leydig cells in the interstitium were normal; however, heterozygote mice displayed abnormal sperm morphology, with numerous spermatozoa displaying a rounded, condensed head (Fig. 2EndashG), and the condensed nucleus frequently displayed a small rounded vacuole measuring about 1–2 µm (Fig. 2G, arrowhead). Nonetheless, sperm were subsequently released into the lumen and attained normal flagellar movement. The epididymis of the Elovl2+/− mice showed the presence of numerous spermatozoa, which, as in the seminiferous tubules, displayed rounded and condensed heads (Fig. 2H). No obvious differences were noticed in total number of sperm, which is in line with the normal testis weight found in heterozygote mice. Together, these data show that the impaired fertility in Elovl2−/− and Elovl2+/− male mice is due to developmental blockage of spermatide formation and occurs at the stage of sperm head packaging during spermatogenesis, respectively (Fig. 2M).
Gene expression of spermatogenesis markers
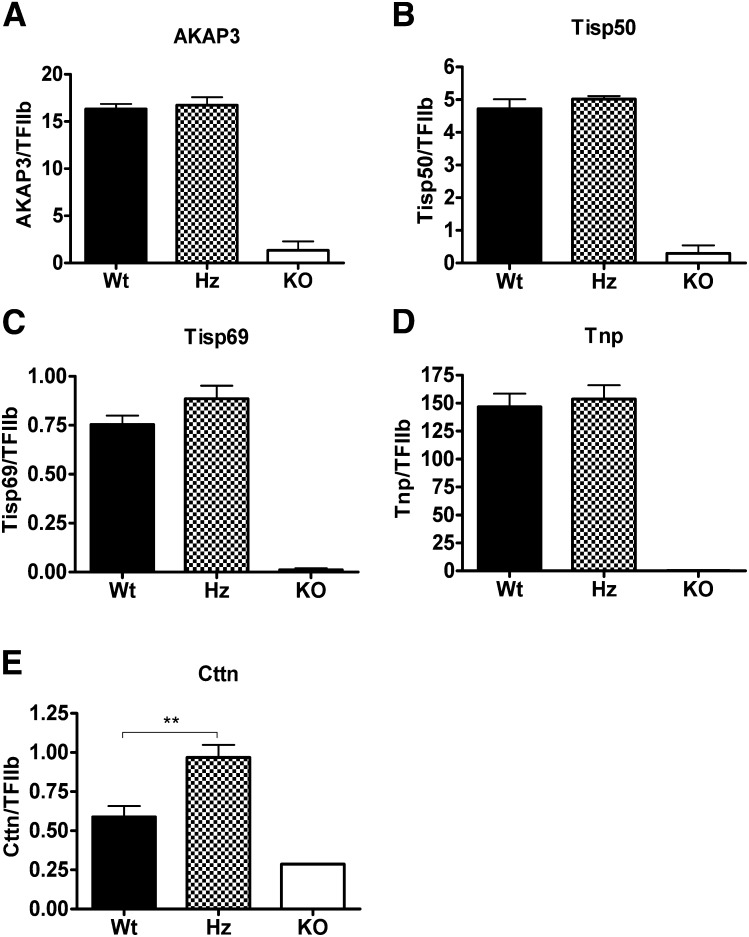
As expected, the levels of mRNA expression of several stage-specific gene products, which are controlled by the bZip-type cAMP-responsive element modulator (CREM) transcription factor during late spermiogenesis (14), were dramatically downregulated in the Elovl2−/− mice, including the A-kinase anchoring protein-3 (AKAP3), a scaffolding protein for regulatory proteins in the flagella; transcript induced in spermiogenesis 50 (TISP50), a sperm flagellar protein; TISP69, an E3 ligase promoting proteasome-mediated degradation of spermatid proteins in the late spermatid stage; and transition protein-1 (Tnp1), which is involved in sperm head compaction (Fig. ). However, the expression levels of these same encoding genes were unchanged in the Elovl2+/− mice, which is consistent with the fact that it is the formation and extrusion of the head components of the spermatids during their release into the lumen of the seminiferous tubules and not the formation of the flagellum and mitochondrial sheath that is impaired in the heterozygote Elovl2+/− mice. Of the genes investigated, only the Cttn (cortactin) gene expression, which is expressed in the apical membrane of Sertoli cells facing germ cells and is involved in a complex of spermatid-Sertoli cell interactions during spermatid head shaping (15, 16), was changed in Elovl2+/− male mice (Fig. 3E).
Fig. 3.
Testicular gene expression. Analysis of the AKAP3 (A), TISP50 (B), TISP69 (C), Tnp (D) and Cttn (E) genes were examined by real-time PCR in wild-type (Wt), Elovl2 +/−(Hz [heterozygous]), and Elovl2−/− (KO [knockout]) mice. Results shown are the means ± SEM of 7 Wt and Hz animals and 2 KO animals. *, P < 0.05; **, P < 0.01.
ELOVL2 controls the synthesis of 28:5 and 30:5n-6 PUFA in testis
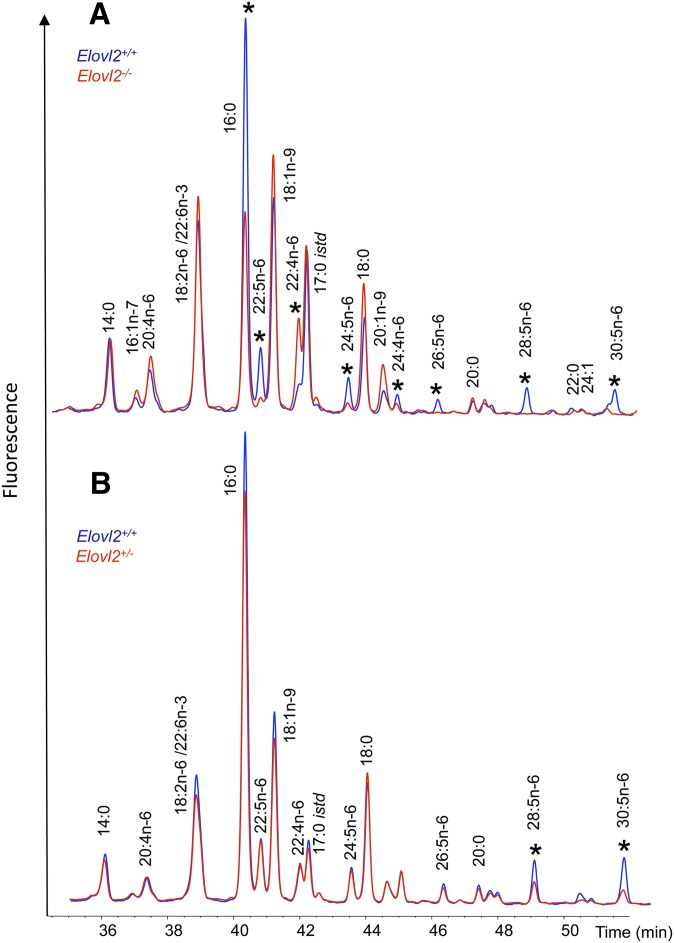
In order to directly link PUFA synthesis in the testis with the impaired spermatogenesis seen in our mutant mice, acyl-CoA profiling of testis was performed in the Elovl2−/− and Elovl2+/− mice and compared with that in age-matched littermates. When we looked at the metabolic pool in which the Elovl enzymes work, except for the C22:6n-3 (DHA) PUFA, the levels of n-3 PUFA-CoAs were too low to be identified within the testis; however, the Elovl2−/− mice exhibited increased levels of C22:4n-6-CoA, accompanied by significantly reduced levels of C22:5n-6-CoA (Fig. 4A). Intriguingly, the mice exhibited an almost complete loss of the acyl-CoA elongation products of C24:5n-6, that is, C26:5n-6, C28:5n-6, and C30:5n-6. These results describe a novel role for ELOVL2 in the formation of VLC-PUFAs and show that n-6 PUFA elongation beyond 22:4 in testis is exclusively dependent on ELOVL2 activity (Fig. 5, gray area).
Fig. 4.
Biosynthesis of VLC-PUFAs in wild-type, Elovl2+/−, and Elovl2−/− mice. The composition of the testicular acyl-CoA pool was determined by extraction, derivatization, and HPLC analysis of acyl-etheno-CoAs. Carbon 17:0 istd is the internal standard acyl-CoA. A: Acyl-CoA composition of wild-type and Elovl2−/− testes. Blue trace corresponds to Elovl2+/+, and the red trace corresponds to Elovl2−/− mice. *, indicates differences between the genotypes. B: Acyl-CoA composition of wild-type and Elovl2+/− testes. Blue trace corresponds to Elovl2+/+ mice, and the red trace corresponds to Elovl2+/− mice. *, indicates differences between the genotypes. Results shown are pooled samples from 7 wild-type and heterozygous mice and 2 knockout animals.
Fig. 5.
In vivo VLC-PUFA biosynthesis by ELOVL2, ELOVL4, and ELOVL5. The 18:3n-3 and 18:2n-6 PUFAs were obtained from the diet and were subsequently elongated and desaturated by the indicated enzymes into longer PUFAs. ELOVL5 is involved in the elongation of PUFAs up to 22 carbons in length. ELOVL2 elongates from 20:4n-6 and 20:5n-3 series. Specifically, in the testis, ELOVL2 is involved in the biosynthesis of VLC-PUFA (≥C24) up to 30 carbons in length of the n-6 family (gray area). ELOVL4 elongates up to C38 in length. C26FADS, fatty acid desaturase.
To further assess the consequences of impaired Elovl2 expression in the heterozygote mice, it was clearly shown that as in Elovl2−/− mice, the acyl-CoA levels of C28:5n-6 and C30:5n-6 PUFA were significantly reduced in the testis of Elovl2+/− mice as well (Fig. 4B). In addition to the changes observed in PUFA levels, we noted a decrease in total fatty acids (FAME mass) and triglycerides in the Elovl2+/− mice (Table 2). Despite this, there were no differences in the composition of triacylglycerols among the mouse strains. The levels of cholesterol, which is the substrate for testosterone synthesis in Leydig cells, and cholesterol esters were not different between wild-type and Elovl2+/− mice (Table 2), indicating that testosterone production is normal in the Elovl2+/− mice. In addition, there were no differences in the appearance of the vesicula seminalis, again, implying that the androgen levels are normal in the Elovl2+/− mice.
TABLE 2.
Neutral lipid composition in testis of wild-type and Elovl2+/− male mice
| Lipid class and length | Wild-type mice | Elovl2+/− mice |
|---|---|---|
| Triglyceride C49 | 2.5 ± 0.9a | 3.5 ± 0.7 |
| Triglyceride C51 | 5.5 ± 0.5a | 6.0 ± 0.6 |
| Triglyceride C53 | 25.5 ± 0.9a | 25.6 ± 0.7 |
| Triglyceride C55 | 38.8 ± 1.7a | 36.8 ± 1.4 |
| Triglyceride C57 | 23.0 ± 0.7a | 22.5 ± 0.7 |
| Triglyceride C59 | 4.7 ± 1.7a | 5.7 ± 1.2 |
| Cholesterol | 15.3 ± 0.9b | 13.5 ± 0.9 |
| Cholesterol esters | 2.7 ± 0.4b | 2.3 ± 0.3 |
| Cholesterol esters C16 | 36.5 ± 2.0a | 37.2 ± 1.2 |
| Cholesterol esters C18 | 55.1 ± 2.4a | 54.7 ± 1.2 |
| Cholesterol esters C20:4 (n-6) | 8.5 ± 1.7a | 8.1 ± 1.8 |
| Triglycerides | 32.4 ± 10.8b | 14.4 ± 5.4 |
| Relative FAME mass | 63.9 ± 11.7 | 22.0 ± 6.0** |
Units are percentage of total ± SEM.
Units are µg/mg.
, P < 0.01.
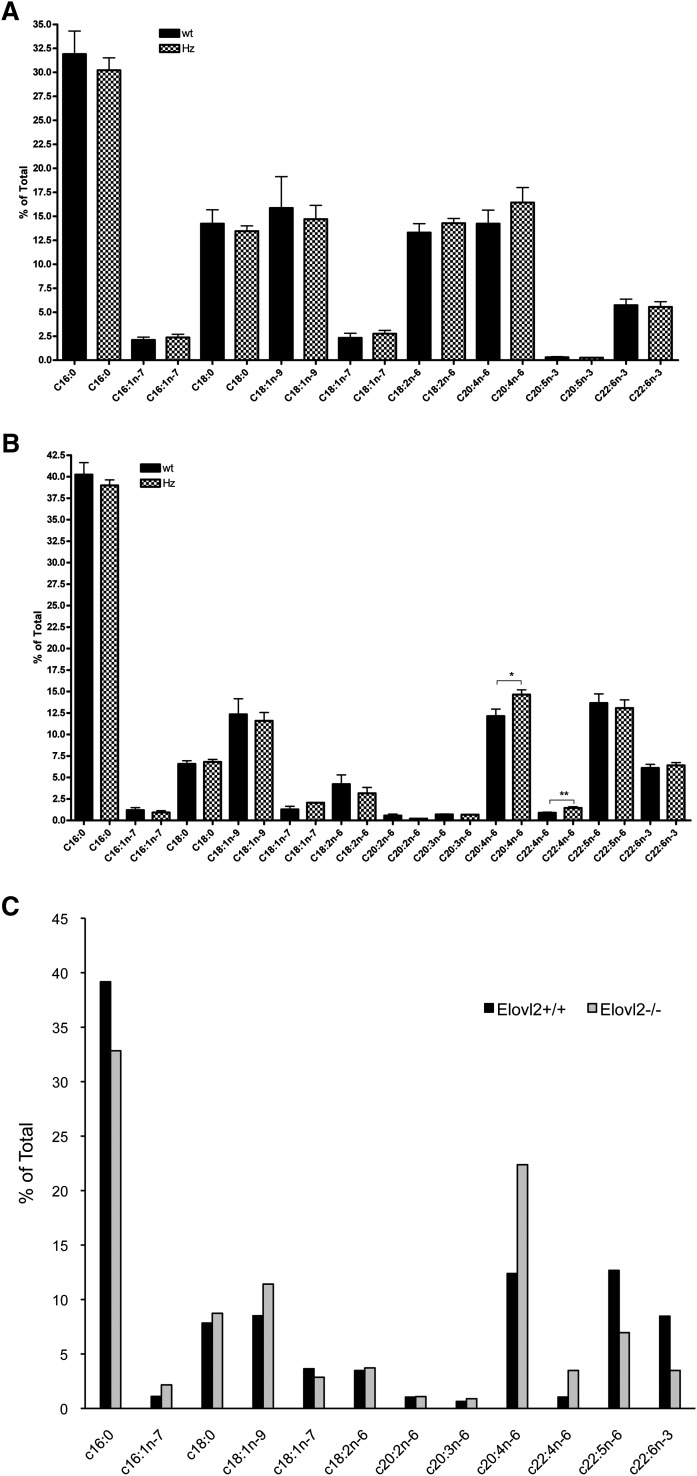
Because Elovl2 expression was reduced in the liver of heterozygote Elovl2+/− male mice to a level that was similar to that in testis, we also performed total lipid profiling by gas chromatography analysis of liver fatty acids and compared those results with levels in testis. Although we were unable to detect any fatty acids longer than C24 in liver, no major changes were seen in PUFA levels in Elovl2+/− mice (Fig. 6A). However, in testis, the level of C20:4n-6 was increased to 25% of total fatty acids compared with that in wild-type mice testis (Fig. 6B), implying a differential mechanism in the control of PUFA levels in different tissues. Furthermore, the elongated product of C20:4n-6 (i.e., C22:4n-6) was increased by almost 70%. Surprisingly, in contrast to the Elovl2−/− mice (Fig. 6C), no reduction was seen in the levels of C22:5n-6 and C22:6n-3, suggesting that the activity of Δ6-desaturase is very sensitive to the levels of these fatty acids in testis.
Fig. 6.
Total fatty acid composition of liver and testis. Fatty acid composition up to C24, analyzed by gas chromatography of liver (A) and testis (B) of wild-type (wt) and Elovl2+/− (Hz [heterozygous]) male mice and testis of two pooled wild-type and Elovl2−/− male mice (C). Bars indicate means ± SEM from 6 animals. *, P < 0.05; **, P < 0.01.
Deficiencies in serum levels of n-3 and n-6 fatty acids in Elovl2−/− mice
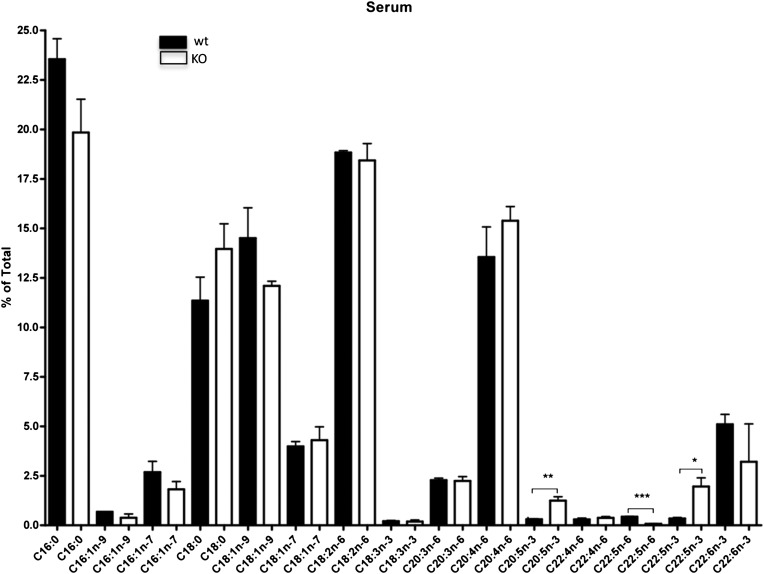
When we analyzed serum lipid composition, we found that C20:5n-3 and C22:5n-3 levels in Elovl2−/− mice were increased compared with that in age-matched littermates (Fig. 7), which suggests that Elovl2 ablation causes a general blockage in the elongation of C22:5n-3 for the formation of C24:5n-3, which is the substrate for Δ6-desaturase and precursor for C22:6n-3 synthesis (Fig. 5). In addition, finding that the level of C22:5n-6, which is the final product of C22:4n-6 elongation and Δ6-desaturation, also was reduced implies that ELOVL2 is important for C22 elongation within both the n-3 and the n-6 series (Fig. 5).
Fig. 7.
Differences in total PUFA composition of serum. Fatty acid composition up to C24, by gas chromatography analysis of serum in wild-type and Elovl2−/− male mice. Bars indicate means ± SEM from 3 animals per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DHA supplementation does not restore fertility in male Elovl2+/− mice
Recently, two different mouse strains lacking Δ6-desaturase (FADS2) have been associated with impaired reproduction. As in the case of Elovl2+/− mice, Fads2-ablated mice show abnormal sperm head conformation (17, 18) and are characterized by a reduction in levels of C20:4n-6, C22:5n-6, and C22:6n-3. Interestingly, dietary supplementation of C22:6 n-3 (DHA) completely restored sperm morphology and subsequently male fertility of these mice (19), suggesting that 22:5n-6 PUFA are dispensable if 22:6n-3 is supplemented in the diet. In contrast, despite distorted levels of C22 PUFAs in testis of Elovl2+/− mice, dietary supplementation of DHA for 3 months (Tables 3 and 4) could not restore male fertility of these mice, emphasizing the importance of ELOVL2 and the generation of testis-specific VLC-PUFA required for male fertility.
TABLE 4.
DHA content in male liver after dietary supplementation
| Measure | Elovl2+/+ fed chow | Elovl2+/− fed chow | Elovl2+/− fed DHA supplement |
|---|---|---|---|
| % DHA ± SEM in liver | 5.7 ± 0.6 | 5.5 ± 0.6 | 11.3 ± 3.5 |
DISCUSSION
Here we report a novel mechanism involving ELOVL2-derived fatty acids in mammalian spermatogenesis. Using homologous recombination, we developed Elovl2−/− mice, which resulted in a complete arrest of spermatogenesis, with seminiferous tubules displaying only spermatogonia and primary spermatocytes, without further germinal cells in male mice. The main reason for this result is the complete absence of C24–C30 VLC-PUFAs of the n-6 family in the testis, of which C24:4 n-6 is essential for the production of C22:5 n-6. Even more surprising is that heterozygote Elovl2+/− mice exhibited haploinsufficiency with reduced levels of n-6 C28:5 and C30:5 PUFAs, which gives rise to impaired packaging of haploid spermatides and infertility in almost all male mice, implying that even minor changes in Elovl2 expression and testicular omega-6 PUFA content may have severe effects on male reproduction ability. This implies that C28–C30 n-6 PUFAs are formed by Elovl2 and are crucial for normal spermatogenesis, although we cannot rule out the possibility that the effect seen in Elovl2−/− mice at an earlier stage of spermatogenesis may be a direct consequence of impaired levels of C22–C26 PUFAs as well.
The histological phenotype of the Elovl2−/− mice testis resembles in part the morphology seen in two mouse strains with disruption of the epimorphin gene, a member of the SNARE family, which plays an essential role in membrane fusion at the terminal step of cytokinesis and vesicle fusion during exocytosis (20–22). As in the case of Elovl2 ablation, those germ cells failed to complete the meiotic division to generate haploid cells, thereby resulting in the formation of multinucleated cells in the lumens of the seminiferous tubules. In addition, mutations within the bond gene, an Elovl homolog in Drosophila, with unknown fatty acid specificity, block cleavage-furrow ingression during early telophase in dividing spermatocytes, which is due to collapse of the contractile rings as a consequence of impaired interaction between cytoskeleton and plasma membrane at this stage (23). Those findings together with our data presented here imply that ELOVL2-synthesized VLC PUFAs are essential membrane components for normal completion of spermatocyte cytokinesis in mammals.
With maturation, the spermatides accumulate significant amounts of unusual sphingolipids containing VLC PUFAs (24, 25), which are principally localized to the sperm head (26) where they may stabilize cellular membranes with high curvature, such as the rims of the sperm head. In addition, during acrosomal biogenesis, proacrosomal vesicles derived from the Golgi apparatus dock and fuse along a cytoskeletal plate, which then reorganizes to modulate exogenous shear forces exerted by the Sertoli cells to guide nuclear elongation (27). Our data for heterozygote Elovl2+/− mice imply that Elovl2-derived C28:5 and C30:5 n-6 fatty acids are essential membrane components in circumstances when exceptionally strong forces are applied to the cell.
A distinguishing feature between the Elovl2+/− mice and the Fads2- and Elovl5-ablated mice (Fig. 5) is that fertility is already severely affected at a heterozygous stage in Elovl2+/− male mice, suggesting a dominant negative effect, which cannot be rescued by any other metabolic pathway. In addition, mice heterozygous for both Elovl5 and Fads2 ablation produce offspring in a normal Mendelian ratio, and male Elovl5−/− mice are fertile (18, 28).
Because ELOVL4 has been shown to be the enzyme responsible for VLC-PUFA synthesis in retina (3), the enzyme has been suggested to perform the same action in testis. As both ELOVL2 and ELOVL4 are expressed at significant levels in testis and retina, our data clearly imply that ELOVL2 and ELOVL4 have two distinct functions in these tissues: ELOVL2 preferentially elongates C22 PUFA up to C30 of the n-6 series, and ELOVL4 preferentially elongates C26 PUFA up to C36 of the n-3 series (Fig. 5). In addition, several mutations and deletions in the Elvol4 gene have been shown to be associated with the pathogenesis of dominant macular dystrophies in mice and humans (29–33). However, diabetes-induced retinopathy has recently been shown to be associated with reduced expression of both Elovl2 and Elovl4, which is paralleled by a downregulation of DHA and n-3 VLC-PUFAs in retina (34), suggesting that ELOVL2 and ELOVL4 may operate at different levels also within the same elongation pathway.
It is noteworthy that in addition to its expression in retina and testis, Elovl4 is also expressed in skin (3). Elovl4-ablated mice die within a few hours after birth due to dehydration caused by a reduction of specific ceramides and saturated VLCFA longer than C26 in the epidermis of mutant mice (35). In contrast, Elovl2 knockout mice do not show any obvious skin phenotype, which is in accordance with undetectable levels of Elovl2 expression in skin of normal mice. Overall, these data emphasize ELOVL2 as the critical enzyme for the conversion of C22 to C26 PUFA, which may then act as a substrate for ELOVL4.
In summary, from our data here, it is apparent that de novo synthesis of n-6 VLC-PUFAs with C28 and C30 carbon atoms is a prerequisite of normal sperm maturation and that any interference with this process has severe consequences for male reproduction. Considering recent findings for the relationship between ELOVL2 expression and metabolic syndromes found in humans (36, 37), our results presented here may therefore give new insights into the field and show how nutritional interventions, and/or modulation of ELOVL2 expression, could represent a potential therapeutic target for male infertility caused by impaired lipid metabolism.
Acknowledgments
The authors thank Dr. R. Feinstein, Dr. A. Asadi, Dr. O. Söder, B. Leksell, and S. Sundberg for technical assistance.
Footnotes
Abbreviations:
- DHA
- docosahexaenoic acid
- dNTP
- deoxynucleoside triphosphate
- ELOVL
- elongation of very-long-chain fatty acid (protein)
- ES
- embryonic stem (cells)
- FAME
- fatty acid methyl ester
- Hz
- heterozygous
- KO
- knockout
- TK
- thymidine kinase
- VLCFA
- very-long-chain FA
This work was supported by grants from the Swedish Research Council and Cancer Foundation (A.J.), Sven och Dagmar Saléns stiftelse (D.Z.), Institut National de la Recherche Agronomique, and Formas (H.G. and A.J.). Rothamsted Research receives grant support from the Biotechnology and Biological Sciences Research Council, United Kingdom (J.N.).
REFERENCES
- 1.Burr G. O., Burr M. M. 1973. Nutrition classics from The Journal of Biological Chemistry 82:345–67, 1929. A new deficiency disease produced by the rigid exclusion of fat from the diet. Nutr. Rev. 31: 248–249. [DOI] [PubMed] [Google Scholar]
- 2.Guillou H., Zadravec D., Martin P. G., Jacobsson A. 2010. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 49: 186–199. [DOI] [PubMed] [Google Scholar]
- 3.Mandal M. N., Ambasudhan R., Wong P. W., Gage P. J., Sieving P. A., Ayyagari R. 2004. Characterization of mouse orthologue of ELOVL4: genomic organization and spatial and temporal expression. Genomics. 83: 626–635. [DOI] [PubMed] [Google Scholar]
- 4.Agbaga M. P., Brush R. S., Mandal M. N., Henry K., Elliott M. H., Anderson R. E. 2008. Role of Stargardt-3 macular dystrophy protein (ELOVL4) in the biosynthesis of very long chain fatty acids. Proc. Natl. Acad. Sci. U S A. 105: 12843–12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonard A. E., Bobik E. G., Dorado J., Kroeger P. E., Chuang L. T., Thurmond J. M., Parker-Barnes J. M., Das T., Huang Y. S., Mukerji P. 2000. Cloning of a human cDNA encoding a novel enzyme involved in the elongation of long-chain polyunsaturated fatty acids. Biochem. J. 350: 765–770. [PMC free article] [PubMed] [Google Scholar]
- 6.Moon Y. A., Shah N. A., Mohapatra S., Warrington J. A., Horton J. D. 2001. Identification of a mammalian long chain fatty acyl elongase regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 276: 45358–45366. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Torres-Gonzalez M., Tripathy S., Botolin D., Christian B., Jump D. B. 2008. Elevated hepatic fatty acid elongase-5 activity affects multiple pathways controlling hepatic lipid and carbohydrate composition. J. Lipid Res. 49: 1538–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprecher H. 2000. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim. Biophys. Acta. 1486: 219–231. [DOI] [PubMed] [Google Scholar]
- 9.Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 90: 8424–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebouissou S., Imbeaud S., Balabaud C., Boulanger V., Bertrand-Michel J., Terce F., Auffray C., Bioulac-Sage P., Zucman-Rossi J. 2007. HNF1alpha inactivation promotes lipogenesis in human hepatocellular adenoma independently of SREBP-1 and carbohydrate-response element-binding protein (ChREBP) activation. J. Biol. Chem. 282: 14437–14446. [DOI] [PubMed] [Google Scholar]
- 11.Larson T. R., Edgell T., Byrne J., Dehesh K., Graham I. A. 2002. Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. Plant J. 32: 519–527. [DOI] [PubMed] [Google Scholar]
- 12.Larson T. R., Graham I. A. 2001. Technical advance: a novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant J. 25: 115–125. [DOI] [PubMed] [Google Scholar]
- 13.Sayanova O., Haslam R., Venegas Caleron M., Napier J. A. 2007. Cloning and characterization of unusual fatty acid desaturases from Anemone leveillei: identification of an acyl-coenzyme A C20 Delta5-desaturase responsible for the synthesis of sciadonic acid. Plant Physiol. 144: 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagamori I., Yomogida K., Adams P. D., Sassone-Corsi P., Nojima H. 2006. Transcription factors, cAMP-responsive element modulator (CREM) and Tisp40, act in concert in postmeiotic transcriptional regulation. J. Biol. Chem. 281: 15073–15081. [DOI] [PubMed] [Google Scholar]
- 15.Kai M., Irie M., Okutsu T., Inoue K., Ogonuki N., Miki H., Yokoyama M., Migishima R., Muguruma K., Fujimura H., et al. 2004. The novel dominant mutation Dspd leads to a severe spermiogenesis defect in mice. Biol. Reprod. 70: 1213–1221. [DOI] [PubMed] [Google Scholar]
- 16.Vitale M. L., Akpovi C. D., Pelletier R. M. 2009. Cortactin/tyrosine-phosphorylated cortactin interaction with connexin 43 in mouse seminiferous tubules. Microsc. Res. Tech. 72: 856–867. [DOI] [PubMed] [Google Scholar]
- 17.Stoffel W., Holz B., Jenke B., Binczek E., Gunter R. H., Kiss C., Karakesisoglou I., Thevis M., Weber A. A., Arnhold S., et al. 2008. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 27: 2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stroud C. K., Nara T. Y., Roqueta-Rivera M., Radlowski E. C., Lawrence P., Zhang Y., Cho B. H., Segre M., Hess R. A., Brenna J. T., Haschek W. M., Nakamura M. T. 2009. Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 50: 1870–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roqueta-Rivera M., Stroud C. K., Haschek W. M., Akare S. J., Segre M., Brush R. S., Agbaga M. P., Anderson R. E., Hess R. A., Nakamura M. T. 2010. Docosahexaenoic acid supplementation fully restores fertility and spermatogenesis in male delta-6 desaturase knockout mice. J. Lipid Res. 51: 360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Wang L., Iordanov H., Swietlicki E. A., Zheng Q., Jiang S., Tang Y., Levin M. S., Rubin D. C. 2006. Epimorphin(−/−) mice have increased intestinal growth, decreased susceptibility to dextran sodium sulfate colitis, and impaired spermatogenesis. J. Clin. Invest. 116: 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama K., Akimaru S., Asano Y., Khalaj M., Kiyosu C., Masoudi A. A., Takahashi S., Katayama K., Tsuji T., Noguchi J., et al. 2008. A new ENU-induced mutant mouse with defective spermatogenesis caused by a nonsense mutation of the syntaxin 2/epimorphin (Stx2/Epim) gene. J. Reprod. Dev. 54: 122–128. [DOI] [PubMed] [Google Scholar]
- 22.Low S. H., Li X., Miura M., Kudo N., Quinones B., Weimbs T. 2003. Syntaxin 2 and endobrevin are required for the terminal step of cytokinesis in mammalian cells. Dev. Cell. 4: 753–759. [DOI] [PubMed] [Google Scholar]
- 23.Szafer-Glusman E., Giansanti M. G., Nishihama R., Bolival B., Pringle J., Gatti M., Fuller M. T. 2008. A role for very-long-chain fatty acids in furrow ingression during cytokinesis in Drosophila spermatocytes. Curr. Biol. 18: 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aveldano M. I., Sprecher H. 1987. Very long chain (C24 to C36) polyenoic fatty acids of the n-3 and n-6 series in dipolyunsaturated phosphatidylcholines from bovine retina. J. Biol. Chem. 262: 1180–1186. [PubMed] [Google Scholar]
- 25.Rabionet M., van der Spoel A. C., Chuang C. C., von Tumpling-Radosta B., Litjens M., Bouwmeester D., Hellbusch C. C., Korner C., Wiegandt H., Gorgas K., et al. 2008. Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. J. Biol. Chem. 283: 13357–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furland N. E., Oresti G. M., Antollini S. S., Venturino A., Maldonado E. N., Aveldano M. I. 2007. Very long-chain polyunsaturated fatty acids are the major acyl groups of sphingomyelins and ceramides in the head of mammalian spermatozoa. J. Biol. Chem. 282: 18151–18161. [DOI] [PubMed] [Google Scholar]
- 27.Kierszenbaum A. L., Rivkin E., Tres L. L. 2007. Molecular biology of sperm head shaping. Soc. Reprod. Fertil. Suppl. 65: 33–43. [PubMed] [Google Scholar]
- 28.Moon Y. A., Hammer R. E., Horton J. D. 2009. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J. Lipid Res. 50: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards A. O., Donoso L. A., Ritter R., 3rd 2001. A novel gene for autosomal dominant Stargardt-like macular dystrophy with homology to the SUR4 protein family. Invest. Ophthalmol. Vis. Sci. 42: 2652–2663. [PubMed] [Google Scholar]
- 30.Zhang K., Kniazeva M., Han M., Li W., Yu Z., Yang Z., Li Y., Metzker M. L., Allikmets R., Zack D. J., et al. 2001. A 5-bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat. Genet. 27: 89–93. [DOI] [PubMed] [Google Scholar]
- 31.Karan G., Yang Z., Howes K., Zhao Y., Chen Y., Cameron D. J., Lin Y., Pearson E., Zhang K. 2005. Loss of ER retention and sequestration of the wild-type ELOVL4 by Stargardt disease dominant negative mutants. Mol. Vis. 11: 657–664. [PubMed] [Google Scholar]
- 32.Grayson C., Molday R. S. 2005. Dominant negative mechanism underlies autosomal dominant Stargardt-like macular dystrophy linked to mutations in ELOVL4. J. Biol. Chem. 280: 32521–32530. [DOI] [PubMed] [Google Scholar]
- 33.Bernstein P. S., Tammur J., Singh N., Hutchinson A., Dixon M., Pappas C. M., Zabriskie N. A., Zhang K., Petrukhin K., Leppert M., et al. 2001. Diverse macular dystrophy phenotype caused by a novel complex mutation in the ELOVL4 gene. Invest. Ophthalmol. Vis. Sci. 42: 3331–3336. [PubMed] [Google Scholar]
- 34.Tikhonenko M., Lydic T. A., Wang Y., Chen W., Opreanu M., Sochacki A., McSorley K. M., Renis R. L., Kern T., Jump D. B., et al. 2009. Remodeling of retinal fatty acids in an animal model of diabetes: a decrease in long chain polyunsaturated fatty acids is associated with a decrease in fatty acid elongases Elovl2 and Elovl4. Diabetes. 59:219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cameron D. J., Tong Z., Yang Z., Kaminoh J., Kamiyah S., Chen H., Zeng J., Chen Y., Luo L., Zhang K. 2007. Essential role of Elovl4 in very long chain fatty acid synthesis, skin permeability barrier function, and neonatal survival. Int. J. Biol. Sci. 3: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka T., Shen J., Abecasis G. R., Kisialiou A., Ordovas J. M., Guralnik J. M., Singleton A., Bandinelli S., Cherubini A., Arnett D., et al. 2009. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI study. PLoS Genet. 5: e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Illig T., Gieger C., Zhai G., Romisch-Margl W., Wang-Sattler R., Prehn C., Altmaier E., Kastenmuller G., Kato B. S., Mewes H. W., et al. 2010. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 42: 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]