Abstract
Neutral glycosphingolipids containing one to six sugars in their oligosaccharide chains have been isolated from cysts of the brine shrimp Artemia franciscana. The structures of these glycolipids were identified by methylation analysis, partial acid hydrolysis, gas-liquid chromatography, combined gas-liquid chromatography-mass spectrometry, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, and proton nuclear magnetic resonance spectroscopy to be Glcβ1-Cer, Manβ1-4Glcβ1-Cer, Fucα1-3Manβ1-4Glcβ1-Cer, GlcNAcβ1-3Manβ1-4Glcβ1-Cer, GlcNAcα1-2Fucα1-3Manβ1-4Glcβ1-Cer, GalNAcβ1-4GlcNAcβ1-3Manβ1-4Glcβ1-Cer, GalNAcβ1-4(Fucα1-3)GlcNAcβ1-3Manβ1-4Glcβ1-Cer (CPS), and GalNAcβ1-4(GlcNAcα1-2Fucα1-3)GlcNAcβ1-3Manβ1-4Glcβ1-Cer (CHS). Two glycosphingolipids, CPS and CHS, were characterized as novel structures. Because Artemia contains a certain series of glycosphingolipids (-Fucα3Manβ4GlcβCer), which differ from the core sugar sequences reported thus far, we tentatively designated the glycosphingolipids characterized as nonarthro-series ones. Furthermore, CHS exhibited a hybrid structure of arthro-series and nonarthro-series sugar chain. Two novel glycosphingolipids were characterized from the brine shrimp Artemia franciscana; one was composed of arthrotetraose and a branching fucose attached to N-acetylglucosamine residue, and the other was composed of CPS with an additional N-acetylglucosamine residue attached to the branching fucose.
Keywords: sphingolipid, sphingosine, long-chain base, oligosaccharide, fucomannolipid, terminal alpha-N-acetylglucosamine residue, structure characterization, conventional destructive analysis, Branchiopoda, dormant cyst
Glycosphingolipid (GSL) is composed of sugar chain and ceramide, the latter of which consists of a fatty acid and a sphingoid. GSLs, which are expressed on the outer side of the lipid bilayer in animal cells, form patches with sphingomyelin (microdomains) and play a foundational role in intercellular adhesion, cellular recognition, differentiation/growth, and immune response (1–3). However, a comprehensive understanding of GSL function has not yet been attained because of the structural complexity in the sugar chain of GSLs even in vertebrates and invertebrates (4, 5). We have analyzed the GSL structures in lower animals on the assumption that their complexity plays an important role in maintaining the biological activity of multicellular organisms.
In invertebrates, structural analyses of GSLs have been performed in several phyla and are well known in Arthropoda, Mollusca, and Nematoda. The immune response by α-galactosylceramide from a marine sponge (3) and the induction of cytokine secretion by zwitterionic GSLs from a parasitic nematode (6) are interesting in terms of GSL function, and investigation of invertebrate GSL structures could therefore be a productive research area. In Arthropoda, structural analyses of GSLs have begun for flies (Diptera Insecta) (7, 8), and a characteristic arthro-series sugar chain (GlcNAcβ3Manβ4GlcβCer; At3Cer) has been identified. In Drosophila, mactosyl ceramide (MacCer) and arthrotriaosylceramide (At3Cer) are biosynthesized by catalytic β4-mannosyltransferase (egghead, egh) and β3-N-acetylglucosaminyltransferase (brainiac, brn), respectively (9–12). It was reported that the mutant of brn was shown to be a lethal phenotype at the pupal stage (9); therefore, the arthro-series trisaccharide appears to be essential for development in insects.
In this report, we analyzed GSL structures in cysts of the brine shrimp Artemia franciscana, a crustacean arthropod. This species is a kind of plankton inhabiting saline environments such as the Great Salt Lake in the USA. Analytical reports on this species have mainly related to nutritional analyses in aquaculture using the species as instant live food (13). There have only been two reports regarding sphingolipid characterization: one on a ceramide tetrasaccharide (CTeS) in newly hatched nauplii (14) and the other on sphingomyelin in diapausing eggs (15). Here, we present novel nonarthro-series and arthro-series GSLs, as well as hybrid structure GSL composed of core arthro-series sugar chains with a branching nonarthro-series disaccharide in the brine shrimp.
MATERIALS AND METHODS
Isolation of neutral GSLs
Great Salt Lake brine shrimp cysts (1.8kg) purchased from A and A Marine LLC (Salt Lake City, UT) were ground to powder by using automatic mortars. Lipids were extracted once with 7.2 liter of chloroform-methanol (2:1, v/v), once with 5.3 liter of chloroform-methanol (2:1, v/v) and once with 4.4 liter of chloroform-methanol (1:1, v/v). The combined chloroform-methanol extracts were concentrated by a rotary evaporator and subjected to mild alkaline hydrolysis with 0.5 M potassium hydroxide (KOH) in methanol. The hydrolyzate was acidified (pH 1) with several drops of concentrated hydrochloride (HCl), kept in an ice bath for 1 h, and dialyzed against tap water for 2 days. The inner fluid was concentrated to near dryness in vacuo at 40°C, and precipitated by addition of cold acetone (yield: 8.6 g, alkaline-stable product). The alkaline-stable product was dissolved in chloroform-methanol-water (30:60:8, v/v/v) and applied to a column (ø3.5 × 48 cm) packed with quaternary ammonium ethyl (QAE)-Sephadex A-25, OH- form (GE Healthcare Co.) equilibrated with the same solvent. The column was successively eluted with the same solvent (5 column volumes) and pure methanol (1 vol) as neutral solvents, and with 0.45 M ammonium acetate in methanol (5 vols) as a polar solvent. The separation of neutral and acidic glycolipids was monitored by thin-layer chromatography (TLC) as described below. The eluates obtained from this column using the neutral solvents were pooled and evaporated to dryness (yield: 610 mg, crude neutral GSL fraction). The crude neutral GSL fraction was acetylated and then fractionated on a column (column size 2.0 × 57 cm) packed with Florisil, 60 ∼100 mesh (Nacalai Tesque, Inc.) by slightly modifying the method of Saito and Hakomori (16). The column was successively eluted with 3 column volumes of n-hexane-dichloroethane (1:4, v/v), 3 vols of pure dichloroethane, 3 vols of dichloroethane-acetone (1:1, v/v), 6 vols of dichloroethane-methanol (9:1, v/v), 3 vols of dichloroethane-methanol (3:1, v/v), 3 vols of dichloroethane-methanol-water (2:8:1, v/v/v), 6 vols of chloroform-methanol-water (6:4:1, v/v/v), and 3 vols of chloroform-methanol-water (2:8:1, v/v/v). In the course of this fractionation, different species of neutral GSLs were eluted with the various mixtures of dichloroethane-acetone and dichloroethane-methanol solvents. The solutions of acetylated GSLs were each evaporated to dryness, deacetylated with 0.5 M KOH in methanol at 37°C for 6 h, and dialyzed against tap water for 2 days. The inner fluids were concentrated to near dryness in vacuo at 40°C. The concentrated lipid fractions were tested by TLC as described below and combined (yield: 102 mg, neutral GSL fraction). For isolation of each neutral GSL, the neutral GSL fraction (97 mg) was applied to an Iatrobeads (6RS-8060, Mitsubishi Kagaku Iatron Inc., Tokyo, Japan) column (column size 1.0 × 111cm). The neutral GSLs were eluted with two linear gradient elution systems of chloroform-methanol-water with compositions of [80:20:1 (v/v/v) 255 ml ∼50:50:5 (v/v/v) 325 ml] and [50:50:5 (v/v/v) 252 ml ∼20:80:10 (v/v/v) 330ml], respectively. Fractions of 3 ml were collected in each tube and aliquots from every three tubes were tested by HPTLC.
Solvent system for TLC
The following solvent system was used: chloroform-methanol-water (60:40:10, v/v/v). In QAE-Sephadex and Florisil column chromatography, the eluates on TLC plates of silica gel 60 (Merck KGaA) were visualized by spraying with orcinol-sulfuric acid reagent (17) followed by heating at 110°C and by spraying Dittmer-Lester reagent (18) to detect sugar and phosphate groups, respectively. In Iatrobeads column chromatography, GSLs on HPTLC plates of silica gel 60 (Merck KGaA) were visualized by spraying with orcinol-sulfuric acid reagent followed by heating at 110°C.
Analysis of fatty acid and sugar components
For determination of the composition of fatty acids and sugars, 0.1–0.2 mg of each GSL was methanolyzed with 1 M anhydrous methanolic HCl at 100°C for 3 h. The produced fatty acid methyl esters were extracted with n-hexane and analyzed by GC and GC-MS. The remaining methanolic phase was neutralized by adding silver carbonate and evaporated to dryness after removal of silver chloride. The residue containing methyl glycosides was N-acetylated by adding 10 μl of pyridine and 50 μl of anhydrous acetate in 0.5 ml of methanol for 30 min (19), evaporated under a nitrogen stream, and dried in a vacuum desiccator with a water aspirator. The N-acetylated residue was trimethylsilylated (20), and subjected to GC.
Methylation analysis
About 0.2 mg of each GSL was permethylated according to the method of Ciucanu and Kerek (21). In brief, each GSL was dried in a screw-capped glass test tube and dissolved in 0.2 ml of dimethylsulfoxide while sonicating for 5 min. Immediately after, about 20 mg of fine powdered sodium hydroxide (NaOH) and 0.2 ml of methyl iodide were added, the test tube was capped tightly and vigorously stirred for 2 min. The methylated GSL was extracted with chloroform, washed with water six times, dried under a nitrogen stream, and hydrolyzed with 0.3 ml of acetic acid-HCl-water (16:1:3, v/v/v) using a microwave oven (22, 23). The acetolyzate was dried under a nitrogen stream using a few drops of toluene, dried in a desiccator with a water aspirator for 1 h, and then reduced by adding 0.25 ml of 0.01 M NaOH followed by 0.25 ml of 0.01 M NaOH solution containing 2% sodium borohydride (final concentration: 1% sodium borohydride, 0.01 M NaOH) at room temperature overnight. The reduction was stopped by adding a few drops of acetic acid. The reduced and partially methylated alditols were dried under a nitrogen stream while adding ∼1 ml of methanol a few times, and then dried in a desiccator with a water aspirator for 1 h. The residue was acetylated by adding 0.25 ml of pyridine followed by 0.25 ml of acetic anhydride and then incubated in boiling water for 12 min. The partially methylated alditol acetates were extracted with chloroform, washed with water six times, and then subjected to GC and GC-MS.
Analysis of partial acid hydrolysis
About 4 mg of ceramide hexasaccharide (CHS) was hydrolyzed with 1 ml of 0.1 M HCl in boiling water for 60 min. The hydrolyzate was extracted into a lower phase by adding 5 ml of chloroform-methanol (2:1, v/v), evaporated to dryness under a nitrogen stream, and dissolved in 2 ml of chloroform-methanol (2:1, v/v). The GSL fragments produced were separated on a silica gel 60 TLC plate, developed in the solvent mixture of chloroform-methanol-water (60:40:10, v/v/v) for 20 min, and dried. After slight exposure to iodine vapor, the spots corresponding to mono-, di-, tri-, and tetraglycosyl ceramide were scraped off, extracted with chloroform-methanol-water (2:1:0.1, v/v/v), and subjected to sugar composition analysis.
Analysis of sphingoids
Sphingoid composition was determined by the method of Gaver and Sweeley (24). In brief, about 0.2 –0.3 mg of each GSL was measured into a screw-capped glass test tube and dried under a nitrogen stream. After adding 0.2 ml of aqueous HCl-methanol reagent (methanolic reagent containing 8.6% concentrated HCl and 9.4% water), the mixture was hydrolyzed at 70°C in the oven for 18 h. The hydrolyzate was washed with n-hexane to remove fatty acids. The residual methanolic phase was dried under a nitrogen stream, alkalized with 0.6 ml of methanol-1 M NaOH solution (4:3, v/v), extracted with 0.72 ml of chloroform, and washed twice with 0.4 ml of methanol-water (1:1, v/v). The residual chloroform solution was evaporated under a nitrogen stream, dried in a vacuum desiccator with a water aspirator, trimethylsilylated, and subjected to GC and GC-MS.
GC and GC-MS
Compositional analyses of methylation, fatty acids, sugars, and sphingoids were carried out using a Shimadzu GC-18A gas chromatograph with a Shimadzu HiCap-CBP 5 capillary column (0.22 mm × 25 m). The temperature increase was programmed at 2°C/min from 140 to 230°C for sugar component analysis, 4°C/min from 140 to 230°C for the methylation study, 4°C/min from 170 to 230°C for fatty acid analysis, and 2°C/min from 210 to 230°C for sphingoid analysis. Electron impact ionization mass spectra were obtained using a Shimadzu GCMS-QP5050 gas chromatograph-mass spectrometer with the same capillary column under the following conditions: an interface temperature of 250°C, an injection port temperature of 240°C, a helium gas pressure of 100kPa, and an ionizing voltage of 70eV. The oven temperatures for GC-MS analyses were 80 (2 min) to 180 (20°C/min) to 240°C (4°C/min) for the methylation study, 80 (2min) to 170 (20°C/min) to 240°C (4°C/min) for fatty acid analysis, and 80 (2min) to 220 (20°C/min) to 280°C (6°C/min) for sphingoid analysis, respectively.
MALDI-TOF MS
MALDI-time-of-flight (TOF) MS analysis was performed using an Applied Biosystems/Voyager-DE STR™ Biospectrometer with a nitrogen laser (337 nm) operating in the reflector positive-ion mode at an acceleration voltage of 20 kV. The matrix used was α-cyano-4-hydroxycinnamic acid (Proteomics Grade, Wako Chemical Co.). External mass calibration was provided by the [M+Na]+ ions of angiotensin I (1296.96 mass units; Sigma Chemical Co.) and bradykinin fragment I-V (573.31 mass units; Sigma Chemical Co.).
Proton-nuclear magnetic resonance (1H-NMR) spectroscopy
NMR spectra were obtained using a JEOL-ECS400 400MHz NMR spectrometer at an operating temperature of 60°C. The purified GSL was dissolved in 0.5 ml of d6-dimethylsulfoxide containing 2% D2O. The chemical shift was referenced to the solvent signals (δH 2.49 ppm) in d6-dimethylsulfoxide as the internal standard.
RESULTS
Purified neutral GSLs
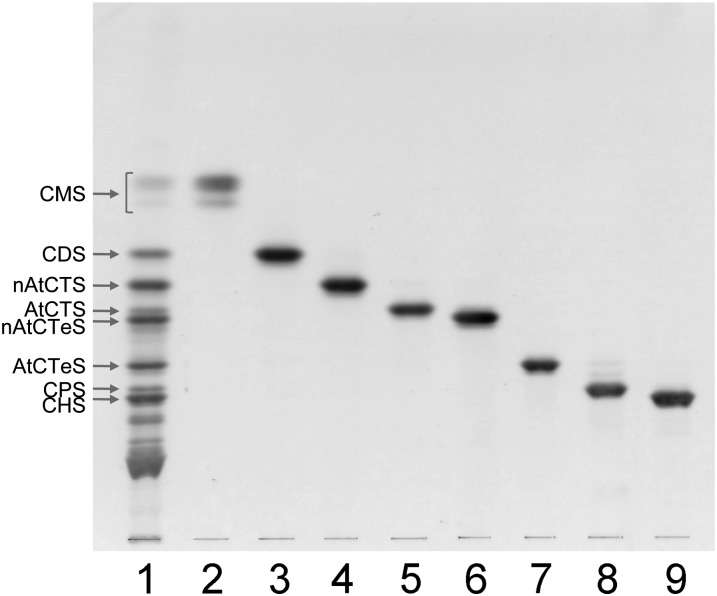
Crude sphingolipids (8.6 g) were obtained from chloroform-methanol extracts (285 g) of brine shrimp (Artemia franciscana) cysts (1.8 kg) by alkaline- and acid-treatment and chromatographed on a QAE-Sephadex column using a solvent mixture of chloroform-methanol-water (30:60:8, v/v/v). In the TLC analysis of the resultant fraction (610 mg), GSLs (102 mg) and sphingomyelin (359 mg) were detected with orcinol-sulfuric acid and Dittmer-Lester reagent, respectively. Zwitterionic GSL could not be detected. GSLs were separated using Florisil and silicic acid column chromatography into 26 fractions. GSLs with one to six sugar residues were developed by TLC (Fig. 1) . The yields of the purified GSLs obtained from 1.8 kg of brine shrimp cysts were 2.8 (CMS), 2.7 (CDS), 5.1 (nonarthro CTS, nAtCTS), 2.1 (arthro CTS, AtCTS), 5.1 (nonarthro CTeS, nAtCTeS), 6.9 (arthro CTeS, AtCTeS), 1.9 (CPS), and 5.3 mg (CHS). GSLs with more than six sugar residues are currently under investigation.
Fig. 1.
Thin-layer chromatogram of neutral glycosphingolipids isolated from the brine shrimp Artemia franciscana. Lane 1, total neutral glycosphingolipid fraction obtained by QAE-Sephadex A-25 and Florisil column chromatographies; lanes 2 to 9, isolated CMS, CDS, nAtCTS, AtCTS, nAtCTeS, AtCTeS, CPS, and CHS, respectively. The HPTLC plate was developed in chloroform-methanol-water (60:40:10, v/v/v). The spots were visualized by orcinol-sulfuric acid reagent.
Sugar composition
Each GSL was methanolyzed and the methylglycosides were converted to N-acetyl-O-trimethylsilyl derivatives for GC analysis. Their gas chromatograms defined the sugar composition of CMS as glucose; CDS as glucose and mannose (molar ratio, 1:1); nAtCTS as glucose, mannose, and fucose (1:1:1); AtCTS as glucose, mannose, and N-acetylglucosamine (1:1:1); nAtCTeS as glucose, mannose, fucose, and N-acetylglucosamine (1:1:1:1); AtCTeS as glucose, mannose, N-acetylglucosamine, and N-acetylgalactosamine (1:1:1:1); CPS as glucose, mannose, N-acetylglucosamine, N-acetylgalactosamine, and fucose (1:1:1:1:1); and CHS as glucose, mannose, N-acetylglucosamine, N-acetylgalactosamine, and fucose (1:1:2:1:1) (see Table 1). Sugar compositional analysis of AtCTS indicated 0.1 mol of fucose corresponding to the impurities in nAtCTeS, which could not be separated by the Iatrobeads column chromatography.
TABLE 1.
Sugar components and their molar ratios in the neutral glycosphingolipids purified from the brine shrimp Artemia franciscana
| Glc | Man | Fuc | GlcNAc | GalNAc | |
|---|---|---|---|---|---|
| CMS | + | − | − | − | − |
| CDS | 1 | 0.8 | − | − | − |
| nAtCTS | 1 | 0.9 | 0.8 | − | − |
| AtCTS | 1 | 0.9 | 0.1 | 1.1 | − |
| nAtCTeS | 1 | 0.9 | 0.8 | 1.0 | − |
| AtCTeS | 1 | 1.0 | − | 1.0 | 1.1 |
| CPS | 1 | 1.0 | 0.8 | 1.1 | 1.0 |
| CHS | 1 | 0.9 | 0.8 | 2.0 | 1.2 |
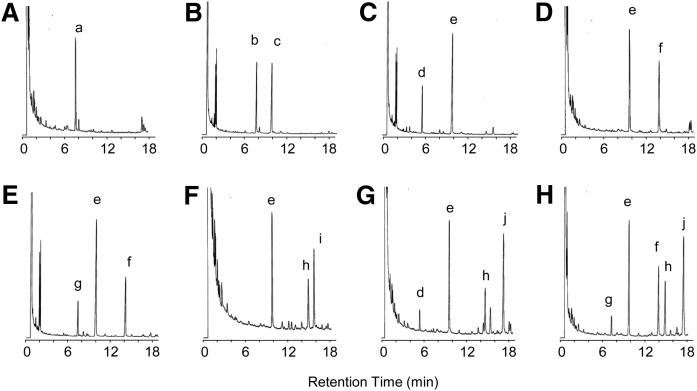
Methylation analysis
The partially methylated alditol acetates derived from the obtained GSLs were separated by GC as shown in Fig. 2. The identification was confirmed by GC-MS analysis according to the data of Jansson et al. (25) and Tai et al. (26). The methylation analysis revealed 1,5-di-O-acetyl-2,3,4,6-tetra-O-methylglucitol (1Glc) from CMS; 1,4,5-tri-O-acetyl-2,3,6-tri-O-methylglucitol (1,4Glc) and 1,5-di-O-acetyl-2,3,4,6-tetra-O-methylmannitol (1Man) from CDS; 1,4Glc, 1,3,5-tri-O-acetyl-2,4,6-tri-O-methylmannitol (1,3Man), and 1,5-di-O-acetyl-2,3,4-tri-O-methylfucitol (1Fuc) from nAtCTS; 1,4Glc, 1,3Man, and 1,5-di-O-acetyl-3,4,6-tri-O-methyl-N-acetylglucosaminitol (1GlcNAc) from AtCTS; 1,4Glc, 1,3Man, 1,2,5-tri-O-acetyl-3,4-di-O-methylfucitol (1,2Fuc), and 1GlcNAc from nAtCTeS; 1,4Glc, 1,3Man, 1,4,5-tri-O-acetyl-3,6-di-O-methyl-N-acetylglucosaminitol (1,4GlcNAc), and 1,5-di-O-acetyl-3,4,6-tri-O-methyl-N-acetylgalactosaminitol (1GalNAc) from AtCTeS; 1,4Glc, 1,3Man, 1,3,4,5-tetra-O-acetyl-6-O-methyl-N-acetylglucosaminitol (1,3,4GlcNAc), 1GalNAc, and 1Fuc from CPS; and 1,4Glc, 1,3Man, 1,3,4GlcNAc, 1GalNAc, 1,2Fuc, and 1GlcNAc from CHS. Although 1,4Glc and 1,3Man are not separated by GC analysis using the Shimadzu HiCap-CBP 5 capillary column, their identities were determined from their electron impact ionization mass spectra. Their identities were subsequently confirmed by repeating the analysis using a column of DB-210 capillary column (0.25mm × 30 m, Agilent Technology) that provided separation and characteristic retention times for both 1,4Glc and 1,3Man (data not shown).
Fig. 2.
Gas chromatograms of partially methylated alditol acetates derived from the isolated GSLs. A: CMS; B: CDS; C: nAtCTS; D: AtCTS; E: nAtCTeS; F: AtCTeS; G: CPS; H: CHS; a: 1Glc; b: 1Man; c: 1,4Glc; d: 1Fuc; e: 1,4Glc and 1,3Man; f: 1GlcNAc; g: 1,2Fuc; h: 1GalNAc; i: 1,4GlcNAc; j: 1,3,4GlcNAc.
Position of glycosidic substitution on the branching saccharide by partial acid hydrolysis
The methylation analysis described above for CHS indicated the presence of a saccharide branch on the N-acetylglucosamine residue. To determine the position of glycosidic substitution on N-acetylglucosamine to N-acetylgalactosamine residue, CHS was hydrolyzed with 0.1 M HCl at 100°C for 60 min, and the resulting products were separated with preparative TLC. From the hydrolysis, four products corresponding to GSLs with one to four sugar residues (CHS-M, -D, -T, and -Q) were obtained (Fig. 3), methanolyzed, trimethylsilylated, and subjected to GC. The gas chromatogram revealed the sugar composition of CHS-M as Glc, CHS-D as Glc and Man (molar ratio, 1:1); CHS-T as Glc, Man, and GlcNAc (1:1:1); CHS-Q as Glc, Man, GlcNAc, and GalNAc (1:1:1:1). This result indicates the presence of the sugar chain, GalNAc-GlcNAc-Man-Glc, in the CHS structure as a core sequence. Therefore, we concluded that the branching Fuc residue attaches to the 3-position on GlcNAc residue in the core AtCTeS, GalNAc1-4GlcNAc1-3Man1-4Glc.
Fig. 3.
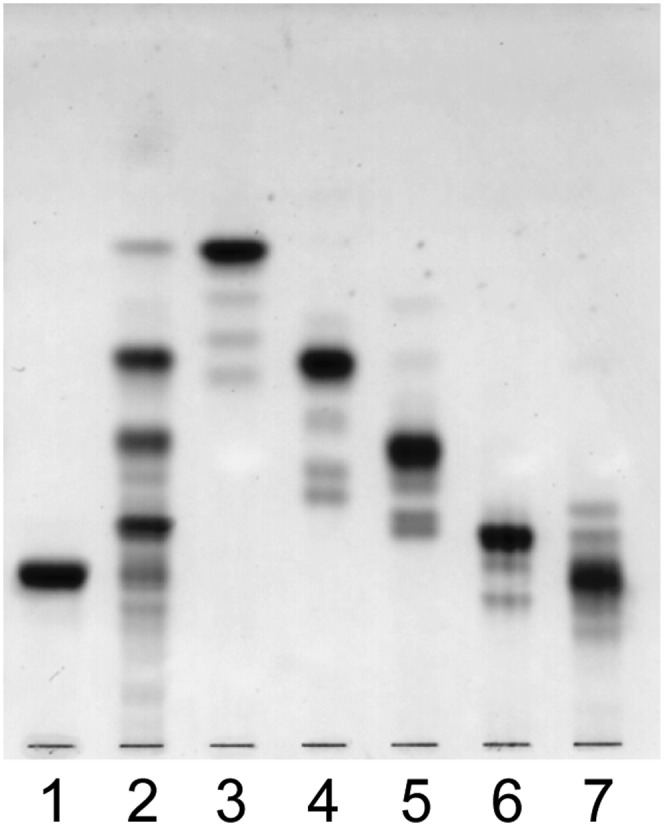
Thin-layer chromatogram of hydrolyzates of CHS with HCl. 1, intact CHS; 2, hydrolyzates before separation; 3, product with mono-saccharide residue (CHS-M); 4, product with di-saccharide residue (CHS-D); 5, product with tri-saccharide residue (CHS-T); 6, product with tetra-saccharide residue (CHS-Q); 7, remaining CHS. The plate was developed with chloroform-methanol-water (60:40:10, v/v/v). Spots were visualized with orcinol-sulfuric acid reagent.
Aliphatic components
The compositions of fatty acid and sphingoid in the obtained GSLs are given in Table 2. The fatty acids were mainly normal saturated acids ranging in length from C16 to C24, of which C22 acid was the most predominant. In some GSLs, odd-numbered saturated acids ranging in length from C19 to C23 were also detected in low amounts. The monoenoic acids of C22 and C24 were common in all GSLs. CMS contained 2-hydroxy acids as minor components. The sphingoids of the GSLs were composed of hexadeca- and heptadeca-4-sphingenines. In each case, the amount of the shorter base accounted for more than 57.5% of the total amount.
TABLE 2.
Ceramide composition of the neutral glycosphingolipids purified from Artemia franciscana
| FA (%) | CMS | CDS | nAtCTS | ArCTS | nAtCTeS | AtCTeS | CPS | CHS |
|---|---|---|---|---|---|---|---|---|
| 16:0 | 1.4 | 1.3 | tr. | tr. | tr. | tr. | 1.8 | tr. |
| 18:0 | 14.5 | 8.7 | 5.4 | 2.4 | 7.9 | 5.4 | 3.8 | 4.9 |
| 19:0 | tr. | — | tr. | — | tr. | — | — | — |
| 20:0 | 1.9 | 2.6 | 1.6 | 1.5 | 2.0 | 1.6 | 1.5 | 1.7 |
| 21:0 | tr. | 1.2 | tr. | 1.0 | 1.0 | tr. | tr. | tr. |
| 22:1 | 1.4 | 2.7 | 8.3 | 3.5 | 5.9 | 3.0 | 3.0 | 2.7 |
| 22:0 | 66.4 | 83.5 | 77.4 | 86.4 | 80.7 | 87.4 | 87.2 | 88.0 |
| 23:0 | tr. | tr. | tr. | 1.3 | tr. | tr. | 1.3 | tr. |
| 24:1 | 1.7 | tr. | 6.0 | 1.6 | 2.5 | 1.2 | tr. | 1.1 |
| 24:0 | 1.1 | tr. | 1.3 | 2.3 | tr. | 1.4 | 1.4 | 1.6 |
| 2 h16:0 | 1.0 | — | — | — | — | — | — | — |
| 2 h18:0 | 9.2 | — | — | — | — | — | — | — |
| 2 h20:0 | tr. | — | — | — | — | — | — | — |
| 2 h22:0 | 1.4 | — | — | — | — | — | — | — |
| LCB (%) | ||||||||
| d16:1 | 75.8 | 64.2 | 57.5 | 65.3 | 61.2 | 60.6 | 58.7 | 64.0 |
| d17:1 | 24.2 | 35.8 | 42.5 | 34.7 | 38.8 | 39.4 | 41.3 | 36.0 |
2 h, 2-hydroxy; d, dihydroxy sphingoid; tr., trace. —, not detected.
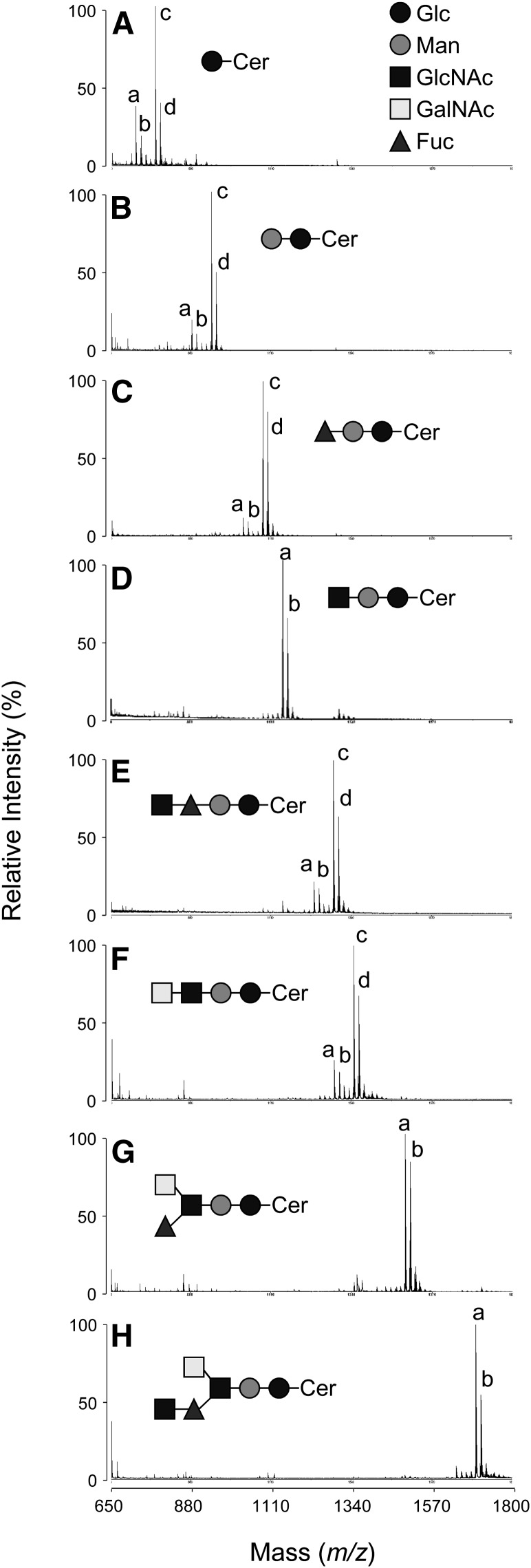
MALDI-TOF MS analysis
The putative structures of the eight purified GSLs were confirmed by the positive-ion reflector mode of MALDI-TOF MS analysis, as shown in Fig. 4 and summarized in the supplemental table. Their mass spectra have two to four sodium adducted ion species, [M+Na]+ ion species, the mass difference of which ranged from −319 to +463 ppm against the values calculated from the proposed structures, throughout the whole measurement period; that is, for CMS, [M+Na]+ ions at m/z 722.78, 738.77, 778.86, and 792.87 with one mole each of glucose, fatty acid (assigned as 18:0, h18:0 or 22:0), and sphingoid (assigned as d16:1 or d17:1); for CDS, [M+Na]+ ions at m/z 884.62, 898.64, 940.69, and 954.70 with one mole each of glucose, mannose, fatty acid (assigned as 18:0 or 22:0), and sphingoid (assigned as above); for nAtCTS, [M+Na]+ ions at m/z 1030.67, 1044.68, 1086.73, and 1100.74 with one mole each of glucose, mannose, fucose, fatty acid (assigned as above), and sphingoid (assigned as above); for AtCTS, [M+Na]+ ions at m/z 1143.69 and 1157.69 with one mole each of glucose, mannose, N-acetylglucosamine, fatty acid (assigned as 22:0), and sphingoid (assigned as above); for nAtCTeS, [M+Na]+ ions at m/z 1233.88, 1247.89, 1289.95, and 1303.96 with one mole each of glucose, mannose, fucose, N-acetylglucosamine, fatty acid (assigned as 18:0 or 22:0), and sphingoid (assigned as above); for AtCTeS, [M+Na]+ ions at m/z 1290.78, 1304.80, 1346.84, and 1360.85 with one mole each of glucose, mannose, N-acetylglucosamine, N-acetylgalactosamine, fatty acid (assigned as above), and sphingoid (assigned as above); for CPS, [M+Na]+ ion at m/z 1493.18 and 1507.16 with one mole each of glucose, mannose, N-acetylglucosamine, N-acetylgalactosamine, fucose, fatty acid (assigned as 22:0), and sphingoid (assigned as above); and for CHS, [M+Na]+ ion at m/z 1695.54 and 1709.55 with one mole each of glucose, mannose, N-acetylgalactosamine, fucose, two moles of N-acetylglucosamine, and one mole each of fatty acid (assigned as 22:0) and sphingoid (assigned as above), respectively.
Fig. 4.
Positive-ion reflector mode MALDI-TOF MS spectra of the isolated GSLs. A: CMS; a: [M+Na]+ ion at m/z 722.78; b: [M+Na]+ ion at m/z 738.77; c: [M+Na]+ ion at m/z 778.86; d: [M+Na]+ ion at m/z 792.87; B: CDS; a: [M+Na]+ ion at m/z 884.62; b: [M+Na]+ ion at m/z 898.64; c: [M+Na]+ ion at m/z 940.69; d: [M+Na]+ ion at m/z 954.70; C: nAtCTS; a: [M+Na]+ ion at m/z 1030.67; b: [M+Na]+ ion at m/z 1044.68; c: [M+Na]+ ion at m/z 1086.73; d: [M+Na]+ ion at m/z 1100.74; D: AtCTS; a: [M+Na]+ ion at m/z 1143.69; b: [M+Na]+ ion at m/z 1157.69; E: nAtCTeS; a: [M+Na]+ ion at m/z 1233.88; b: [M+Na]+ ion at m/z 1247.89; c: [M+Na]+ ion at m/z 1289.95; d: [M+Na]+ ion at m/z 1303.96; F: AtCTeS; a: [M+Na]+ ion at m/z 1290.78; b: [M+Na]+ ion at m/z 1304.80; c: [M+Na]+ ion at m/z 1346.84; d: [M+Na]+ ion at m/z 1360.85; G: CPS; a: [M+Na]+ ion at m/z 1493.18; b: [M+Na]+ ion at m/z 1507.16; H: CHS; a: [M+Na]+ ion at m/z 1695.54; b: [M+Na]+ ion at m/z 1709.55.
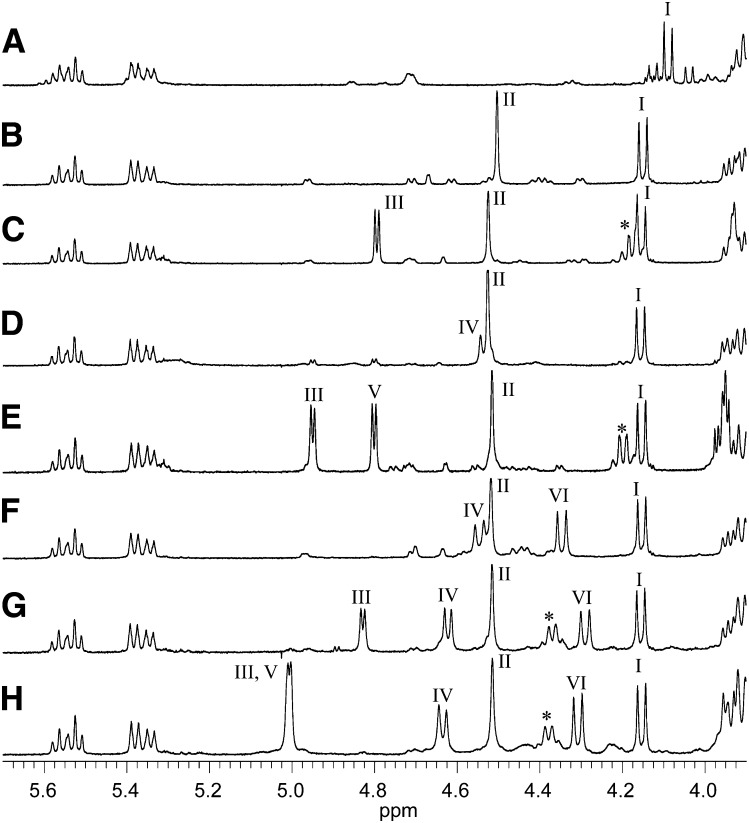
Anomeric configurations of the sugar residues
Anomeric configurations of the sugar residue in GSLs were determined by a 1H-NMR spectrometer (Fig. 5). Configurations were assigned by reference to the data of MacCer and At3Cer (11), that of At5Cer (27), of Fuc (28), and of α-GlcNAc (29, 30) in terms of chemical shifts and coupling constants, these assignments of which are listed in Table 3. The anomeric assignments of α-Fuc and α-GlcNAc in nAtCTeS were determined by downfield shift of glycosyl-substituted fucose compared with the data of nAtCTS, which was consistent with the data of Xu et al. (14). In the anomeric region of the spectrum for each GSL, the following anomeric proton resonances were observed: at 4.09 ppm (J1,2=7.8Hz) for β-Glc (Fig. 5A, CMS); at 4.15 ppm (J1,2=7.8Hz) for β-Glc and at 4.50 ppm (J1,2= ∼1Hz) for β-Man (Fig. 5B, CDS); at 4.15 ppm (J1,2=7.8Hz) for β-Glc, at 4.52 ppm (J1,2= ∼1Hz) for β-Man and at 4.79 ppm (J1,2=3.7Hz) for α-Fuc (Fig. 5C, nAtCTS); at 4.16 ppm (J1,2=7.8Hz) for β-Glc, at 4.53 ppm (J1,2= ∼1Hz) for β-Man, at 4.53 ppm (J1,2=7.3Hz) for β-GlcNAc (Fig. 5D, AtCTS); at 4.15 ppm (J1,2=7.8Hz) for β-Glc, at 4.52 ppm (J1,2= ∼1Hz) for β-Man, at 4.95 ppm (J1,2=3.7Hz) for α-Fuc, and at 4.80 ppm (J1,2=3.7Hz) for α-GlcNAc (Fig. 5E, nAtCTeS); at 4.15 ppm (J1,2=7.8Hz) for β-Glc, at 4.52 ppm (J1,2= ∼1Hz) for β-Man, at 4.55 ppm (J1,2=8.2Hz) for β-GlcNAc and at 4.35 ppm (J1,2=8.7Hz) for β-GalNAc (Fig. 5F, AtCTeS); at 4.16 ppm (J1,2=7.8Hz) for β-Glc, at 4.52 ppm (J1,2= ∼1Hz) for β-Man, at 4.62 ppm (J1,2=6.4Hz) for β-GlcNAc, at 4.29 ppm (J1,2=8.7Hz) for β-GalNAc, and at 4.83 ppm (J1,2=3.7Hz) for α-Fuc (Fig. 5G, CPS); at 4.15 ppm (J1,2=7.8Hz) for β-Glc, at 4.52 ppm (J1,2= ∼1Hz) for β-Man, at 4.64 ppm (J1,2=7.3Hz) for β-GlcNAc, at 4.31 ppm (J1,2=8.2Hz) for β-GalNAc, and at 5.01 ppm (J1,2=2.7Hz) for overlapping α-Fuc and α-GlcNAc (Fig. 5H, CHS).
Fig. 5.
Anomeric proton regions of the 1H-NMR spectra of the isolated GSLs. A: CMS; B: CDS; C: nAtCTS; D: AtCTS; E: nAtCTeS; F: AtCTeS; G: CPS; H: CHS; I: Glcβ; II: Manβ; III: Fucα; IV: GlcNAcβ; V: GlcNAcα; VI: GalNAcβ; *, Fuc5H.
TABLE 3.
Chemical shifts and J1,2 coupling constants of the protons of the isolated glycosphingolipids in the anomeric regions
| CMS | Glc1 | |||||
|---|---|---|---|---|---|---|
| I | ||||||
| Chemical shifts (ppm) | 4.09 | |||||
| Coupling constants (Hz) | 7.8 | |||||
| CDS | Man1- | 4Glc1 | ||||
| II | I | |||||
| Chemical shifts (ppm) | 4.50 | 4.15 | ||||
| Coupling constants (Hz) | ∼1 | 7.8 | ||||
| nAtCTS | Fuc1- | 3Man1- | 4Glc1 | |||
| III | II | I | ||||
| Chemical shifts (ppm) | 4.79 | 4.52 | 4.15 | |||
| Coupling constants (Hz) | 3.7 | ∼1 | 7.8 | |||
| AtCTS | GlcNAc1- | 3Man1- | 4Glc1 | |||
| IV | II | I | ||||
| Chemical shifts (ppm) | 4.53 | 4.53 | 4.16 | |||
| Coupling constants (Hz) | 7.3 | ∼1 | 7.8 | |||
| nAtCTeS | GlcNAc1- | 2Fuc1- | 3Man1- | 4Glc1 | ||
| V | III | II | I | |||
| Chemical shifts (ppm) | 4.80 | 4.95 | 4.52 | 4.15 | ||
| Coupling constants (Hz) | 3.7 | 3.7 | ∼1 | 7.8 | ||
| AtCTeS | GalNAc1- | 4GlcNAc1- | 3Man1- | 4Glc1 | ||
| VI | IV | II | I | |||
| Chemical shifts (ppm) | 4.35 | 4.55 | 4.52 | 4.15 | ||
| Coupling constants (Hz) | 8.7 | 8.2 | ∼1 | 7.8 | ||
| CPS | GalNAc1-4 | (Fuc1-3) | GlcNAc1- | 3Man1- | 4Glc1 | |
| VI | III | IV | II | I | ||
| Chemical shifts (ppm) | 4.29 | 4.83 | 4.62 | 4.52 | 4.16 | |
| Coupling constants (Hz) | 8.7 | 3.7 | 6.4 | ∼1 | 7.8 | |
| CHS | GalNAc1-4 | (GlcNAc1- | 2Fuc1-3) | GlcNAc1- | 3Man1- | 4Glc1 |
| VI | V | III | IV | II | I | |
| Chemical shifts (ppm) | 4.31 | 5.01 | 5.01 | 4.64 | 4.52 | 4.15 |
| Coupling constants (Hz) | 8.2 | 2.7 | 2.7 | 7.3 | ∼1 | 7.8 |
DISCUSSION
This study shows the structures of cerebroside, mannolipid, and fucomannolipids as neutral GSLs in the cysts of the brine shrimp Artemia franciscana. There are some differences in the sphingolipids between the brine shrimp and insect culture cell/flies; the differences are that the dominant sphingoids are mainly hexadeca-4-sphingenine/heptadeca-4-sphingenine and tetradeca-4-sphingenine/hexadeca-4-sphingenine, respectively, and that the dominant fatty acids are behenic acid and arachidic acid/stearic acid/behenic acid, respectively (7, 31). Compared with the dominant sphingoids of the CTeS (BSG-I and II) in newly hatched nauplii of the brine shrimp (14), those of nAtCTeS are similar, whereas the dominant fatty acids are quite different between the nauplii and the cysts. In the CTeS isolated from the hatched shrimp, the dominant fatty acid was 2-hydroxy behenic acid, followed by behenic acid, as inferred from the density of BSG-I and BSG-II on the TLC plate (14). On the other hand, in the CTeS isolated from the cysts, behenic acid was largely dominant and 2-hydroxy behenic acid was below detection limits. There are many sibling Artemia species, which cannot mate with each other despite being indistinguishably similar in morphology among Artemia species: A. franciscana, A. monica, A. parthenogenetica, A. persimilis, A. salina, A. sinica, A. tibetiana, A. tunisiana, and A. urmiana. It would be interesting to know whether the difference due to sibling species or stages is due to the difference in the fatty acid composition of CTeS in two stages. Apart from CTeS, it appears that the hatched brine shrimp contains GSLs, such as CMS, CDS, CTS, CPS, and CHS, as indicated by a previous report on the CTeS in the hatched shrimp by Horibata et al. (32). The dominant GSLs are CTS and CTeS; of these, CTS appears to be Fucα3Manβ4GlcβCer because CTeS is a nonarthro-series GSL (GlcNAcα2Fucα3Manβ4GlcβCer). As for CPS and CHS, the cysts apparently contain a higher amount than the hatched shrimp; this could be due to the fact that Horibata et al. used only the solvent of chloroform-methanol 2:1 (v/v) as the solvent in the lipid extraction and recovered only the lower-phase extract after a Folch partition in the purification of lipids. Compared with the GSLs in the hatched shrimp, those in the cysts differ considerably regarding the amount of CDS. Although only a small difference exists in the simple GSLs between the cysts and the hatched shrimp, further studies are required in the future to determine whether there are further differences in the complex GSLs between the two developmental stages and the other stages. In addition, it is also interesting to note that identical dominant ceramide components of sphingomyelin and GSLs have been detected in the cysts of the brine shrimp Artemia franciscana (15). It seems likely that in the case of neutral GSLs as well as the sphingomyelin ceramide, biosynthesis in this species utilizes a common ceramide pool.
It was assumed that the arthro-series GSL would be detected in the brine shrimp as a major GSL because this species belongs to the phylum Arthropoda. Structural analyses of neutral GSLs with some sugar residues have been performed in several Arthropodae: the green-bottle fly Lucilia caesar (7) and the blowfly Calliphora vicina (8), the fruit fly Drosophila melanogaster (33), the insect culture cell line High Five Cell derived from the cabbage looper Trichoplusia ni (31), the moth Manduca sexta (34), and the millipede Parafontaria laminata armigera (28), all of which have been verified to contain At3Cer. From the brine shrimp, arthro-series GSL (At3Cer) was characterized as a minor CTS, but on the other hand, and surprisingly, a nonarthro-series GSL (nAtCTS) was characterized as a major CTS. However, nonarthro-series GSLs account for ∼10% of the total 102 mg of neutral GSLs. Both CTSes are elongated from MacCer as the common substrate by the two enzymes involved, β3-N-acetylglucosaminyltransferase and α3-fucosyltransferase (see Scheme 1), and both CTSes independently elongate a saccharide to synthesize each series of CTeS. Interestingly, not only nAtCTeS but also At4Cer were abundant as CTeS in the brine shrimp Artemia franciscana, despite the fact that there is no report that has detected At4Cer to be abundant except for reports of flies’ GSLs (5, 7, 8, 33). It has been shown in Arthropoda that the nonarthro-series sugar chain was dominant in a cultured insect cell line, namely High-Five cells (31). The report discussed the following: 1) the existence of an alternative nonarthro CTS (Galβ3Manβ4Glcβ1Cer) to At3Cer in High Five GSLs implies that there is potential for the generation of multiple pathways with a possibility for core switching as observed in mammalian cells, 2) the fact that the dominant CTS of the cell is itself a substrate for further glycosylation supports the premise that the potential for sequence variability is inherent in the nature of the machinery for glycosylation, and 3) although the potential existence of a Galβ3Manβ4Glcβ1Cer core pathway is not sufficient to rescue the brn mutation, the observation that brn is essential for development does not rule out the existence of one or more alternative glycosylation pathways operating during the life cycle of a normal fly. It was also indicated that more complex GSLs such as At4Cer and At5Cer were necessary for normal development in Drosophila (27). In the current report on Artemia, we show that AtCTeS was abundant in amount (6.9 mg) whereas AtCTS was minor (2.1 mg), and we propose that the likely source of GSL with more than five sugar residues is the core arthro-series sugar chain, elongated by transfer of the sugars occurring at the terminus of the nonarthro-series, as a branch (see ). In Artemia, although both arthro-series and nonarthro-series GSLs were detected as both CTS and CTeS, the only CPS detected was derived from AtCTeS. Thus, CPS is composed of only the core arthro-series sugar chain plus a branching fucose, whereas CHS is characterized by the additional presence of an N-acetylglucosamine residue on this branching fucose. CHS is therefore a hybrid structure, as it were, composed of the core arthro-series sugar chain plus the sugars from the nonreducing end of the nonarthro-series chain as a branch. We speculate that Artemia contains two pools (arthro and nonarthro) of CTS and CTeS, each pool serving individual biological functions, and that the organism may divert either form of CTS and CTeS as needed for the biosynthesis of more complex GSLs, such as a sequence variability.
Dominant pathways for glycosphingolipid glycosylation in Artemia franciscana.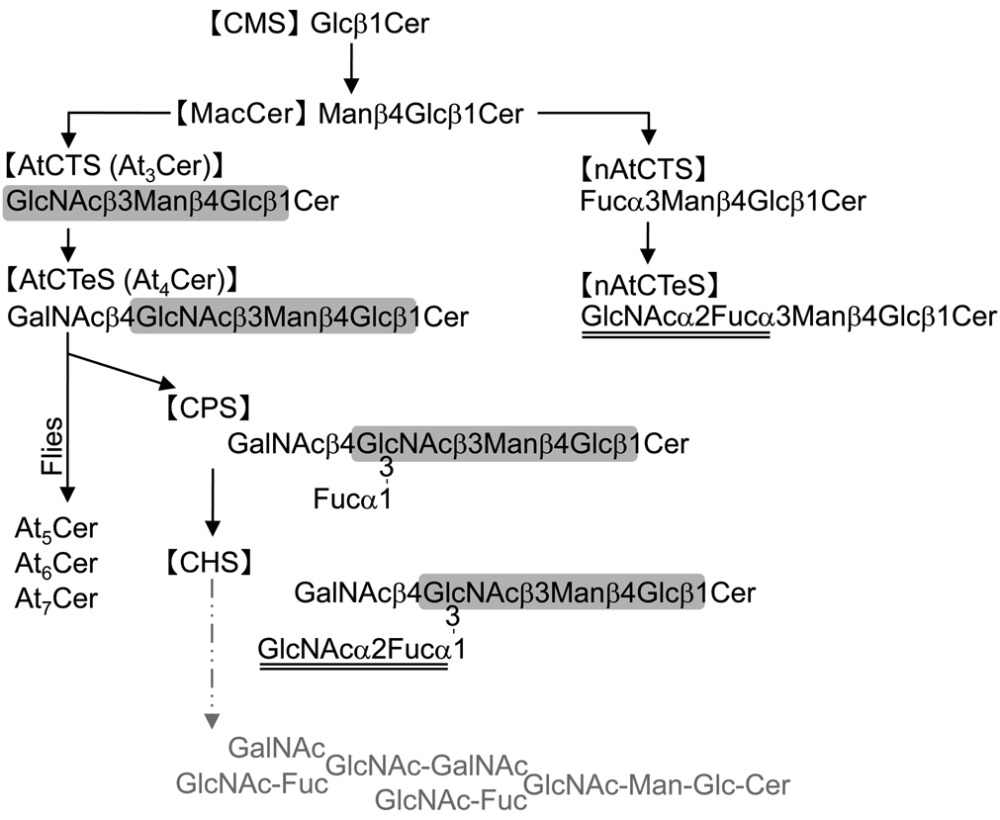
|
Scheme 1. |
The observations reported herein that a fucosylated LacdiNAc trisaccharide (GalNAcβ1-4[Fucα1-3]GlcNAcβ) determinant has been found in CPS and internally in CHS is interesting in several respects. This trisaccharide is a Lewis X (LeX) analog previously reported in the parasite Schistosoma mansoni (35, 36), albeit on different aglycone structures. It has also been found as a carbohydrate terminus on a human immunosuppressive glycoprotein, glycodelin (37). An enzyme α1,3-fucosyltransferase, which is capable of producing fucosylated LacdiNAc trisaccharide from LacdiNAc, has been cloned from the nematode Caenorhabditis elegans as CEFT-1 (38) and from the honeybee Apis mellifera (39). Although the presence of this enzyme has not been studied, it might be found in the brine shrimp. As far as we know, there is no report about whether terminal αGlcNAc, terminal αFuc, or the disaccharide of GlcNAcα2Fucα3 in GSL are involved in brine shrimp development. Because CPS and CHS contain fucosylated LacdiNAc structure, a LeX analog, it is possible that these novel GSLs interact with each other and aggregate to form patches of microdomain in which developmental signals are transduced according to GSL-GSL interaction, such as on Lex-Lex, Ley-H, and Gb4-Gg3 (40).
The structure of CHS consisted of core arthro-series GSLs with a branching nonarthro-series disaccharide (GlcNAcα2Fucα-). In the structures of nAtCTeS and CHS, “α-anomeric” GlcNAc was attached at the nonreducing end. This type of sugar chain in GSL structure is reported in only a few cases: GlcNAcα4GalαCer from the marine sponge Axinella damicornis (29), GlcNAcα1-HPO3-6GalCer from the liver fluke Fasciola hepatica (30), and CTeS (BSG-I and II) from newly hatched nauplii in the brine shrimp (14). Mucin-type O-glycan with terminal αGlcNAc is also reported in only two cases: gastric mucins from pig [Fucα2Galβ4GlcNAcβ6(GlcNAcα4Galβ3)GalNAc-ol and GlcNAcα4Galβ4GlcNAcβ6(GlcNAcα4Galβ3)GalNAc-ol] (41) and a human gastric mucin [GlcNAcα4Galβ4GlcNAcβ6(GlcNAcα4Galβ3)GalNAcαSer/Thr] (42). Interestingly, it was reported that these mucins arrested proliferation of the spiral-shaped gram-negative bacterium Helicobacter pylori, which causes stomach upsets and peptic ulcers (43). It will be interesting to examine whether nAtCTeS and CHS from the brine shrimp arrest the growth of H. pylori as the mucins do and to investigate the presence of further unique sugar chains in more complex GSLs.
This report on Artemia shows the existence of a sugar chain with rare terminal αGlcNAc. To the best of our knowledge, although the sugar addition appears to be catalyzed by α1,2-N-acetylglucosaminyltransferase (α2GnT), it has not been reported among animals to date. Its homologous transferase, α1,4-N-acetylglucosaminyltransferase (α4GnT) has been characterized in human (42). By using the data of nucleic or amino acid sequences of α4GnT, homology searches were performed via BLAST searches (blastn, tblastn, and tblastx) with the filter of “invertebrate” on the DDBJ web server. As a result, only two homologous sequences were found: α1,4-N-acetylgalactosaminyltransferase (GalNAcT) from the fruit fly Drosophila melanogaster and lactosylceramide 4-α-galactosyltransferase putative mRNA from the salmon lice Lepeophtheirus salmonis. Further homology searches were performed according to the method mentioned above by using the data of nucleic or amino acid sequences of the fly GalNAcT, and again only two records from the same organisms as mentioned above were found. Thus, α2GnT appears rare. This warrants further study in organisms in which sugar chain with the terminal αGlcNAc is detected.
Supplementary Material
Acknowledgments
The authors thank Dr. John T. Dulaney who kindly read this manuscript while it was in preparation and for providing helpful comments.
Footnotes
Abbreviations:
- At
- arthro
- At3Cer
- arthrotriaosylceramide
- At4Cer
- arthrotetraosylceramide
- At5Cer
- arthropentaosylceramide
- At6Cer
- arthrohexaosylceramide
- At7Cer
- arthroheptaosylceramide
- CDS
- ceramide disaccharide
- CHS
- ceramide hexasaccharide
- CMS
- ceramide monosaccharide
- CPS
- ceramide pentasaccharide
- CTS
- ceramide trisaccharide
- CTeS
- ceramide tetrasaccharide
- GSL
- glycosphingolipid
- LeX
- Lewis X
- 1H-NMR
- proton nuclear magnetic resonance
- HPTLC
- high performance thin-layer chromatography
- MacCer
- mactosyl ceramide
- nAt
- nonarthro
- TOF
- time-of-flight
This work was supported in part by the Grant-in-Aid for Scientific Research from the Japanese Ministry for Education, Science, Culture and Sports under the Grant No. 10010417, and a scholarship from the Society for the Advancement of Science and Technology at Ritsumeikan University.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of a table.
REFERENCES
- 1.Hakomori S. 2000. Cell adhesion/recognition and signal transduction through glycosphingolipid microdomain. Glycoconj. J. 17: 143–151. [DOI] [PubMed] [Google Scholar]
- 2.Yu R. K., Nakatani Y., Yanagisawa M. 2009. The role of glycosphingolipid metabolism in the developing brain. J. Lipid Res. 50: S440–S445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E., et al. 1997. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 278: 1626–1629. [DOI] [PubMed] [Google Scholar]
- 4.Yu R. K., Yanagisawa M., Ariga T. 2007. Glycosphingolipid structures. Comprehensive Glycoscience 1: Introduction to Glycoscience Synthesis of Carbohydrates. Kamerling J. P. editor Elsevier, Oxford: 73–122. [Google Scholar]
- 5.Itonori S., Sugita M. 2007. Glycophylogenetic aspects of lower animals. Comprehensive Glycoscience 3: Biochemistry of Glycoconjugate Glycans Carbohydrate-mediated Interactions. Kamerling J. P., editor Elsevier, Oxford. U.K: 253–284. [Google Scholar]
- 6.Lochnit G., Dennis R. D., Ulmer A. J., Geyer R. 1998. Structural elucidation and monokine-inducing activity of two biologically active zwitterionic glycosphingolipids derived from the porcine parasitic nematode Ascaris suum. J. Biol. Chem. 273: 466–474. [DOI] [PubMed] [Google Scholar]
- 7.Sugita M., Nishida M., Hori T. 1982. Studies on glycosphingolipids of larvae of the green-bottle fly, Lucilia caesar. I. Isolation and characterization of glycosphingolipids having novel sugar sequences. J. Biochem. 92: 327–334. [DOI] [PubMed] [Google Scholar]
- 8.Dennis R., Geyer R., Egge H., Peter-Katalinic J., Li S., Stirm S., Wiegandt H. 1985. Glycosphingolipids in insects; chemical structures of ceramide tetra-, penta-, hexa-, and heptasaccharide from Calliphora vicina pupae (Insecta: Diptera). J. Biol. Chem. 260: 5370–5375. [PubMed] [Google Scholar]
- 9.Müller R., Altmann F., Zhou D., Hennet T. 2002. The Drosophila melanogaster brainiac protein is a glycolipid-specific β1,3N-acetylglucosaminyltransferase. J. Biol. Chem. 277: 32417–32420. [DOI] [PubMed] [Google Scholar]
- 10.Wandall H. H., Pedersen J. W., Park C., Levery S. B., Pizette S., Cohen S. M., Schwientek T., Clausen H. 2003. Drosophila egghead encodes a β1,4-mannosyltransferase predicted to form the immediate precursor glycosphingolipid substrate for brainiac. J. Biol. Chem. 278: 1411–1414. [DOI] [PubMed] [Google Scholar]
- 11.Schwientek T., Keck B., Levery S. B., Jensen M. A., Pedersen J. W., Wandall H. H., Stroud M., Cohen S. M., Amado M., Clausen H. 2002. The drosophila gene brainiac encodes a glycosyltransferase putatively involved in glycosphingolipid synthesis. J. Biol. Chem. 277: 32421–32429. [DOI] [PubMed] [Google Scholar]
- 12.Wandall H. H., Pizette S., Pedersen J. W., Eichert H., Levery S. B., Mandel U., Cohen S. M., Clausen H. 2005. Egghead and brainiac are essential for glycosphingolipid biosynthesis in vivo. J. Biol. Chem. 280: 4858–4863. [DOI] [PubMed] [Google Scholar]
- 13.Dhont J., Sorgeloos P. 2002. Artemia and aquaculture. Artemia: Basic and Applied Biology (Biology of Aquatic Organisms). Abatzopoulos T. J., Beardmore J. A., Clegg J. S., Sorgeloos P, Kluwer Academic Publishers, Dordrecht, The Netherlands. 259–269. [Google Scholar]
- 14.Xu X., Horibata Y., Inagaki M., Hama Y., Sakaguchi K., Goda H. M., Okino N., Ito M. 2009. A novel fucosyl glycosphingolipid of brine shrimp that is highly sensitive to endoglycoceramidase. Glycobiology. 19: 1446–1451. [DOI] [PubMed] [Google Scholar]
- 15.Kojima H., Inoue T., Sugita M., Itonori S., Ito M. 2010. Biochemical studies on sphingolipid of Artemia franciscana (I) isolation and characterization of sphingomyelin. Lipids. 45: 635–643. [DOI] [PubMed] [Google Scholar]
- 16.Saito T., Hakomori S. 1971. Quantitative isolation of total glycosphingolipids from animal cells. J. Lipid Res. 12: 257–259. [PubMed] [Google Scholar]
- 17.Svennerholm L. 1956. The quantitative estimation of cerebrosides in nervous tissue. J. Neurochem. 1: 42–53. [DOI] [PubMed] [Google Scholar]
- 18.Dittmer J. C., Lester R. L. 1964. A simple, specific spray for the detection of phospholipids of thin-layer chromatograms. J. Lipid Res. 5: 126–127. [PubMed] [Google Scholar]
- 19.Kozulić B., Ries B., Mildner P. 1979. N-acetylation of amino sugar methyl glycosides for gas-liquid chromatographic analysis. Anal. Biochem. 94: 36–39. [DOI] [PubMed] [Google Scholar]
- 20.Kishimoto Y., Hoshi M. 1972. Isolation, purification, and assay of fatty acids and steroids from the nervous system. Methods of Neurochemistry. Fried R., editor Marcel Dekker, New York: 75–152. [Google Scholar]
- 21.Ciucanu I., Kerek F. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131: 209–217. [Google Scholar]
- 22.Itonori S., Takahashi M., Sugita M. 2000. Application of microwave-mediated reaction to lipid analysis. Japan Oil Chemists’ Society American Oil Chemists’ Society World Congress 2000, Kyoto, October 22–27, 2000. [Google Scholar]
- 23.Itonori S., Takahashi M., Aoki K., Sugita M. 2001. Microwave-mediated acetolysis of glycosphingolipids. Shiga Daigaku Kyoikugakubu Kiyo, III: Shizen Kagaku, Volume Date 2000, 50, 17–24. Shiga Daigaku Kyoikugakubu, CODEN: SDKKFY, ISSN: 1342–9272. In Japanese. CAN 136:147387, AN 2001:364705. [DOI] [PubMed] [Google Scholar]
- 24.Gaver R. C., Sweeley C. C. 1965. Methods for methanolysis of sphingolipids and direct determination of long-chain bases by gas chromatography. J. Am. Oil Chem. Soc. 42: 294–298. [DOI] [PubMed] [Google Scholar]
- 25.Jansson P. E., Kenne L., Liedgren H., Lindberg B., Lönngren J. 1976. A practical guide to the methylation analysis of carbohydrates. A practical guide to the methylation analysis of carbohydrates. Chemical Communications (Stockholm University), 8, 1–75, CODEN: CCUSBN, ISSN: 0366–5607. General Review in English. CAN 87:136153, AN 1977:536153. [Google Scholar]
- 26.Tai T., Yamashita K., Kobata A. 1975. Synthesis and mass fragmentographic analysis of partially O-methylated 2-N-methylglucosamines. J. Biochem. 78: 679–686. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y., Pedersen J. W., Wandall H. H., Levery S. B., Pizette S., Clausen H., Cohen S. M. 2007. Glycosphingolipids with extended sugar chain have specialized functions in development and behavior of Drosophila. Dev. Biol. 306: 736–749. [DOI] [PubMed] [Google Scholar]
- 28.Sugita M., Hayata C., Yoshida T., Suzuki M., Suzuki A., Takeda T., Hori T., Nakatani F. 1994. A novel fucosylated glycosphingolipid from the millipede Parafontaria laminata armigera. Biochim. Biophys. Acta. 1215: 163–169. [DOI] [PubMed] [Google Scholar]
- 29.Costantino V., D'Esposito M., Fattorusso E., Mangoni A., Basilico N., Parapini S., Taramelli D. 2005. Damicoside from Axinella damicornis: the influence of a glycosylated galactose 4-OH group on the immunostimulatory activity of α-galactoglycosphingolipids. J. Med. Chem. 48: 7411–7417. [DOI] [PubMed] [Google Scholar]
- 30.Wuhrer M., Grimm C., Zähringer U., Dennis R. D., Berkefeld C. M., Idris M. A., Geyer R. 2003. A novel GlcNAcα1-HPO3-6Gal(1–1)ceramide antigen and alkylated inositol-phosphoglycerolipids expressed by the liver fluke Fasciola hepatica. Glycobiology. 13: 129–137. [DOI] [PubMed] [Google Scholar]
- 31.Fuller M. D., Schwientek T., Wandall H. H., Pedersen J. W., Clausen H., Levery S. B. 2005. Structure elucidation of neutral, di-, tri-, and tetraglycosylceramides from High Five cells: identification of a novel (non-arthro-series) glycosphingolipid pathway. Glycobiology. 15: 1286–1301. [DOI] [PubMed] [Google Scholar]
- 32.Horibata Y., Sakaguchi K., Okino N., Iida H., Inagaki M., Fujisawa T., Hama Y., Ito M. 2004. Unique catabolic pathway of glycosphingolipids in a hydrozoan, Hydra magnipapillata, involving endoglycoceramidase. J. Biol. Chem. 279: 33379–33389. [DOI] [PubMed] [Google Scholar]
- 33.Seppo A., Moreland M., Schweingruber H., Tiemeyer M. 2000. Zwitterionic and acidic glycosphingolipids of the Drosophila melanogaster embryo. Eur. J. Biochem. 267: 3549–3558. [DOI] [PubMed] [Google Scholar]
- 34.Abeytunga D. T. U., Oland L., Somogyi A., Polt R. 2008. Structural studies on the neutral glycosphingolipids of Manduca sexta. Bioorg. Chem. 36: 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khoo K-H., Chatterjee D., Caulfield J. P., Morris H. R., Dell A. 1997. Structural mapping of the glycans from the egg glycoproteins of Schistosoma mansoni and Schistosoma japonicum: identification of novel core structures and terminal sequences. Glycobiology. 7: 663–677. [DOI] [PubMed] [Google Scholar]
- 36.Nyame A. K., Yoshino T. P., Cummings R. D. 2002. Differential expression of LacdiNAc, fucosylated LacdiNAc, and Lewis X glycan antigens in intramolluscan stages of Schistosoma mansoni. J. Parasitol. 88: 890–897. [DOI] [PubMed] [Google Scholar]
- 37.Dell A., Morris H. R., Easton R. L., Panico M., Patankar M., Oehninger S., Koistinen R., Koistinen H., Seppala M., Clark G. F. 1995. Structural analysis of the oligosaccharides derived from glycodelin, a human glycoprotein with potent immunosuppressive and contraceptive activities. J. Biol. Chem. 270: 24116–24126. [DOI] [PubMed] [Google Scholar]
- 38.DeBose-Boyd R. A., Nyame A. K., Cummings R. D. 1998. Molecular cloning and characterization of an α1,3 fucosyltransferase, CEFT-1, from Caenorhabditis elegans. Glycobiology. 8: 905–917. [DOI] [PubMed] [Google Scholar]
- 39.Rendi D., Klaudiny J., Stemmer U., Schmidt J., Paschinger K., Wilson I. B. H. 2007. Towards abolition of immunogenic structures in insect cells: characterization of a honey-bee (Apis mellifera) multi-gene family reveals both an allergy-related core α1,3-fucosyltransferase and the first insect Lewis-histo-blood-group-related antigen-synthesizing enzyme. Biochem. J. 402: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakomori S. 2008. Structure and function of glycosphingolipids and sphingolipids: recollections and future trends. Biochim. Biophys. Acta. 1780: 325–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishihara K., Kurihara M., Goso Y., Urata T., Ota H., Katsuyama T., Hotta K. 1996. Peripheral α-linked N-acetylglucosamine on the carbohydrate moiety of mucin derived from mammalian gastric gland mucous cells: epitope recognized by a newly characterized monoclonal antibody. Biochem. J. 318: 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama J., Yen J. C., Misra A. K., Ito S., Katsuyama T., Fukuda M. 1999. Expression cloning of a human α1,4-N-acetylglucosaminyltransferase that forms GlcNAcα1→4Galß→R, a glycan specifically expressed in the gastric gland mucous cell-type mucin. Proc. Natl. Acad. Sci. USA. 96: 8991–8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawakubo M., Ito Y., Okimura Y., Kobayashi M., Sakura K., Kasama S., Fukuda N. M., Fukuda M., Katsuyama T., Nakayama J. 2004. Natural antibiotic function of a human gastric mucin against Helicobacter pylori infection. Science. 305: 1003–1006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.