Abstract
Endothelial lipase (EL) is a phospholipase A1 (PLA1) enzyme that hydrolyzes phospholipids at the sn-1 position to produce lysophospholipids and free fatty acids. Measurement of the PLA1 activity of EL is usually accomplished by the use of substrates that are also hydrolyzed by lipases in other subfamilies such as PLA2 enzymes. In order to distinguish PLA1 activity of EL from PLA2 enzymatic activity in cell-based assays, cell supernatants, and other nonhomogeneous systems, a novel fluorogenic substrate with selectivity toward PLA1 hydrolysis was conceived and characterized. This substrate was preferred by PLA1 enzymes, such as EL and hepatic lipase, and was cleaved with much lower efficiency by lipases that exhibit primarily triglyceride lipase activity, such as LPL or a lipase with PLA2 activity. The phospholipase activity detected by the PLA1 substrate could be inhibited with the small molecule esterase inhibitor ebelactone B. Furthermore, the PLA1 substrate was able to detect EL activity in human umbilical vein endothelial cells in a cell-based assay. This substrate is a useful reagent for identifying modulators of PLA1 enzymes, such as EL, and aiding in characterizing their mechanisms of action.
Keywords: phospholipase A1
Endothelial lipase (EL), a phospholipase A1 (PLA1) subfamily lipase, has gained attention as a potential therapeutic target for raising HDL and protecting against atherosclerotic plaque formation (1, 2). EL is a secreted enzyme that subsequently becomes associated with the surface of endothelial cells, where it retains its enzymatic activity (1, 2). Therefore, EL resides in the proximity of the plasma compartment, where it alters lipoproteins and regulates lipoprotein metabolism. Its primary function is the hydrolysis of HDL phospholipids, which increases HDL turnover and decreases HDL cholesterol (HDL-C) and apolipoprotein A-1 (apoA-1) content (3–5), resulting in an overall decrease in plasma levels of HDL particles. In addition to reducing plasma HDL-C, EL elicits other proatherogenic effects, including increased free fatty acid and lyso-phosphatidycholine, monocyte adhesion to the vessel wall (6), and increased levels of plasma small dense LDL (7). EL is expressed in mouse aorta (8), and its expression is upregulated in response to various inflammatory mediators such as interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α) (9), and IFN-γ (10). EL expression is also upregulated in the aortas of apoE−/− mice, a model for atherosclerosis research (8). Thus, the role of EL in lipoprotein metabolism is highly responsive to the proinflammatory conditions found in atherosclerotic plaques.
Inhibition of EL with an anti-EL antibody increases HDL in several mouse strains, with no increase in triglyceride (TG), LDL, or VLDL (11). In addition, HDL particles were increased approximately 60% in EL knockout mice (12, 13). More significantly, the atherosclerotic lesion area in EL/apoE double-knockout mice was reduced by 70% (6) and was accompanied by increased plasma HDL and reduced vessel macrophage content. However, a more recent study of both EL/apoE and EL/LDLR double-knockout mice did not show significant improvement in the lesion area, despite clear increases in HDL (14). The reason for this discrepancy is not known but may be the result of differences in environmental factors or stage of development of the atherosclerotic lesions at the time of measurement.
Five separate studies have reported mutations in human EL associated with increased plasma HDL-C (13, 15–18). In particular, two nonsynonymous mutations in human subjects were shown to promote elevated HDL either by reduced EL secretion (19) or by reduced EL catalytic activity (20). In addition, EL expression was evident in infiltrating cells, macrophages, and smooth muscle cells within human atheromatous plaques (21). Intriguingly, high levels of plasma EL are correlated with low levels of HDL-C, high levels of small dense LDL, and metabolic syndrome (22). Elevated EL is also associated with increased visceral adiposity (23) and plasma inflammatory markers (24). Thus, increases in EL activity may contribute to the low HDL levels observed in patients with obesity and metabolic syndrome. Overall, data indicate that EL plays an important role in HDL metabolism in humans and that inhibition of EL should increase plasma HDL, especially in patients with low levels of HDL-C. Given the association of low levels of HDL with increased risk of atherosclerosis-induced cardiovascular events in humans (2, 25–27), EL represents a promising target for potential inhibition of atherosclerotic disease.
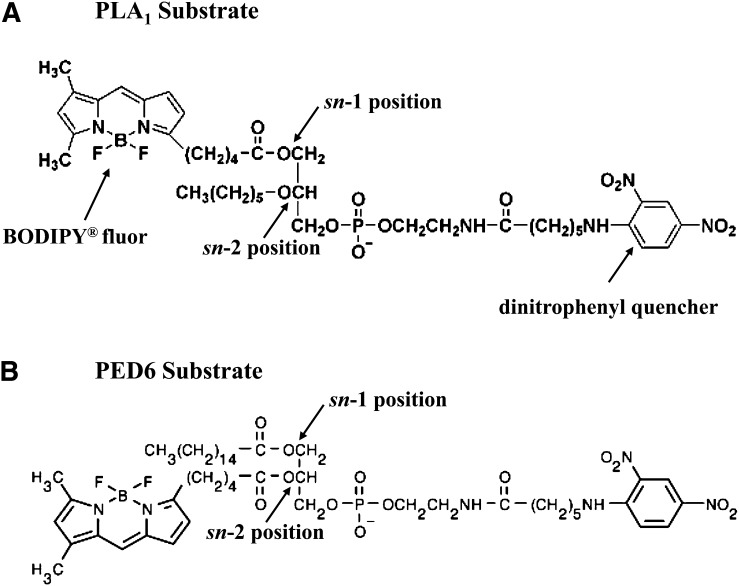
To identify inhibitors of EL as potential cardiovascular disease therapeutic agents, we developed an assay that can be used to reliably detect cell-based EL enzymatic activity and that is also adaptable to a high-throughput compound screening format. Although EL is a secreted enzyme, it remains associated with the cell surface via interactions with proteoglycans through a conserved heparin binding site (28, 29), similar to the related PLA1 enzymes LPL and HL. Thus, cells expressing EL are amenable to monitoring EL enzymatic activity with a suitable substrate. In order to distinguish PLA1 activity of EL from nonspecific PLA2 activity in cell-based assays, we designed and characterized a novel fluorogenic substrate with selectivity for PLA1-specific hydrolysis. This unique substrate has a 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene fluor-conjugated to the fatty acyl chain at the sn-1 position (BODIPY FL-C5). In addition, it has an ether bond at the sn-2 position to preclude hydrolysis by PLA2 enzymes. Finally, the PLA1 substrate has a dinitrophenyl group on the phospholipid head. This chemical moiety quenches the BODIPY fluor until hydrolysis of the substrate takes place, creating distance between the fluor and the quencher, allowing fluorescence to occur. Use of this substrate in PLA1 assays allows continuous monitoring of enzymatic activity over time.
MATERIALS AND METHODS
Materials and reagents
We conceived the PLA1-specific substrate (N-{[6-(2,4-dinitrophenyl) amino] hexanoyl}-1-{4,4-difluoro-5,7-dimethyl-4-bora-3a,-4a-diaza-s-indacene-3-pentanoyl}-2-hexyl-sn-glycero-3-phosphoethanolamine; molecular weight [mw] = 879) (Fig. 1A), based on our need for a PLA1-sensitive substrate for a high-throughput screen for endothelial lipase inhibitors. Because this substrate was not commercially available at the time (2005), we contracted with Molecular Probes to synthesize a customized compound. Following its synthesis for our internal use, Invitrogen made the substrate available for sale as PED-A1 in its 2007 catalog. Compound PED6 (N-{[6-(2,4-dinitrophenyl) amino]hexanoyl}-2-[4,4-difluoro-5,7-dimethyl-4-bora-3a,-4a-diaza-s-indacene-3-pentanoyl}-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine; mw = 1,136) (Fig. 1B) and the cleavage products of both the PED6 and the PLA1 substrate (4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoic acid [BODIPY FL-C5]) (Fig. 2A) were purchased from Molecular Probes (Invitrogen, Eugene, OR). Bovine lipoprotein lipase, ebelactone B, TNF-α, and IL-1β were purchased from Sigma (St. Louis, MO), while bee venom PLA2 was purchased from Cayman Chemical Co. (Ann Arbor, MI).
Fig. 1.
Structures of the fluorescent substrates used. A: Chemical structure of PLA1-specific substrate (N-{[6-(2,4-dinitrophenyl) amino] hexanoyl}-1-{4,4-difluoro-5,7-dimethyl-4-bora-3a,-4a-diaza-s-indacene-3-pentanoyl}-2-hexyl-sn-glycero-3-phosphoethanolamine; mw = 849). B: Chemical structure of PED6 substrate (N-{[6-(2,4-dinitrophenyl) amino]hexanoyl}-2-[4,4-difluoro-5,7-dimethyl-4-bora-3a,-4a-diaza-s-indacene-3-pentanoyl}-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine; mw = 1136). Positions of the sn-1 and sn-2 cleavage sites are indicated for both substrates, and the BODIPY fluor and dinotrophenyl quencher is indicated on the PLA1 substrate.
Fig. 2.
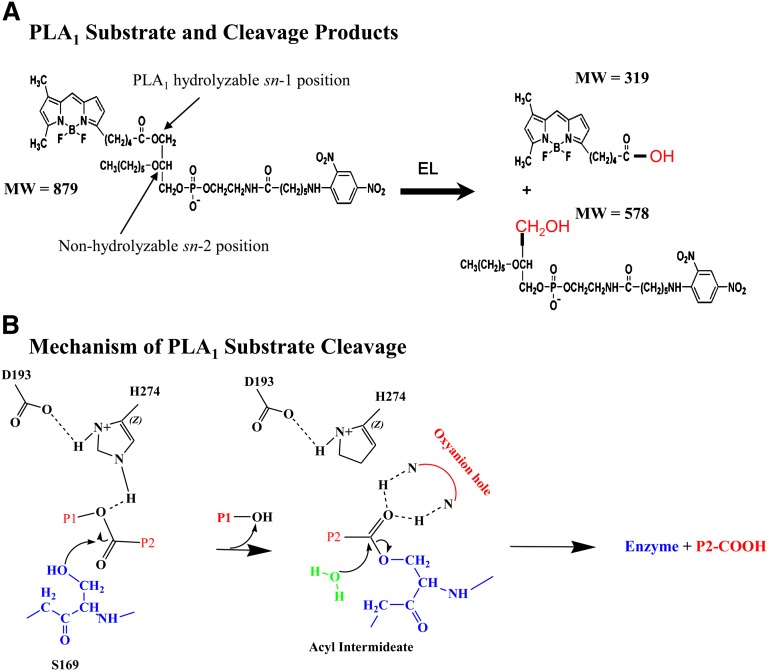
PLA1 substrate cleavage products and the mechanism of catalysis. A: Reaction shows how the PLA1 substrate is hydrolyzed into its two cleavage products by a PLA1 enzyme such as EL. Molecular weights (MW) of the substrate and predicted products are shown. B: The mechanism by which EL catalyzes the cleavage of the ester bond via an sn-1 reaction is shown. The amino acids (Ser169, Asp 193, and His274) of the catalytic triad reported to be at the active site of murine EL are shown. P1, P2, products 1 and 2.
Expression and purification of mouse EL
The cDNA from mouse EL was modified by changing most of the nucleotide bases at the wobble position of the amino acid codons. The resulting cDNA retains the ability to encode full-length wild-type mouse EL polypeptide; however, it is unable to hybridize with the human EL cDNA sequence under high stringency conditions. The modified mouse EL cDNA was synthesized by DNA2.0 (Menlo Park, CA) with both six-histidine (6xHis) and 3xFLAG tags at the C terminus and was then subcloned, using unique BamHI and XbaI sites, into pVL1393. The final construct sequence was confirmed (Seqwright, Houston, TX). Spodoptera frugiperda Sf9 insect cells were routinely maintained between 0.8 × 106 and 4.0 × 106 cells/ml in ESF921 medium (Expression Systems, Woodland, CA) at 27°C. To generate recombinant EL baculovirus, the pVL1393/EL vector was transfected into Sf9 insect cells by using a BaculoGold transfection kit (BD Biosciences, San Diego, CA) according to the manufacturer's protocol. Five days after transfection, passage 0, virus was harvested and stored at 4°C. High-titered virus (passage 3) was obtained through two sequential rounds of amplification and used for large-scale production of EL. Trichoplusia ni High Five cells were used for large-scale production. During growth, cells were maintained between 0.3 × 106 and 4 × 106 cells/ml in ESF921 medium at 27°C. Cells were infected with recombinant P3 EL baculovirus at a density of ∼1.6 × 106 cells/ml, and a multiplicity of infection of 1 at 27°C. Following 72 h of infection, the medium was cleared by centrifugation at 800 g for 10 min at 4°C. Further clarification was achieved with an additional centrifugation at 6,000 g for 10 min at 4°C. Expression was validated by immunoblotting using anti-FLAG (M2) antibody (Sigma).
Medium (6 liters) was concentrated to 0.7 liter by using a Kvick Lab system (GE Healthcare, Piscataway, NJ). Concentrated medium underwent buffer exchange by adding 4 liters of 50 mM Tris, pH 7.4, 150 mM NaCl (buffer A) and was reconcentrated to 0.7 of a liter. Buffer-exchanged medium was clarified by centrifugation at 6,000 g for 1 h. Cleared medium was loaded onto a 10-ml anti-FLAG M2 affinity gel (Sigma) at 1 ml/min at 4°C, using an AKTA Explorer System (GE Healthcare, Piscataway, NJ). The column was washed with 15–20 column volumes of buffer A. Mouse EL was eluted with 5 column volumes of buffer A containing 0.1 mg/ml 3xFLAG peptide (Sigma). Fractions were analyzed by SDS-PAGE, and those fractions containing mouse EL were pooled. Expression yields following purification were ∼1 mg/l as determined by Bradford assay using a protein assay kit from Bio-Rad (Hercules, CA) according to the manufacturer's instructions with BSA as the standard.
Assays with purified lipases
Analysis of EL activity, using purified EL protein, 15 µl of assay buffer (HBSS without calcium, magnesium, or phenol red, with 25 mM HEPES [Mediatech, Manassas, VA]) was placed in a 384-well PCR plate (Abgene, Epsom, UK). Three microliters of PLA1 or PED6 substrate (50 μM) dissolved in DMSO was added using a Multidrop reagent dispenser (Thermo Fisher, Waltham, MA) for a final substrate concentration of 5 μM or as indicated in the figure. The plate was incubated for 10 min at 37°C to avoid the lag phase. Purified mouse EL protein (12 µl; for a final concentration of 0.4 μM) was added for a final assay volume of 30 µl. Fluorescence signal was monitored for 40 min at 37°C with a Safire II plate reader (Tecan, Raleigh, NC) in kinetic mode (80 cycles; kinetic interval, 30 s) with an excitation wavelength of 490 nm and an emission wavelength of 515 nm. Linear regression of the fluorescence intensity values collected from 400 to 1,500 s was used to calculate the reaction rate (the slope), and slopes were used to calculate IC50 values, where appropriate. Conversion of relative fluorescence units (RFU) to moles of substrate turnover was accomplished by determining the specific activity (RFU × M−1) of BODIPY FL-C5 following linear regression of 0.05 to 200 nM (serial dilution) under the exact assay conditions used for each experiment. This was completed at multiple substrate concentrations (each substrate concentration was used in all experiments) and multiple instrument gain settings to account for interfilter effects from the substrate. This must be determined empirically for experiments because the RFU × M−1 value is dependent upon the instrument, instrument settings, assay conditions (of buffer and other components), temperature, and the assay plate.
To analyze for purified bovine LPL (catalog no. L2254; Sigma) and bee venom PLA2 (catalog no. 765001; Cayman) activities, 80 µl of assay buffer (DMEM with no glutamine and no phenol red but with 25 mM HEPES, pH 7.2, [Mediatech]) was placed in a Corning 96-well black plate with clear bottom (catalog no. 3340; Corning, NY). PLA1 or PED6 substrate (100 µl diluted in assay buffer for a series of final concentrations between 0.25 and 10 μM) was added using a Multidrop reagent dispenser. Purified bovine LPL or bee venom PLA2 protein was added to a final concentration of 2.5 μg/ml in a 200 µl assay volume. Fluorescence signal was monitored for 90 min at 37°C with a Safire II plate reader (Tecan, Raleigh, NC), at an excitation wavelength of 490 nm and an emission wavelength of 515 nm. RFU were converted to nM × min−1 of free BODIPY FL-C5 liberated over time from a BODIPY FL-C5 (Molecular Probes) standard curve.
Analysis of PLA1 substrate hydrolysis by EL, using LC/MS
Purified mouse EL (200 nM) was incubated with PLA1 substrate (10 µM), and reactions were quenched with MeOH at a final concentration of 25%. Samples were subjected to LC/MS analysis (using negative-ion mode) monitoring A220, A503, and m/z mass to charge ratio 319 for selective ion monitoring (SIM) or scanning the m/z range 300–350 and 879 (SIM, scan 800–900 mass units) and excitation/emission at 490 nm/525 nm (8 nm band pass) using an Agilent 1100 LC unit coupled to an Agilent MSD system (Agilent Technologies, Santa Clara, CA). Stationary and mobile phases consisted of 10 mM NH4OAc (pH 7.5; Sigma Ultra) and 10 mM NH4OAc plus 95% MeOH (LC/MS grade; EMD), respectively. Chromatography was accomplished with a 20%-to-95% MeOH gradient over 3 min, where the mobile phase was maintained at 100% for an additional 3 min, and the column was equilibrated for 4 min with 20% MeOH at 0.5 ml × min−1 using a Jupiter C5 50 × 3 mm column (Phenomenex, Torrance, CA). Substrate hydrolysis was quantified by using the equation for A505 data, [S] × AUCproduct/(AUCproduct + AUCsubstrate), and the equation for LC/MS SIM, AUCproduct/7.53 × 104 × uM−1, where 7.53 × 104 is the area under the curve (AUC) units × uM−1 of purified BODIPY FL-C5, as determined empirically from standard curve analysis.
Assays with HEK293 cells stably expressing lipases
The murine EL gene (LIPG [NM_010720]) cDNA sequence was modified, optimized for codon preference, and fused to an in-frame C-terminal 6xHis 3xFLAG affinity epitope tag (as described above). Resulting cDNA was subcloned into pcDNA3.1(+) with neomycin resistance (Invitrogen, Carlsbad, CA). The human HL (hHL) (LIPC [NM_000236]) and human LPL (hLPL) (NM_000237) genes were obtained from OriGene Technologies, TrueClone™ collection (catalog nos. SC120025 and SC120026, respectively). Full-length HL cDNA, isolated as a NotI fragment, and full-length LPL cDNA, isolated as an EcoRI fragment, were subcloned into the mammalian expression vector pcDNA3Neo (Invitrogen, Carlsbad, CA). HEK293 cells were transfected with either EL, HL, or LPL cDNA by using SuperFect transfection reagent (Qiagen, Valencia, CA) as directed by the manufacturer, and selected for stable expression of the desired plasmid with 1,200 mg/ml G418 solution (Mediatech). Stable HEK293 cell lines were maintained in DMEM (Mediatech), 10% fetal bovine serum (HyClone, Logan, UT), 2 mM l-glutamine (Mediatech), 50 U/ml penicillin and 50 µg/ml streptomycin (GIBCO/Invitrogen, Grand Island, NY), and 1,200 mg/ml G418. Immunocytochemistry assays was performed using 1) FLAG M2 MAb (Sigma) to identify clones that expressed EL; 2) anti-hHL MAb (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) to identify clones that expressed HL; and 3) anti-LPL MAb (Abcam, Cambridge, MA) to identify clones that expressed LPL.
PLA1 or PED6 substrate stock solution consisted of 5 mM substrate in DMSO, stored at −80°C. Stock was diluted 1:625 in HBSS with 25 mM HEPES to achieve an 8 µM working solution. To analyze for cell surface lipase activity, cells were plated in CellBIND® 384-well plates (Corning, Lowell, MA) in 25 µl of serum-free medium at a density of 2,000 cells/well. After medium was incubated for 18–24 h at 37°C, it was removed and replaced with 15 µl of working solution (containing either PLA1 or PED6 substrate) and 15 µl of HBSS with 25 mM HEPES (1:2 dilution) to achieve a final concentration of 4 µM (or the final concentrations given in the figure legends). Substrate was dispensed using a Multidrop reagent dispenser. Fluorescence signal was monitored for 30 min at 37°C with a Safire II plate reader in kinetic mode (60 cycles; kinetic interval, 30 s) with an excitation wavelength of 490 nm and an emission wavelength of 515 nm. The linear regression of fluorescence intensity collected from 480 to 1,500 s was used to calculate the reaction rate (the slope), and slopes were used to calculate IC50 values where appropriate. The amount of BODIPY-labeled product generated was calculated at the 30 min time point as determined from standard curve analysis of purified BODIPY FL-C5. In all studies using the inhibitor ebelactone B, consistent results were obtained when it was dissolved as a stock solution in DMSO immediately before use.
Human umbilical vein endothelial cell assay
Pooled human umbilical vein endothelial cells (HUVEC) (Cambrex/Lonza, Walkersville, MD) were cultured with EBM2 medium containing 5% fetal bovine serum and Endothelial Growth Medium supplements (Cambrex/Lonza) and plated for assays at a density of 2,000 cells/well in BD collagen-coated 384-well plates (BD Biosciences, San Jose, CA) and incubated for 24 h at 37°C. Cells were then washed with phosphate-buffered saline and incubated with serum-free medium containing 10 ng/ml TNF-α and 1 ng/ml IL-1β for an additional 24 h at 37°C. Medium was removed and replaced with 15 µl of assay buffer and assayed as described for the stable HEK293 cell lines.
RESULTS
In order to develop a cell-based assay that would distinguish the activity of PLA1 from that of PLA2 enzyme, we conceived and designed a phospholipid substrate with a BODIPY fluor affixed at the sn-1 position and an alkyl chain attached to the sn-2 position with an ether bond (Fig. 1A). Since this molecule was not commercially available at the time (2005), we contracted to have it custom synthesized at Invitrogen (subsequently, Invitrogen added the novel substrate to its regular catalog [2007]). The potentially fluorogenic phospholipid, herein referred to as the PLA1 substrate, is similar in structure to PED6 (Fig. 1B), a commercially available substrate for PLA2 enzymes. Although PLA1 enzymes are able to hydrolyze PED6, they do so with much lower efficiency than PLA2 lipases. Therefore, the PED6 substrate is not favorable for the detection of PLA1 enzymatic activity in a cell-based assay. We engineered the PLA1 substrate to have an ether bond at the sn-2 position. Therefore, this substrate should not be readily cleaved by PLA2 enzymes and would be a preferred substrate for PLA1 lipases. In addition to the BODIPY fluor, the PLA1 substrate also contains a dinitrophenyl quencher on the phospholipid head group, allowing for continuous measurement of fluorescence increase associated with PLA1 hydrolysis.
Confirmation of activity and appropriate products, using purified EL
A depiction of how the PLA1 substrate is predicted to be hydrolyzed by a PLA1 enzyme, along with its products, is presented in Fig. 2A. The prescribed mechanism, an acid/base catalysis with an acyl-enzyme intermediate following the release of product 1 (Fig. 2B, P1), has been described previously (Fig. 2B) (7, 29–33). An oxyanion hole formed by amide backbone nitrogen atoms within the active site stabilizes enzyme product 2 (Fig. 2B, P2) complex. A catalytic water molecule then enters the active site, which allows for completion of the cleavage reaction releasing P2, and the free enzyme is ready for another round of catalysis.
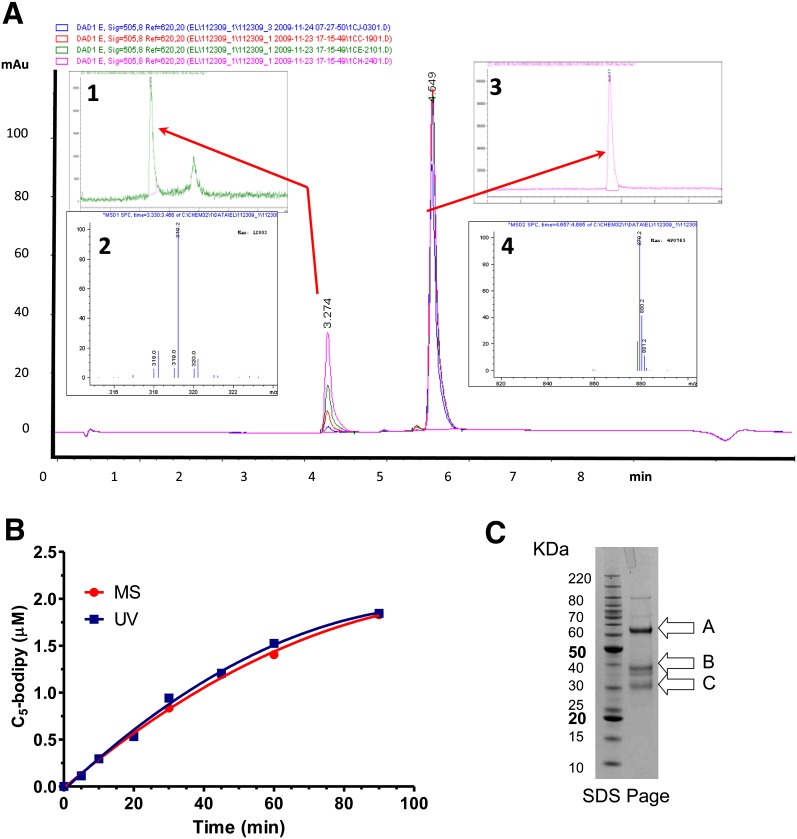
LC/MS analysis confirmed that the PLA1 substrate was cleaved appropriately as a function of time, using 200 nM mouse EL enzyme. Data in Fig. 3A show that the masses of the substrate (Fig. 3A, inset 4) and the BODIPY FL-C5 product (Fig. 3A, inset 3) are consistent with cleavage at the sn-1 position (Fig. 1). The production of the appropriate product accumulates as a function of time and is consistent with its predicted molecular weight throughout the time course of the experiment. The proportionate reduction in substrate signal and BODIPY FL-C5 product accumulation as a function of time and the absence of additional peaks at A505 or at A220 in the chromatogram (data not shown) indicate the absence of substrate hydrolysis at alternative sites. As shown in Fig. 3B, the increase in fluorescence of the PLA1 substrate turnover monitored by A505 corresponded with its quantification by MS as a function of time. Thus, the cleavage of the PLA1 substrate by EL is occurring exclusively at the appropriate sn-1 position. Examination by SDS-PAGE of the purified murine EL used in our analysis indicates that our preparation is predominantly intact as shown in Fig. 3C. However, it also contained inactive cleavage products that we confirmed by N-terminal sequence analysis, due to partial processing by proprotein convertases, as previously identified by others (34, 35). With the PLA1 substrate, this purified mouse EL had an average Km of 10.0 ± 3.8 µM and a Kcat/Km of 84 ± 4.5 M−1 s−1 as calculated from a total of seven independent determinations.
Fig. 3.
Cleavage of the PLA1 substrate by purified mouse EL as a function of time and confirmation of the products by LC/MS are shown. Purified mouse EL (200 nM) was incubated with the PLA1 substrate (10 µM) and monitored for cleavage as a function of time. A: Chromatograms for the 10, 30, 60, and 90 min samples: inset 1, LC/MS chromatogram (h/z 300–350) of the 60 min time point; inset 2, corresponding mass (M) signature of the 3.28 min peak from inset 1; inset 3, LC/MS chromatogram (h/z 800–900) of the 60 min time point; inset 4, corresponding mass signature of the 4.6 min peak from inset 3. B: Quantitation of the PLA1 substrate turnover as a function time monitored by (ultraviolet [UV]) A505 or by MS. C: A Coomassie blue-stained NuPage bis-Tris 4-12%-gradient gel showing purified mouse EL; (A) Full-length mouse EL with cleaved signal sequence amino acids (21–482); (B) Proprotein convertase cleaved mouse EL, N-terminal fragment (amino acids 21–330); (C) Proprotein convertase cleaved mouse EL, C-terminal fragment (amino acids 331–482). Bands were confirmed by N-terminal sequencing.
PLA1 enzymes demonstrate preferential cleavage of the PLA1 substrate
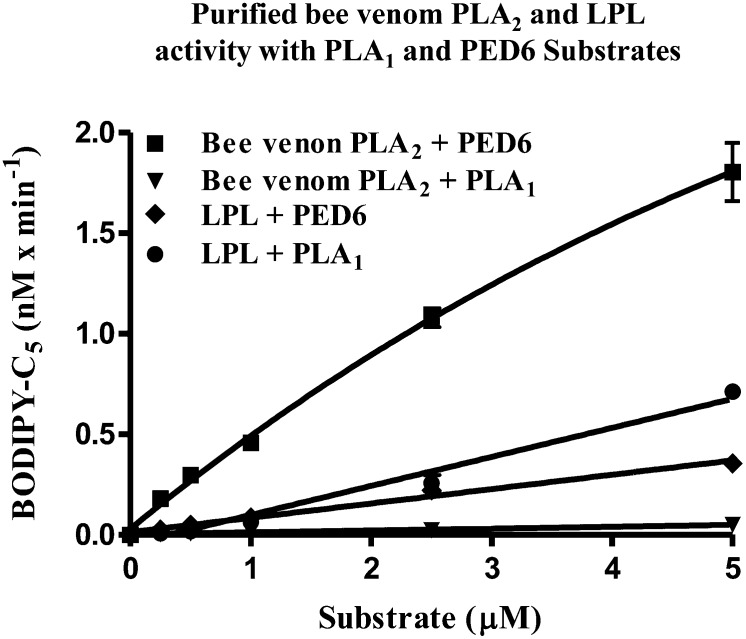
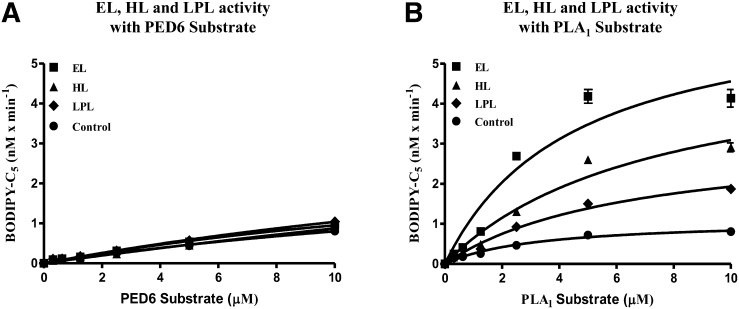
We investigated the preference for cleavage of the PLA1 substrate initially by using a standard PLA2 enzyme, bee venom PLA2. As shown in Fig. 4,, purified bee venom PLA2 displayed robust enzymatic activity when incubated with the PED6 substrate, but under identical conditions, it displayed negligible enzymatic activity when incubated with the PLA1 substrate. This reflected the inability of bee venom PLA2 to cleave the PLA1 substrate at the sn-1 position. Purified bovine LPL, which has been reported to more readily cleave TG than phospholipid substrates (3–5), demonstrated limited enzymatic activity on the PLA1 substrate. The PLA1 substrate was then directly compared with PED6 in a cell-based assay using cells stably expressing murine EL, hHL, or hLPL. As shown in Fig. 5A,, under the assay conditions used, the PED6 substrate showed no significant cleavage in cells expressing EL, HL, or LPL greater than that of nontransfected control cells at all substrate concentrations tested. In contrast to PED6, the PLA1 substrate demonstrated significant cleavage with these three lipases (Fig. 5B). The rank order of cleavage was observed to be EL > HL > LPL > untransfected control cells. Moreover, these stably expressing cell lines were determined to have comparable levels of lipase expression (data not shown). EL demonstrated the highest activity on the PLA1 substrate. This result is consistent with its phospholipid preference and strong bias for the sn-1 acyl ester bond (5, 28, 29).
Fig. 4.
PLA1 and PED6 substrate turnover by purified LPL and bee venom PLA2. Comparison demonstrates substrate preferences between purified bovine LPL and bee venom PLA2, using either the PLA1 or PED6 substrate. Standard 90 min assays were run with increasing substrate concentrations. This triplicate assay represents one of two different experiments yielding identical results.
Fig. 5.
PED6 substrate versus PLA1 substrate turnover by EL, HL and LPL in stably expressing HEK293 cell lines. A and B: Cells stably expressing either murine EL, hHL or hLPL were assayed using either the PED6 (A) or PLA1 (B) substrate. For both substrates, 30 min cell-based assays were run with increasing substrate concentrations. The level of expression for the three lipases in each cell line was comparable, based on qualitative Western blotting (data not shown). These triplicate assays represent one of three different experiments yielding identical results.
Inhibition of the PLA1 substrate cleavage with ebelactone B
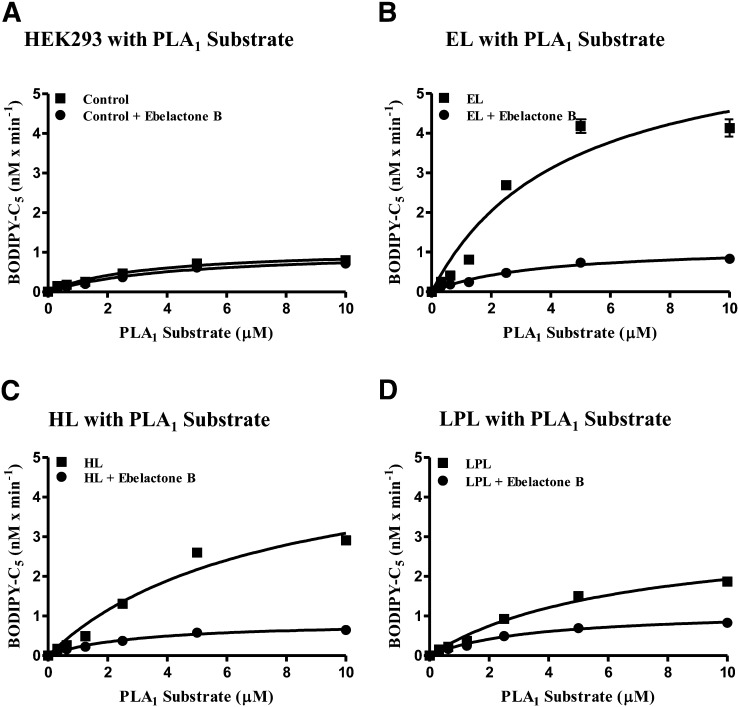
We next examined the potential of a small molecule inhibitor, ebelactone B, to block the hydrolytic activity of the PLA1 lipases in our cell-based assays. Ebelactone B is known to be a promiscuous, covalent inhibitor of lipases (36). Using HEK293 cells stably expressing EL, HL, LPL, or nontransfected HEK293 control cells, we assayed the PLA1 substrate in the absence or presence of 10 µM ebelactone B. As shown in Fig. 6,, with the exception of nontransfected HEK293 cells, PLA1 substrate hydrolysis was decreased significantly when ebelactone B was present. As determined by subsequent ebelactone B dose-response curves in these cell-based assays, the IC50 values for the three lipases were determined to be 0.15 ± 0.06 µM (n = 28) for murine EL; 0.05 ± 0.01 µM (n = 24) for hHL; and 0.12 ± 0.07 µM (n = 24) for hLPL. This indicates that use of the PLA1 substrate can help identify and characterize small molecule inhibitors of EL as well as determine selectivity in counter-screen assays of related lipases. Furthermore, the murine EL HEK293 cell line was used for a high-throughput screen of 200,000 compounds in order to detect inhibitors of EL activity. Statistical parameters for this high throughput were as follows: 12.9 ± 1.5 RFU/s; coefficient of variation % = 11.9; and z′ = 0.50. When used as an assay to screen compounds, generate dose-response curves, and determine their IC50 values, the z′ factor per 384-well plate ranged between 0.5 and 0.75.
Fig. 6.
Ebelactone B inhibition of EL, HL, and LPL in cell-based assays. Cell-based assays with increasing PLA1 substrate concentrations, using HEK293 untransfected control cells (A) or transfected HEK293 expressing either murine EL (B), hHL (C), or hLPL (D) were run in the absence or presence of the nonselective esterase inhibitor ebelactone B (10 µM). These assays, run in triplicate for 30 min, represent one of three different experiments yielding identical results.
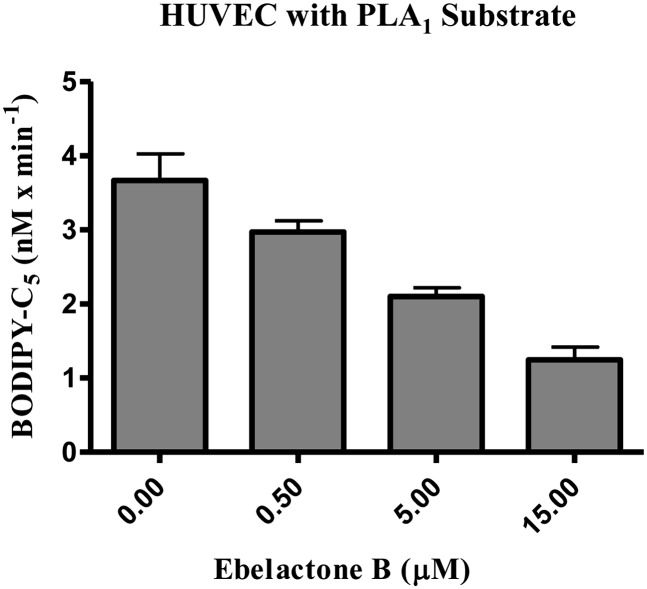
To further explore how the PLA1 substrate can be used to measure EL activity in a cell-based assay, we used HUVECs in which EL expression is known to be upregulated by cytokines (9, 37). Following stimulation with TNF-α and IL-1β, HUVECs were used in a cell-based assay with the PLA1 substrate and increasing concentrations of ebelactone B. As shown in Fig. 7, hydrolysis of the PLA1 substrate by cells endogenously expressing EL was inhibited in a dose-dependent manner by increasing concentrations of ebelactone B. Using this HUVEC assay, we determined the IC50 for ebelactone B inhibition to be 0.15 ± 0.03 µM (n = 18). This value agrees with the IC50 value obtained with ebelactone B, using EL-expressing HEK293 cells. When used as an assay to screen compounds, generate dose-response curves, and determine their IC50 values, the statistical parameters of the assay were as follows: 125.6 ± 18.7 RFU/min; coefficient of variation % = 14.9; and z′ factor ranging between 0.46 and 0.70.
Fig. 7.
PLA1 substrate turnover by HUVECs is inhibited by ebelactone B. Cell-based assays of HUVECs were run for 120 min, using 2.5 µM PLA1 substrate and increasing concentrations of ebelactone B. This triplicate assay represents one of two different experiments yielding identical results.
DISCUSSION
We conceived of and hypothesized the specificity of a novel BODIPY-labeled, self-quenched PLA1 substrate that could be used in homogeneous high-throughput kinetic assays to measure PLA1-specific phospholipase activities in order to identify small molecular inhibitors of EL as potential therapeutic agents. Because it was not commercially available at the time, we contracted with Invitrogen to custom synthesize this substrate. EL showed robust phospholipase activity, using this substrate to monitor its hydrolytic activity in both cell-free and cell-based assays. We have routinely run the cell-based PLA1 substrate assay in a 384-well format. This novel PLA1 substrate was compared with the widely used PED6 substrate and found to have superior characteristics, making it a more favorable substrate for monitoring EL activity. In addition, the PLA1 substrate can be used to analyze the hydrolytic activity of other lipases in counter-screens for monitoring compound selectivity.
Like PED6, the PLA1 substrate has a BODIPY fluor coupled to a fatty acyl chain and a dinitrophenyl quencher on the phospholipid head group. Unlike PED6, the PLA1 substrate has the BODIPY fluor coupled to the fatty acyl chain at the sn-1 position. Further selectivity is designed into the novel PLA1 substrate by placing a nonhydrolyzable ether bond at the sn-2 position, which prevents hydrolysis at this site by nonspecific PLA2 enzymes.
Other similar fluorescent substrates have been described and used with phospholipid or TG micelles and with synthetic HDL particles in cell-free assays (38). We saw no particular requirement to incorporate the PLA1 substrate characterized here into micelles as it could be used directly in either cell-free or cell-based assays. The critical micelle concentration for the PLA1 substrate was determined to be approximately 6 µM by fluor titration (unpublished results). Cleavage products were confirmed by LC/MS in purified mouse EL as well as in a cell-based assay of HEK293 cells expressing human EL (data not shown).
Recently, an indirect assay for EL activity has been described using HDL as a substrate by monitoring the liberation of free fatty acids in coupled enzymatic and detection reactions (39). Even though this assay incorporates HDL, which is the more physiologically relevant substrate for EL, the complex coupled reactions it requires would make analyses of the mode of inhibition difficult and thus inappropriate for guiding chemical optimization of small molecule inhibitors of EL. In addition, although this published assay was used in a high-throughput fashion, the complex nature of the substrate preparation would make it less convenient and more prone to interassay variability than the assays presented here.
We attempted to support the physiological relevance of these assays by incorporating the substrate into HDL or HDL-like particles (in collaboration with Roar Biomedical) but were unsuccessful due to dissociation of the substrate from the particles (unpublished observations). We hypothesize that the rapid dissociation was the result of the significantly shorter chain length at the sn-2 position compared with that of PED6 (Fig. 1), which has been shown to be useful in an assay incorporated into HDL (40). To mimic the normal plasma environment for EL activity, we also added plasma to the assay but found the binding of the substrate by BSA resulted in a significant release of self-quenching (unpublished observations).
Like LPL and HL, EL is secreted and appears to predominantly remain bound to the cell surface by proteoglycan binding. Although it remains unproven, the cell surface-associated form of EL may constitute the bulk of active EL within the plasma compartment in vivo. Another consideration for this enzyme regarding in vivo activity is the recent finding that EL exists as a head-to-tail dimer. It is possible that this conformation results in a quaternary structure whereby the monomeric units are situated side-by-side (41). This dimerization occurs intracellularly and is likely present in this form on the cell surface. Since the improved PLA1 substrate was shown to be amenable to monitoring EL enzymatic activity in cells expressing EL, the assay is able to assess inhibitory activity of compounds against the native form of the enzyme. We used the nonspecific esterase inhibitor ebelactone B as an example of how a small molecule inhibitor of lipases can be detected in cell-based assays by using the PLA1 substrate. More recently, a large screening campaign using the above-described assays resulted in the identification of potent and selective inhibitors of EL, which, when administered to animals, resulted in significant increases in HDL-C (unpublished data), which is the expected physiological and therapeutic consequences of in vivo EL inhibition. These recent findings provide important validation for the relevance of the above-described assays in identifying selective and effective therapeutic inhibitors of EL.
In summary, we have conceived, characterized, and validated an improved PLA1 substrate for use in the screening and characterization of specific inhibitors of EL in both purified enzyme and cell-based activity assay formats. Given that inhibition of EL is a potential therapeutic target for raising HDL-C and inhibiting atherosclerotic disease progression in addition to statin therapy (2, 42), the use of these described assays using the improved PLA1 substrate may facilitate the development of selective EL inhibitors for the eventual potential evaluation in human disease.
Acknowledgments
The authors thank Eric Devine, HongChang Ma, Diane Maguire, and Wenfeng Sun for technical contributions to this work. We also thank Kathy Free and Devon Larson, Molecular Probes custom synthesis division, and Michelle Steel, Invitrogen, for facilitating the special order for this unique substrate. In addition, we gratefully acknowledge chemists HeeChol Kang and Nabi Malekzadeh, responsible for the custom synthesis.
Footnotes
Abbreviations:
- BODIPY FL-C5
- 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene fluor-conjugated to the fatty acyl chain at the sn-1 position
- EL
- endothelial lipase
- apoA-1
- apolipoprotein A-1
- AUC
- area under the curve
- HDL-C
- HDL-cholesterol
- hHL
- human HL
- hLPL
- human LPL
- HUVEC
- human umbilical vein endothelial cells
- IL-1β
- interleukin-1β
- PLA1
- phospholipase A1
- RFU
- relative fluorescence units
- SIM
- selective ion monitoring
- TG
- triglyceride
- TNF-α
- tumor necrosis factor-α
REFERENCES
- 1.Brown R. J., Rader D. J. 2007. Lipases as modulators of atherosclerosis in murine models. Curr. Drug Targets. 8: 1307–1319. [DOI] [PubMed] [Google Scholar]
- 2.Duffy D., Rader D. J. 2009. Update on strategies to increase HDL quantity and function. Nat. Rev. Cardiol. 6: 455–463. [DOI] [PubMed] [Google Scholar]
- 3.Choi S. Y., Hirata K., Ishida T., Quertermous T., Cooper A. D. 2002. Endothelial lipase: a new lipase on the block. J. Lipid Res. 43: 1763–1769. [DOI] [PubMed] [Google Scholar]
- 4.Duong M., Psaltis M., Rader D. J., Marchadier D., Barter P. J., Rye K. A. 2003. Evidence that hepatic lipase and endothelial lipase have different substrate specificities for high-density lipoprotein phospholipids. Biochemistry. 42: 13778–13785. [DOI] [PubMed] [Google Scholar]
- 5.McCoy M. G., Sun G. S., Marchadier D., Maugeais C., Glick J. M., Rader D. J. 2002. Characterization of the lipolytic activity of endothelial lipase. J. Lipid Res. 43: 921–929. [PubMed] [Google Scholar]
- 6.Ishida T., Choi S. Y., Kundu R. K., Spin J., Yamashita T., Hirata K., Kojima Y., Yokoyama M., Cooper A. D., Quertermous T. 2004. Endothelial lipase modulates susceptibility to atherosclerosis in apolipoprotein-E-deficient mice. J. Biol. Chem. 279: 45085–45092. [DOI] [PubMed] [Google Scholar]
- 7.Broedl U. C., Maugeais C., Millar J. S., Jin W., Moore R. E., Fuki I. V., Marchadier D., Glick J. M., Rader D. J. 2004. Endothelial lipase promotes the catabolism of ApoB-containing lipoproteins. Circ. Res. 94: 1554–1561. [DOI] [PubMed] [Google Scholar]
- 8.Yu K. C., David C., Kadambi S., Stahl A., Hirata K., Ishida T., Quertermous T., Cooper A. D., Choi S. Y. 2004. Endothelial lipase is synthesized by hepatic and aorta endothelial cells and its expression is altered in apoE-deficient mice. J. Lipid Res. 45: 1614–1623. [DOI] [PubMed] [Google Scholar]
- 9.Hirata K., Ishida T., Matsushita H., Tsao P. S., Quertermous T. 2000. Regulated expression of endothelial cell-derived lipase. Biochem. Biophys. Res. Commun. 272: 90–93. [DOI] [PubMed] [Google Scholar]
- 10.Broedl U. C., Jin W., Rader D. J. 2004. Endothelial lipase: a modulator of lipoprotein metabolism upregulated by inflammation. Trends Cardiovasc. Med. 14: 202–206. [DOI] [PubMed] [Google Scholar]
- 11.Jin W., Millar J. S., Broedl U., Glick J. M., Rader D. J. 2003. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. J. Clin. Invest. 111: 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishida T., Choi S., Kundu R. K., Hirata K., Rubin E. M., Cooper A. D., Quertermous T. 2003. Endothelial lipase is a major determinant of HDL level. J. Clin. Invest. 111: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma K., Cilingiroglu M., Otvos J. D., Ballantyne C. M., Marian A. J., Chan L. 2003. Endothelial lipase is a major genetic determinant for high-density lipoprotein concentration, structure, and metabolism. Proc. Natl. Acad. Sci. U S A. 100: 2748–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko K. W., Paul A., Ma K., Li L., Chan L. 2005. Endothelial lipase modulates HDL but has no effect on atherosclerosis development in apoE−/− and LDLR−/− mice. J. Lipid Res. 46: 2586–2594. [DOI] [PubMed] [Google Scholar]
- 15.deLemos A. S., Wolfe M. L., Long C. J., Sivapackianathan R., Rader D. J. 2002. Identification of genetic variants in endothelial lipase in persons with elevated high-density lipoprotein cholesterol. Circulation. 106: 1321–1326. [DOI] [PubMed] [Google Scholar]
- 16.Mank-Seymour A. R., Durham K. L., Thompson J. F., Seymour A. B., Milos P. M. 2004. Association between single-nucleotide polymorphisms in the endothelial lipase (LIPG) gene and high-density lipoprotein cholesterol levels. Biochim. Biophys. Acta. 1636: 40–46. [DOI] [PubMed] [Google Scholar]
- 17.Paradis M. E., Couture P., Bosse Y., Despres J. P., Perusse L., Bouchard C., Vohl M. C., Lamarche B. 2003. The T111I mutation in the EL gene modulates the impact of dietary fat on the HDL profile in women. J. Lipid Res. 44: 1902–1908. [DOI] [PubMed] [Google Scholar]
- 18.Yamakawa-Kobayashi K., Yanagi H., Endo K., Arinami T., Hamaguchi H. 2003. Relationship between serum HDL-C levels and common genetic variants of the endothelial lipase gene in Japanese school-aged children. Hum. Genet. 113: 311–315. [DOI] [PubMed] [Google Scholar]
- 19.Brown R. J., Edmondson A. C., Griffon N., Hill T. B., Fuki I. V., Badellino K. O., Li M., Wolfe M. L., Reilly M. P., Rader D. J. 2009. A naturally occurring variant of endothelial lipase associated with elevated HDL exhibits impaired synthesis. J. Lipid Res. 50: 1910–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edmondson A. C., Brown R. J., Kathiresan S., Cupples L. A., Demissie S., Manning A. K., Jensen M. K., Rimm E. B., Wang J., Rodrigues A., et al. 2009. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J. Clin. Invest. 119: 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azumi H., Hirata K., Ishida T., Kojima Y., Rikitake Y., Takeuchi S., Inoue N., Kawashima S., Hayashi Y., Itoh H., et al. 2003. Immunohistochemical localization of endothelial cell-derived lipase in atherosclerotic human coronary arteries. Cardiovasc. Res. 58: 647–654. [DOI] [PubMed] [Google Scholar]
- 22.Badellino K. O., Wolfe M. L., Reilly M. P., Rader D. J. 2006. Endothelial lipase concentrations are increased in metabolic syndrome and associated with coronary atherosclerosis. PLoS Med. 3: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paradis M. E., Badellino K. O., Rader D. J., Tchernof A., Richard C., Luu-The V., Deshaies Y., Bergeron J., Archer W. R., Couture P., et al. 2006. Visceral adiposity and endothelial lipase. J. Clin. Endocrinol. Metab. 91: 3538–3543. [DOI] [PubMed] [Google Scholar]
- 24.Badellino K. O., Wolfe M. L., Reilly M. P., Rader D. J. 2008. Endothelial lipase is increased in vivo by inflammation in humans. Circulation. 117: 678–685. [DOI] [PubMed] [Google Scholar]
- 25.Barter P., Kastelein J., Nunn A., Hobbs R. 2003. High density lipoproteins (HDLs) and atherosclerosis; the unanswered questions. Atherosclerosis. 168: 195–211. [DOI] [PubMed] [Google Scholar]
- 26.Melnikova I. 2005. Raising HDL cholesterol. Nat. Rev. Drug Discov. 4: 185–186. [DOI] [PubMed] [Google Scholar]
- 27.Rader D. J. 2006. Molecular regulation of HDL metabolism and function: implications for novel therapies. J. Clin. Invest. 116: 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirata K., Dichek H. L., Cioffi J. A., Choi S. Y., Leeper N. J., Quintana L., Kronmal G. S., Cooper A. D., Quertermous T. 1999. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J. Biol. Chem. 274: 14170–14175. [DOI] [PubMed] [Google Scholar]
- 29.Jaye M., Lynch K. J., Krawiec J., Marchadier D., Maugeais C., Doan K., South V., Amin D., Perrone M., Rader D. J. 1999. A novel endothelial-derived lipase that modulates HDL metabolism. Nat. Genet. 21: 424–428. [DOI] [PubMed] [Google Scholar]
- 30.Schrag J. D., Cygler M. 1997. Lipases and alpha/beta hydrolase fold. Methods Enzymol. 284: 85–107. [DOI] [PubMed] [Google Scholar]
- 31.Li J. J., Bugg T. D. 2007. Investigation of a general base mechanism for ester hydrolysis in C–C hydrolase enzymes of the alpha/beta-hydrolase superfamily: a novel mechanism for the serine catalytic triad. Org. Biomol. Chem. 5: 507–513. [DOI] [PubMed] [Google Scholar]
- 32.van Tilbeurgh H., Roussel A., Lalouel J. M., Cambillau C. 1994. Lipoprotein lipase. Molecular model based on the pancreatic lipase x-ray structure: consequences for heparin binding and catalysis. J. Biol. Chem. 269: 4626–4633. [PubMed] [Google Scholar]
- 33.Egloff M. P., Marguet F., Buono G., Verger R., Cambillau C., van Tilbeurgh H. 1995. The 2.46 A resolution structure of the pancreatic lipase-colipase complex inhibited by a C11 alkyl phosphonate. Biochemistry. 34: 2751–2762. [DOI] [PubMed] [Google Scholar]
- 34.Gauster M., Oskolkova O., Innerlohinger J., Glatter O., Knipping G., Frank S. 2004. Endothelial lipase-modified high-density lipoprotein exhibits diminished ability to mediate SR-BI (scavenger receptor B type I)-dependent free-cholesterol efflux. Biochem. J. 382: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin W., Fuki I. V., Seidah N. G., Benjannet S., Glick J. M., Rader D. J. 2005. Proprotein convertases [corrected] are responsible for proteolysis and inactivation of endothelial lipase. J. Biol. Chem. 280: 36551–36559. [DOI] [PubMed] [Google Scholar]
- 36.Nonaka Y., Ohtaki H., Ohtsuka E., Kocha T., Fukuda T., Takeuchi T., Aoyagi T. 1996. Effects of ebelactone B, a lipase inhibitor, on intestinal fat absorption in the rat. J. Enzyme Inhib. 10: 57–63. [DOI] [PubMed] [Google Scholar]
- 37.Jin W., Sun G. S., Marchadier D., Octtaviani E., Glick J. M., Rader D. J. 2003. Endothelial cells secrete triglyceride lipase and phospholipase activities in response to cytokines as a result of endothelial lipase. Circ. Res. 92: 644–650. [DOI] [PubMed] [Google Scholar]
- 38.Mitnaul L. J., Tian J., Burton C., Lam M. H., Zhu Y., Olson S. H., Schneeweis J. E., Zuck P., Pandit S., Anderson M., et al. 2007. Fluorogenic substrates for high-throughput measurements of endothelial lipase activity. J. Lipid Res. 48: 472–482. [DOI] [PubMed] [Google Scholar]
- 39.Keller P. M., Rust T., Murphy D. J., Matico R., Trill J. J., Krawiec J. A., Jurewicz A., Jaye M., Harpel M., Thrall S., et al. 2008. A high-throughput screen for endothelial lipase using HDL as substrate. J. Biomol. Screen. 13: 468–475. [DOI] [PubMed] [Google Scholar]
- 40.Schwartz B., Sweitzer T., Krawiec J. 2007. Development of a plasma based assay for endothelial lipase. Presented at the 13th Annual Meeting of the Society for Biomolecular Screening, Montréal, Canada, April 15–19. [Google Scholar]
- 41.Griffon N., Jin W., Petty T. J., Millar J., Badellino K. O., Saven J. G., Marchadier D. H., Kempner E. S., Billheimer J., Glick J. M., et al. 2009. Identification of the active form of endothelial lipase, a homodimer in a head-to-tail conformation. J. Biol. Chem. 284: 23322–23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paradis M. E., Lamarche B. 2006. Endothelial lipase: Its role in cardiovascular disease. Can. J. Cardiol. 22: 31B–34B. [DOI] [PMC free article] [PubMed] [Google Scholar]