Abstract
Protein S-acylation is a major posttranslational modification whereby a cysteine thiol is converted to a thioester. A prototype is S-palmitoylation (fatty acylation), in which a protein undergoes acylation with a hydrophobic 16 carbon lipid chain. Although this modification is a well-recognized determinant of protein function and localization, current techniques to study cellular S-acylation are cumbersome and/or technically demanding. We recently described a simple and robust methodology to rapidly identify S-nitrosylation sites in proteins via resin-assisted capture (RAC) and provided an initial description of the applicability of the technique to S-acylated proteins (acyl-RAC). Here we expand on the acyl-RAC assay, coupled with mass spectrometry-based proteomics, to characterize both previously reported and novel sites of endogenous S-acylation. Acyl-RAC should therefore find general applicability in studies of both global and individual protein S-acylation in mammalian cells.
Keywords: acylation, H-Ras, lipid, palmitoylation, proteomics
Protein cysteine residues undergo a wide variety of chemical reactions owing to thiol nucleophilicity and redox reactivity. These reactions include S-nitrosylation (1, 2), S-prenylation (3, 4), and S-acylation (5, 6), which involve the adduction of nitroso, isoprenyl (thioether), and acyl (thioester) moieties, respectively. Within mammalian cells, an important type of S-acylation involves S-palmitoylation (the attachment of a 16 carbon fully saturated lipid moiety). S-palmitoylation has been shown to significantly impact protein function and localization, largely via modulating membrane affinity and protein stability (7–9). In contrast to the stable thioether linkage of S-prenylation, the thioester linkage of S-acylation confers a reversible and dynamic nature on this modification, and many recent efforts are shedding light on how this modification is regulated (8–11).
There are a variety of methodologies to detect protein S-acylation/palmitoylation in intact cells. A well-established method involves incubating cells with 3H-labeled palmitate, followed by autoradiography to visualize the degree of isotopic incorporation. However, this approach requires high levels of [3H]palmitate (as many as several mCi per sample) and exposure times on the order of weeks (12, 13). More recent methods have cleverly circumvented these issues by using nonradioactive derivatives of palmitate, which can be enriched or detected via cycloaddition reactions ( “click chemistry”) (14–17). Nonetheless, these “palmitate-centric” approaches are encumbered by i) the need for radioactive or chemically modified palmitate analogs; ii) the likely bias for proteins that undergo rapid palmitate turnover versus proteins that are more stably palmitoylated; iii) difficulty in detecting individual S-acylated proteins or their specific sites of S-acylation; and iv) the inability to detect proteins that are acylated with moieties other than palmitate (e.g., shorter, longer, or unsaturated lipid chains).
Recently, a “cysteine-centric” approach to identify S-acylated proteins was introduced that uses the conversion of the protein thioester to a disulfide-linked biotin (18, 19). This assay, known as acyl-biotin exchange (ABE), is readily adapted to immunoblotting techniques and is also adaptable to mass spectrometric-based identification of individual S-acylated proteins (19–22). However, the detection of biotinylated proteins requires expensive reagents and complicated procedures (e.g., repeated protein precipitations, SDS neutralization, and avidin pull down). We recently provided an initial description of a simple and robust alternative to ABE that uses the detection of S-acylated species via resin-assisted capture (acyl-RAC) in lieu of biotinylation (23). The method is rapid (the entire procedure can be completed in several hours) and is readily adapted to mass spectrometry techniques for identifying sites of S-acylation. Here we provide a detailed validation and expansion of the acyl-RAC method and demonstrate its efficacy in detecting S-acylated protein substrates and sites of modification.
EXPERIMENTAL PROCEDURES
Materials and reagents
All materials were obtained from Sigma Chemicals (St. Louis, MO), unless otherwise indicated. Sources of antibodies were mouse MAb α-HA (code 2367; Cell Signaling Technology); and rabbit polyclonal antibody α-H-Ras (code sc-520; Santa Cruz Biotechnology). Bovine brain membranes were isolated as described previously (24).
Mammalian cell culture and transfection
All cells were cultured at 37°C in a 5% Co2 atmosphere. Cell lines were obtained from the Duke Cell Culture Facility and grown in DMEM (HEK293 cells) or McCoy's 5A medium (T24 cells) supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were transfected with Superfect (Qiagen) per the manufacturer's instructions. In general, HEK293 cells were grown in 10 cm dishes to 70%–80% confluency and transfected with 12 μg of the indicated DNA and 48 μl of Superfect (Qiagen). Approximately 24 h later, cells were harvested with cold PBS and used immediately.
Cloning and DNA manipulation
All PCR procedures were performed with Advantage Taq DNA polymerase (Clontech), and products were verified by DNA sequencing (Duke DNA Sequencing Facility). The pCDNA3.1-3xHA-H-Ras construct was acquired from Missouri S and T cDNA Resource Center (product no. RASH00TN00). Integrated Molecular Analysis of Genomics and Expression (IMAGE) clones containing cDNAs for human Sec61B (BC001734), Rps11 (BC070224), and MGST3 (BC005964) were acquired from OpenBiosystems (shown in parentheses are the corresponding Genbank accession numbers). These three cDNAs were amplified by PCR and subcloned into pIRES-puro3 (Clontech) at the 5′-NheI and 3′-EcoRI sites to generate mammalian expression vectors containing an N- or C-terminal hemagglutinin (HA)-tagged cDNA. Primers used for generating pIRES-puro3-Sec61B-HA were 5′-TATTAGCTAGCACCATGGCTGGTCCGACCCCCAGTG-3′ and 5′-TTAAGAATTCTTAAGCGTAGTCTGGGACGTCGTATGGGTACGAACGAGTGTACTTGCCCCAAATG-3′; primers for pIRES-puro3-Rps11-HA were 5′-TATTAGCTAGCACCATGGCGGACATTCAGACTGAG-3′ and 5′-TTAAGAATTCTTAAGCGTAGTCTGGGACGTCGTATGGGTAGAACTTCTGGAACTGCTTCTTGGTGCC-3′; and primers for pIRES-puro3-HA-MGST3 were 5′-TATTAGCTAGCACCATGGTTTACCCATACGACGTCCCAGACTACGCTGCTGTCCTCTCTAAGG-3′ and 5′-TTAAGAATTCTTAATGGCAGCATTTGGGTCC-3′. Point mutations in MGST3 were generated via PCR as above, except reverse 3′- primer 5′-ATTAGAATTCTTAATGGCAGCTTTTGGGTCCACTGC-3′ was used for C150S; primer 5′-ATTAGAATTCTTAATGGCTGCATTTGGGTCCACTGC-3′ was used for C151S; and primer 5′-ATTAGAATTCTTAATGGCTGCTTTTGGGTCCACTGC-3′ was used for C150/151S. Other Cys-to-Ser point mutations were generated with a QuikChange XL II kit (Stratagene) according to the manufacturer's instructions, using primers 5′-CGGCAGCATGAGCAGCAAGTGTG’3- and 5′-CACACTTGCTGCTCATGCTGCCG-3′ for pDNA3.1-3xHA-H-Ras C181/184S; primers 5′-AATGCCAGCAGTGGGACAAGGAGTGC-3′ and 5′-GCACTCCTTGTCCCACTGCTGGCATT-3′ for Sec61B C39S; and primers 5′-CATTGACAAGAAAAGCCCCTTCACTGG-3′ and 5′-CCAGTGAAGGGGCTTTTCTTGTCAATG-3′ for Rps11 C60S.
Detection of S-acylated proteins by acyl-RAC
Following the indicated treatments/transfections, cells were collected and washed in cold PBS. After undergoing a freeze-thaw cycle, cells were lysed in lysis buffer (25 mM HEPES, 25 mM NaCl, 1 mM EDTA, pH 7.5) containing protease inhibitor cocktail (Roche). Lysis was improved by repeated passaging through a 28 gauge needle. For enrichment of membranes, lysates were depleted of nuclei via centrifugation at 800 g for 5 min. The supernatant was then centrifuged at 20,000 g for 30 min, and the pellet was resuspended in lysis buffer containing 0.5% Triton X-100. Total protein was quantified with a bicinchononic acid (BCA) assay (Pierce) using BSA as the standard. Methodology for acyl-RAC, including blocking of free thiols with methyl methanethiosulfonate (MMTS), cleavage of thioester linkages, and capture of nascent thiols on thiopropyl Sepharose, was carried out essentially as described previously (23). In particular, equal amounts of protein (0.5–2.0 mg for immunoblot experiments and 10–20 mg for mass spectrometry experiments) were diluted to a concentration of 2 mg/ml in blocking buffer (100 mM HEPES, 1.0 mM EDTA, 2.5% SDS, 0.1% MMTS, pH 7.5) and incubated at 40°C for 10 min with frequent vortexing. Three volumes of cold acetone were added, and proteins were allowed to precipitate at −20°C for 20 min. Following centrifugation of the solution at 5,000 g for 10 min, the pellet was extensively washed with 70% acetone, resuspended in 300 μl of binding buffer (100 mM HEPES, 1.0 mM EDTA, 1% SDS, pH 7.5) and added to ∼40 μl of prewashed thiopropyl Sepharose (GE-Amersham). To this mixture was added 40 μl of either 2 M NH2OH (freshly prepared in H2O from HCl salt and brought to pH 7.5 with concentrated NaOH) or 2 M NaCl. Binding reactions were carried out on a rotator at room temperature for 2–4 h. Approximately 20 μl of each supernatant was saved as the “total input.” Resins were washed at least five times with binding buffer. For immunoblot analysis, elution was performed using 60 μl of binding buffer containing 50 mM DTT at room temperature for 20 min. Supernatants were removed and mixed with Laemmli loading buffer, heated to 95°C for 5 min, and separated via SDS-PAGE on a Mini-Gel apparatus (Bio-Rad).
On-resin trypsinization and mass spectrometric analysis of S-acylated sites
This procedure was performed essentially as described previously (23) but is fully detailed in the supplementary information (available at http://www.jlr.org).
RESULTS
Application of the acyl-RAC technique using purified bovine brain membranes
The acyl-RAC assay is chemically analogous to the ABE assay, although it replaces the biotinylation/avidin pull-down step with the use of direct conjugation to resin containing thiol-reactive thiopyridinyl groups (Fig. 1). This strategy is advantageous for examining cysteine-based modifications because it is rapid and economical, and it allows the resin-immobilized proteins to be processed conveniently with virtually any chemical or enzyme treatment, except reductants (which would drive elution). As shown in supplementary Fig. I, acyl-RAC was applied to examine S-acylated proteins in bovine brain membranes, which are known to be rich in S-palmitoylated proteins. A number of proteins were readily detected by acyl-RAC in a hydroxylamine-dependent manner via Coomassie staining of eluted proteins resolved by SDS-PAGE. In addition, two S-palmitoylated proteins known to be present in brain, Gαz (25) and GAP-43 (26), were readily detected by immunoblot analysis of acyl-RAC proteins, and only if the samples had been treated with hydroxylamine to cleave endogenous thioesters. In contrast, synaptophysin, which is not a substrate for S-acylation, was not detected by acyl-RAC. Thus, the acyl-RAC technique can be applied to the isolation and identification of S-acylated proteins in complex biological samples.
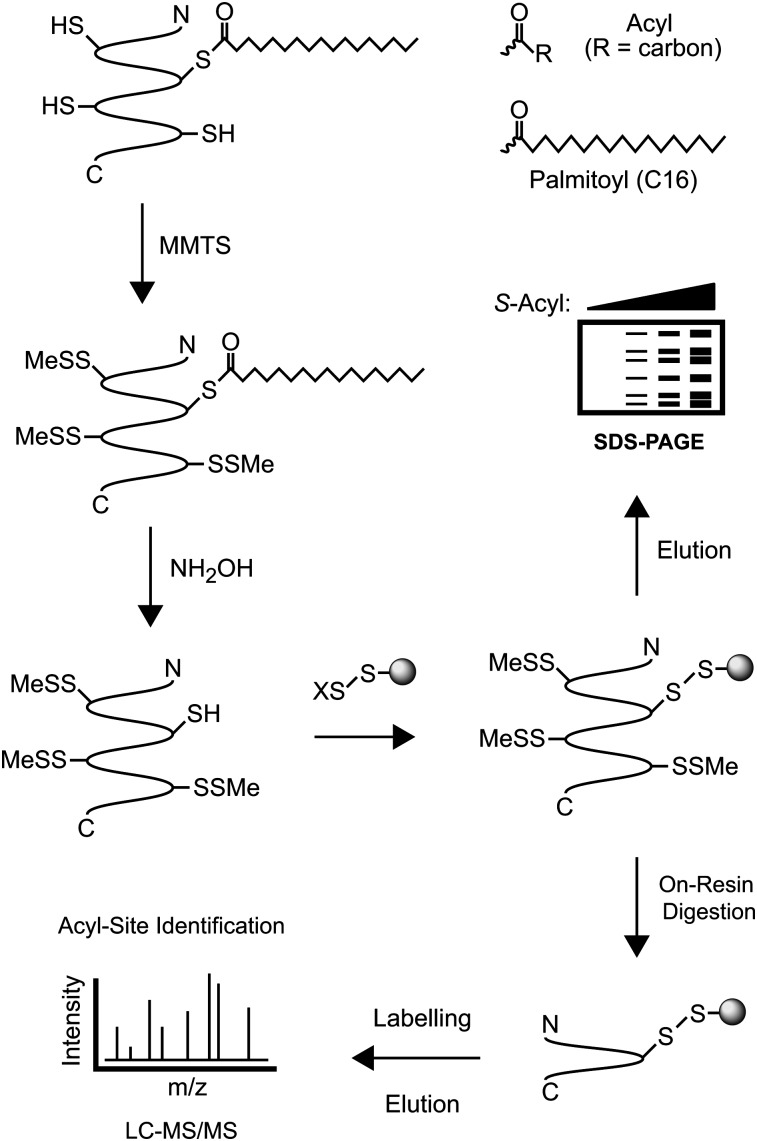
Fig. 1.
A schematic overview of the acyl-RAC assay. Free thiols are first blocked with MMTS. Thioesters are then cleaved with neutral hydroxylamine (NH2OH), and the newly liberated thiols are captured with thiol-reactive Sepharose resin. After being washed, captured proteins are eluted with reductant and analyzed by SDS-PAGE with either protein staining or immunoblotting. To identify individual sites of S-acylation, captured proteins are subjected to “on-resin” proteolysis (typically with trypsin), and resulting peptides are eluted and analyzed by mass spectrometry (LC-MS/MS). X, 2-thiopyridyl.
Application of acyl-RAC to analysis of H-Ras, a model S-palmitoylated protein
To further explore the utility of acyl-RAC to detect S-acylation in an intact mammalian cell culture system, HEK293 cells were transfected with vectors encoding H-Ras, which is known to undergo S-palmitoylation on Cys181 and Cys184 (27) and S-farnesylation on Cys186 (3, 4). The highly modified C terminus of human H-Ras is shown in Fig. 2A. As shown in Fig. 2B, acyl-RAC readily detected S-palmitoylation of H-Ras in a hydroxylamine-dependent manner. Importantly, the C181/184S double mutant, which cannot undergo S-acylation, was not detected. Furthermore, because the C181/184S mutant continues to undergo S-farnesylation on Cys186 (28), these results confirm the expected result that acyl-RAC does not detect S-prenylated proteins (because the thioether linkage is not susceptible to hydroxylamine cleavage). Further confirmation that the protein species identified by acyl-RAC are indeed S-acylated was provided by the observation that the degree of H-Ras S-palmitoylation was attenuated by incubation with 2-bromopalmitate, a known inhibitor of S-palmitoylation (Fig. 2C). Endogenous S-palmitoylated H-Ras could also be readily detected in the T24 bladder carcinoma cell line (supplementary Fig. IIA), in which the oncogenic G12V variant of H-Ras is known to drive the tumorigenic phenotype (29).
Fig. 2.
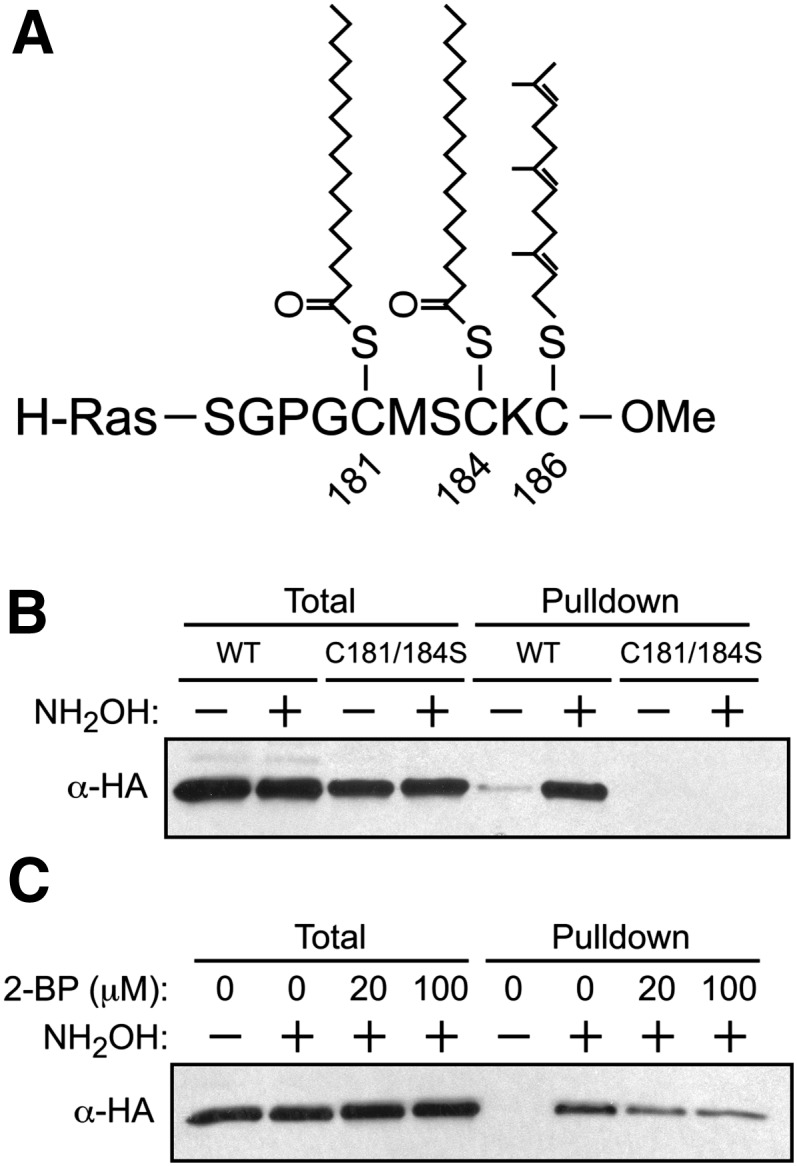
Detection of S-acylated H-Ras by acyl-RAC. A: H-Ras C terminus and associated posttranslational modifications. H-Ras undergoes S-acylation at Cys181 and Cys184 (via thioester linkages), as well as S-prenylation at Cys186 (via a thioether linkage). B: HEK293 cells were transfected with wild-type (WT) HA-H-Ras or the Cys181/184Ser mutant of HA-H-Ras and subjected to acyl-RAC, and captured proteins were analyzed by immunoblotting for HA. C: HEK293 cells were transfected with WT HA-H-Ras for 18 h and then treated with 2-bromopalmitate (2-BP)for another 18 h. Cells were subjected to acyl-RAC and analyzed by immunoblotting for HA.
Although H-Ras is a highly studied prototype of S-acylated proteins, a more complex system was desired to verify the general applicability of acyl-RAC. To that end, a membrane-enriched fraction from HEK293 cells was pretreated with either buffer or palmitoyl-CoA, followed by analysis via acyl-RAC and direct visualization of captured proteins via SDS-PAGE and Coomassie staining (supplementary Fig. IIB). Capture of cellular proteins was both augmented by palmitoyl-CoA pretreatment and dependent on NH2OH during the assay, demonstrating that acyl-RAC can detect a range of S-palmitoylated proteins.
Mass spectrometry-coupled acyl-RAC for identification of S-acylation sites
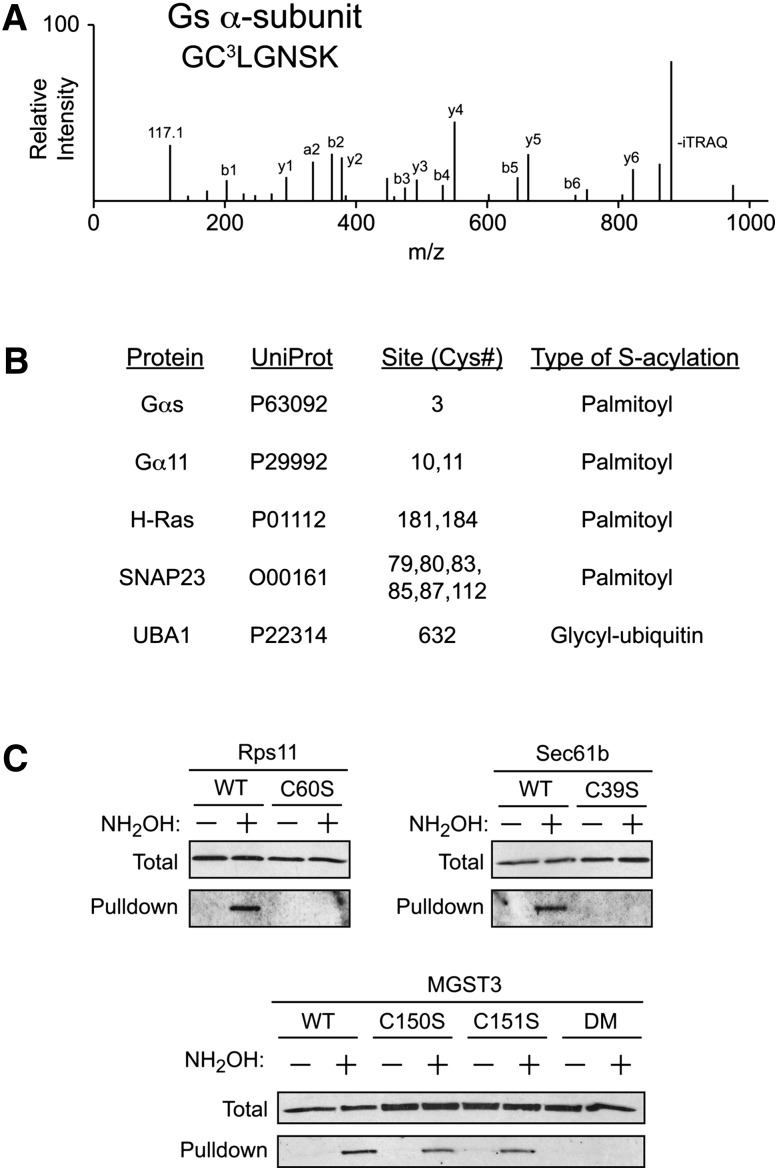
We also assessed the utility of acyl-RAC in identification of specific sites of S-acylation on captured proteins, by using isobaric labeling and LC-MS/MS. Samples of a membrane-enriched fraction from HEK293 cells were subjected to the acyl-RAC procedure in the presence and absence of NH2OH, followed by on-resin trypsinization of captured proteins and isobaric labeling with either iTRAQ-114 atomic mass units (amu) (control) or iTRAQ-117 amu (plus NH2OH) reporter tags. Resins containing the proteins captured from both conditions were combined, and the resulting eluants were analyzed by LC-MS/MS. From a search of the human Swiss-Prot database, 93 putative sites of S-acylation on 88 peptides were identified (supplementary Table I), including a number of sites previously known to undergo S-palmitoylation (Fig. 3B). Of the 88 identified peptides, 84 peptides contained at least one Cys residue (the database search was not restricted to Cys-containing peptides and therefore provided another internal control). As an example, data obtained from the α-subunit of the heterotrimeric G-protein Gs, which is palmitoylated on the N-terminal Cys3 (30, 31), are shown (Fig. 3A). This N-terminal peptide containing Cys3 was identified by MS-coupled acyl-RAC, whereas none of the 7 other potential Cys-containing peptides from Gs were identified in the analysis. These data further validate the utility of acyl-RAC for identifying sites of S-palmitoylation in intact cells.
Fig. 3.
Identification of S-acylation sites by acyl-RAC coupled with mass spectrometry. A: Representative MS/MS spectrum of the N-terminal peptide from the heterotrimeric GTPase Gαs containing Cys3, the known site of S-palmitoylation. The 117 amu peak corresponds to the reporter ion from the NH2OH+ sample, whereas the 114 amu peak (control) was not detected. B: Representative list of known S-acylated sites identified by MS-coupled acyl-RAC (see supplementary Table I for the complete listing of sites identified). C: Validation of MS data by transfection of HEK293 cells with putative S-acylated proteins followed by acyl-RAC and immunoblotting for the specified individual proteins. In each case, the identified sites of S-acylation were mutated to serine as noted. For MGST3, the identified peptide contained two Cys residues (Cys150 and Cys151); therefore, both single and double mutants (DM) were subjected to acyl-RAC.
Several other established sites of S-acylation that were identified by using acyl-RAC are shown in Fig. 3B. These sites include Cys9 and Cys10 within the α-subunit of the heterotrimeric G-protein, G11 (32), Cys181 and Cys184 of H-Ras (Fig. 2A, B, and see reference [27]), as well as 6 Cys residues within SNAP23, of which 4 are conserved in its better-studied homolog SNAP25, a known S-palmitoylated protein (33). Also identified was the active site cysteine, Cys632, of E1 ubiquitin activating enzyme 1 (UBA1). This protein is known to form a thioester with the C-terminal glycine of ubiquitin at Cys632 (34), which is required for ubiquitin transfer to downstream E2 proteins. Although UBA1 contains 19 Cys residues, acyl-RAC detected only the Cys-containing peptide from the active site, demonstrating that acyl-RAC is capable of identifying diverse types of S-acylation.
Validation of novel S-acylated targets identified via MS-coupled acyl-RAC
To examine the fidelity of MS-based identification of S-acylation sites from acyl-RAC-identified proteins, three candidate proteins that were not previously studied in the context of S-acylation were selected for further analysis: the β-subunit of the protein translocating system (Sec61b), ribosomal protein S11 (Rps11), and microsomal glutathione-S-transferase 3 (MGST3). These three proteins, and mutations of each in which the identified S-acylated Cys had been changed to a Ser residue, were expressed in HEK293 cells. Cells were transfected with the respective HA-tagged constructs and then analyzed by acyl-RAC with anti-HA immunoblotting. As shown in Fig. 3C, acyl-RAC detected all three proteins in a hydroxylamine-dependent fashion, whereas mutations of the putative acylation sites abrogated their detection by acyl-RAC. These findings confirm the fidelity of acyl-RAC in detecting both known and novel sites of S-acylation in intact cells.
DISCUSSION
Fatty acylation of proteins is increasingly recognized as a regulator of protein localization and a facilitator of signaling from cellular membranes (8–11, 35). Given the broad range of proteins that are known to undergo S-acylation (e.g., heterotrimeric Gα isoforms, H-Ras, N-Ras, Src family members, and G-protein-coupled receptors [GPCR]), substantial effort has been focused on understanding the mechanisms and biological consequences of S-acylation. These efforts will be aided significantly by the development of efficient tools for detecting S-acylated proteins and sites of modification. Here we have described in detail and validated the acyl-RAC methodology, which efficaciously detects endogenous S-acylation. Notably, the acyl-RAC procedure can be completed in an afternoon and is fully compatible with modern proteomic methodologies to unambiguously identify proteins and their sites of modification.
The acyl-RAC methodology should help provide new insight into the dynamics of S-acylation. The major advantage of acyl-RAC, however, is the simplicity with which it can be performed. Compared with the ABE assay, acyl-RAC uses far fewer procedures, thus minimizing the number of steps at which mistakes might inadvertently occur. Insofar as acyl-RAC is analogous to the detection of S-nitrosylated proteins by resin-assisted capture (SNO-RAC), one would expect acyl-RAC to be similar with regard to sensitivity to the ABE assay (except for high-molecular-weight proteins, where resin-based approaches like acyl-RAC and SNO-RAC are likely superior) (23).
In combination with metabolic labeling approaches (e.g., [3H]palmitate, palmitate-based click chemistry), acyl-RAC can provide a powerful approach to the analysis of dynamic fatty acylation. It should be emphasized that acyl-RAC (as in the case of ABE) detects all types of S-acylation, (i.e., the presence of thioesterified Cys residues) and cannot characterize the nature of the endogenous acyl group. For example, the active site, Cys632, of UBA1 undergoes S-acylation with a glycyl ubiquitin moiety, but UBA1 is apparently not S-palmitoylated. When questions regarding the acyl group arise, metabolic labeling with radiolabeled palmitate or chemically derivatized palmitate analogs (14–17), as well as inhibitors of palmitoylation (e.g., 2-bromopalmitate), can help distinguish the nature of the acyl moiety. The acyl-RAC method should facilitate analysis of cellular protein S-acylation under physiological and pathophysiological conditions.
Supplementary Material
Acknowledgments
The authors are grateful to Matthew W. Foster, Ian Cushman, Michelle E. Kimple, and Christine E. Eyler for helpful discussions and advice.
Footnotes
- ABE
- acyl-biotin exchange
- acyl-RAC
- S-acylation by resin-assisted capture
- MMTS
- S-methylmethanethiosulfonate
- NH2OH
- hydroxylamine
- SNO-RAC
- detection of S-nitrosylated proteins by resin-assisted capture
- UBA1
- E1 ubiquitin activating enzyme
This work was funded in part by National Institutes of Health Grant RO1-DK-076488 (P.J.C.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three methods, two figures, and one table.
REFERENCES
- 1.Benhar M., Forrester M. T., Stamler J. S. 2009. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10: 721–732. [DOI] [PubMed] [Google Scholar]
- 2.Foster M. W., Hess D. T., Stamler J. S. 2009. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med. 15: 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright L. P., Philips M. R. 2006. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47: 883–891. [DOI] [PubMed] [Google Scholar]
- 4.Zhang F. L., Casey P. J. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65: 241–269. [DOI] [PubMed] [Google Scholar]
- 5.Schultz A. M., Henderson L. E., Oroszlan S. 1988. Fatty acylation of proteins. Annu. Rev. Cell Biol. 4: 611–647. [DOI] [PubMed] [Google Scholar]
- 6.Smotrys J. E., Linder M. E. 2004. Palmitoylation of intracellular signaling proteins: regulation and function. Annu. Rev. Biochem. 73: 559–587. [DOI] [PubMed] [Google Scholar]
- 7.Nadolski M. J., Linder M. E. 2007. Protein lipidation. FEBS J. 274: 5202–5210. [DOI] [PubMed] [Google Scholar]
- 8.Linder M. E., Deschenes R. J. 2007. Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8: 74–84. [DOI] [PubMed] [Google Scholar]
- 9.Resh M. D. 2006. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2: 584–590. [DOI] [PubMed] [Google Scholar]
- 10.Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Scholermann B., et al. 2010. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6: 449–456. [DOI] [PubMed] [Google Scholar]
- 11.Rocks O., Gerauer M., Vartak N., Koch S., Huang Z. P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., et al. 2010. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell. 141: 458–471. [DOI] [PubMed] [Google Scholar]
- 12.Magee A. I., Wootton J., de Bony J. 1995. Detecting radiolabeled lipid-modified proteins in polyacrylamide gels. Methods Enzymol. 250: 330–336. [DOI] [PubMed] [Google Scholar]
- 13.Bizzozero O. A. 1995. Chemical analysis of acylation sites and species. Methods Enzymol. 250: 361–379. [DOI] [PubMed] [Google Scholar]
- 14.Charron G., Zhang M. M., Yount J. S., Wilson J., Raghavan A. S., Shamir E., Hang H. C. 2009. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131: 4967–4975. [DOI] [PubMed] [Google Scholar]
- 15.Martin B. R., Cravatt B. F. 2009. Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods. 6: 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yap M. C., Kostiuk M. A., Martin D. D., Perinpanayagam M. A., Hak P. C., Siddam A., Majjigapu J. R., Rajaiah G., Keller B. O., Prescher J. A., Wu P., Bertozzi C. R., Falck J. R., Berthiaume L. G. 2010. Rapid and selective detection of fatty acylated proteins using ω-alkynyl-fatty acids and click chemistry. J. Lipid Res. 51: 1566–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannoush R. N., Arenas-Ramirez N. 2009. Imaging the lipidome: omega-alkynyl fatty acids for detection and cellular visualization of lipid-modified proteins. ACS Chem. Biol. 4: 581–587. [DOI] [PubMed] [Google Scholar]
- 18.Drisdel R. C., Green W. N. 2004. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 36: 276–285. [DOI] [PubMed] [Google Scholar]
- 19.Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., III, Davis N. G. 2006. Global analysis of protein palmitoylation in yeast. Cell. 125: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M. R., El-Husseini A. 2009. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 23: 2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., et al. 2008. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 456: 904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. 2010. Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteomics. 9: 54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., Stamler J. S. 2009. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27: 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sternweis P. C., Robishaw J. D. 1984. Isolation of two proteins with high affinity for guanine nucleotides from membranes of bovine brain. J. Biol. Chem. 259: 13806–13813. [PubMed] [Google Scholar]
- 25.Morales J., Fishburn C. S., Wilson P. T., Bourne H. R. 1998. Plasma membrane localization of G alpha z requires two signals. Mol. Biol. Cell. 9: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudo Y., Valenzuela D., Beck-Sickinger A. G., Fishman M. C., Strittmatter S. M. 1992. Palmitoylation alters protein activity: blockade of G(o) stimulation by GAP-43. EMBO J. 11: 2095–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. 1989. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 57: 1167–1177. [DOI] [PubMed] [Google Scholar]
- 28.Schroeder H., Leventis R., Rex S., Schelhaas M., Nagele E., Waldmann H., Silvius J. R. 1997. S-Acylation and plasma membrane targeting of the farnesylated carboxyl-terminal peptide of N-ras in mammalian fibroblasts. Biochemistry. 36: 13102–13109. [DOI] [PubMed] [Google Scholar]
- 29.Taparowsky E., Suard Y., Fasano O., Shimizu K., Goldfarb M., Wigler M. 1982. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature. 300: 762–765. [DOI] [PubMed] [Google Scholar]
- 30.Linder M. E., Middleton P., Hepler J. R., Taussig R., Gilman A. G., Mumby S. M. 1993. Lipid modifications of G proteins: alpha subunits are palmitoylated. Proc. Natl. Acad. Sci. U S A. 90: 3675–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mumby S. M., Kleuss C., Gilman A. G. 1994. Receptor regulation of G-protein palmitoylation. Proc. Natl. Acad. Sci. U S A. 91: 2800–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCallum J. F., Wise A., Grassie M. A., Magee A. I., Guzzi F., Parenti M., Milligan G. 1995. The role of palmitoylation of the guanine nucleotide binding protein G11 alpha in defining interaction with the plasma membrane. Biochem. J. 310: 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess D. T., Slater T. M., Wilson M. C., Skene J. H. 1992. The 25 kDa synaptosomal-associated protein SNAP-25 is the major methionine-rich polypeptide in rapid axonal transport and a major substrate for palmitoylation in adult CNS. J. Neurosci. 12: 4634–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee I., Schindelin H. 2008. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 134: 268–278. [DOI] [PubMed] [Google Scholar]
- 35.Hancock J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4: 373–384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.