Abstract
Immortalized human lymphoblastoid cell lines have been used to demonstrate that it is possible to use an in vitro model system to identify genetic factors that affect responses to xenobiotics. To extend the application of such studies to investigative toxicology by assessing interindividual and population-wide variability and heritability of chemical-induced toxicity phenotypes, we have used cell lines from the Centre d'Etude du Polymorphisme Humain (CEPH) trios assembled by the HapMap Consortium. Our goal is to aid in the development of predictive in vitro genetics-anchored models of chemical-induced toxicity. Cell lines from the CEPH trios were exposed to three concentrations of 14 environmental chemicals. We assessed ATP production and caspase-3/7 activity 24 h after treatment. Replicate analyses were used to evaluate experimental variability and classify responses. We show that variability of response across the cell lines exists for some, but not all, chemicals, with perfluorooctanoic acid (PFOA) and phenobarbital eliciting the greatest degree of interindividual variability. Although the data for the chemicals used here do not show evidence for broad-sense heritability of toxicity response phenotypes, substantial cell line variation was found, and candidate genetic factors contributing to the variability in response to PFOA were investigated using genome-wide association analysis. The approach of screening chemicals for toxicity in a genetically defined yet diverse in vitro human cell-based system is potentially useful for identification of chemicals that may pose a highest risk, the extent of within-species variability in the population, and genetic loci of interest that potentially contribute to chemical susceptibility.
Keywords: chemical toxicity, human, lymphoblasts, population variability
Environmental and public health agencies are faced with the overwhelming task of assessing the human risk from tens of thousands of chemicals that are released in the environment each year (Judson et al., 2009). Traditional approaches to toxicity characterization rely on extensive animal testing, cost millions of dollars, can take many years per chemical, and have significant uncertainty factors. Because of the time consuming and costly nature of in vivo testing, data currently exist for only a small fraction of the chemicals of potential concern (Judson et al., 2009). The National Research Council report (NRC, 2007) argues for a transition from in vivo models to in vitro/in silico methods in order to capitalize on the advances in technology and computational biology to more efficiently and effectively predict human health risk and reduce the time and cost of testing. Governmental agencies in the United States have responded by initiating large-scale high-throughput screening programs, devising methods to coordinate data, and considering modified approaches to risk assessment (Collins et al., 2008).
To address the pressures for more efficient risk assessment and prioritization strategies, there is a need for in vitro toxicity testing in population-based models (NRC, 2008). Although genetic information can play a key role in understanding and quantifying human susceptibility, an essential step in many of the risk assessments used to shape policy (Cullen et al., 2008), traditional toxicity testing paradigms do not address this critical need. Toxicity data derived from studies on lower organisms (such as yeast), only a single cell line, or rodent strain cannot be used to understand how constitutional genetic variation between individuals in a human population may affect toxicity (Rusyn et al., 2010). In the absence of an understanding of population genetic variation, default uncertainty factors, rather than scientific data, drive risk assessment decisions on most chemicals. Better population-based, genetically defined models are needed to fill these data gaps and to develop more meaningful uncertainty factors.
Recent studies using immortalized human lymphoblastoid cell lines (LCLs) from the International HapMap Consortium show promise in filling this gap with meaningful data. This in vitro population-based model has been used successfully to evaluate interindividual and gender-specific differences in responses to drugs (Dolan et al., 2004; Watters et al., 2004) and to identify genes in the transcriptional response to radiation, including genes involved in cell cycle control, DNA repair, and cell death (Jen and Cheung, 2003). HapMap cell lines of Northern and Western European ancestry (CEU) are arranged in family trios, have been densely genotyped (Frazer et al., 2007), and provide large, renewable sample sets for a wide range of studies (Meucci et al., 2005). Although HapMap cell lines have important limitations (Choy et al., 2008), we hypothesized that a population-based human in vitro model may be used for assessment of interindividual and population-wide variability in chemical-induced toxicity phenotypes and provide information for the biological interpretation of the variability.
The Centre d'Etude du Polymorphisme Humain (CEPH) panel of LCL trios was treated with 14 model environmental toxicants, and two widely used markers (Huang et al., 2008; Xia et al., 2008) of cell death/viability (ATP production and caspase-3/7 activity) were used to assess toxicity. We show that this in vitro genetics-anchored human model system can be used to demonstrate (1) the utility of a population-based approach to screening for chemical toxicity and (2) the potential to identify the genetic susceptibility factors that can be used as candidates in genotype-phenotype studies.
MATERIALS AND METHODS
Chemicals.
A set of 14 chemicals includes model chemicals representing a wide range of classes of toxicants. Chemicals were obtained from the U.S. Environmental Protection Agency National Center for Computational Toxicology and dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 20mM, aliquoted to a 96-well plate, and stored at −20°C. Chemical concentrations for in vitro experiments were chosen from the reports in the literature. Dilutions (100×) were prepared using dimethyl sulfoxide according to the necessary final concentration, and a 96-well working plate was kept at room temperature for no longer than 10 days. The 96-well treatment plates were prepared by aliquoting 1 μl each of 14 chemicals at three concentrations (with a replicate of each concentration) using Biomek 3000 (Beckman Coulter, Fullerton, CA) robot. Wells containing vehicle alone (1% final concentration), cells in vehicle, and a positive control (tetraoctyl ammonium bromide) were also included. Final chemical concentrations and plate design are shown in Supplementary figure 1.
Cell lines and culture conditions.
A panel of 87 immortalized human lymphoblast cell lines from the CEPH trios assembled by the HapMap Consortium was purchased from Coriell Cell Repository (Camden, NJ). Each of the CEU trios consists of a family of two parents and one child. Cell lines were cultured as a suspension in RPMI 1640 media (Gibco, Carlsbad, CA) supplemented with 15% Fetal Bovine Serum (HyClone, South Logan, UT) and 1% Pen-Strep (Gibco) as recommended by Coriell and maintained at 37°C and 5% CO2. Cell density and viability were assessed prior to treatment using Cellometer Auto T4 Plus (Nexcelcom Bioscience, Lawrence, MA). Cells were grown to a concentration of 106 cells per milliliter, volume of at least 12 ml, and viability of at least 85% before treatment. Cells (100 μl containing 104 cells) were aliquoted to each well in a 96-well treatment plate (following addition of the chemicals, see above) and mixed using the Biomek 3000 robot. Plates were incubated for 24 h after treatment at 37°C and 0.5% CO2.
Cell viability and caspase-3/7 assays.
Following a 24-h incubation period, Cell-Titer-Glo Luminescent Cell Viability and Caspase-Glo 3/7 (Promega Corporation, Madison, WI) assays were used according to manufacturer’s protocol. These assays assess ATP production, a marker of cell viability, and caspase-3/7 activity, a measure of apoptosis. Luminescence was measured and recorded using DTX880 (Beckman Coulter) plate reader. Replicate wells at each dose were averaged, and raw values were normalized relative to the baseline (cells in vehicle) and background (vehicle only) at each dose. These normalized percentage of control values were used in all subsequent analyses. A subset of replicate cell viability (18 cell lines) and caspase-3/7 (31 cell lines) experiments were performed independently to evaluate experimental variability. Raw data are publicly available from PubChem (AID: 1976).
Receiver operating characteristic curve analysis and response classification.
Values from the replicate experiments were used to determine experimental variability of chemical-elicited responses for each assay. Although the data are inherently quantitative, it is useful to develop thresholds for classification of individual cell lines as “responders” and “nonresponders.” The interplate replicated values (“first run” and “second run”) provide a useful comparison to establish thresholds to maximize reproducibility. Briefly, for each of the 14 chemicals, a threshold was applied to the quantitative response for the second run and used to provisionally classify individuals as responders/nonresponders. By comparing these provisional dichotomized values to the original quantitative response values for the first run, a receiver operating characteristic (ROC) curve analysis can be used as a measure of correspondence. Here the dichotomized response values serve the role of “true negative/positive” classification in the ROC calculation, although it should be recognized that our purpose is different than for typical ROC analysis. Each threshold resulted in a different ROC curve and an accompanying area underneath the curve. By examining various possible thresholds, we found the assay response value that gave the largest area underneath the ROC curve, and this value was used as the response/nonresponse threshold applied to the plate- and replicate-averaged response (see Supplementary table 1).
Estimation and testing of heritability.
Heritability estimates for parent-child trios follow from a standard quantitative trait model: y = β0 + β1 (am + ap) + ϵ for normalized ATP or caspase-3/7 phenotypes y as an average across plate and replicate measurements (see, e.g., similar expressions in Roy-Gagnon et al., 2008), where am and ap represent the separate maternal and paternal allelic contributions across all contributing loci, i.e., the polygenic effects. The normal error terms ϵ were assumed to be independent across individuals. We did not include an extra family-specific environmental term, reasoning that the above model would potentially slightly overestimate heritability, and any apparently significant results would be subject to additional scrutiny and modeling. A straightforward variance component approach implemented in R (v. 2.10.1) fits each family separately as multivariate normal, with fixed effect mean β0, overall variance (the genetic and error variances), covariances 0 between the unrelated parents, and covariances between parents and children. A likelihood ratio (LR) statistic with 1 degree of freedom was formed for the term , equivalent to testing . The positive constraint on the variance term implies that 2log(LR) is approximately distributed as a mixture of 0 and , and this approximation was used to obtain p values. The suitability of this approximation for a sample size of 30 trios was confirmed via additional simulations (data not shown).
Modeling of cell line, plate, and replicate effects.
The availability of multiple plate measurements on a subset of individuals enabled dissection of the variance components because of each of the cell line, plate, and replicate effects for each chemical and each of the normalized ATP and caspase measurements. For the ATP assay, 16 cell lines and 34 plates were used for this modeling, with 2 plate replications for 14 cell lines and 3 plate replications for 2 cell lines. For the caspase-3/7 assay, 20 cell lines and 50 plates were used for this modeling, with 2 plate replications for 13 cell lines, 3 plate replications for 4 cell lines, and 4 plate replications for 3 cell lines. Using the terminology “north” and “south” to represent the upper and lower positions of replicates on the same plate (Supplementary fig. 1), we used the lme4 function in R v. 2.10.1 to fit the hierarchical model where and , for the ith replicate within plate (i = 2 corresponding to south), the jth plate, and the kth cell line. Here β0 is a fixed intercept term, and β1 is a fixed position effect, to account for possible systematic position bias. The terms correspond to variance contributions from (replicate) error, plate, and cell line, respectively. Although the β1 term was significant for many chemicals, removal of the term did not appreciably affect the variance components (not shown). The models did not use family relationship information, as the heritability values were not significant, and the inclusion of the relationship information (which largely affects correlation structure in the error terms) would not appreciably change the estimates.
For the genome-wide scans, a single phenotype measurement was used by averaging across the plates and north/south replicates. The modeling approach described above can then be used to estimate the proportion of variability due to cell line for each assay and chemical. We have the new model for the kth cell line , where yk is the average assay value across north/south replicates and available plates and b0k is a random intercept with variance estimable from the previous variance component modeling. We have , regardless of plate replication, and for cell lines with r plate replicates (r = 1, 2, 3, or 4), we have . The proportion of variation due to cell line for the averaged phenotype is, finally, var(b0k)/var(yk), where the denominator is averaged over the number of replications.
Genome-wide association study.
Genotypes for the cell lines were obtained from phase II genotypes of the International HapMap Project (Frazer et al., 2007). Quantitative transmission disequilibrium testing was applied to evaluate the association between genetic markers and chemical-elicited ATP production and caspase-3/7 activation response phenotypes using PLINK (Purcell et al., 2007). For this analysis, quantile normalization was used on the assay response values to reduce the influence of extreme observations. The University of California-San Cruz Genome Browser (http://genome.ucsc.edu/index.html) (Kent et al., 2002) NCBI Build 36.1 was utilized for examination of genomic loci of interest. To investigate networks related to genes of interest, we used Ingenuity Pathway Analysis Software (http://www.ingenuity.com; Ingenuity Systems, Mountain View, CA).
RESULTS
Interindividual Variability
We successfully screened 85 cell lines for cell viability and 83 cell lines for caspase-3/7 activity endpoints. These assays were selected based on their utility for in vitro screening of cytotoxicity in cell type- (Xia et al., 2008) and individual-independent manner (Choy et al., 2008). A combination of the two assays compensates for the limitations of the individual assays and therefore can provide sensitive and reliable estimation of cytotoxicity (Shi et al., 2010).
Using “on the plate” duplicates and a subset of independent replicate experiments (containing duplicate wells for each chemical/concentration), we determined the extent of experimental variability. The thresholds established using this analysis (see Materials and Methods section and Supplementary table 1) were used to classify responder/nonresponder cell lines for each chemical and assay. Among the chemicals screened in this study, there was a range of interindividual variability in cytotoxicity across the population of cell lines. Even though three concentrations were tested for each chemical in all cell lines and assays, only the highest concentrations elicited robust responses (data not shown), and further analyses were performed on top concentrations. For cell viability assay, the intraplate correlations between responses to 14 chemicals (rPearson = 0.80 ± 0.17, rSpearman = 0.68 ± 0.22) were generally higher than interplate correlations (rPearson = 0.65 ± 0.28, rSpearman = 0.55 ± 0.23). For caspase-3/7 assay, the intraplate correlations (rPearson = 0.89 ± 0.17, rSpearman = 0.64 ± 0.14) were as high as or even lower than those in interplate comparisons (rPearson = 0.88 ± 0.13, rSpearman = 0.73 ± 0.16).
Interindividual (i.e., between cell lines) variability in responses was observed for some, but not all, chemicals in the panel of 14. For each chemical/assay, we used random-effect modeling to determine the portions of the variability due to cell line, plate replicates, and within-plate replicates (see Materials and Methods section and Supplementary tables 2A and 2B). The results indicate substantial portions of variability because of cell line for some chemicals, keeping in mind that the initial modeling reflected portions of variability for each measurement, not considering the impact of averaging across replicates. When the impact of replicate averaging is considered (Supplementary tables 2A and 2B), the proportion of variation because of cell line is even higher, ranging from 15 to 40% for several assay/chemical combinations. The averaged phenotype values are reflective of the individual “phenotype” value used for later association analysis, and such fractions of explained variability compare quite favorably to the cumulative effect size distributions for modern genome-wide association study data sets (Park et al., 2010), although additional heritability investigation is warranted. Basal values for each of the assays tended to be correlated with the response values at the highest dose; thus, the analysis using normalized values was key to eliminate potentially spurious correlation because of the baseline differences between cell lines.
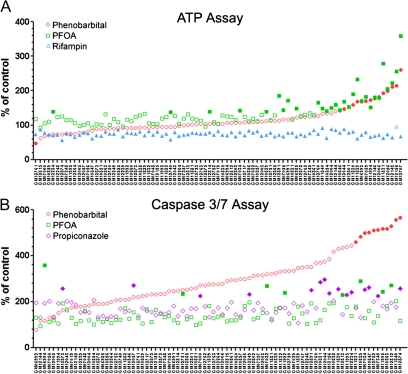
Representative examples of trends in responses that were seen across the population of cell lines are shown in Figure 1. The cell viability (ATP production) response (Fig. 1A) was most variable for perfluorooctanoic acid (PFOA) (100μM) and phenobarbital (3mM), and there is correlation between the two on the individual cell line level. At the same time, the loss of cell viability induced by rifampin (100μM) was very consistent across the panel. Induction of apoptosis (activation of caspase-3/7) phenotype (Fig. 1B) was most variable with phenobarbital; however, PFOA and propiconazole (100μM) did exhibit some but not as wide a range of responses.
FIG. 1.
Interindividual variability in response to toxicants in a panel of human lymphoblast cell lines. Production of ATP (A) and activation of caspase-3/7 (B) were assessed 24 h after treatment with phenobarbital (○, 3000μM), PFOA (□, 100mM), rifampin (▴, 100μM), or propiconazole (◊, 100μM). Percent change over the baseline (vehicle, 1% DMSO) in each cell line is shown. Cell lines are ordered (in each panel separately) according to the response to phenobarbital. Cell lines were considered responders (filled symbols) if percent change was exceeding the significance threshold (see Materials and Methods section ).
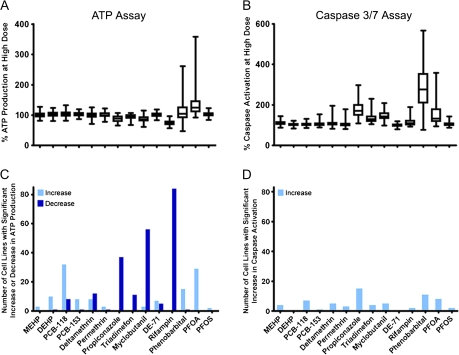
FIG. 2.
Population-wide variability in response to 14 model toxicants. Production of ATP (A and C) and activation of caspase-3/7 (B and D) in a panel of human lymphoblast cells were assessed 24 h after treatment with the highest dose (see Supplementary fig. 1) of each compound. Box and whiskers plots (A and B) were used to exhibit the variability in responses across the population. Bar graphs (C and D) demonstrate the number of cell lines with a significant response above (light bars) or below (dark bars) the vehicle control.
Population-Wide Variability
Next, we assessed the ranges of variability in the population that were exhibited by the chemicals tested. Varying degrees of population-wide variability were exhibited in both phenotypes in response to the panel of 14 chemicals (Figs. 2A and 2B). Most of the chemicals tested showed rather narrow ranges of variability and little overall effect, except for PFOA and phenobarbital. The number of responder cell lines (see Materials and Methods section for a description of the classification) is shown in Figures 2C and 2D. Rifampin, myclobutanil (100μM), and propiconazole caused a robust decrease in ATP production (loss of cell viability) in half or more of the cell lines even though there was little population-wide variability. Polychlorinated biphenyl-118, PFOA, and phenobarbital had an opposite effect on the ATP production phenotype. It has been reported previously that some cytotoxic treatments may lead to a transient increase in the intracellular ATP, an effect attributed to enhancement in energy production when cells are undergoing self-repair in response to toxic challenge shortly after exposure (Shi et al., 2010). Propiconazole, and phenobarbital also caused the most increases in activation of caspase-3/7, even though the fraction of responder cell lines was much smaller, only 10–20%. Interestingly, whereas rifampin exhibited a uniform effect of loss of ATP production in this panel of cell lines and thus produced the highest number of responders, chemicals that showed great variability in responses (e.g., PFOA, phenobarbital, and propiconazole) also elicited a number of robust responses in both assays. It should be noted that cytotoxicity was observed in at least one individual in response to most of the chemicals (except for mono-ethyl-hexyl phthalate, PFOA, and perfluorooctanesulfonic acid for the ATP assay, and di-ethyl-hexyl phthalate and diethyl ether for the caspase-3/7 assay), further arguing for the utility of screening in a genetically diverse panel of cells whereby both the population-wide and the individual's responses can be evaluated.
Comparison of Chemical-Elicited Responses
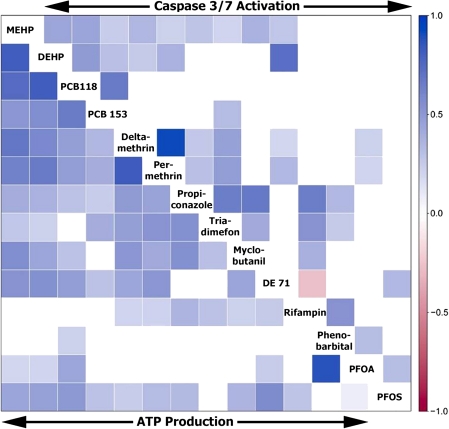
Similarities and/or differences in response to chemicals in two assays probing divergent modes of cell death in a population may provide additional clues for mode of action analysis. We compared compound effects both within and between assays by calculating correlations between compounds with respect to their population-wide effects. A heat map (Fig. 3) of Spearman correlation coefficients (see sidebar scale for reference) shows that strong concordance of responses across a population exists for some chemicals known to have similar modes of action (or a parent-metabolite pair) within each assay. However, some closely related chemicals, such as PFOA and perfluorooctanesulfonic acid, did not elicit highly similar responses within either assay. In addition (as is also shown on Fig. 1), responses to PFOA and phenobarbital were highly correlated in the ATP production assay, but not in the caspase-3/7 assay. Correlation of compound's activity between the two assays was also calculated; yet, no significant correlations between chemical-elicited responses in ATP production and caspase-3/7 activity were found (data not shown).
FIG. 3.
Within-assay correlation map of compound activity across the population. Correlations between compounds with respect to their population-wide effects in the ATP production (bottom left) or caspase-3/7 activation (upper right) assays are shown as a heat map using Spearman correlation coefficient values (see sidebar scale for reference). Only DE 71 and Rifampin (Caspase 3/7 activation) showed negative correlation.
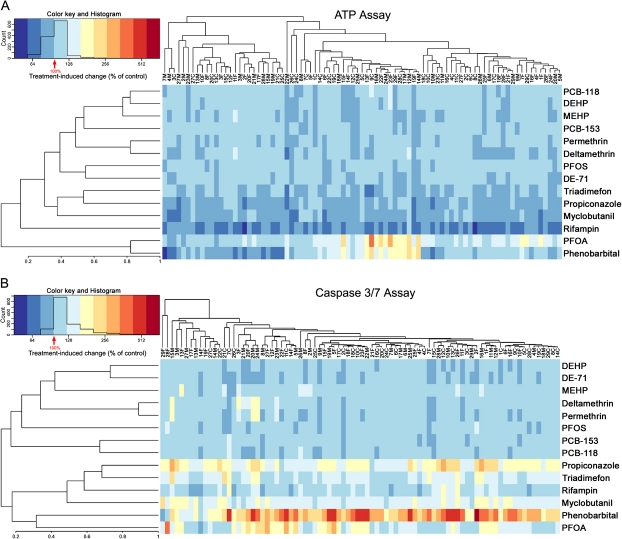
In addition to evaluating similarities in responses across chemicals, we assessed similarities among cell lines in the population. Correlations between responses to the 14 compounds in these cell lines were evaluated and unsupervised clustering performed (average linkage hierarchical clustering using the correlation metric) using the data from ATP production (Fig. 4A) and caspase-3/7 activation (Fig. 4B) assays. Similar to the observations in Figure 3, chemicals of the same class showed similar responses and clustered together. Across the individual cell lines, subclusters of response are apparent; however, no familial clustering of cell lines was evident for responses to this panel of chemicals.
FIG. 4.
Correlation of responses between chemicals and individuals. Heat maps were generated by clustering (average linkage on the correlation metric) the data on percent ATP production and percent caspase activation at highest dose across all chemicals and individuals. The heat maps are ordered both vertically and horizontally with the most correlated participants and chemicals being displayed near to one another. Data were mean centered and transformed as indicated in the corresponding histograms.
As a more formal approach to assess the impact of familial relationships, we performed variance components heritability testing in the trios for each chemical/assay (see Materials and Methods section and Supplementary table 3). None of these results was significant, an observation which is consistent with informal examination of mid-parent assay values compared with those of the offspring (data not shown).
Phenotype-Genotype Association Mapping
The cell-based model system used in this study is not only genetically diverse but is also genetically defined, with dense genotyping information (several million single nucleotide polymorphisms [SNPs]) publicly available (Frazer et al., 2007). Thus, it is possible to examine associations between the chemical-induced phenotypes (ATP production and caspase-3/7 activity) and the genetic markers, investigating potential genetic contributions to the observed variability in responses. Although phenotype-genotype relationships were assessed using genome-wide association scans (see Materials and Methods section ) on all chemical/assay combinations, we report here a detailed analysis of the effect of PFOA in ATP production (cell viability) assay. PFOA elicited the widest range of interindividual variability in this phenotype, with close to 35% of cell lines classified as responders, across the population of cells. Both pharmacokinetics and toxicity of PFOA are known to be highly variable in many species, as evidenced, e.g., by studies with controlled exposures in nonhuman primates (Butenhoff et al., 2002). In addition, PFOA was estimated to exhibit approximately 20% variation due to cell line for the averaged response (Supplementary table 2), although as described earlier there was no significant evidence of heritability. In such circumstances, the power to detect significant associations is likely to be limited, but a genome scan for these samples can nonetheless be performed as the genotypes of these cell lines are publicly available, potentially identifying candidate regions for follow-up analyses.
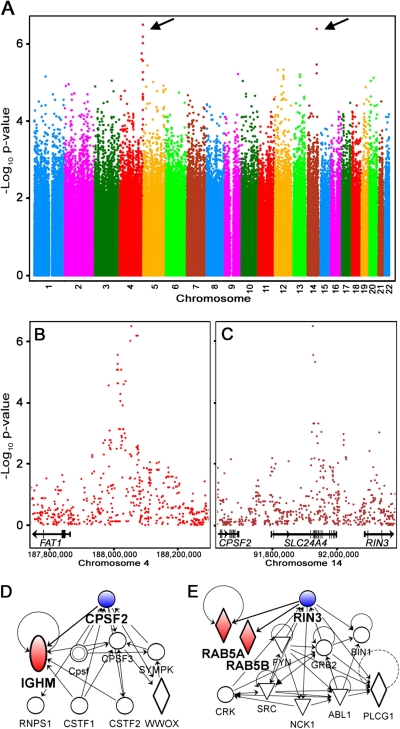
The genome-wide plot in Figure 5A shows ∼2.2 million SNPs and their strengths of association with PFOA-elicited ATP production phenotype. Using a genome-wide threshold of p < 10−6 as suggestive evidence identifies loci on chromosomes 4 (Fig. 5B) and 14 (Fig. 5C). Within the 500-kb loci flanking SNPs with highest association, there are several potential candidate genes for susceptibility to PFOA. On chromosome 4, FAT tumor suppressor homolog 1 (FAT1) is a human gene whose homolog in the rat has been shown to be responsive to PFOA treatment (in the liver) (Guruge et al., 2006). On chromosome 14, three genes were located in the candidate quantitative trait locus (QTL): solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 (SLC24A4); cleavage and polyadenylation specific factor 2 (CPSF2); and Ras and Rab interactor 3 (RIN3). SLC24A4 is a sodium ion carrier, and numerous genes from the solute ion carrier family have been identified as responsive to PFOA treatment in rat liver, rat kidney, and mouse liver (Guruge et al., 2006; Kudo et al., 2002; Rosen et al., 2007). Although CPSF2 and RIN3 have not been shown in previous studies to be responsive to treatment with PFOA, they are tightly linked through a gene network to genes that have been observed as responsive to PFOA treatment in other species. Networks for CPSF2 (Fig. 5D) and RIN3 (Fig. 5E) show the interactions with immunoglobulin heavy constant mu and RAB5A/B, member of RAS oncogene family (RAB5A and RAB5B), respectively, which are responsive to PFOA in rat and chicken liver (Guruge et al., 2006; Yeung et al., 2007).
FIG. 5.
Genome-wide association scan of ATP production response to PFOA. (A) Genome-wide plot of SNPs and their degree of association with PFOA-elicited ATP production phenotype. Arrows indicate the loci with highest association. (B and C) Loci (500 kb) on chromosomes 4 and 14 flanking SNPs that are highly associated (p < 0.000001) with PFOA-elicited phenotype. Genes located in these loci are shown as well as gene networks for CPSF2 (D) and RIN3 (E). Shaded symbols (IGHM, RAB5A and RAB5B) indicate genes that have been identified as responsive to PFOA treatment in rat and chicken liver (Guruge et al., 2006; Yeung et al., 2007).
DISCUSSION
Although the utility and relevance of the cell-based models with regard to species, metabolism capacity, concentration selection, and other important factors are still under intense debate (Coecke et al., 2006; Hartung and Daston, 2009), high-throughput in vitro screening methods are destined to become the major means for addressing numerous gaps in chemical toxicity data (Andersen and Krewski, 2009). One important dimension that is yet to be comprehensively considered in chemical testing is the role of the genetic variability that plays a major role in differential susceptibility to chemicals. Indeed, an in vitro model system that is genetically defined, representative of a diverse population, and amenable to high-throughput screening could significantly aid in chemical prioritization, identification of susceptibility factors, population-wide uncertainty analysis, and predictive toxicity testing.
The human population-based in vitro resource provided by the HapMap Consortium allows generation of phenotype data in a cell-based model system, which contains genetic information representative of a diverse human population (Meucci et al., 2005). The toxicity data obtained in this model system can potentially be combined with high-density genotype information to enable the discovery of genetic causes of susceptibility and variability in response (Watters et al., 2004). Combining genotype and phenotype information for both molecular profiles and complex traits is a promising strategy for understanding which genes, pathways, and biological processes are also under the influence of a given QTL (Harrill and Rusyn, 2008). Additional advantages of using HapMap LCLs are ease of experimental manipulation and ability to control the experimental environment, allowing the genetic contributions toward a specific chemical-induced phenotype to be tested (Huang et al., 2007).
Some (Choy et al., 2008) have argued that nongenetic factors for the LCL cell lines could offer alternative explanations for cell line variability, hampering efforts to elucidate genetic variation. Although we have taken steps to minimize alternate sources of variation, the question will remain open until clear genetic susceptibility variants have been identified. The establishment of apparent cell line variation indicates the need for caution in interpreting a potential population-wide risk for a chemical, both because of the potential for underlying genetic susceptibility as well as the possibility that genomic assays performed by other researchers on much more limited sample sets have also been subject to such variation. Nonetheless, we point out clear differences in the scientific goals of toxicogenetic testing approaches compared with that of other genetic subdisciplines (e.g., pharmacogenomics). In toxicology, merely establishing ranges of variability is important, both because such ranges may reflect true underlying genetic variability but also because they may reflect other sources of variability, the extent of which may have gone unrecognized.
The freshly isolated lymphocytes (generally described as peripheral blood mononucleated cells) are widely used as a surrogate tissue for toxicity studies and as a source of DNA for genotyping. Numerous reports in the literature have examined the metabolic capacity of the total lymphocyte preparations, as well as fractions of T and B lymphocytes (Sempoux et al., 1999; Siest et al., 2008; Vanden Heuvel et al., 1993), and their capacity for induction of the metabolism genes by xenobiotics (Krovat et al., 2000). Whereas lymphocytes do not have the metabolic capacity of the liver, or even that of freshly isolated hepatocytes, they do express a number of nuclear receptors, as well as most genes of the phase I and II metabolism, and transporters (Siest et al., 2008). Thus, even though the metabolism-dependent toxicity may not be detected in cultured lymphocytes and lymphoblasts, this model system is comparable to many other transformed cells; yet, it offers a critical advantage of the defined genetic variability.
This study shows that data produced by testing a set of chemicals in a genetically defined yet diverse human population-based in vitro system can be useful in many ways. The examination of interindividual variability shows that for some chemicals, such as phenobarbital and PFOA, there is an appreciable interindividual variability in response across the population, whereas for others, such as rifampin, toxicity response is uniform across the population. This suggests that there is utility of using this population-based in vitro system for both prioritization of chemicals for more in-depth screening (for those exhibiting great variability across the population) and understanding the potential genetic causes of individual susceptibility. In addition, these data represent a measure of the degree of variability in toxicity-relevant phenotypes, information which may be useful in defining the extent of a population-wide uncertainty for risk assessment.
Identifying chemicals with the widest variability in response in a genetically diverse population signifies the need to further examine the mechanisms behind interindividual responses. First, the comparisons of the adverse effects for a panel of chemicals and across a population can generate hypotheses concerning modes of action that may explain the similarities and/or differences within the population and between chemicals. For example, responses to PFOA and phenobarbital were highly correlated in the ATP production assay, but not in the caspase-3/7 assay, indicating similarities in their modes of action with regard to the effect on general cell metabolism and cell proliferation, but not on the activation of apoptosis. Second, the lack of significant correlations between chemical-elicited responses in ATP production and caspase-3/7 activity may indicate that none of the chemicals that we tested reduces cell viability through the apoptotic pathway at the concentrations used. Third, comparing responses to chemicals across the population is another important assessment that provides information on population-wide effects. Lack of familial clustering may indicate that ATP production and caspase-3/7 activation responses to chemicals in this panel of cells are not heritable, but because of the complexity of the traits that we observed and the small number of chemicals, concentrations, and time points tested, further studies may be needed.
In addition, this study identified several potential candidate susceptibility genes for PFOA-induced adverse effects. The genes in the susceptibility loci can be linked either directly or indirectly to the known effects of PFOA in other tissues and species (Guruge et al., 2006; Kudo et al., 2002; Rosen et al., 2007). Existence of previous reports from the in vivo model systems provides biological plausibility to the discovered associations and serves as an illustrative example supporting the potential of using high-throughput phenotype and genotype data from an in vitro human population-based resource to discover genetic loci that may be relevant to observable trends in phenotypes across the population. In addition, the candidate regions can be further elucidated using existing expression QTL (eQTL) data under the hypothesis that regional SNP variants may act on susceptibility via expression-based regulation. Whereas a search of the eQTL browser (eqtl.uchicago.edu) found no cis-eQTL associations in the FAT1 region with the requisite evidence for display (cis-eQTL evidence with p < 10−3), two SNPs (rs12586368 and rs7159431) in intron 1 of SLC24A4 were reported as having evidence of cis-eQTL action on transcript CCDC88 and one SNP (rs17783660) in the exon 2 of RIN3 was reported as having evidence of cis-eQTL action on BTBD7. All the SNPs and transcripts are in the same 11q32 region based on the RNA-seq data (Montgomery et al., 2010). Although the tissue sources for examination of eQTL evidence are currently limited, larger and richer eQTL data sets will soon be available, with organ-specific evidence of genetic regulation of transcription. Further studies are needed to firmly address the potential biological role of these genes in PFOA-induced adverse effects and to test these and other candidates in human or animal studies to verify their mechanistic relevance to interindividual susceptibility.
It is important to consider that in vitro systems have many limitations. Culture conditions of many cell-based in vitro systems do not mimic physiological conditions, are not homeostatic, and are often oxygen deprived. Cell densities are often lower than tissue densities, impairing normal intracellular signaling, and multiple cell types usually are not represented, limiting cell-cell interactions. Many in vitro cell-based systems are of cancer origin, which is problematic because it is known that cancerous cells have thousands of mutations and even chromosomal aberrations which can affect phenotypes (Hartung and Daston, 2009), and these limitations make prediction of human toxicity based on in vitro toxicity results very challenging. The HapMap cell lines used in this study are not devoid of many of these limitations. In addition, the CEPH panel consists of cell lines derived from a human population that have been immortalized via Epstein-Barr virus transformation to induce long-term growth (Tosato and Cohen, 2007). A concern was expressed that Epstein-Barr virus copy number as well as cellular growth rate and ATP levels play a confounding role in studies using these cell lines to detect genetic contributions to phenotypic traits (Choy et al., 2008).
Although the HapMap cell lines have important limitations, we were able use these cell lines to generate data that can be used for hazard identification in a population context. Our study was limited in size and throughput but serves as an important proof of concept that may be scaled to larger samples. In addition, high-throughput studies should be done to explore a wider range of chemicals and time points to fully explore the ability of this system to provide a measure of population-based dose-response for risk assessment purposes. Nevertheless, our work does demonstrate that a genetically defined, diverse in vitro model system can be used to generate data that are useful for exploring population-wide interindividual variability in response, chemical mode of action characteristics, and genetic contributions to observed phenotypes.
CONCLUSIONS
This study aimed to investigate the utility of using a genetically defined yet diverse human in vitro model system to screen for adverse effects of environmental chemicals. A population of cell lines was exposed to a panel of model toxicants, and cell viability and caspase activation were assessed. The interpretation of the data from in vitro screening in the population-based cell culture models, rather than collections of unrelated cell lines from multiple species and tissues, is affording several advantages. First, our results show that some, but not all, chemicals elicit interindividual variation in response to treatment. Chemicals that vary in their effects across the population may need to be prioritized for further testing using additional in vitro or in vivo approaches. Second, this screening method provides a population-wide measure of uncertainty, information which may be crucial for the future risk assessment approaches that will rely heavily on in vitro data (Andersen and Krewski, 2009). Third, such data may be used to explore potential differences/similarities in modes of action between chemicals on the population-wide level. Lastly, by combining the toxicity data and publicly available genetic information, it is possible to probe the contribution of genetics to adverse phenotypes and select the candidate genes and regulatory networks for further studies to verify their mechanistic relevance with regard to differences in susceptibility to chemical treatment.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health (R01 ES015241); United States Environmental Protection Agency (RD832720 and RD833825).
Acknowledgments
We thank Dr Raymond R. Tice (National Toxicology Program, National Institute of Environmental Health Sciences) for his interest in this study and valuable comments on the study design and data analysis approaches and Dr David Dix (National Center for Computational Toxicology, United States Environmental Protection Agency) for contribution of the chemicals that were used in this study. Conflict of interest: The research described in this article has not been subjected to each funding agency's peer review and policy review and therefore does not necessarily reflect their views and no official endorsement should be inferred. The authors declare no competing financial interests.
References
- Andersen ME, Krewski D. Toxicity testing in the 21st century: bringing the vision to life. Toxicol. Sci. 2009;107:324–330. doi: 10.1093/toxsci/kfn255. [DOI] [PubMed] [Google Scholar]
- Butenhoff J, Costa G, Elcombe C, Farrar D, Hansen K, Iwai H, Jung R, Kennedy G, Jr, Lieder P, Olsen G, et al. Toxicity of ammonium perfluorooctanoate in male cynomolgus monkeys after oral dosing for 6 months. Toxicol. Sci. 2002;69:244–257. doi: 10.1093/toxsci/69.1.244. [DOI] [PubMed] [Google Scholar]
- Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coecke S, Ahr H, Blaauboer BJ, Bremer S, Casati S, Castell J, Combes R, Corvi R, Crespi CL, Cunningham ML, et al. Metabolism: a bottleneck in in vitro toxicological test development. The report and recommendations of ECVAM workshop 54. Altern. Lab. Anim. 2006;34:49–84. doi: 10.1177/026119290603400113. [DOI] [PubMed] [Google Scholar]
- Collins FS, Gray GM, Bucher JR. Toxicology. Transforming environmental health protection. Science. 2008;319:906–907. doi: 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen AC, Corrales MA, Kramer CB, Faustman EM. The application of genetic information for regulatory standard setting under the clean air act: a decision-analytic approach. Risk Anal. 2008;28:877–890. doi: 10.1111/j.1539-6924.2008.01084.x. [DOI] [PubMed] [Google Scholar]
- Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr, Badner JA. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruge KS, Yeung LW, Yamanaka N, Miyazaki S, Lam PK, Giesy JP, Jones PD, Yamashita N. Gene expression profiles in rat liver treated with perfluorooctanoic acid (PFOA) Toxicol. Sci. 2006;89:93–107. doi: 10.1093/toxsci/kfj011. [DOI] [PubMed] [Google Scholar]
- Harrill AH, Rusyn I. Systems biology and functional genomics approaches for the identification of cellular responses to drug toxicity. Expert Opin. Drug Metab. Toxicol. 2008;4:1379–1389. doi: 10.1517/17425255.4.11.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T, Daston G. Are in vitro tests suitable for regulatory use? Toxicol. Sci. 2009;111:233–237. doi: 10.1093/toxsci/kfp149. [DOI] [PubMed] [Google Scholar]
- Huang R, Southall N, Cho MH, Xia M, Inglese J, Austin CP. Characterization of diversity in toxicity mechanism using in vitro cytotoxicity assays in quantitative high throughput screening. Chem. Res. Toxicol. 2008;21:659–667. doi: 10.1021/tx700365e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen KY, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res. 2003;13:2092–2100. doi: 10.1101/gr.1240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P, et al. The toxicity data landscape for environmental chemicals. Environ. Health Perspect. 2009;117:685–695. doi: 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krovat BC, Tracy JH, Omiecinski CJ. Fingerprinting of cytochrome P450 and microsomal epoxide hydrolase gene expression in human blood cells. Toxicol. Sci. 2000;55:352–360. doi: 10.1093/toxsci/55.2.352. [DOI] [PubMed] [Google Scholar]
- Kudo N, Katakura M, Sato Y, Kawashima Y. Sex hormone-regulated renal transport of perfluorooctanoic acid. Chem. Biol. Interact. 2002;139:301–316. doi: 10.1016/s0009-2797(02)00006-6. [DOI] [PubMed] [Google Scholar]
- Meucci MA, Marsh S, Watters JW, McLeod HL. CEPH individuals are representative of the European American population: implications for pharmacogenetics. Pharmacogenomics. 2005;6:59–63. doi: 10.1517/14622416.6.1.59. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (NRC) Toxicity Testing in the 21st Century: A Vision and a Strategy. Washington, DC: National Academies Press; 2007. [DOI] [PubMed] [Google Scholar]
- National Research Council (NRC) Science and Decisions: Advancing Risk Assessment. Washington, DC: National Academies Press; 2008. [PubMed] [Google Scholar]
- Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MB, Thibodeaux JR, Wood CR, Zehr RD, Schmid JE, Lau C. Gene expression profiling in the lung and liver of PFOA-exposed mouse fetuses. Toxicology. 2007;239:15–33. doi: 10.1016/j.tox.2007.06.095. [DOI] [PubMed] [Google Scholar]
- Roy-Gagnon MH, Mathias RA, Fallin MD, Jee SH, Broman KW, Wilson AF. An extension of the regression of offspring on mid-parent to test for association and estimate locus-specific heritability: the revised ROMP method. Ann. Hum. Genet. 2008;72:115–125. doi: 10.1111/j.1469-1809.2007.00401.x. [DOI] [PubMed] [Google Scholar]
- Rusyn I, Gatti DM, Wilshire T, Kleeberger SR, Threadgill DW. Toxicogenetics: population-based testing of drug and chemical safety in mouse models. Pharmacogenomics. 2010;11:1127–1136. doi: 10.2217/pgs.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempoux C, Starkel P, Stevens M, Van Den Berge V, Horsmans Y. Cytochrome P450 3A proteins are expressed in B lymphocytes but not in T lymphocytes. Pharmacogenetics. 1999;9:263–265. [PubMed] [Google Scholar]
- Shi J, Springer S, Escobar P. Coupling cytotoxicity biomarkers with DNA damage assessment in TK6 human lymphoblast cells. Mutat. Res. 2010;696:167–178. doi: 10.1016/j.mrgentox.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Siest G, Jeannesson E, Marteau JB, Samara A, Marie B, Pfister M, Visvikis-Siest S. Transcription factor and drug-metabolizing enzyme gene expression in lymphocytes from healthy human subjects. Drug Metab. Dispos. 2008;36:182–189. doi: 10.1124/dmd.107.017228. [DOI] [PubMed] [Google Scholar]
- Tosato G, Cohen JI. Generation of Epstein-Barr Virus (EBV)-immortalized B cell lines. Curr. Protoc. Immunol. 2007 doi: 10.1002/0471142735.im0722s76. Chapter 7, Unit 7.22. [DOI] [PubMed] [Google Scholar]
- Vanden Heuvel JP, Clark GC, Thompson CL, McCoy Z, Miller CR, Lucier GW, Bell DA. CYP1A1 mRNA levels as a human exposure biomarker: use of quantitative polymerase chain reaction to measure CYP1A1 expression in human peripheral blood lymphocytes. Carcinogenesis. 1993;14:2003–2006. doi: 10.1093/carcin/14.10.2003. [DOI] [PubMed] [Google Scholar]
- Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, et al. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ. Health Perspect. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung LW, Guruge KS, Yamanaka N, Miyazaki S, Lam PK. Differential expression of chicken hepatic genes responsive to PFOA and PFOS. Toxicology. 2007;237:111–125. doi: 10.1016/j.tox.2007.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.