Abstract
By virtue of its ability to regulate both protein turnover and nonproteolytic signalling functions, ubiquitin protein conjugation has been implicated in the control of multiple cellular processes, including protein localization, cell cycle control, transcription regulation, DNA damage repair and endocytosis. Ubiquitin metabolism enzymes have been identified as either oncogenes or tumor suppressors in a variety of cancers. Given that ubiquitin metabolism is governed by enzymes—E1, E2, E3, E4, deubiquitinases (DUBs) and the proteasome-the system as a whole is ripe for target and drug discovery in cancer. Of the ubiquitin/proteasome system components, the E3s and DUBs can recognize substrates with the most specificity, and are thus of key interest as drug targets in cancer. This review examines the molecular role in cancer, relevant substrates and potential for pharmacologic development, of E3s and DUBs that have been associated thus far with human malignancies as oncogenes or tumor suppressors.
Key words: ubiquitin, E3, cancer, DUB, ligase, proteasome
Introduction
Ubiquitin, a small, highly conserved regulatory protein that is covalently tagged to proteins as a signal for proteasome degradation, has been implicated in the control of multiple cellular processes including protein localization, cell cycle control, transcription regulation, DNA damage repair and endocytosis.1,2 As the addition and removal of ubiquitin chains is a fundamental biologic process in all eukaryotic cells, it is not surprising that ubiquitin metabolism enzymes feature prominently as either oncogenes or tumor suppressors in a variety of cancers and many signaling/regulatory pathways relevant to cancer. More importantly, given that ubiquitin metabolism is governed by enzymes—E1, E2, E3, E4, deubiquitinases (DUBs) and the proteasome- the system as a whole is ripe for target and drug discovery in cancer as well as many other human diseases. Though only one drug targeting the ubiquitin/proteasome system, bortezomib (a proteasome inhibitor), is currently FDA approved in cancer, many are in development.3 Of the ubiquitin/proteasome system components, the E3s and DUBs can recognize substrates with the most specificity and are thus of key interest as drug targets in cancer. This review will therefore examine the molecular role in cancer, relevant substrates and potential for pharmacologic development, of E3s and DUBs that have been associated thus far with human malignancies as oncogenes or tumor suppressors.
The mechanics of ubiquitin conjugation.
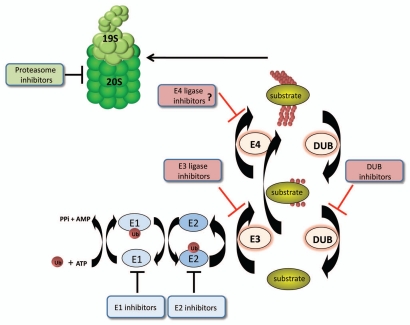
Protein ubiquitination is an ATP-dependent and highly ordered multistep enzymatic process (Fig. 1) that requires the sequential action of three enzymes. The E1 activating enzyme forms a thioester bond with the C-terminal glycine of ubiquitin through its active site cysteine.4 Ubiquitin is then transferred from the E1 to the active site cysteine of an E2 conjugating enzyme.4 An E3 ubiquitin ligase next facilitates transfer of ubiquitin to the protein substrate, catalyzing either the polyubiquitination of protein substrates that can lead to rapid proteasome degradation or monoubiquitination of protein substrates, which is often associated with signaling or vesicle trafficking events.4 The extension of polyubiquitin chains on certain monoubiquitinated protein substrates requires the cooperation of an additional “E4” ubiquitin ligase, which might encode an independent ligase activity or enhance the processivity of the E3.5–7 In addition, polyubiquitin chains can be formed using any of the 7 lysines found in ubiquitin and these different chain topologies lead to different functional outcomes. Of the most well studied linkages, K48-linked polyubiquitin chains generally target substrates in vivo for proteasomal degradation, while K63-linked chains are often involved in non-proteolytic signal transduction or DNA damage repair processes.8,9
Figure 1.
The enzyme cascade of the ubiquitin/proteasome system. Catalytic ubiquitin conjugating and deconjugating enzymes are pictured. Inhibitors have been characterized for all the Ub metabolism enzymes and the proteasome, as noted, except for E4s. The separation of monoubiquitination and polyubiquitination into two steps requiring two enzymes occurs for certain substrates, but as indicated, many substrates are effectively polyubiquitinated by a single E3 enzyme.
The E3 ubiquitin ligases, unlike E1 and E2, are specific to the protein substrate. E3s provide substrate selectivity through a specific substrate recognition domain or via other cofactors in the E3 ubiquitin complex. In accordance, more than 600 proteins have been identified bearing E3 signatures, and they can be grouped into four major classes, based on their structure and biochemical mechanism in the ubiquitination reaction. The HECT (Homolog of E6-AP C Terminus) E3s are usually monomeric and facilitate ubiquitin transfer onto protein substrates via formation of a thioester intermediate with ubiquitin.4 RING (Really Interesting New Gene) domain E3s can function individually as “single-subunit” E3s or as components of multisubunit E3s. In both configurations, the RING domain recruits the E2 within proximity of the substrate, facilitating direct ubiquitin transfer from the E2 to protein substrate.4 A hallmark of RING domain E3s is autoubiquitination by the E2/E3 complex. PHD domain and U-box-type E3s are more recently discovered E3 families, and their enzymatic domain structures closely resemble the RING motif, despite disparate primary sequence signatures.10 Other “atypical” E3 enzymes have also been recently characterized that share the property of coordinating zinc, but are distinct from RING E3s in structure.11
Ubiquitin deconjugation.
Ubiquitin conjugation, like other protein modifications, can be reversed by deubiquitinating enzymes (DUBs; Fig. 1). Over 100 DUBs have been identified in the human genome and they have been linked to the physiologic regulation of diverse cellular functions.12 DUBs are able to regenerate monoubiquitin for reuse by the cell by disassembling polyubiquitin chains that are either free or attached to target proteins.12 DUBs also function in ubiquitin maturation via the modification of the ubiquitin pro-protein, which is usually fused to ribosomal proteins.12 Whereas some DUBs act non-specifically to disassemble Ub chains without regard to their conjugated substrate, many DUBs are now known to trim or remove Ub moieties from specific substrates, with the same level of specificity as encoded in the E3s that attach Ub chains to substrates. Thus, DUBs play a role in the Ub/proteasome system akin to phosphatases in the protein kinase/phosphatase regulatory network.
DUBs are categorized into five conserved families. Except for the JAB1/MPN/Mov34 metalloenzyme (JAMM) domain family, which are zinc metalloproteases, all other DUB families are papain-like cysteine proteases.12 The four classes of cysteine protease DUBs are: ubiquitin C-terminal hydrolase (UCH); ubiquitin specific protease (USP); ovarian tumor domain (OTU); and Machado Josephin domain (MJD). Of these DUB families, the USP family is by far the largest, with over 50 predicted members.12
E3 Ubiquitin Ligases in Cancer
E3 ligases regulate diverse cellular processes such as cell proliferation, cell cycle arrest and apoptosis. The dysregulation of these E3s and their ubiquitin network is often linked with human diseases, particularly neurodegenerative disease and cancer,3 and E3 ligases are the second most prevalent cancer-related functional gene family next to protein kinases.13 E3 ubiquitin ligases can be either tumor suppressors or oncogenes, depending, in turn, on their ability to trigger degradation of either oncogenes or tumor suppressor proteins, respectively (Table 1). As E3 ligases are either intrinsic enzymes (e.g., HECT E3s) or incorporate an E2 enzyme as part of a multisubunit E3, oncogenic E3s have also generated intense interest as potential therapeutic targets for cancer therapy, and a limited number of E3 inhibitors have been described, with many more under development.3 The following are among the most well understood E3 ubiquitin ligases with a role in cancer development or suppression:
Table 1.
E3's and cancer
| E3 | Substrate | Category | Associated cancer | Biological function(s) impacted by dysregulation | Outcome of substrate ubiquitination | Alteration in tumors |
| E6-AP | p53,15 p27131 | Oncogene | Infection by high-risk HPv in cervical, anal, genital, head and neck carcinomas15,16 | Cell cycle control, apoptosis | Proteasomal degradation | No link of mutation to non-HPv tumors; co-factor for HPV E6 at physiologic levels15,16 |
| MDM2 | p53132 | Oncogene | One third of sarcomas,23 breast24 and lung cancer22 | Cell cycle control, apoptosis | Proteasomal degradation, N/C translocation | Gene amplification and overexpression,22 missense, non-sense, frameshift mutation in Zinc-finger domain133 |
| SCFskp2 | p2736,39 | Oncogene | Lung, glioma, gastric, prostate, ovarian,36 breast47 and colon cancer46 | Cell cycle control | Proteasomal degradation | Gene amplification and overexpression134 |
| SCFfbw7 | c-Myc,49 c-jun,50 Notch,51 mTOr52 and cyclin E53 | Tumor suppressor | Cholangiocarcinomas, T-cell acute lymphocytic leukemia, endometrium, colon and stomach cancer135 | Cell cycle control, cell growth, cell division and cell differentiation | Proteasomal degradation | A wide spectrum of mutations as high as 6%, including missense, non-sense, and 75% are point mutations in substrate recognition domain48,135 |
| VHL | HIF-1α57 | Tumor suppressor | Renal cell carcinoma of the clear cell type, Pancreatic cancer, cerebellar and retinal hemangioblastomas55 | Tumor vascularization | Proteasomal degradation | 20–37% large or partial germline deletion, 30–38% missense mutation, 23–27% nonsense or frameshift mutations59 |
| BrCA1 | CtIP,68 NPM,66 RNA polymerase II67 | Tumor suppressor | Breast and ovarian cancer62 | DNA damage response, cell cycle control | Chromatin association, DNA damage foci formation and protein stabilization | Genomic rearrangement (deletion, exon amplification),136 single point mutations, hypermethylation of promoter137 |
| Nedd4-1 | PTeN?70,74 viral gag proteins138 | Oncogene? | Prostate and bladder cancer,70 colorectal and gastric cancer139 | Apoptosis. cell transformation | Proteasomal degradation, N/C translocation | Overexpression70,139 |
| Smurf2 | Smad1,75 Smad2,77 TGFβ receptor 176 | Oncogene? | Esophageal squamous cell cancer79 | Cell growth, cell migration, metastasis? | Proteasomal degradation | Overexpression79 |
| WWP1 | KLF5,140 p53,85 p63,141 ErbB4142 | Oncogene | Breast and prostate cancer143,144 | Transcription activity, apoptosis, cell cycle control | Proteasomal degradation, N/C translocation | Gene amplification and overexpression81,144 |
p53 and its E3s: MDM2, E6AP and Arf-BP1.
p53, known as the “guardian of the genome”, is perhaps the most well studied tumor suppressor gene. p53 is a transcription factor that marshals the cellular response to a wide variety of oncogenic and non-oncogenic stress signals. The pivotal role of p53 in tumor suppression is to prevent genome instability and mutation, by either halting DNA replication to allow for genome repair or activating apoptotic programs in the face of catastrophic DNA damage. p53 activity is restrained and kept silent in unstressed conditions due to highly active ubiquitination and proteasome degradation. Ubiquitination plays a key role in regulating p53 not only due to signaling p53 degradation by the 26S proteasome, but also by regulating the activity and localization of p53.14
E6-Associated Protein (E6-AP) was the first E3 ubiquitin ligase found to target p53 for degradation.15 It is the prototype member of the HECT E3 ubiquitin ligase family, encoding a 350 amino acid residue C-terminal ubiquitin ligase domain. High-risk human papillomaviruses (HPV) 16 and 18 have been etiologically associated with malignant lesions, most notably with cervical cancer, but also with anal, genital and head and neck cancers.16–19 E6-AP was initially discovered as an interaction partner of the E6 oncoproteins of HPV 16 or 18 that mediates their interaction with p53 protein, and is required for E6 mediated ubiquitination and degradation of p53.15 Therefore, E6-AP catalyzes ubiquitin conjugation and subsequent proteasome degradation of p53 upon virus infection by high risk HPV's, but not in uninfected cells where HPV E6 protein is not present.15 HPV infected cells, as a result, are released from p53-induced cell cycle arrest to allow viral genome replication.15
Subsequent to the discovery of E6/E6AP as a specific p53 E3, MDM2 was discovered as the principal physiologic E3 ubiquitin ligase of mammalian p53. Mouse genetic studies revealed that MDM2 is the principal cellular negative regulator of p53.20 The MDM2-null mouse is lethal due to uncontrolled p53 activity, and the lethality can be rescued by concurrent p53 deletion.21 Similar to E6/E6-AP, MDM2 is an oncogenic E3 ligase when overexpressed, due to its ability to maintain p53 instability and prevent p53 activation in the face of genotoxic stress.14 MDM2 gene amplification and protein overexpression are present in more than one third of human sarcomas, breast cancer, lung cancers and many other tumor types.22–24
The prevalence of MDM2 dysregulation in tumorigenesis, and our deep understanding of the p53-MDM2 regulatory loop have generated great interest in MDM2 as a bone fide target in cancer therapy. Several different strategies have been used to design MDM2 inhibitors: (1) Detaching MDM2 from p53 by interfering with their interaction; (2) Targeting MDM2 intrinsic E3 ligase activity; (3) Reducing MDM2 expression.25 Nutlin-3a, a small chemical inhibitor that interferes with p53-MDM2 binding,26 can induce cell cycle arrest or apoptosis in tumor cells expressing wild type p53, and exhibits modest antitumor activity in mouse models.27 The pre-clinical investigation and clinical trials of other anti-MDM2 cancer therapeutic compounds RITA, MI-63 and syl-155 is ongoing, and all of these therapeutics, like Nutlin, are based on targeting the MDM2-p53 interaction.28–30
Another p53 E3 ligase, ARF-BP1, originally purified in a complex with p14ARF, can directly interact with p53 and induce p53 ubiquitination and degradation.31 ARF-BP1 is highly expressed in 80% of breast cancer cell lines suggesting a potential role of ARF-BP1 in breast cancer tumorigenesis.32 COP1, Pirh2 and synoviolin have also been characterized as E3 ubiquitin ligases for p53 in various contexts.33–35 Their physiological significance in tumorigenesis, however, remains to be determined.
p27 and SCFskp2 E3 ligase complex.
SCF (Skp1-Cullin-F-box) type multisubunit E3 ubiquitin ligases play multiple roles in cell proliferation, cell survival and cell size control.36 SCF ligases are composed of invariable subunits Skp1 (bridging/adaptor protein), Cul1 (scaffold) and the enzymatic RING finger ligase Rbx1/Roc1, along with a variable F-box subunit protein that serves as a recognition site for protein substrates.37 Sixty-nine F-box proteins have been identified in the human genome and nine F-box proteins have been paired with specific substrates.36 In contrast to other ubiquitin ligases, SCF F-box proteins generally recognize protein substrates only after post-translational modification, most often phosphorylation.38
Skp2 and Fbw7 are the two most well studied F-box proteins and the dysregulation of their SCF complexes, SCFskp2 and SCFFBW7 has been linked to cell cycle dysregulation and tumorigenesis.36 SCFskp2 promotes entry into S phase by regulating the ubiquitination of several cell cycle control proteins, notably p27, a CDK (Cyclin Dependent Kinase) inhibitor and tumor suppressor gene.39 Indeed, several independent studies found accelerated p27 degradation coupled with lower protein levels of p27 in many aggressive human tumors.40 p27 knockout (-/-) mice spontaneously develop pituitary tumors and show increased susceptibility to carcinogen induced tumor formation.41–43 p27 haploinsufficiency is also associated with accelerated mammary and prostate tumorigenesis in transgenic oncogene-driven mouse models.44 As the E3 ligase of p27, SCFskp2 was predicted to have tumorigenic activity, and forced overexpression of SCFskp2 in mice was found to promote tumor formation.45 In addition, SCFskp2 overexpression correlates with lower p27 expression in breast and colon cancer specimens.46,47 Given the mouse tumorigenesis data on Skp2 overexpression and correlations of SKP2 overexpression with reduced p27 levels in human tumors, SCFskp2 is a clearly attractive therapeutic target for development of an E3 inhibitor—whether directed to the catalytic subunit, which may be problematic given the participation of Rbx1 in many different SCF complexes, vs. targeting of SKP2, which may offer a better therapeutic index.
Another SCF ubiquitin ligase, SCFFBW7 is believed to act as a tumor suppressor complex.48 SCFFBW7 targets multiple oncogenic protein substrates such as c-Myc,49 c-jun,50 Notch,51 mTOR52 and cyclin E.53 Fbw7 is mutated in a wide spectrum of tumors, suggesting its loss is positively selected for in tumorigenesis. In the mouse, Fbw7 functions as a p53-dependent haploinsufficient tumor suppressor.54 The identity of which Fbw7 targets are most critical for tumor suppressor activity, however, remains incompletely understood.48
HIF-1 and VHL ligase complex.
The von Hippel-Lindau (VHL) gene is named after a hereditary cancer syndrome first described over 100 years ago. Patients with VHL disease are characterized by the development of a variety of highly angiogenic tumors, including renal cell carcinoma of the clear cell type (CC-RCC), pancreatic cysts and tumors, as well as hemangioblastomas of the retina and central nervous system.55 After identification of the VHL gene by positional cloning using cells from VHL patients,56 VHL protein was discovered to serve as a substrate recognition subunit in the VCB-Cul2-VHL ubiquitin ligase complex. VHL, together with Cul2, Rbx1 and elongins B and C, compose the VCB-Cul2-VHL complex and Elongin C and Cul2 resemble Skp1 and Cdc53/Cul1 in the SCF ubiquitin complex.57
One of the best known substrates of the VHL ligase is HIF-1α (Hypoxia-inducible factor-1α), a key mediator of oxygen homeostasis and regulator of genes in energy metabolism and angiogenesis.57 Under normoxic conditions, HIF-1α is hydroxylated at a conserved proline, leading to its recognition by VHL, ubiquitination by the VCB complex and rapid proteasome degradation. Under hypoxic conditions, HIF-1α escapes from VHL-induced degradation and induces such genes as Vascular Endothelial Growth Factor and Glucose Transporter 1, which promote angiogenesis and boost metabolism in favor of tumor vascularization.58 Mutations in VHL prevent degradation of HIF-1α under normal oxygen conditions, leading to the upregulation of HIF-1α-induced genes, which enable high levels of vascularization of tumors.59,60 The tumor suppression function of VHL therefore becomes manifest through the functional link with HIF-1α. The restoration of VHL ligase function, should it prove technically feasible, would be a promising strategy to cure or treat VHL-associated—sporadic or inherited—tumors.
BRCA1/BARD1.
The BRCA1 (Breast cancer susceptibility gene 1) tumor suppressor gene behaves as a classic autosomal dominantly inherited tumor suppressor gene, and is mutated in more than 50% of inherited breast cancers.61 BRCA1 mutation results in predisposition to early-onset breast and ovarian cancers, due to the development of genomic instability in the epithelial precursor cells for breast and ovarian cancers, leading to the development of a malignant phenotype.62 Accordingly, BRCA1 has been implicated as an important factor involved in both DNA damage repair and the regulation of cell cycle checkpoint control.
BRCA1 encodes an N-terminal RING E3 ubiquitin ligase,63 and acquires significantly increased ubiquitin ligase activity when heterodimerized with another RING finger protein, BRCA1 Associated Ring Domain 1 (BARD1).64 It is clear that the E3 ligase activity of BRCA1 is of critical functional importance for the tumor suppressor function of BRCA1, since tumor-derived BRCA1 alleles are frequently deficient in E3 ubiquitin ligase activity.65 The bona fide protein substrate of BRCA1/BARD1 E3 ligase activity has not yet been identified, though there have been a number of candidates proposed over the past few years.66–68 One could imagine that if the target of BRCA1 E3 activity must be inactivated for tumor suppression to be maintained, then the physiologic target of BRCA1 E3 activity might serve as a drug target for the prevention or treatment of inherited as well as certain BRCA1-deficient sporadic cases of breast or ovarian cancer.
Nedd4-like E3s.
The Nedd4-like subgroup of HECT E3 ligases contain three conserved functional domains: a C-terminal HECT domain for ubiquitin protein ligation, an N-terminal C2 domain for membrane binding, and a WW2 domain for protein- protein interaction.69 Nedd4-like E3s regulate monoubiquitin-mediated trafficking, proteasome degradation and nuclear translocation of many protein targets. Nedd4-like E3s include nine members, and among them, Smurf2, WWP1 and Nedd4-1 have been genetically linked to tumorigenesis.
Nedd4-1. Nedd4-1 has been proposed as an oncogene, as it negatively regulates the tumor suppressor Phosphatase and Tensin Homolog (PTEN) by mediating PTEN ubiquitination and degradation.70 PTEN is a lipid phosphatase that inactivates PI3-kinase, and is the one of the most frequently mutated tumor suppressor genes in human tumors.71 Overexpression of Nedd4-1 was correlated with lower PTEN protein levels in a mouse prostate tumor model and multiple human cancer samples.70 Furthermore, the depletion of Nedd4-1 inhibited xenograft tumor growth, and this inhibition of tumor growth was PTEN dependent.70 However, Nedd4-1 mediated PTEN ubiquitination may serve other functions beyond signaling degradation.72 Nedd4-1 dependent PTEN ubiquitination on K289 led to PTEN translocation into the nucleus and mono-ubiquitinated PTEN accumulated in the nucleus when Nedd4-1 was overexpressed.72 Nuclear localization of PTEN has been reported to correlate with its tumor suppressor function.73 Taking all of the current data together, Nedd4-1 E3 ligase regulates the proteasome degradation and subcellular localization of PTEN, and thereby, modulates PTEN tumor suppression function.70,72
In contrast to all of the above data, a Nedd4-1 knockout mouse demonstrated no dysregulation of PTEN protein level or cellular distribution, arguing against a critical role for Nedd4-1 in PTEN regulation.74 A caveat to understanding the Nedd4-1 mouse knockout data and phenotype interpretation is the possibility of a compensating epigenetic change that suppresses the phenotype occurring during development of the Nedd4-1 knockout mouse. Further research is therefore necessary to resolve the disparities among the various experimental systems, but there is no doubt that a clearer understanding of how PTEN stability and localization is regulated—whether by-Nedd4-1 or not—is important to understanding the etiology and progression of a multitude of malignancies.
Smurf2. Smad ubiquitination regulatory factor 2 (Smurf2) is a Nedd4-like E3 ligase that regulates the protein stability of Smad2, Smad1 and TGFβ (transforming growth factor β) receptor 1, the key signal mediators of TGFβ signaling cascades.75–77 Smad2 becomes activated upon TGFβ receptor activation and translocates into the nucleus to trigger the expression of target genes in the TGFβ pathway. Smurf2 mediated ubiquitination of Smad2, which is induced by TGFβ,75 decreases the cellular levels of Smad2 protein and attenuates the cellular response to TGFβ.75 Notably, the TGFβ pathway can either inhibit cancer cell proliferation or promote tumor progression depending on the cellular and tissue context.78
Smurf2 upregulation and decreased Smad2 protein level have been observed in esophageal squamous cell carcinoma.79 Increased Smurf2 expression is also associated with higher invasiveness and metastatic potential in esophageal squamous cell cancer.79 In this case, the TGFβ pathway is likely operating in a tumor suppressive mode and loss of Smad2 and TGFβ signaling in these cancers enhances tumor progression—though the data remains purely correlative at this time, and lacking in dispositive animal data to confirm or refute this hypothesis.
WWP1. WWP1 is another Nedd4-like E3 ubiquitin ligase aberrantly regulated in human cancers. The frequent amplification of WWP1, and the overexpression of its gene product in prostate and breast cancer samples, suggests that WWP1 is a potential oncogene in these specific cancer types.80,81 WWP1 has been proposed to target the KLF5 (Krüppel-like factor 5) transcription factor for ubiquitination, inducing KLF5 proteasome degradation.82 KLF5 is a putative tumor suppressor, as evidenced by frequent downregulation in breast cancer cell lines, and its ability to generally suppress cell growth in cancer cells.82–84 Thus, it is highly possible that WWP1 could act as an oncogene by inhibiting KLF5. The initial work on WWP1/KLF5 requires further animal model experiments to provide physiologic support to this mechanism.
Interestingly, WWP1 has been reported to regulate p53 ubiquitination and leads to subsequent p53 translocation to the cytoplasm and diminished p53 transcriptional activity.85 In theory, the inactivation of WWP1 will facilitate p53 transcriptional induction. This reverse correlation of WWP1 and p53 provides a new avenue for understanding regulation of p53 transcription activity, and thus WWP1 may have additional value as a cancer drug target if its inhibition can activate p53, beyond its effects on KLF5.
DUBs and Cancer
Like E3s, DUBs have been implicated in the regulation of proteasome dependent degradation, protein localization, transcription activities and endocytosis. The dysregulation of DUBs is highly related to human diseases, especially human cancers, with examples of both oncogenic and tumor suppressor DUBs that are discussed below (Table 2). Moreover, the growing interest in the role of oncogenic DUBs in cancer is linked to the idea that DUBs are evolutionarily linked to a well-studied pharmaceutical target class—proteases—and DUBs may serve as equally or more useful drug targets within the ubiquitin/proteasome system alongside E3s and the proteasome.
Table 2.
DUBs and cancer
| DUB | Substrate | Category | Associated cancer | Biological function | Outcome of substrate Deubiquitination | Alteration in tumors |
| HAUSP (USP7) | p53,88 MDM2,89 MDMX,91 PTEN,93 FOXO492 | Oncogene/Tumor suppressor? | Prostate cancer;93 tumor suppressor in colon cancer, non-small cell lung cancer?145,146 | Apoptosis, cell cycle arrest, cell proliferation | Protein stabilization, N/C translocation | Overexpression,93 downregulation145, 146 |
| USP2a | FAS,97 MDM2,99 MDMX100 | Oncogene | Prostate cancer101 | Apoptosis | Protein stabilization | Overexpression101 |
| USP28 | c-MYC,105 CHK2, 53BP1106 | Oncogene | Colon and breast cancers105 | Cell cycle control, apoptosis, DNA damage response | Protein stabilization | Overexpression, somatic mutation in lobular breast cancer114 |
| CYLD | TRAF2, TRAF6, TRAF7, NEMO, RIP1 and TAK1108–113 | Tumor suppressor | Familial cylindromatosis (skin tumors), multiple myeloma, B-cell malignancy, hepatocellular carcinoma, uterine cervix carcinoma, kidney cancer, colon cancer and melanomas115 | Immune response, apoptosis, cell proliferation, cell migration and spermatogenesis | Signaling, N/C translocation | Somatic mutation in cylindromatosis, reduced expression, gene mutation and gene copy number reduction in multiple cancers115 |
| A20 | RIP1, RIP2, TRAF2, TRAF6, NEMO and MALT11,117–122, 124 | Tumor suppressor | Several lymphomas, including B cell lymphomas, Hodgkin lymphoma, mantle cell lymphoma, MALT lymphoma, marginal zone lymphoma125,126 | Immune response, apoptosis | Signaling, proteasomal degradation | Genomic deletion and mutations, promoter methylation147–149 |
| USP9x | β-catenin?128 SMAD4,129 MCL1130 | Oncogene | Follicular lymphomas, diffuse large B-cell lymphomas, colon cancers, small cell lung carcinoma, breast cancer130–150 | Apoptosis, cell migration | Protein stabilization, signaling | Overexpression130 |
HAUSP (USP7).
Originally identified in complexes with herpes viral proteins vmw 110 (also known as infected cell protein 0 ICP0) and Epstein-Barr nuclear antigen 1,86,87 and termed Herpes Associated Ubiquitin Specific Protease (HAUSP), HAUSP subsequently was shown to be a critical regulator of p53 stability through a complicated mechanism of targeting both p53 and its E3 ubiquitin ligase, MDM2.88–90 HAUSP independently interacts with, and catalyzes deubiquitination of, both p53 and MDM2; therefore, HAUSP counteracts ubiquitin-dependent proteasome degradation and stabilizes both p53 and MDM2.88,89 The complete depletion of HAUSP has a net stabilization effect on p53 in cells where MDM2 is the major negative regulator of p53 stability.89,90 Upon DNA damage stress, ATM-dependent regulation causes reduced interaction between HAUSP and MDM2 and results in destabilized MDM2, therefore allowing proper p53 activation after DNA damage.91 The HAUSP role in DNA damage regulation of p53 is further supported by its deubiquitination and destabilization of MDM2's dimerization partner and co-regulator of p53 activity, MDM-X, upon DNA damage.91 This dynamic role of HAUSP in the p53-MDM2 pathway is the first clue to a likely role for this DUB in tumorigenesis.
More recently, HAUSP was discovered to deubiquitinate two tumor suppressor proteins—Forkhead box O4 (FOXO4) and PTEN. FOXO4 was shown to interact with HAUSP upon oxidative stress.92 HAUSP deubiquitination does not influence the stability of FOXO4, but instead decreases the nuclear targeting and transcriptional activity of FOXO4.92 HAUSP-mediated deubiquitination of PTEN favors PTEN cytoplasmic accumulation, as monoubiquination is a nuclear localization signal for PTEN.93 HAUSP overexpression and associated PTEN nuclear exclusion are found in human prostate tumors and correlate with tumor aggressiveness.93
With its multiplicity of tumor suppressor targets, HAUSP could be a therapeutic target in human cancers due to its negative regulation of these tumor suppressor genes. A genome wide RNAi screen of the active DUBs in cancer models identified inhibition of HAUSP as a promising DUB to target in cancer treatment.94 The small molecule HAUSP inhibitor HBX 41108 (Hybrigenics) has been reported to effectively activate p53 in a non-genotoxic manner, leading to p53-dependent apoptosis.95 Ongoing basic research on HAUSP, especially animal models of HAUSP inactivation, will clarify the net outcome of HAUSP positive and negative dysregulation, and provide more comprehensive and precise knowledge for the use of HAUSP inhibitors in cancer therapy.
USP2a.
The USP2 gene encodes two ubiquitin-specific proteases, USP2a and USP2b, that arise due to alternative splicing of the 5′ USP2 exon.96,97 USP2a mRNA abundance in the mammalian brain is regulated by the circadian regulator protein, CLOCK.98 Another CLOCK-regulated gene in mouse liver is fatty acid synthase (FAS), a deubiquitination substrate of USP2a.97,98 FAS is overexpressed in many cancers, including breast and prostate cancers.97 Growing evidence suggests that FAS is a metabolic oncogene, facilitating de novo lipid biogenesis required for cancer cell growth and tumor progression.97 Significantly, USP2a deubiquitinates, and subsequently stabilizes FAS and the depletion of USP2a destabilizes FAS and results in apoptosis due to loss of FAS anti-apoptotic activity.97
Additionally, USP2a interacts with and deubiquitinates, MDM2. Ectopic USP2a expression stabilizes MDM2 and promotes p53 degradation.99 MDMX, another MDM2-like p53 repressor, is also a USP2a substrate.100 USP2a, therefore, is a significant repressor of p53 activity, adding to its potential value as a therapeutic target, especially in wild type p53 expressing cancers.
USP2a exhibits oncogenic behavior in cultured cells, in that its overexpression protects human tumor cell lines from apoptosis.101 Increased expression of USP2a was found in 44% of prostate tumor samples, and the overexpression of USP2a was also correlated with increased fatty acid synthesis.101 The bona fide physiological contribution of USP2a to tumorigenesis will require further animal and human tumor studies. However, biochemical studies characterizing the targets of its DUB activity do support further investigation of USP2a as a therapeutic target, at least in those cancers where it is overexpressed.
Usp28.
C-Myc, as the first identified and extensively studied oncogene, is a central regulator of cell growth, proliferation and apoptosis.102 It was traditionally thought to be regulated mainly at the transcriptional level.103 However, like most transcription factors, c-Myc is a fundamentally unstable protein and two independent groups have identified the F-box ubiquitin ligase SCFFBW7 as capable of inducing ubiquitin dependent degradation of c-Myc.49,104 Linking ubiquitin dependent regulation to its oncogenic functions, the c-Myc hot spot mutation at Thr58 found in Burkitt lymphoma converts the protein to a form resistant to SCFFBW7 induced degradation and induces an elevation in protein level of c-Myc.49,104
A recent genome-wide shRNA screen for genes required for c-Myc induced apoptosis identified USP28.105 USP28 does not directly interact with c-Myc. Instead, USP28 is recruited to c-Myc through its binding to SCFFBW7, the E3 ligase that targets c-Myc.105 SCFFBW7 thus ubiquitinates and USP28 deubiquitinates, c-Myc, with both requiring Thr58 phosphorylation of c-Myc for their activities.105 Adding complexity, USP28 stability is increased when SCFFBW7 is depleted.105 The ability of USP28 to deubiquitinate and stabilize c-Myc suggests that USP28 has oncogenic potential. In fact, USP28 is overexpressed in colon and breast carcinomas.105 Furthermore, USP28 is essential for c-Myc induced tumor cell proliferation in colon carcinoma cells.105 C-Myc dysregulation is nearly universal in human malignancy and cancer cells require c-Myc activity for growth.102 However, c-Myc has proved difficult to practically and efficiently target in cancer cells with any type of pharmacologic strategy. The inhibition of USP28 may therefore provide a means of targeting malignancies via indirect interference with c-Myc function, by accelerating its degradation.
As is the case for most other DUBs when considering their inactivation as a therapeutic strategy, the cellular functions of USP28 are pleiotropic. USP28 is also involved in DNA damage responses, and is required for 53BP1 and CHK2 stabilization after DNA damage.106 In the absence of USP28, DNA damage signals are attenuated and p53-induced apoptosis is abrogated.106 Therefore, USP28 positively regulates the CHK2-p53 repair pathway, the critical component of the DNA damage pathway that responds to ionizing radiation and many genotoxic chemotherapeutics.106 Taken together, USP28 warrants further investigation as a therapeutic target in cancer, if agents can be developed that selectively block its ability to stabilize c-Myc, but do not interfere with its functions that contribute to p53 activation after DNA damage. Alternatively, USP28 may serve as a useful target specifically in p53 mutated/deleted tumors.
CYLD.
CYLD is a UCH-family DUB that is mutated in the autosomally dominant inherited disease Familial Cylindromatosis, characterized by the development of multiple skin tumors.107 The full extent of substrates and pathways regulated by CYLD remains unclear, but CYLD's regulation of the NFκB pathway has been well studied. NFκB is a crucial mediator of immune responses and cell survival. Multiple reports have shown that CYLD negatively regulates NFκB by deubiquitinating K63-linked polyubiquitin chains from TNF Receptor Associated Factors 2, 6 and 7 (TRAF2, TRAF6 and TRAF7), Nuclear factor kappa B Essential Modifier (NEMO), Receptor Interacting Protein 1 (RIP1) and TGFβ-activated kinase 1 (TAK1),108–113 all of which are critical mediators of I kappa kinase (IKK) and NFκB activation. CYLD impacts the NFκB pathway also through the deubiquitination of Bcl-3 protein,114 which is a critical transcription co-activator for NFκB. Bcl-3 is inactive in the cytoplasm and polyubiquitination triggers its translocation into the nucleus, where it forms a complex with the NFκB dimer p50/p52 to switch p50/p52 function from repression to activation. In turn, Bcl-3 activates NFκB transcription activity and promotes cell survival.114 The CYLD-Bcl-3 association removes K63-linked polyubiquitin chains from Bcl-3 and blocks its nuclear translocation, therefore inhibiting NFκB activation.114 CYLD-/- knockout mice recapitulate the phenotype of familial cylindromatosis patients.114 These CYLD deficient mice are highly sensitive to chemically induced skin tumors, in accordance with increased NFκB activity.114
Reduced expression or mutation of CYLD has been characterized in multiple myeloma, B-cell malignancies, hepatocellular carcinoma, uterine cervix carcinoma, kidney cancer, colon cancer and melanoma.115 CYLD is also involved in the regulation of other cellular processes, such as cell migration and spermatogenesis.116 Though not a therapeutic target due to its tumor suppressor status, a better understanding of CYLD regulation may be of value in developing strategies to reactivate its expression, and/or lead to its use as a biomarker for prognostic or therapeutic use related to the activity of the NFκB pathway in various cancers.
A20.
A20 is a second DUB functioning in the restriction of NFκB activity. A20 bears unique biochemical characteristics, in that it is both an E3 ubiquitin ligase and a deubiquitinating enzyme, regulating the ubiquitination status of multiple protein mediators in the NFκB signaling pathway. A20 contains an OTU domain at the N-terminus, which removes activating K63-linked polyubiquitin chains from RIP1, RIP2, TRAF6, NEMO and Mucosa-associated lymphoid tissue lymphoma translocation protein 1 (MALT1).11,117–122 A20 contains seven zinc fingers at its C-terminus that function as an E3 ubiquitin ligase, conjugating K48-linked polyubiquitin chains to RIP1 and TRAF2, which then target RIP1 and TRAF2 for degradation.123 A20 therefore inhibits NFκB activation both by disassembly of K63 ubiquitin chains, as well as by K48 polyubiquitination and degradation of NFκB signaling intermediates. The interference of A20 with the NFκB pathway, and thus immune responses, was confirmed in A20 deficient mice, which exhibit spontaneous inflammation and premature death.122
Recent studies have, not surprisingly, identified A20 as a crucial tumor suppressor gene. A20 protein is often inactivated by deletion, bi-allelic mutation or promoter methylation in several B cell lymphomas and Hodgkin's lymphoma.125 Among B cell malignancies, A20 inactivation has been reported in mantle cell lymphoma, MALT lymphoma and marginal zone lymphoma.126 The study of A20 has therefore linked ubiquitination, inflammation and tumorigenesis in mouse models and human disease. Like CYLD, A20 is not an obvious drug target, but further understanding of its action could again lead to uses as a biomarker or lead to proteins that act as A20 negative regulators that themselves might be useful drug targets.
USP9x.
Originally cloned from Drosophila where its mutation leads to erroneously developed eyes, USP9x (FAM in Drosophila) is a DUB belonging to the USP family.127 USP9x was first linked to cancer with the discovery of its ability to stabilize β-catenin, given that β-catenin is well known for promoting colon cancer cell growth.128 Later, it was established that USP9x positively regulates the TGFβ signaling pathway by targeting Smad4 for deubiquitination.129 Monoubiquitination of Smad4 impedes its association with phosphorylated Smad2, which is a decisive step in the activation of the TGFβ signaling pathway.129 USP9x removes the inhibitory monoubiquitination signal from Smad4 allowing Smad4 to positively regulate the TGFβ pathway.129 The exact relationship of USP9x with cancer development based on its regulation of Smad4 remains unclear, however, given the consideration of the dual roles of TGFβ signaling in tumor initiation vs. progression which is highly dependent on signal strength and duration, as well as distinctive cellular/tissue contexts.
A more recent discovery, which established USP9x as the specific deubiquitinase of MCL1, provides a stronger link between USP9x and its potential role as an oncogene.130 MCL1 is a powerfully anti-apoptotic BCL-2 homolog.130 High levels of MCL1 in some subtypes of lymphomas, leukemias and myelomas contributes to chemotherapy resistance.130 USP9x deubiquitination of MCL1 blocks MCL1 proteasome degradation, providing a new mechanism for aberrant MCL1 stabilization in certain tumors.130 USP9x overexpression is coupled with MCL1 upregulation in follicular lymphomas, diffuse large B-cell lymphomas, breast ductal adenocarcinomas, colon adenocarcinomas and small lung cancer carcinomas. MCL1 overexpression is also often associated with poor prognostic outcome.130 Inhibition of USP9x destabilizes MCL1 and can sensitize resistant cancer cells to chemotherapy.130 USP9x is thus an exciting new therapeutic target in diseases where its expression is upregulated-perhaps with particular use as a chemosensitizer for resistant hematologic malignancies that also overexpress MCL1.
Concluding Remarks
E3 ubiquitin ligases clearly contribute to either cancer initiation and development or tumor suppression. E3s that degrade tumor suppressor genes, such as MDM2, are oncogenes. E3s that degrade oncogenic genes are tumor suppressors, for example the VHL ligase complex. Distinctive E3s function in different types of cancers. The large body of evidence from cell culture work, primary tumor sample studies and mouse models provides, in many cases, a molecular explanation of individual E3 action in tumorigenesis. The interference with protein-protein interaction between E3s and their substrates may lead to the development of cancer therapeutics, though the drug development process has been hampered by technical difficulties of specificity and potency of inhibitors. However, in theory, E3 ligase-based drugs might be expected to generate less toxic cancer drugs compared with 26S proteasome inhibitors, which carry substantial toxicity due to the lack of target specificity.
The targeting of DUBs by small molecules has been technically simpler and may yield more practical cancer therapeutics than anti-E3 drugs. Though DUBs have only very recently been linked directly to oncogenic or tumor suppressor phenotypes, there appears to be in most substrates a symmetry of DUB and E3 action not unlike that seen with kinases and phosphatases. The number and diversity of DUBs, at least for now, is well below that of E3s, but it is quite likely that additional DUB domains or DUB complexes will be discovered that will add to the diversity and specificity of substrate activity, and therefore add to the potential value of DUBs as specific and useful drug targets in cancer.
Acknowledgements
The authors would like to thank R. Kulikov and M. Straza for providing comments on the manuscript. S.R.G. was supported by CA107532 from NCI. The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH.
Abbreviations
- DUB
deubiquitinating enzyme
- HECT
homolog of E6-AP C terminus
- RING
really interesting new gene
- JAMM
JAB1/MPN/Mov34 metalloenzyme
- UCH
ubiquitin C-terminal hydrolase
- USP
ubiquitin specific protease
- OTU
ovarian tumor domain
- MJD
machado josephin domain
- HPV
human papillomavirus
- SCF
Skp1-Cullin-F-box
- CDK
cyclin dependent kinase
- VHL
von hippel-lindau
- HIF-1a
hypoxia-inducible factor-1 a
- BRCA1
breast cancer susceptibility gene 1
- BARD1
BRCA1 associated ring domain 1
- PTEN
phosphatase and tensin homolog
- Smurf2
smad ubiquitination regulatory factor 2
- TGFβ
transforming growth factorβ
- KLF5
krüppel-like factor 5
- HAUSP
herpes associated ubiquitin specific protease
- FOXO4
forkhead box O4
- FAS
fatty acid synthase
- TRAF
TNF receptor associated factors
- NEMO
nuclear factor kappaB essential modifier
- RIP1
receptor interacting protein 1
- IKK
I-kappa kinase
- MALT
mucosa-associated lymphoid tissue
Footnotes
Previously published online: www.landesbioscience.com/journals/cbt/article/13417
References
- 1.Ciechanover A, Elias S, Heller H, Ferber S, Hershko A. Characterization of the heat-stable polypeptide of the ATP-dependent proteolytic system from reticulocytes. J Biol Chem. 1980;255:7525–7528. [PubMed] [Google Scholar]
- 2.Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc Natl Acad Sci USA. 1980;77:1365–1368. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 5.Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci USA. 2009;106:16275–16280. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoppe T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn't fit all. Trends Biochem Sci. 2005;30:183–187. doi: 10.1016/j.tibs.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 8.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 9.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 10.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 11.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NFkappaB signalling. Nature. 2004;430:694–679. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 12.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 16.Scheffner M. Ubiquitin, E6-AP and their role in p53 inactivation. Pharmacol Ther. 1998;78:129–139. doi: 10.1016/s0163-7258(98)00003-5. [DOI] [PubMed] [Google Scholar]
- 17.Giordano G, Azzoni C, D'Adda T, Rocco A, Gnetti L, Froio E, et al. Human papilloma virus (HPV) status, p16INK4a and p53 overexpression in epithelial malignant and borderline ovarian neoplasms. Pathol Res Pract. 2008;204:163–174. doi: 10.1016/j.prp.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 18.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 19.Galani E, Christodoulou C. Human papilloma viruses and cancer in the post-vaccine era. Clin Microbiol Infect. 2009;15:977–981. doi: 10.1111/j.1469-0691.2009.03032.x. [DOI] [PubMed] [Google Scholar]
- 20.de Rozieres S, Maya R, Oren M, Lozano G. The loss of mdm2 induces p53-mediated apoptosis. Oncogene. 2000;19:1691–1697. doi: 10.1038/sj.onc.1203468. [DOI] [PubMed] [Google Scholar]
- 21.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 22.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- 24.Quesnel B, Preudhomme C, Fournier J, Fenaux P, Peyrat JP. MDM2 gene amplification in human breast cancer. Eur J Cancer. 1994;30:982–984. doi: 10.1016/0959-8049(94)90128-7. [DOI] [PubMed] [Google Scholar]
- 25.Allende-Vega N, Saville MK. Targeting the ubiquitin-proteasome system to activate wild-type p53 for cancer therapy. Semin Cancer Biol. 2010;20:29–39. doi: 10.1016/j.semcancer.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Cheok CF, Dey A, Lane DP. Cyclin-dependent kinase inhibitors sensitize tumor cells to nutlin-induced apoptosis: a potent drug combination. Mol Cancer Res. 2007;5:1133–1145. doi: 10.1158/1541-7786.MCR-07-0161. [DOI] [PubMed] [Google Scholar]
- 27.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Canner JA, Sobo M, Ball S, Hutzen B, DeAngelis S, Willis W, et al. MI-63: a novel small-molecule inhibitor targets MDM2 and induces apoptosis in embryonal and alveolar rhabdomyosarcoma cells with wild-type p53. Br J Cancer. 2009;101:774–781. doi: 10.1038/sj.bjc.6605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 30.Li WD, Wang MJ, Ding F, Yin DL, Liu ZH. Cytotoxic effect of a non-peptidic small molecular inhibitor of the p53-HDM2 interaction on tumor cells. World J Gastroenterol. 2005;11:2927–2931. doi: 10.3748/wjg.v11.i19.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 32.Chen D, Brooks CL, Gu W. ARF-BP1 as a potential therapeutic target. Br J Cancer. 2006;94:1555–1558. doi: 10.1038/sj.bjc.6603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dornan D, Bheddah S, Newton K, Ince W, Frantz GD, Dowd P, et al. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64:7226–7230. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 34.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 35.Yamasaki S, Yagishita N, Sasaki T, Nakazawa M, Kato Y, Yamadera T, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2007;26:113–122. doi: 10.1038/sj.emboj.7601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:3002. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 39.Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, et al. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 40.Slingerland J, Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J Cell Physiol. 2000;183:10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 41.Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, et al. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, et al. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 43.Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 44.Muraoka RS, Lenferink AE, Law B, Hamilton E, Brantley DM, Roebuck LR, et al. ErbB2/Neu-induced, cyclin D1-dependent transformation is accelerated in p27-haploinsufficient mammary epithelial cells but impaired in p27-null cells. Mol Cell Biol. 2002;22:2204–2219. doi: 10.1128/MCB.22.7.2204-2219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Latres E, Chiarle R, Schulman BA, Pavletich NP, Pellicer A, Inghirami G, et al. Role of the F-box protein Skp2 in lymphomagenesis. Proc Natl Acad Sci USA. 2001;98:2515–2520. doi: 10.1073/pnas.041475098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapira M, Ben-Izhak O, Linn S, Futerman B, Minkov I, Hershko DD. The prognostic impact of the ubiquitin ligase subunits Skp2 and Cks1 in colorectal carcinoma. Cancer. 2005;103:1336–1346. doi: 10.1002/cncr.20917. [DOI] [PubMed] [Google Scholar]
- 47.Traub F, Mengel M, Luck HJ, Kreipe HH, von Wasielewski R. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res Treat. 2006;99:185–191. doi: 10.1007/s10549-006-9202-3. [DOI] [PubMed] [Google Scholar]
- 48.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 49.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 51.Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 52.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–1502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 54.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 55.Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, et al. von Hippel-Lindau disease. Lancet. 2003;361:2059–2067. doi: 10.1016/S0140-6736(03)13643-4. [DOI] [PubMed] [Google Scholar]
- 56.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 57.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 58.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 59.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol. 2004;22:4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 60.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med. 2002;8:62–67. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- 61.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scully R. Role of BRCA gene dysfunction in breast and ovarian cancer predisposition. Breast Cancer Res. 2000;2:324–330. doi: 10.1186/bcr76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 64.Ohta T, Fukuda M. Ubiquitin and breast cancer. Oncogene. 2004;23:2079–2088. doi: 10.1038/sj.onc.1207371. [DOI] [PubMed] [Google Scholar]
- 65.Boulton SJ. BRCA1-mediated ubiquitylation. Cell Cycle. 2006;5:1481–1486. doi: 10.4161/cc.5.14.2930. [DOI] [PubMed] [Google Scholar]
- 66.Sato K, Hayami R, Wu W, Nishikawa T, Nishikawa H, Okuda Y, et al. Nucleophosmin/B23 is a candidate substrate for the BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279:30919–30922. doi: 10.1074/jbc.C400169200. [DOI] [PubMed] [Google Scholar]
- 67.Starita LM, Horwitz AA, Keogh MC, Ishioka C, Parvin JD, Chiba N. BRCA1/BARD1 ubiquitinate phosphorylated RNA polymerase II. J Biol Chem. 2005;280:24498–24505. doi: 10.1074/jbc.M414020200. [DOI] [PubMed] [Google Scholar]
- 68.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen C, Matesic LE. The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev. 2007;26:587–604. doi: 10.1007/s10555-007-9091-x. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 72.Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lian Z, Di Cristofano A. Class reunion: PTEN joins the nuclear crew. Oncogene. 2005;24:7394–7400. doi: 10.1038/sj.onc.1209089. [DOI] [PubMed] [Google Scholar]
- 74.Fouladkou F, Landry T, Kawabe H, Neeb A, Lu C, Brose N, et al. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci USA. 2008;105:8585–8590. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGFbeta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 77.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 78.Ikushima H, Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 79.Fukuchi M, Fukai Y, Masuda N, Miyazaki T, Nakajima M, Sohda M, et al. High-level expression of the Smad ubiquitin ligase Smurf2 correlates with poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Res. 2002;62:7162–7165. [PubMed] [Google Scholar]
- 80.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, et al. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene. 2005;24:3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 81.Chen C, Zhou Z, Sheehan CE, Slodkowska E, Sheehan CB, Boguniewicz A, et al. Overexpression of WWP1 is associated with the estrogen receptor and insulin-like growth factor receptor 1 in breast carcinoma. Int J Cancer. 2009;124:2829–2836. doi: 10.1002/ijc.24266. [DOI] [PubMed] [Google Scholar]
- 82.Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 83.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and downregulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 84.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 85.Laine A, Ronai Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene. 2007;26:1477–1483. doi: 10.1038/sj.onc.1209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, et al. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- 87.Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135 kDa cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 88.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 89.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 90.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:486. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 91.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemensen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 92.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 93.Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Colland F. Ubiquitin and cancer: from molecular targets and mechanisms to the clinic—AACR Special Conference. IDrugs. 2006;9:179–181. [PubMed] [Google Scholar]
- 95.Colland F, Formstecher E, Jacq X, Reverdy C, Planquette C, Conrath S, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol Cancer Ther. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 96.Gousseva N, Baker RT. Gene structure, alternate splicing, tissue distribution, cellular localization and developmental expression pattern of mouse deubiquitinating enzyme isoforms Usp2-45 and Usp2-69. Gene Expr. 2003;11:163–179. doi: 10.3727/000000003108749053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Graner E, Tang D, Rossi S, Baron A, Migita T, Weinstein LJ, et al. The isopeptidase USP2a regulates the stability of fatty acid synthase in prostate cancer. Cancer Cell. 2004;5:253–261. doi: 10.1016/s1535-6108(04)00055-8. [DOI] [PubMed] [Google Scholar]
- 98.Oishi K, Miyazaki K, Kadota K, Kikuno R, Nagase T, Atsumi G, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 99.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Allende-Vega N, Sparks A, Lane DP, Saville MK. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2010;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- 101.Priolo C, Tang D, Brahamandan M, Benassi B, Sicinska E, Ogino S, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 2006;66:8625–8632. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- 102.Albihn A, Johnsen JI, Henriksson MA. MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res. 2010;107:163–224. doi: 10.1016/S0065-230X(10)07006-5. [DOI] [PubMed] [Google Scholar]
- 103.Amati B, Sanchez-Arevalo Lobo VJ. MYC degradation: deubiquitinating enzymes enter the dance. Nat Cell Biol. 2007;9:729–731. doi: 10.1038/ncb0707-729. [DOI] [PubMed] [Google Scholar]
- 104.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Popov N, Wanzel M, Madiredjo M, Zhang D, Beijersbergen R, Bernards R, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 106.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 107.Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 108.Yoshida H, Jono H, Kai H, Li JD. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative crosstalk with TRAF6 AND TRAF7. J Biol Chem. 2005;280:41111–41121. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- 109.Jono H, Lim JH, Chen LF, Xu H, Trompouki E, Pan ZK, et al. NFkappaB is essential for induction of CYLD, the negative regulator of NFkappaB: evidence for a novel inducible autoregulatory feedback pathway. J Biol Chem. 2004;279:36171–36174. doi: 10.1074/jbc.M406638200. [DOI] [PubMed] [Google Scholar]
- 110.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 111.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NFkappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 112.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NFkappaB signalling by deubiquitination. Nature. 2003;424:801–815. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 113.Reiley WW, Jin W, Lee AJ, Wright A, Wu X, Tewalt EF, et al. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J Exp Med. 2007;204:1475–1485. doi: 10.1084/jem.20062694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NFkappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 115.Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NFkappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sacco JJ, Coulson JM, Clague MJ, Urbe S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, et al. Homeostatic MyD88-dependent signals cause lethal inflamMation in the absence of A20. J Exp Med. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Duwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, et al. A20 negatively regulates T cell receptor signaling to NFkappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 120.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 121.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NFkappaB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 122.Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, et al. Failure to regulate TNF-induced NFkappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Song HY, Rothe M, Goeddel DV. The tumor necrosis factor-inducible zinc finger protein A20 interacts with TRAF1/TRAF2 and inhibits NFkappaB activation. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li L, Soetandyo N, Wang Q, Ye Y. The zinc finger protein A20 targets TRAF2 to the lysosomes for degradation. Biochim Biophys Acta. 2009;1793:346–353. doi: 10.1016/j.bbamcr.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med. 2009;206:981–989. doi: 10.1084/jem.20090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 127.Huang Y, Baker RT, Fischer-Vize JA. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 128.Murray RZ, Jolly LA, Wood SA. The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and beta-catenin. Mol Biol Cell. 2004;15:1591–1599. doi: 10.1091/mbc.E03-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 130.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 131.Mishra A, Godavarthi SK, Jana NR. UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol Dis. 2009;36:26–34. doi: 10.1016/j.nbd.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 132.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 133.Schlott T, Reimer S, Jahns A, Ohlenbusch A, Ruschenburg I, Nagel H, et al. Point mutations and nucleotide insertions in the MDM2 zinc finger structure of human tumours. J Pathol. 1997;182:54–61. doi: 10.1002/(SICI)1096-9896(199705)182:1<54::AID-PATH815>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 134.Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–1424. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- 135.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 136.Hogervorst FB, Nederlof PM, Gille JJ, McElgunn CJ, Grippeling M, Pruntel R, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- 137.Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 138.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim SS, Yoo NJ, Jeong EG, Kim MS, Lee SH. Expression of NEDD4-1, a PTEN regulator, in gastric and colorectal carcinomas. APMIS. 2008;116:779–784. doi: 10.1111/j.1600-0463.2008.00999.x. [DOI] [PubMed] [Google Scholar]
- 140.Chen C, Zhou Z, Guo P, Dong JT. Proteasomal degradation of the KLF5 transcription factor through a ubiquitin-independent pathway. FEBS Lett. 2007;581:1124–1130. doi: 10.1016/j.febslet.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 141.Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15:1941–1951. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- 142.Li Y, Zhou Z, Alimandi M, Chen C. WW domain containing E3 ubiquitin protein ligase 1 targets the full-length ErbB4 for ubiquitin-mediated degradation in breast cancer. Oncogene. 2009;28:2948–2958. doi: 10.1038/onc.2009.162. [DOI] [PubMed] [Google Scholar]
- 143.Chen C, Sun X, Guo P, Dong XY, Sethi P, Zhou W, et al. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26:2386–2394. doi: 10.1038/sj.onc.1210021. [DOI] [PubMed] [Google Scholar]
- 144.Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;121:80–87. doi: 10.1002/ijc.22653. [DOI] [PubMed] [Google Scholar]
- 145.Becker K, Marchenko ND, Palacios G, Moll UM. A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle. 2008;7:1205–1213. doi: 10.4161/cc.7.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Masuya D, Huang C, Liu D, Nakashima T, Yokomise H, Ueno M, et al. The HAUSP gene plays an important role in non-small cell lung carcinogenesis through p53-dependent pathways. J Pathol. 2006;208:724–732. doi: 10.1002/path.1931. [DOI] [PubMed] [Google Scholar]
- 147.Novak U, Rinaldi A, Kwee I, Nandula SV, Rancoita PM, Compagno M, et al. The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood. 2009;113:4918–4921. doi: 10.1182/blood-2008-08-174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chanudet E, Huang Y, Ichimura K, Dong G, Hamoudi RA, Radford J, et al. A20 is targeted by promoter methylation, deletion and inactivating mutation in MALT lymphoma. Leukemia. 2010;24:483–487. doi: 10.1038/leu.2009.234. [DOI] [PubMed] [Google Scholar]
- 149.Chanudet E, Ye H, Ferry J, Bacon CM, Adam P, Muller-Hermelink HK, et al. A20 deletion is associated with copy number gain at the TNFA/B/C locus and occurs preferentially in translocation-negative MALT lymphoma of the ocular adnexa and salivary glands. J Pathol. 2009;217:420–430. doi: 10.1002/path.2466. [DOI] [PubMed] [Google Scholar]
- 150.Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, et al. Overexpression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]