Abstract
The general transcription factor TFIIB plays essential, but as yet unclear, roles in transcription initiation by RNA polymerase II. We recently found that phosphorylation of TFIIB is required for productive transcription. We discuss the implications of this work for the functions of TFIIB in promoter escape and gene loop formation.
Key words: transcription, TFIIB, promoter, terminator, RNA polymerase II, gene loops
Introduction
The general transcription factors (GTFs) guide RNA polymerase II (pol II) to the promoter and assist in the initiation of transcription at the correct site.1 Once the preinitiation complex (PIC) is formed, open complex formation facilitates pol II access to the template strand, leading to synthesis of the first phosphodiester bond. Before pol II enters into productive elongation, it passes through a transition, promoter escape, at which a subset of GTFs remain at the promoter forming a scaffold for the formation of the next transcription initiation complex.2,3 Coincident with transcription initiation, the C-terminal domain (CTD) of pol II is phosphorylated at Serine-5, which stimulates the recruitment of mRNA 5′end capping enzymes.4,5 mRNA capping, splicing and polyadenylation take place co-transcriptionally, as the pol II CTD is phosphorylated at Serine-2 and Serine-5 is progressively dephosphorylated as elongation proceeds.6,7 Some genes also require pol II CTD Serine-7 phosphorylation in transcription regulation.8 In living cells, gene loops, which connect promoters and terminators, have been proposed as a potential mechanism to link the multiple events involved in transcription.
TFIIB
TFIIB plays a critical role in the PIC, showing absolute requirement at all promoters for the recruitment of pol II. Human TFIIB is a single peptide of 33 kDa consisting of a zinc ribbon at the N-terminus and a C-terminal core domain composed of α-helices that form two direct repeats.9 The core domain of TFIIB complexes with TBP at the TATA box and interacts with the DNA both upstream and downstream of TATA. The zinc ribbon of TFIIB is required for the recruitment of pol II and interacts with both pol II and TFIIF.9,10 The region between the zinc ribbon and core domain of TFIIB is highly conserved and contains structures termed the B-finger/B-reader. At the promoter, the B-finger/B-reader project into the RNA catalytic center and abuts the initiation site, where it is proposed to play a role in transcription start site selection.11
TFIIB has been proposed as a target of transcriptional activators.9 Interestingly, the N- and C-terminal domains of TFIIB engage in an intramolecular interaction. This involves direct binding of the B-finger/B-reader region of TFIIB to the second direct repeat and results in equilibrium between an “open” form of TFIIB and a “closed” form. We and others have shown that upon interaction with an activator, TFIIB intramolecular interaction is disrupted, thereby stabilizing the “open” form of TFIIB.9 Mutations in TFIIB that stabilize the closed form of TFIIB result in defects in transcriptional activation, suggesting that TFIIB conformational change is important in the activation process.12,13
TFIIB and Transcription Initiation
The B-finger hairpin has been refined as a unidirectional extension of the TFIIB N-terminal ribbon into an a-helix and a mobile loop, reaching into the active center of pol II, where it interacts directly with the DNA-RNA hybrids.11 The C-terminal region of TFIIB is located above the enzyme active center cleft.14 Given the contact with the catalytic center of pol II via the RNA exit channel and the mobility evident from the different structures, the B-finger/B-reader domain likely functions at several stages, including transcription initiation and clearance of pol II from the promoter. It has been proposed that mutations within the B-finger/B-reader alter the interaction of TFIIB with the pol II active site and possibly impair bubble formation around the TSS, leading to failure to expose the path of promoter DNA across the central cleft of pol II and thereby cause aberrant start site selection.15 These defects can be suppressed by the RAP74 subunit of TFIIF and pol II subunits.16
Several studies suggest a role for TFIIB in promoter escape by pol II.11,14,17,18 In early pol II transcription, short DNA-RNA hybrids are unstable and are transiently ejected from the enzyme (abortive transcription initiation). It has been proposed that the TFIIB B-finger/B-reader loop stabilizes the short RNA transcripts with pol II until the growing RNA reaches seven nucleotides and TFIIB is triggered to release from the initiation apparatus as pol II enters the productive elongation complex (promoter escape).19,20 Bubble collapse (reannealing of the upstream edge of DNA strands) is associated with TFIIB release, triggered by a clash of newly synthesized RNA with the B-finger/B-reader and a clash of the upstream DNA duplex with the TFIIB B-linker (the region between B-finger/B-reader and C-terminus of TFIIB).11,20
New Roles for the TFIIB B-finger/B-reader in Transcription Initiation Control
Our recent study demonstrated that human TFIIB is phosphorylated at Serine-65 within the B-finger/B-reader domain and that this modification is required for productive transcription initiation.21 Mutation of TFIIB Serine-65 to Alanine led to failure in productive transcription at several genes and the accumulation of pol II phosphorylated at serine-5 at the gene 5′ end. Our findings confirm the previous reported importance of the TFIIB B-finger/B-reader loop in regulating transcription initiation post-PIC assembly, and in addition suggest that its function in transcription initiation is subject to regulation.
We found that the phosphorylation of TFIIB in vivo was significantly inhibited by the CDK inhibitor 5,6-dichloro-1-h-ribofuranosyl-benzimidazole (DRB).21 200 µM DRB was required for inhibition and 50 µM DRB had minimal effect in vitro. Although DRB is a broad inhibitor, these results raise the possibility that the CDK7 component of TFIIH might be a TFIIB Serine-65 kinase. However, purified TFIIH failed to significantly phosphorylate recombinant TFIIB in vitro, suggesting that, if TFIIB is indeed phosphorylated by TFIIH, then other factor(s) are required to enhance this activity. Indeed, there is precedent for this in the ability of mediator or TFIIE to enhance pol II CTD Serine-5 phosphorylation by TFIIH.22,23 TFIIB Serine-65 is not nested within a sequence (PSR) that is typical of a CDK substrate. However, we note that Serine-7 of the pol II CTD also contains the proline residue N-terminal to the target serine that is phosphorylated by TFIIH.8 Although TFIIH is a prime candidate as the TFIIB Serine-65 kinase, this remains an open question. The presence of TFIIB phosphorylated at Serine-65 at several promoters and the requirement for this modification for productive transcription suggests that phosphorylation of TFIIB likely takes place within the PIC. More work will be necessary to determine the enzymes that are the physiologically relevant kinases of TFIIB Serine-65.
Promoter-Terminator Connections and TFIIB
TFIIB dissociates from the promoter at transcription initiation and does not travel with the elongating pol II or remain as part of the scaffold.9 However, a recent genome-wide search found that TFIIB occupies both the promoter and terminator regions of at least 120 genes in yeast.24 Similar genome-wide mapping in metazoans uncovered TFIIB at the promoter and terminator regions of a subset of protein coding genes.25 TFIIB has been linked with components of the transcription termination and polyadenylation complexes CPSF and CstF genetically and/or by direct protein-protein interactions.26,27 Gene looping, which links promoters and terminators, has been shown to be dependent on pol II CTD Serine-5 phosphorylation and polyadenylation factors Ssu72 and Pta1.28 TFIIB was found to be involved in this link through interaction with Ssu72 directly.27 The role of gene loops in transcription is unclear. However, it has been demonstrated that gene looping accompanies activated transcription and is dependent on a transcriptional activator.29 Significantly, activators that facilitate loop formation do not directly interact with the 3′ end of the genes, but physically interact with TFIIB.
Model for the Mechanism of Action of TFIIB Phosphorylation in Formation of Gene Loops in Mammalian Cells
We found that TFIIB phosphorylation at Serine-65 does not regulate its interaction with Ssu72. However, in the absence of TFIIB phosphorylation, Ssu72 is recruited to the gene terminator, but not the promoter. In contrast, TFIIB phosphorylation at Serine-65 directly regulates its contact with CstF-64. In the absence of TFIIB phosphorylation, CstF-64 is not recruited to the gene promoter or terminator.21 A possible explanation for these observations is that the association of phosphorylated TFIIB with CstF-64 promotes the formation of gene loops, which stabilizes the interaction between TFIIB and Ssu72. Our data also suggest that the occupancy of Ssu72 at the terminator is transcription independent.21 This observation seems to be contrary to the previously reported role of pol II in regulating the association of Ssu72 during productive transcriptional elongation.30 However, Ssu72 has been shown to be dispensable for transcription termination as it may function later in the established cleavage-competent complex after the recognition of polyadenylation signals by the other processing factors.31 In addition, Ssu72 CTD phosphatase activity has been shown to be required immediately after 5′ end capping of mRNA to facilitate the transition of pol II into the elongation state.32 Thus, it is possible that the assembly of Ssu72 at the terminator is auxiliary but not essential for productive transcription regulation or at least the promoter function dominates to regulate transcription start site selection. It has been reported before that the transcription coactivator PC4 (Sub1 in yeast) genetically interacts with CstF-64 and facilitates its recruitment to the promoter.33 It is therefore significant that PC4 can directly interact with TFIIB in vitro, suppresses the effects of TFIIB B-reader mutations, and is proposed to play a role in the release of TFIIB from the transcription complex.26 It will be interesting to determine the effects of TFIIB phosphorylation on PC4 recruitment to promoters and terminators in vivo.
Our data are in agreement with previous findings that the formation of gene loops is dependent on active transcription. However, previous studies found that the role of TFIIB in gene loop formation was transcription-independent.34 The TFIIB B-finger/reader also plays a role in facilitating “bubble collapse” leading to the transition of pol II into a productive transcribing enzyme.20 One possibility is that gene looping and bubble collapse are coupled by TFIIB phosphorylation. However, it is still not clear whether gene loop structure is a cause or a consequence of productive transcription initiation. Localization of the CPSF and CstF complexes close to transcription start sites has been described before, although the function has not been established.30 Ssu72 and PC4, in concert with TFIIB, can both regulate transcription start site selection in vivo.16 It is also possible that their initial recruitment to the promoter is linked to transcription initiation by modulating the release of TFIIB from promoters. Indeed, recent studies have revealed that gene looping is conferred by activator-dependent interactions between transcription initiation and termination.29 Interestingly, activators facilitate the formation of gene loops through direct interaction with TFIIB. As described previously, the B-finger/B-reader plays a role in modulating the conformation of TFIIB during the activation of transcription.12,13 It is therefore possible that phosphorylation of TFIIB Serine-65 modulates the capacity of TFIIB to contact transcriptional activators and thus modulate the formation of gene loops. Alternatively, activator-TFIIB contact may regulate the efficiency of TFIIB phosphorylation.
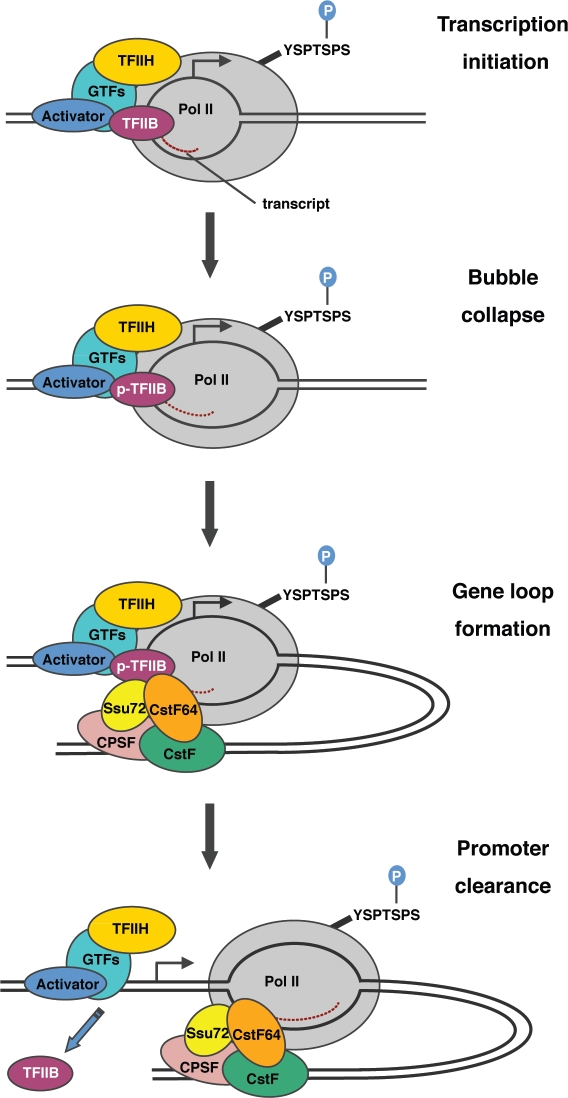
A potential model for the role of TFIIB phosphorylation in transcription-coupled gene loop formation is presented in Figure 1. Following PIC assembly, the melted DNA template strand is exposed to the catalytic center of pol II and TFIIH/CDK7 phosphorylates the pol II CTD at Serine-5. The TFIIB B-finger/reader is projected into the pol II catalytic center and stabilizes short DNA-RNA hybrids. At bubble collapse, the TFIIB N-terminus is released from the catalytic center. It is possible that TFIIB Serine-65 phosphorylation facilitates bubble collapse and the release of TFIIB. Alternatively, TFIIB phosphorylation may occur after the B-finger/reader is released. Phosphorylated TFIIB is then able to associate with CstF-64, which augments the interaction between Ssu72 and TFIIB and drives loop formation. In this case, gene loops may function to facilitate the release of TFIIB and proofread correct initiation before efficient transcription elongation proceeds.
Figure 1.
A potential model for the mechanism by which TFIIB phosphorylation regulates the formation of gene loops. The intermediates involved in the transition from the closed PIC to transcription initiation are shown. TFIIB stabilizes the formation of short DNA-RNA hybrids, but the B-finger/B-reader is ejected at bubble collapse. TFIIB phosphorylation occurs either coincident with or after bubble collapse. Phosphorylated TFIIB then associates with the CstF complex (via CstF-64) and promotes the formation of gene loops, which are further stabilized by interaction between TFIIB and the CPSF complex (via Ssu72). TFIIB is ejected from the complex at promoter clearance.
Future Perspectives
Evidence for a direct role of TFIIB phosphorylation in bubble collapse and/or gene loop formation awaits further studies. It will also be important to determine if TFIIB phosphorylation is universally required for productive transcription initiation. As mentioned above, TFIIB has been proposed as a target of transcriptional activators. The possibility that TFIIB phosphorylation may be important in the process of transcriptional activation, either as a trigger for transcription initiation or in pause release, requires further studies. Clues to the function(s) of TFIIB phosphorylation will also be gained from identification of the kinases/phosphatases responsible and the context(s) in which the modifications occur.
In summary, the finding that TFIIB phosphorylation is required for productive transcription and promoter-terminator contacts offers a potential regulatory mechanism for gene loop formation and a trigger for transcription in eukaryotes.
Abbreviations
- CDK
cyclin-dependent kinase
- CPSF
cleavage and polyadenylation specificity factor
- CstF
cleavage stimulation factor
- CTD
C-terminal domain
- DRB
5,6-dichloro-1-h-ribofuranosyl-benzimidazole
- GTF
general transcription factor
- PIC
preinitiation complex
- Pol II
RNA polymerase II
- TFIIB
transcription factor IIB
- TFIIF
transcription factor IIF
- TFIIH
transcription factor IIH
- TSS
transcription start site
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/12900
References
- 1.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 2.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 3.Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 7.Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, et al. Serine 7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng W, Roberts SGE. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 10.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 11.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 12.Hawkes NA, Evans R, Roberts SGE. The conformation of the transcription factor TFIIB modulates the response to transcriptional activators in vivo. Curr Biol. 2000;10:273–276. doi: 10.1016/s0960-9822(00)00363-8. [DOI] [PubMed] [Google Scholar]
- 13.Glossop JA, Dafforn TR, Roberts SGE. A conformational change in TFIIB is required for activator-mediated assembly of the preinitiation complex. Nucleic Acids Res. 2004;32:1829–1835. doi: 10.1093/nar/gkh504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Bushnell DA, Wang D, Guillermo C, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen HT, Hahn S. Mapping the location of TFIIB within the RNA polymerase II transcription preinitiation complex: a model for the structure of the PIC. Cell. 2004;119:169–180. doi: 10.1016/j.cell.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 16.Sun ZW, Hampsey M. Identification of the gene (SSU72/TFG1) encoding the largest subunit of transcription factor TFIIF as a suppressor of a TFIIB mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr Opin Genet Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilman B, Drullinger LF, Kugel JF, Goodrich JA. TATA-binding protein and transcription factor IIB induce transcript slipping during early transcription by RNA polymerase II. J Biol Chem. 2009;284:9093–9098. doi: 10.1074/jbc.M900019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho EJ, Buratowski S. Evidence that transcription IIB is required for a post-assembly step in transcription initiation. J Biol Chem. 1999;274:25807–25813. doi: 10.1074/jbc.274.36.25807. [DOI] [PubMed] [Google Scholar]
- 20.Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Fairley JA, Roberts SGE. Phosphorylation of TFIIB links transcription initiation and termination. Curr Biol. 2010;20:548–553. doi: 10.1016/j.cub.2010.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohkuma Y, Roeder RG. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex formation. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 23.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 24.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yochum GS, Rajaraman V, Cleland R, McWeeney S. Localization of TFIIB binding regions using serial analysis of chromatin occupancy. MBC Mol Biol. 2007;8:102. doi: 10.1186/1471-2199-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996;15:1933–1940. [PMC free article] [PubMed] [Google Scholar]
- 27.Ansari A, Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genetics. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 29.Kaderi BEI, Medler S, Raghunayakula S, Ansari A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J Biol Chem. 2009;284:25015–25025. doi: 10.1074/jbc.M109.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calvo O, Manley JL. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 2003;17:1321–1327. doi: 10.1101/gad.1093603. [DOI] [PubMed] [Google Scholar]
- 31.He XY, Khan AU, Cheng H. Functional interactions between the transcription and mRNA 3′end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes-Reyes M, Hampsey M. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol Cell Biol. 2007;27:926–936. doi: 10.1128/MCB.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvo O, Manley JL. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation and termination. Mol Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 34.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]