Abstract
In the course of evolution of multi-cellular eukaryotes, paralogs of general transcription factors and RNA polymerase subunits emerged. Paralogs of transcription factors and of the RPC32 subunit of RNA polymerase III play important roles in cell type- and promoter-specific transcription. Here we discuss their respective functions.
Key words: RNA polymerase III, TFIIIB, TFIIIC, PTF, TBP, RPC32, transcription
Three distinct DNA-dependent RNA polymerases (Pol I, Pol II and Pol III) have been identified in eukaryotes more than 40 years ago.1 The presence of several RNA polymerases in eukaryotes versus one single RNA polymerase in prokaryotes2 reflects increased complexity of the transcription systems that may have been necessary to accommodate the transcription of an increased number of genes. Such a gain in complexity may have allowed the evolution of novel genes or groups of genes that require the presence of more than one polymerase for individual regulation.
A further level of complexity has been added to transcription systems in archebacteria and in eukaryotic cells by the evolution of general transcription factors, replacing as an entity eubacterial sigma factors. In addition, in multicellular eukaryotes, including insects and vertebrates, paralogs of some of the general transcription factors have evolved that fulfill cell type or gene-specific functions. Furthermore, paralogs of RNA polymerase subunits arose during evolution. As a consequence, higher eukaryotes, including plants and vertebrates, contain more than three individual DNA-dependent RNA polymerases.3,4 Here, we review our current knowledge of the existence and function of paralogous components of general transcription machineries and particularly concentrate on components that have been shown to function in Pol III transcription or that may contribute to Pol III transcription under certain circumstances (Table 1).
Table 1.
Components of the general Saccharomyces cerevisiae Pol III transcription machinery and their paralogs in Drosophila
| Transcription factor/polymerase subunit | Orthologous and/or paralogous transcription factors | |
| Saccharomyces cerevisiae | Drosophila | Vertebrates |
| TFIIIC:TFC7 (τ55) | XP_002134843 and XP_001360915 in Dpp. | TFIIIC35 |
| XP_002021117 and XP_002017815 in Dp. | ||
| TFIIIB:BRF1 | BRF1 | BRF1, BRF2 |
| TBP | TBP, TRF1, TRF2 | TBP, TRF2, TRF3 |
| Pol III:RPC31 | RPC31 | POLR3G, RPC32α |
| (NP_788522.1 in Dm) | POLR3GL, RPC32β | |
The Dpp and Dp proteins XP_001360915 and XP_002017815 are 98% identical and 99% homologous. The proteins XP_002134843 and XP_002021117 are identical. Dp, Drosophila persimilis; Dpp, Drosophila pseudoobscura pseudoobscura; Dm, Drosophila melanogaster and vertebrates.
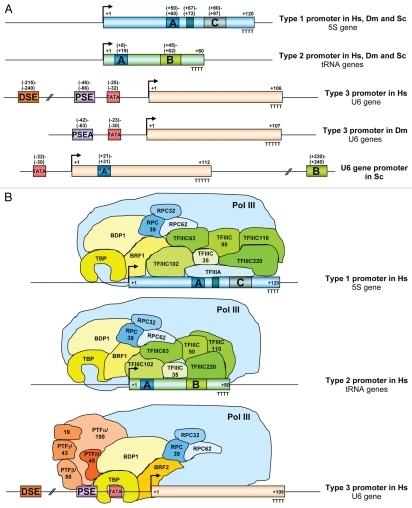
Pol III transcribes small untranslated RNAs that are required for essential cellular processes including the regulation of transcription (7SK RNA, Alu RNAs), RNA processing (U6 snRNA, RNase P RNA, RNase MRP RNA) and translation (5S rRNA, tRNAs). Distinct types of promoters (Fig. 1A) are recognized by promoter-specific sets of transcription factors (Fig. 1B). In mammalian cells, the gene-internal type 1 promoter is recognized by the gene-specific transcription factor TFIIIA, which in turn recruits TFIIIC, TFIIIB-β and finally Pol III itself to the 5S gene. Gene-internal type 2 promoters (e.g., tRNA-, Alu-, adenoviral VA1- and VA2-genes) are directly recognized by TFIIIC, allowing the recruitment of TFIIIB-β and subsequently of Pol III. Type 3 promoters (U6-, 7SK-, RNase P-, RNase MRP-genes) are recognized by the PSE-binding transcription factor (PTF; also known as SNAPc and PBP), followed by the recruitment of TFIIIB-α and of Pol III (Fig. 1B).5,6
Figure 1.
(A) RNA polymerase III promoter types in humans (Hs), Drosophila melanogaster (Dm) and Saccharomyces cerevisiae (Sc). Type 1 and 2 promoters are similar, but U6 promoters are different in the three species. The complete set of genes that utilize the distinct promoter types have been determined.5,6 (B) Promoters and transcription factors of the human (vertebrate) Pol III transcription machinery. Subunits of the PSE-binding transcription factor (PTF) are indicated by their PTF nomenclature and the molecular masses of the corresponding SNAPc subunits. SNAPc19 has not been reported in PTF.
Paralogs of TFIIIC Subunits
TFC7/t55/TFIIIC35-related factors.
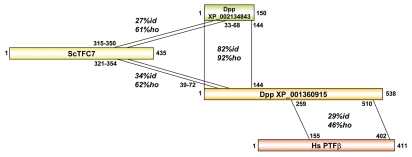
For more than 10 years, Saccharomyces cerevisiae (Sc) TFIIIC has been known to be composed of six subunits (TFC4, TFC1, TFC7, TFC8, TFC3 and TFC6), whereas only five subunits had been described in humans (TFIIIC102, 63, 90, 220 and 110; Fig. 1B). Recently, we identified and characterized the sixth subunit of human (Hs) TFIIIC (TFIIIC35; Fig. 1B), which is orthologous to Saccharomyces cerevisiae (Sc) TFC7 (τ55). We identified TFIIIC35 through a Psi-Blast search by using a 34 amino acid sequence of a yeast Schizosaccharomyces pombe (Sp) protein as a seed, which showed modest sequence conservation with amino acids 317–350 of ScTFC7.7 This 34 amino acid sequence also overlaps with the Pfam 10419:TFIIIC_subunit signature.8 In order to identify novel proteins that may represent paralogs of TFIIIC subunits, we now performed a Psi-Blast search (BLOSUM 62; exclude Saccharomyceta from search) with amino acids 321–364 of ScTFC7. Upon the second iteration, we identified two proteins in two distinct drosophilidae (XP_001360915.2 in Drosophila pseudoobscura pseudoobscura [Dpp] and XP_002017815.1 in Drosophila persimilis [Dp]) that exhibited 34% sequence identity and 62% sequence homology of amino acids 39 to 72 with the amino acids 321–354 of Sc TFC7 (Fig. 2). Surprisingly, amino acids 259 to 510 of the two Drosophila proteins contained a second signature that has been found to be conserved in PTFβ/SNAP50 subunits of PTF/SNAPc/PBP (29% identity and 46% homology to amino acids 155 to 402 of human PTFβ); pfam12251: zf-SNAP50_C; Fig. 2). The presence of a protein in Dp and in Dpp that contains regions of homology to both a subunit of TFIIIC and a subunit of PTF raises the possibility that at least one subunit of TFIIIC and of PTF may have evolved from a common ancestor protein. Interestingly, genomes of Dpp and Dp encode in addition a smaller protein of 150 amino acids (XP_002134843 [Dpp]; XP_002021117 [Dp]) that is highly related (82% identical and 92% homologous) to the N-terminal 144 amino acids of the two larger proteins (537 amino acids [Dp] or 538 amino acids [Dpp]) and that may represent the TFC7 subunit of TFIIIC in the respective Drosophila (Fig. 2). Importantly, the proteins of Dpp and Dp, containing the TFIIIC- and the zf-SNAP_C-signature, were the only proteins related to Drosophila melanogaster (Dm)PSEA-binding protein 49 kD, suggesting that these proteins participate in UsnRNA transcription in these species. Thus, these proteins may be both considered as orthologs of the Dm PSEA-binding protein 49 kD and as paralogs of TFC7.
Figure 2.
Schematic representation of the amino acid homology of a Drosophila pseudoobscura pseudoobscura protein (Genbank accession # XP_001360915) with ScTFC7, with HsPTFβ and with a Drosophila pseudoobscura pseudoobscura protein (Genbank accession # XP_002134843). The percentage of identical amino acids is indicated by “id” and of homologous amino acids by “ho.” The numbers of amino acids of the individual proteins, as well as the regions showing homology in between individual proteins are appropriately indicated. The Pfam signature 12251:zf-SNAP50_C is found in HsPTFβ (amino acids 201–407) and in Dpp XP_001360915 (amino acids 309–515). The Pfam signature 10419:TFIIIC_subunit is contained in ScTFC7 (amino acids 317–351) and the Dpp proteins XP_001360915 and XP_002134843 (amino acids 35–69 in both proteins).
Paralogs of TFIIIB Subunits
ScTFIIIB is composed of three subunits, the TATA-binding protein (TBP), TFIIB-related factor 1 (BRF1) and B double prime 1 (BDP1). Together with TFIIIA, TFIIIC and Pol III, these three components are required and sufficient for reconstituting transcription from all Pol III promoters in Sc.9 ScBRF1 is a paralog of TFIIB and it possesses a structure that is similar to that of TFIIB. It contains an N-terminal zinc ribbon and two direct imperfect repeats, each of which consists of 5 alpha-helices that together adopt a cyclin fold.10 In vertebrate cells, two distinct TFIIIB-activities have been described that both comprise TBP and BDP1, but that differ by either containing TFIIB-related factor 1 (BRF1; component of TFIIIB-β; transcription of type 1 and 2 promoters; Fig. 1B) or TFIIB-related factor 2 (BRF2; component of TFIIIB-α).6,11,12 TFIIIB-α is active in transcription of Pol III genes with gene regulatory elements that are entirely located upstream of the transcription initiation site (type 3 promoter; Fig. 1B) and the BRF2 component of TFIIIB-α has also been cross-linked to genes that contain a combination of gene-internal and gene-external promoter elements.13,14 The evolution of a second TFIIB-related factor at the emergence of vertebrates may have permitted or may have been tolerated by the co-evolution of a novel Pol III promoter type. With respect to the question of whether the evolution of BRF2 led to the co-evolution of a novel promoter type it is noteworthy to mention that promoter elements upstream of the transcription initiation site have also been identified for the U6 and 7SK genes in Drosophila melanogaster (Dm; Fig. 1A).15 Despite the gene regulatory elements being located upstream of the transcription initiation site, only one isoform of a DmTFIIB-related factor has been described. These results indicate that the evolution of upstream promoters in the Pol III transcription system, at least in insects, may not have been driven by the evolution of a second isoform of a TFIIB-related factor, although we cannot exclude that BRF2 sequences may have been lost in the course of evolution in these species. If ever the appearance of type 3 promoters may not be attributable to the evolution of BRF2 it may be asked whether BRF1 or BRF2 possess other gene- or cell type-specific functions. Today, we only know a single gene in vertebrates (coding for BC200 RNA in humans or the functionally analogous BC1 RNA in rodents) that is transcribed by Pol III and that shows neuronal-specific expression.5 ChIP sequencing and ChIP-on-CHIP experiments demonstrated that the BC200 gene is in physical contact with Pol III.13,16 ChIP sequencing further showed the presence of BDP1 at the BC200 genomic locus. Interestingly, however, neither BRF1, nor BRF2 could be detected at the BC200 gene locus.13,16 This could merely be a technical problem, but it could also indicate that transcription initiation of the BC200 gene may be independent of BRF1 and BRF2. In this case, another, hitherto unidentified protein may replace BRF1/BRF2 for transcription of the BC200 gene in neurons. Taken together, the emergence of BRF1 and BRF2 during evolution led to the appearance of two isoforms of TFIIIB with promoter-specific functions.
TBP-related factors.
TBP was once assumed to be a universal transcription factor.17 This point of view was well understandable at that time, because the discovery that TBP participates in transcription of all three RNA polymerases would not have been anticipated some years earlier and TBP was thought to be generally indispensable for transcription. However since then, several paralogs of TBP have been identified (TRF1; TRF2; TRF3) and their discovery indicated that transcription of certain genes may not depend on TBP.
Although first described as a cell type-specific factor, TRF1 turned later out to be widely expressed in Dm and to replace TBP for transcription by Pol III,18 which was confirmed by ChIP-on-CHIP analyses using affinity-purified anti-TRF1 antibodies and Drosophila genome tiling arrays.19 In addition to its essential functions in Dm Pol III transcription, TRF1 has also been implicated in the transcription of a small subset of Pol II genes.20 No ortholog of TRF1 could hitherto be identified in species other than insects.
Several years later, a second paralog of TBP was identified in mammals, Drosophila melanogaster, Caenorhabditis elegans and other metazoans.20 TRF2 was shown to be involved in transcription by Pol II,20 but it was demonstrated that recombinant HsTRF2 was inactive in Pol III transcription in vitro.21
The third and latest member of the TBP-family that has been identified is TRF3/TBP2. A gene encoding TRF3 was found in a variety of metazoans, including humans, mice, frogs and fish. Specific functions for TRF3 in differentiation of mouse myoblasts, in zebrafish hematopoiesis and in Xenopus or mouse oocytes have been suggested.20,22 TRF3/TBP2 knockout studies showed that mice had no apparent phenotype except females being sterile due to defective folliculogenesis.23 A possible involvement of TRF3/TBP2 in Pol III transcription has not yet been reported. However, the high degree of conservation between the C-terminal part of human TBP (residues 141–337) and TRF3 (residues 184–374; 92% identity and 95% homology) makes it likely that TRF3 may be functional in Pol III transcription. Moreover, residues in ScTBP that have been reported to be critical for the interaction with ScBRF1 (S261; D263; S282; E284; E286; L287; R299; V306) or with ScBdp1 (H277) have been conserved in both, HsTBP and HsTRF3/TBP2.24 In line with the possibility that TRF3/TBP2 may be able to replace TBP in Pol III transcription, it has been suggested that TRF3/TBP2 may be a TBP replacement factor in cells that contain low levels of TBP.22
Paralogs of RNA Polymerase III Subunits
Pol III is composed of 17 subunits. Five of these subunits are shared with Pol I and II, two with Pol I only (with paralogous subunits in Pol II) and another five subunits are paralogous to subunits of Pol I and Pol II. However, five subunits are Pol III-specific and no paralogous subunits have been identified in Pols I and II. Recently, it has been proposed that four of the five Pol III-specific subunits (POLR3E/HsRPC80, POLR3D/HsRPC53, POLR3C/HsRPC62, POLR3F/HsRPC39) exhibit sequence/structure homology to the heterodimeric general Pol II transcription factors TFIIE and TFIIF and that they may be considered as permanently recruited forms of these transcription factors.25,26 POLR3G/HsRPC32 is the only Pol III subunit for which no homologous polypeptide has been identified within the Pol I and Pol II transcription machineries.
RPC32-related factors.
As pointed out above, the emergence of three DNA-dependent RNA polymerases in eukaryotes may have allowed a more sophisticated regulation of individual sets of genes. Recently, two distinct isoforms of human Pol III have been reported, which may allow even further specialization for transcription of individual genes. The two isoforms of human Pol III differ at least in containing either the RPC32α (Pol IIIα) or the RPC32β (Pol IIIβ) subunit, and they exhibit isoform-specific expression patterns in distinct cell types. Northern blot analyses of multiple human tissues and cell lines demonstrated that Pol IIIβ is widely expressed, whereas Pol IIIα expression is only detectable in several lymphoma and leukemia cell lines. As a consequence, RPC32β-containing Pol IIIβ may be considered as the general form of Pol III, whereas RPC32α-containing Pol IIIα may only be required in a subset of cells and it may be important for sustaining cell type-specific functions. Analyses of RPC32α expression by RT-qPCR or western blot confirmed low RPC32α mRNA or protein expression levels in human embryonic IMR90 fibroblasts as well as increased expression levels during tumoral transformation, whereas the RPC32β mRNA expression pattern remained unchanged.4 Functional importance for RPC32α expression was demonstrated by RNAi-mediated knockdown of RPC32α expression. The suppression of RPC32α led to reduced colony formation of HeLa cells in soft-agar assays. Thus, the expression of RPC32α changes cellular characteristics and it will be important to determine the underlying molecular mechanisms of how RPC32α affects soft-agar growth. It is likely that Pol IIIα-mediated transcription of genes, which cannot be expressed by Pol IIIβ represents the molecular key to understanding the cellular functions of RPC32α (Pol IIIα). Overexpression of RPC32α in partially transformed human IMR90 fibroblasts strongly enhances the expression of the 7SK gene. However, it can currently not be assessed to which extent transcription of the 7SK gene by RPC32α (Pol IIIα) contributes to the changes in growth behavior that have been observed.4 7SK RNA participates in the regulation of Pol II transcription through alteration of P-TEFb function.27 However, since Pol IIIα-induced effects are likely to be restricted to a limited number of Pol II genes, rather than being due to a general deregulation of Pol II transcription,4 it seems plausible that Pol IIIα-dependent transcription of other RNAs may also contribute to RPC32α-induced induction or maintenance of cell transformation. The existence of such a gene or group of genes is likely, because the acquisition of a novel function for a duplicated gene product enhances its probability of being preserved.28 If ever such genes exist, does their transcription depend on known or novel promoter structures (types 1–3) and transcription factors (TFIIIA, TFIIIC, TFIIIB, PTF)? Today, these questions cannot be answered but will be of interest for future research. Furthermore, from the expression pattern of RPC32α,4 it may be inferred that Pol IIIα has a role in maintaining embryonic stem cells in an undifferentiated state. Therefore, it will be of interest for future research.
Furthermore, from the expression pattern of RPC32α,4 it may be inferred that Pol IIIα has a role in maintaining embryonic stem cells in an undifferentiated state. Therefore, it will be of interest to determine how RPC32α/Pol IIIα is involved in cellular differentiation and de-differentiation processes and whether the existence of two Pol III isoforms is somehow related to cell type- or stage-specific expression of both classical house-keeping29 and non-canonical regulatory30 Pol III genes.
Acknowledgements
This work has been supported by grants from the Conseil Regional d'Aquitaine and the European Regional Development Fund (to M.T.); by grants from the Agence Nationale de la Recherche (ANR) “REGPOLSTRESS” (to M.T.), the Ligue Contre le Cancer-Comites Gironde and Dordogne (to M.T.), the Italian Ministry of Education, University and Research (PRIN 2007 grant to G.D.), the AICCRE-Regione Emilia Romagna (to G.D.). C.P. was supported by a doctoral fellowship from the “Università Italo-Francese/Universite Franco-Italienne.”
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/13192
References
- 1.Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 2.Cramer P. Structure and function of RNA polymerase II. Adv Protein Chem. 2004;67:1–42. doi: 10.1016/S0065-3233(04)67001-X. [DOI] [PubMed] [Google Scholar]
- 3.Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, et al. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M, et al. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci USA. 2010;107:4176–4181. doi: 10.1073/pnas.0914980107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 7.Dumay-Odelot H, Marck C, Durrieu-Gaillard S, Lefebvre O, Jourdain S, Prochazkova M, et al. Identification, molecular cloning and characterization of the sixth subunit of human transcription factor TFIIIC. J Biol Chem. 2007;282:17179–17189. doi: 10.1074/jbc.M611542200. [DOI] [PubMed] [Google Scholar]
- 8.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiduschek EP, Kassavetis GA. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 10.Juo ZS, Kassavetis GA, Wang J, Geiduschek EP, Sigler PB. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature. 2003;422:534–539. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
- 11.Teichmann M, Seifart KH. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 1995;14:5974–5983. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teichmann M, Wang Z, Roeder RG. A stable complex of a novel transcription factor IIB- related factor, human TFIIIB50 and associated proteins mediate selective transcription by RNA polymerase III of genes with upstream promoter elements. Proc Natl Acad Sci USA. 2000;97:14200–14205. doi: 10.1073/pnas.97.26.14200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canella D, Praz V, Reina JH, Cousin P, Hernandez N. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res. 2010;20:710–721. doi: 10.1101/gr.101337.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen RC, Wang Y, Hardin SB, Stumph WE. The proximal sequence element (PSE) plays a major role in establishing the RNA polymerase specificity of Drosophila U-snRNA genes. Nucleic Acids Res. 1998;26:616–622. doi: 10.1093/nar/26.2.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 18.Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- 19.Isogai Y, Takada S, Tjian R, Keles S. Novel TRF1/BRF target genes revealed by genome-wide analysis of Drosophila Pol III transcription. EMBO J. 2007;26:79–89. doi: 10.1038/sj.emboj.7601448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reina JH, Hernandez N. On a roll for new TRF targets. Genes Dev. 2007;21:2855–2860. doi: 10.1101/gad.1623207. [DOI] [PubMed] [Google Scholar]
- 21.Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, et al. Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc Natl Acad Sci USA. 1999;96:13720–13725. doi: 10.1073/pnas.96.24.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akhtar W, Veenstra GJ. TBP2 is a substitute for TBP in Xenopus oocyte transcription. BMC Biol. 2009;7:45. doi: 10.1186/1741-7007-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gazdag E, Santenard A, Ziegler-Birling C, Altobelli G, Poch O, Tora L, et al. TBP2 is essential for germ cell development by regulating transcription and chromatin condensation in the oocyte. Genes Dev. 2009;23:2210–2223. doi: 10.1101/gad.535209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schröder O, Bryant GO, Geiduschek EP, Berk AJ, Kassavetis GA. A common site on TBP for transcription by RNA polymerases II and III. EMBO J. 2003;22:5115–5124. doi: 10.1093/emboj/cdg476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol. 2010;27:1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- 26.Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 28.Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- 29.Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castelnuovo M, Massone S, Tasso R, Fiorino G, Gatti M, Robello M, et al. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J. 2010;24:4033–4046. doi: 10.1096/fj.10-157032. [DOI] [PubMed] [Google Scholar]