Abstract
We recently identified CpG island promoters driving transcription of human telomeric repeat-containing RNA (TERRA). This discovery has shaped a new concept in telomere biology, where TERRA promoters and downstream telomeric sequences constitute autonomous genic units.
Key words: TERRA, telomeres, transcription, CpG-island promoters, DNMTs, heterochromatin
Telomeres are the heterochromatic nucleoprotein complexes located at the termini of linear eukaryotic chromosomes. They allow cells to distinguish between natural chromosome ends and DNA double-stranded breaks.1,2 Telomeres also set the lifespan of human somatic cells by triggering cellular senescence when they become “critically short” in the absence of lengthening mechanisms upon successive cell division cycles.1–3 In vertebrates, the DNA component of telomeres comprises variably long tracts of 5′-TTAGGG-3′/3′-AATCCC-5′ tandem repeats, with the G-rich strand extending beyond its complement to form a 3′ overhang, termed the G-overhang (Fig. 1A).1,2 The core protein component of mammalian telomeres is referred to as shelterin, and in humans it comprises the six polypeptides TRF1, TRF2, POT1, TPP1, TIN2 and hRap1. Shelterin proteins regulate telomere capping and telomere-length homeostasis.1,2
Figure 1.
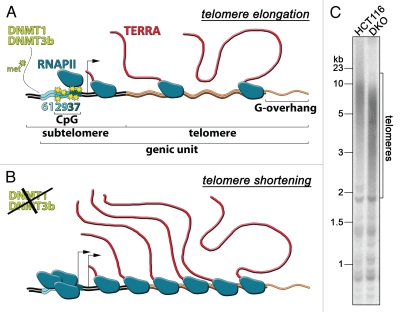
(A) Cartoon sketching of a telomeric genic unit composed of a subtelomeric promoter (61-29-37 repeats) and telomeric repeats. The arrow indicates the TERRA transcription start site. The yellow stars represent methyl groups added to promoter cytosines through concerted action of DNMT1 and DNMT3b. In these settings, telomere extension is favored. (B) Lack of DNMT1 and DNMT3b methyl transferase activities leads to increased telomere transcription, appearance of alternative TERRA start sites and telomere shortening. (C) Genomic DNA from HCT116 and DKO cells was digested with HinfI and RsaI, electrophoresed in 0.7% agarose gels and hybridized to radio-labeled telomeric probes according to standard protocols. Molecular weights are indicated on the left. Note that telomeres in DKO cells are shorter than in parental HCT116 cells.
The longstanding dogma that telomeres are transcriptionally silent genomic loci was overturned by the discovery that long non-coding (nc) RNA molecules named TERRA emanate from chromosome ends in several eukaryotes, including yeasts, birds, fishes, plants and mammals.4–8 Mammalian TERRA comprises telomeric UUAGGG repeats, and ranges in length from approximately 100 bases up to more than 9 kilobases (kb). Moreover, mammalian TERRA localizes preferentially to telomeres throughout the entire cell cycle, suggesting the existence of post-transcriptional mechanisms that regulate the positioning of TERRA at telomeres.4,5 Indeed, depletion of the human proteins UPF1, hEST1A/SMG6 and SMG1, which are effectors of the nonsense mediated mRNA decay (NMD) pathway,9 causes accumulation of telomere-bound TERRA without affecting its half-life or steady-state levels, implying that these factors actively displace TERRA from telomeres. Depletion of the same proteins also leads to stochastic loss of entire telomeric tracts.4 These observations suggest that an excess of chromatin-bound TERRA may compromise telomere integrity or that the telomeric DNA-damage induced by SMG protein depletions might increase TERRA binding to telomeres.4,10 In addition, recent studies show that the shelterin component TRF2 and several heterogeneous ribonucleoproteins (hnRNPs) interact with endogenous TERRA in mammalian protein extracts and promote TERRA association to telomeric heterochromatin in vivo.11,12
While TERRA-associated functions continue to elude researchers, discovery has progressed in TERRA biogenesis. The DNA-dependent RNA polymerase II (RNAPII) uses the C-rich telomeric strand as template to produce TERRA, as demonstrated by substantially decreased TERRA steady-state levels measured in mammalian cells treated with RNAPII inhibitors or in RNAPII-deficient yeast mutants.5,6,13 In addition, RNAPII was found to physically associate with mammalian telomeres in vivo, as well as with the shelterin component TRF1.5 Finally, at least a fraction of mammalian and yeast TERRA is 3′-end polyadenylated, as are the majority of RNAPII products.5,6,13 In budding yeast, the poly-A polymerase Pap1p promotes TERRA polyadenylation and pap1-deficient strains show markedly reduced TERRA levels, implying that poly-A tails could stabilize TERRA molecules.6
Discovery of Human TERRA Promoters
When analyzed, individual human TERRA transcripts contained both a telomeric and a subtelomeric RNA tract,4 suggesting the exciting possibility that TERRA transcription start sites and putative promoters could lie within subtelomeres. With this speculation in mind, we were intrigued by the peculiar sequence organization of the human subtelomeres of chromosomes Xq/Yq and 10q (originally named TelBam3.4 and TelSau2.0, respectively).14 These subtelomeres share a conserved repetitive region that comprises, in a centromere to telomere direction, a 61 bp repeat element, a 29 bp repeat element and a 37 bp repeat element, altogether referred to as 61-29-37 repeats.14,16 The 29 bp and the 37 bp repeats form a DNA island rich in CpG dinucleotides (CpG in Fig. 1A), a feature associated with many mammalian RNAPII promoter sequences.15 Bioinformatics analysis combined with in situ hybridization experiments showed that at least 20 human chromosome ends share 61-29-37 repeats, although different numbers of tandem repeats are present at different subtelomeres.14,16
We developed green fluorescent protein (GFP)-based promoter reporter assays using plasmids comprising progressive 5′ deletions of a 61-29-37 repeat-containing genomic tract from chromosome XqYq and found that the 29 bp and the 37 bp repeat elements alone are sufficient to drive reporter gene expression. In addition, 61-29-37 repeats are bound in vivo by phosphorylation-activated RNAPII. Finally, we found that most XqYq TERRA transcription start sites (TSSs) are located 27 nucleotides downstream of the last 37 bp repeat and about 250 bp upstream of the telomeric tract (Fig. 1A).16 Considering that TERRA molecules are up to more than 9 kb long, we concluded that at least some TERRA transcripts are mainly composed of UUA GGG RNA repeats and that transcription can proceed for several kb through the telomeric tract.
Methylation of cytosines within CpG dinucleotides, the only known methylation-based DNA modification occurring in the human genome, influences gene expression and nuclear architecture.17 CpG methylation is established and maintained by DNA-methyltransferases (DNMTs), and generally leads to transcriptional silencing of genes located within the methylated loci.17 Southern blot-based methylation analysis of genomic DNA from different cancerous and primary human cells revealed that 61-29-37 repeats are embedded within heavily methylated genomic loci, with telomerase-positive cancer cell lines displaying higher degrees of methylation as compared to telomerase-negative cancer cell lines and primary fibroblasts.16
We then used human colon carcinoma HCT116 cell lines knocked-out for either DNMT1 or DNMT3b alone or concomitantly deleted for both enzymes (double KO; DKO).18,19 While a single DNMT gene deletion did not substantially affect the methylation state of TERRA promoters, hypomethylation was observed in DKO cells. This was accompanied by a dramatic increase in cellular TERRA levels, by augmented binding of active RNAPII to TERRA promoter and telomeric DNA, and by appearance of alternative transcription start sites as compared to parental cells (Fig. 1B).16 These data strongly suggest that the methylation of 61-29-37 repeats represses RNAPII-dependent transcription of telomeres and that DNMT1 and DNMT3b cooperatively maintain TERRA promoter methylation and TERRA transcription start site usage, at least in HCT116 cells.
Interestingly, DKO cells have overall shorter telomeres than parental cells (Fig. 1C), indicating that loss of methylation at subtelomeric 61-29-37 promoters and increased TERRA cellular levels might inhibit telomere elongation. Similarly, it has been reported that DNMT3b-deficient cells derived from human patients affected by ICF (immunodeficiency, centromere instability and facial abnormalities type I) syndrome display markedly diminished subtelomeric CpG methylation, shorter telomeres and higher TERRA levels as compared to control cells from healthy individuals.20,21 An apparent contradiction to the effects exerted by DNMTs on TERRA and telomere length homeostasis in human cells is posed by the observations that mouse embryonic stem (ES) cells deficient for DNMT1 or for both DNMT3a and DNMT3b, although displaying hypomethylated subtelomeric CpG islands, have lower TERRA levels and longer telomeres as compared to ES cells derived from wild-type animals.5,22 It seems thus conceivable that DNMTs and CpG methylation might regulate TERRA and telomere length differently in mouse and human cells. It should also be pointed out that TERRA promoters have not yet been identified in mouse cells, leaving open the possibility that murine TERRA promoters are not regulated through CpG methylation.
Another example linking DNA methylation, TERRA expression and telomere stability is represented by the human ATRX (alpha thalassemia/mental retardation syndrome X-linked) protein, which belongs to the SWI2/SNF2 family of chromatin remodeling factors.23 Mutations in ATRX give rise to complex trait syndromes, at least in part due to impairment of ATRX-associated functions in gene transcription regulation.23 ATRX mutations also lead to changes in the CpG methylation pattern of several classes of highly repeated sequences, including subtelomeric repeats corresponding to TelBam3.4 sequences.24 Independent studies have shown that ATRX deficiencies induce accumulation of DNA damage markers at telomeres, as well as increased TERRA steady-state levels. Nevertheless, no obvious effect was observed on telomere length, possibly due to the short experimental time-courses used by the authors.25,26
Conclusions and Future Directions
The existence of regulated promoters dedicated to the transcription of TERRA from independent chromosome ends defines a novel scenario in telomere biology, where subtelomeric TERRA promoters, together with downstream telomeric tracts, constitute autonomous genic units (Fig. 1A).
The identified TERRA promoters could only be mapped to approximately half of the human subtelomeres, while TERRA transcription from chromosome ends apparently devoid of such promoters (for example Xp/Yp and 11q) has been previously documented.4 It will be essential to test whether TERRA transcription from 61-29-37 promoter-less chromosome ends is mediated by alternative promoter types, or rather by run-on transcription of telomeres from upstream genes as is the case in budding yeast mutants lacking Rat1p RNA exonuclease activity.6 Also, the identification of TERRA promoters in non-human organisms will clarify how extensively TERRA biogenesis pathways are conserved throughout evolution.
Another major challenge is the characterization of TERRA-associated functions. Increasing evidence indicates that the vast repertoire of nc transcripts produced by a cell might be involved in regulating epigenetic memory, dosage compensation, heterochromatin formation and gene expression.27 Indeed, transfection of short interfering RNA (siRNA) molecules directed against the UUAGGG repetitive TERRA sequence into human cancer cells induced downregulation of TERRA steady-state levels, diminished density at telomeres of di- and tri-methylated histone H3 lysine 9 (H3K9), telomeric DNA damage and cell cycle arrest.12 It is therefore tempting to speculate that TERRA plays fundamental roles in assuring telomere integrity by depositing and/or maintaining telomeric heterochromatin. Nevertheless, it remains to be carefully evaluated to which extent the scored phenotypes derive from TERRA downregulation rather than from DNA damage induced at telomeres by transfection of telomeric siRNA sequences.
As above mentioned, TERRA might also act as a negative regulator of telomere lengthening mechanisms, as suggested by correlative evidence derived from mammalian and yeast systems, including the telomere shortening observed in cells deficient for DNMTs (Fig. 1C and refs. 5, 6, 20 and 21). TERRA-like short RNA oligonucleotides are able to inhibit telomerase activity in vitro, by base-pairing with the template region of telomerase RNA moiety.5,28 Therefore, TERRA might repress telomerase-mediated extension of telomeres in vivo. It is also possible that the physical action of transcription of telomeres could influence telomere length, both in telomerase-positive and -negative cells, by stripping telomere length regulators off the telomeric sequence.
We believe that a mechanistic characterization of the regulatory circuits governing transcription from TERRA promoters will expand our understanding of TERRA functions and clarify if and how TERRA contributes to heterochromatin establishment and telomere integrity. One first effort to be undertaken is a comprehensive isolation of transcription factors (TFs) that specifically regulate TERRA transcription through direct binding to 61-29-37 repeats. Experimental functional impairment of such TFs will help to unravel TERRA-associated functions.
Landing on “TERRA,” the Latin noun for planet Earth, has marked the beginning of a new era in telomere biology. New avenues are now open towards the characterization of the complex connection between telomeres and crucial aspects of human biology such as cellular senescence, organismal aging and cancer development. In addition, TERRA de-regulation might at least in part contribute to the development of syndromes such as ICF and alpha thalassemia/mental retardation X-linked. Understanding how TERRA integrates into these phenomena might, in the long term, pave the way for the development of new therapeutic approaches.
Acknowledgements
We thank Alicia E. Smith and Vikram G. Panse for critical reading of the manuscript. C.M.A. laboratory is supported by funds from ETHZ (ETH-15 08-1 and ETH-03 08-3), the Swiss National Science Foundation (3100A0-120090 and PP00P3-123356), the European Research Council (BFTERRA) and Fondazione Cariplo (2008-2507). E.G.'s laboratory is supported by funds from Fondazione Cariplo (2008-2507) and Programmi di Ricerca di Rilevante Interesse Nazionale (PRIN 2008).
Abbreviations
- ATRX
alpha-thalassemia/mental retardation syndrome, X-linked
- bp
basepairs
- CpG
cytosine-phosphate-guanine
- DNMT
DNA methyltransferase
- DKO
double knock-out
- ES
embryonic stem
- GFP
green fluorescent protein
- H3K9
histone H3 lysine 9
- ICF
immunodeficiency, centromere instability and facial abnormalities syndrome
- kb
kilobase
- nc
non-coding
- NMD
nonsense mediated mRNA decay
- RNAPII
DNA-dependent RNA polymerase II
- siRNA
short interfering RNA
- TERRA
telomeric repeat-containing RNA
- TF
transcription factor
- TSS
transcription start site
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/13191
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeyapalan JC, Sedivy JM. Cellular senescence and organismal aging. Mech Ageing Dev. 2008;129:467–474. doi: 10.1016/j.mad.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 5.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 6.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 7.Solovei I, Gaginskaya ER, Macgregor HC. The arrangement and transcription of telomere DNA sequences at the ends of lampbrush chromosomes of birds. Chromosome Res. 1994;2:460–470. doi: 10.1007/BF01552869. [DOI] [PubMed] [Google Scholar]
- 8.Vrbsky J, Akimcheva S, Watson JM, Turner TL, Daxinger L, Vyskot B, et al. siRNA-mediated methylation of arabidopsis telomeres. PLoS Genet. 2010;6:1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res. 2008;122:194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- 11.Lopez de Silanes I, Stagno d'Alcontres M, Blasco MA. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nature Communications. 2010 doi: 10.1038/ncomms1032. [DOI] [PubMed] [Google Scholar]
- 12.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azzalin CM, Lingner J. Telomeres: the silence is broken. Cell Cycle. 2008;7:1161–1165. doi: 10.4161/cc.7.9.5836. [DOI] [PubMed] [Google Scholar]
- 14.Brown WR, MacKinnon PJ, Villasanté A, Spurr N, Buckle VJ, Dobson MJ. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 15.Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, et al. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–2194. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng X, Blumenthal RM. Coordinated chromatin control: structural and functional linkage of DNA and histone methylation. Biochemistry. 2010;49:2999–3008. doi: 10.1021/bi100213t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, et al. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 19.Rhee I, Bachman KE, Park BH, Jair KW, Yen RWC, Schuebel KE, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 20.Yehezkel S, Segev Y, Viegas-Péquignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 21.Deng Z, Campbell AE, Lieberman PM. TERRA, CpG methylation and telomere heterochromatin: lessons from ICF syndrome cells. Cell Cycle. 2010;9:69–74. doi: 10.4161/cc.9.1.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 23.Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- 24.Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 25.Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;1;38:5797–5806. doi: 10.1093/nar/gkq296. [DOI] [PMC free article] [PubMed] [Google Scholar]