Abstract
A DNA transposon integrated into -the genome of a primitive mammal some 200 million years ago and, millions of years later, it evolved an essential function in the common ancestor of all placental mammals. This protein, now named ZBED6, was recently discovered because a mutation disrupting one of its binding sites, in an intron of the IGF2 gene, makes pigs grow more muscle. These findings have revealed a new mechanism for regulating muscle growth as well as a novel transcription factor that appears to be of major importance for transcriptional regulation in placental mammals.
Key words: ZBED6, IGF2, transcriptional regulation, evolution, ChIP-seq, muscle growth
The long road towards the identification of the ZBED6 transcription factor started with the mapping of Quantitative Trait Loci (QTL) affecting multifactorial traits in pigs. A paternally expressed QTL with major effects on muscle growth, weight of heart and subcutaneous fat depth was identified in an intercross between the European wild boar and Large White domestic pigs.1 The QTL allele from the domestic pig alters the body composition towards a leaner phenotype (more muscle, less fat), but had no effect on birth weight, general growth, abdominal fat deposition or the weight of the liver.1 In a parallel study using an intercross between Piétrain and Large White pigs, Nezer et al.2 identified the same paternally expressed QTL affecting lean growth. The QTL was localized close to the telomere on the p arm of pig chromosome 2, which immediately revealed IGF2 as the prime positional candidate gene.1,2 The IGF2 gene encodes insulin-like growth factor 2, an important growth factor, and is one of the few paternally expressed genes in this chromosomal region.
This QTL was transformed to a QTN (quantitative trait nucleotide) when the causative mutation was identified as a single nucleotide substitution, a G to A transition, in IGF2 intron 3.3 The mutation is located in an evolutionary conserved CpG island and 16 bp surrounding the mutated site show 100% sequence conservation among 18 out of 18 placental mammals for which sequence data are available; based on combining data presented by Van Laere et al.3 and those available at the UCSC Genome Browser (www.genome.ucsc.edu/, July 4, 2010). The sequence is not conserved in other vertebrates, including platypus, suggesting that the sequence may represent a regulatory element unique to placental mammals.
The mutation was associated with a 3-fold higher IGF2 mRNA expression in postnatal skeletal muscle and higher expression in cardiac muscle, the two tissues where phenotypic effects had been documented. In contrast, no effect on IGF2 mRNA expression was detected in fetal tissues or in postnatal liver, a result consistent with the finding that the mutation has no effect on birth weight, despite IGF2 being a potent fetal growth factor, nor on body weight, weight of liver or circulating levels of IGF2 protein.1,3 Electrophoretic mobility shift assay (EMSA) showed that the mutation disrupts the interaction with an unknown nuclear factor and transient transfection assays using Luciferase reporters showed that the wild-type allele, but not the mutant allele, can recruit a nuclear factor that represses transcription from the endogenous IGF2 P3 promoter. Thus, the IGF2 QTN is a cis-acting regulatory mutation with a tissue- and temporal-specific action. This mutation has been strongly favored in commercial pig breeding programs the past 60 years, since the major breeding goal has been to increase lean growth. This is exactly what this mutation does, but without affecting fetal growth that could have negative effects on the litter size. The mutation has gone through a selective sweep and occurs at a very high allele frequency in major pig populations used for meat production.3,4
Bioinformatics analyses did not reveal any known transcription factor binding motif spanning the QTN. The identification of the unknown transcription factor binding the IGF2 site was therefore accomplished by oligonucleotide capture, using oligonucleotides corresponding to the wild-type and mutant sequences, followed by high-resolution mass spectrometry.5 Nuclear extracts from mouse C2C12 myoblasts labeled with the stable isotope labeling by amino acids in cell culture (SILAC) technology6 were used to identify proteins binding to the wild-type but not the mutant sequence. The protein showing the most significant enrichment with the wild-type oligonucleotide was derived from an open reading frame (ORF) comprising more than 900 codons located in intron 1 of Zc3h11a, a gene encoding a poorly characterized zinc-finger protein. The ORF had previously been annotated as an alternative splice form of Zc3h11a, but a bioinformatics analysis showed that it encodes a novel protein with no significant sequence similarity to the ZC3H11A protein. Recombinant expression of this ORF, combined with EMSA and supershift assays, confirmed that the encoded protein indeed binds oligonucleotides corresponding to the wild-type but not the mutant sequence.5 The identification of this protein as the nuclear factor binding the IGF2 site has been confirmed in a subsequent study by Butter et al.7 also using the SILAC technology.
The protein encoded by the intronic ORF contains two amino-terminal zinc-finger BED8 domains and we therefore decided to name the gene ZBED6, since there are five other annotated genes (ZBED1 to ZBED5) in the mammalian genome that contain at least one BED domain. In the carboxy-terminal, ZBED6 contains a hAT dimerization domain. The overall primary structure therefore places ZBED6 in the hAT transposase family; a family that contains active DNA transposons in many species (e.g., fruit fly and maize) from different kingdoms.9 ZBED6 is not unique in its being originated from a DNA transposon. In the initial analysis of the human genome, Lander et al.10 listed 43 genes as probably derived from DNA transposons. ZBED6 was one of these, but it had received no attention until our discovery that it encodes the nuclear factor binding the QTN site in pigs.
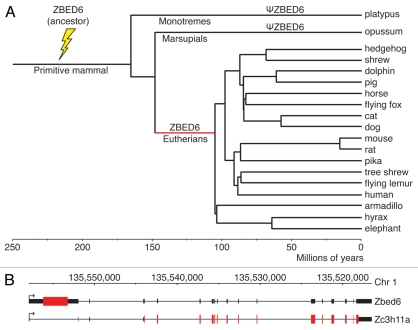
The ancestor of ZBED6 seems to have integrated in the genome of a primitive mammal, prior to the divergence of monotremes and other mammals, since non-functional remnants of ZBED6 were found both in the platypus and the opossum genomes, but not in the sauropsid lineage (birds and reptiles).5 ZBED6 is found at the same position in the genome in all placental mammals for which genome sequence data are presently available (more than 25 species) and the encoded protein sequence, in particular the DNA binding BED domains, is highly conserved within this group. These data demonstrate that ZBED6 is an innovation in placental mammals that has evolved subsequent to the split from the marsupial lineage, but prior to the radiation of different forms of placental mammals (Fig. 1A). Thus, ZBED6 may have played an essential evolutionary role in this group since it may have contributed to the evolution of a more complex regulatory network, a characteristic feature of advanced eukaryotes.
Figure 1.
Genomic integration of ZBED6. (A) Evolutionary tree highlighting the time period (in red) when ZBED6 must have evolved an essential function in a common ancestor of all placental mammals. The ancestor of ZBED6 probably integrated in a primitive mammal, prior to the divergence of monotremes and other mammals, but we cannot rule out the possibility that the integration took place even before that. The placental mammals included in this figure is a subset of those species for which full or partial genome sequence data are available and that represent different forms of placental mammals. The tree has been adapted from Murphy et al.27 and Warren et al.28 (B) Representation of full-length mRNA transcripts of the major splice forms of Zbed6 and Zc3h11a in C2C12 murine cells, as assessed by long-range RT-PCR (for details, see Suppl. 1). These sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers FR682664 and FR682665
Experiments to define the transcription start site (TSS) for ZBED6, including 5′RACE experiments as well as the location of RNA polymerase II binding sites and 5′-cap analysis gene expression tags (CAGE) both generated by the ENCODE project for the human ZBED6/ZC3H11A locus (see UCSC Genome Browser), suggested that ZBED6 hitch-hikes on the ZC3H11A promoter and is expressed as a composite transcript containing its own coding sequence spliced to the exons encoding ZC3H11A.5 Long-range RT-PCR analysis for detection of full-length transcripts performed recently in our lab confirms this. The expression of ZBED6 depends on intron retention; when the ZBED6 sequence is present it will be expressed, as it contains the first initiation codon in the transcript as well as a termination codon, while ZC3H11A is translated from transcripts in which the intron containing ZBED6 has been spliced out (Fig. 1B). In mice, Zbed6 is widely expressed both during development and in adult tissues.5 Furthermore, immunofluorescence analysis using an anti-ZBED6 antibody confirmed a nuclear localization and co-staining with an anti-nucleophosmin antibody revealed that ZBED6 is significantly enriched in the nucleolus. It is possible that the latter localization is relevant for the function of ZBED6, since the nucleolus has an important role for regulating cell growth and proliferation.11 The nucleolus is the site for either coordinated active ribosomal RNA gene (rDNA) transcription during cell growth or repression of rDNA transcription during lineage commitment.12 Recently, a number of transcriptional regulators including JHDM1B, MyoD, Myogenin, TLE1 and Runx2 have been shown to repress rDNA transcription in the nucleolus.13–15 MyoD and Myogenin-dependent transcriptional repression of rDNA transcription have been correlated to commitment of myogenesis of C2C12 cells.14 Interestingly, SRF (serum response factor), a major regulator of MyoD, is a putative target for ZBED6 in C2C12 cells.5
The functional significance of ZBED6 was further explored by RNAi-mediated silencing in mouse C2C12 myoblasts. These experiments, combined with transient transfection assays, confirmed that the binding of ZBED6 to the conserved element derived from the IGF2 intron is able to repress transcription from the endogenous pig IGF2 P3 promoter. Furthermore, Zbed6-silencing resulted in increased Igf2 expression and cell proliferation, and it promoted myotube formation. These phenotypic effects observed in C2C12 cells after Zbed6-silencing mirror remarkably well the phenotype observed in IGF2 mutant pigs. Interestingly, the effect on Igf2 expression was observed at day 6, whereas the effects on proliferation and myotube formation were apparent already at day 3, strongly implicating that the latter two effects were caused by ZBED6 affecting other downstream targets than Igf2.
Chromatin immunoprecipitation using an anti-ZBED6 antibody followed by next generation sequencing (ChIP-seq) was performed using C2C12 cells. The experiment revealed about 2,500 genomic regions with significant enrichment of sequence reads, and ∼1,200 genes had a putative ZBED6 binding site within 5 kb of defined TSS, while ∼300 sites were located far from known protein-coding genes.5 The CpG island corresponding to the mutated site in pig IGF2 was one of the highly enriched sites. Bioinformatics analysis of all the putative binding sites revealed the consensus ZBED6 binding motif as 5′-GCT CGC-3′, in perfect agreement with the wild-type sequence at the IGF2 locus in pigs. Furthermore, ZBED6 binding sites showed a strong association with CpG islands, consistent with the IGF2 site being located in a CpG island. As stated above, ZBED6 binding sites often occurred in the vicinity of TSS, but often downstream of the TSS. A gene ontology analysis revealed a highly significant overrepresentation of genes associated with development, regulation of biological processes and regulation of transcription among the ∼1,200 genes associated with ZBED6 binding sites. As many as 22% of these encoded other transcription factors. Furthermore, an Ingenuity pathway analysis (www.ingenuity.com) showed that the human homologs of putative ZBED6 targets in the mouse were significantly associated with a number of disorders, the most prominent ones being developmental disorders, cancer and cardiovascular disease.
An active DNA transposase physically interacts with DNA and scans the host genome in search for its target sequences. The transposase then uses a cut-and-paste mechanism to move the transposon to another location in the genome. Active DNA transposons can therefore be regarded as parasitic elements for its host and organisms have developed defense mechanisms to combat these hostile elements and their potentially deleterious effects. The host may evolve mechanisms for insulating transposon DNA from host transcription.8 In some instances the host might recruit the transposase for regulation of its own gene transcription and chromatin structure. Indeed, as reviewed by Matsukage et al.16 the Drosophila DREF protein of the hAT superfamily is found in complexes containing NURF chromatin remodeling factors and human DREF/ZBED1 interacts with MI-2 and PC2, a chromatin remodeling factor and a Polycomb group protein involved in heterochromatin formation, respectively.
To obtain further insight into the functional role of ZBED we investigated the association between ZBED6 binding sites and ChIP-seq data for p300, a histone acetyltransferase that is associated with enhancers and activates transcription through chromatin remodeling,17 and E2F1, which is found at promoter regions18 where it is associated with transcriptional repression through, for example, pRB, histone deacetylases and SWI2/SNF complexes. There was a marked association between ZBED6 and E2F1 binding sites, but not between ZBED6 and p300 sites (Fig. 2A and B). This indicates that ZBED6 binds proximal promoters rather than distal enhancers, consistent with our previous data.5 The co-localization of the ZBED6 and E2F1 binding sites may merely reflect the fact that both factors are strongly associated with CpG islands.5,18 The molecular mechanism for how ZBED6 affects gene transcription remains unknown, but a clearer picture would be obtained with the discovery of specific protein-interaction partners. At present we only know that ZBED6 acts as repressor at the IGF2 locus, and we cannot exclude that it acts as an activator at other loci.
Figure 2.
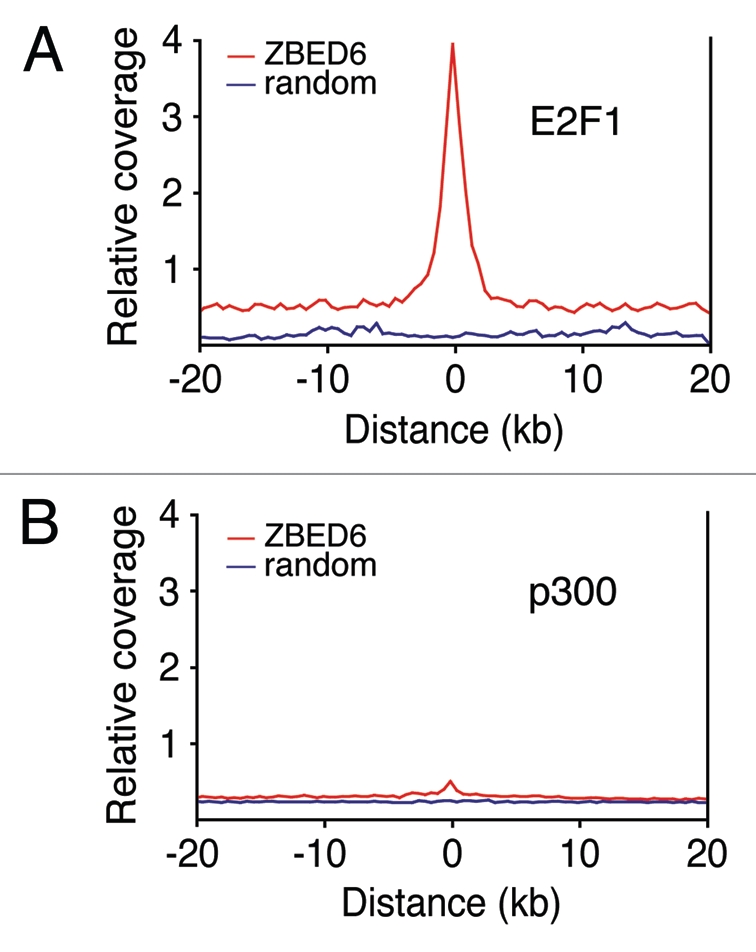
ChIP-seq data29 for E2F1 (A) and p300 (B) in mouse ES cells were realigned to mouse genome (mm9) to assess their correlation with ZBED6 binding sites identified in mouse C2C12 cells. We observed strong enrichment of E2F1 binding in the near vicinity of ZBED6 sites compared with random sites (A), but no significant association to p300 binding sites (B).
The initial characterization of ZBED6 suggests that this gene may be of considerable significance for evolutionary biology as well as for human medical genetics. Cis-regulatory mutations are considered to be a major source for phenotypic evolution.19–21 The importance of cis-acting regulatory mutations for shaping phenotypic diversity is strongly supported by molecular characterization of phenotypic traits in domestic animals,22 and the mutation at the IGF2 locus in pigs is one of the most prominent examples. The loss of only one of the ZBED6 binding sites in the pig, the one in IGF2, has a profound effect on body composition. Therefore, loss and gain of ZBED6 binding sites may have contributed to phenotypic evolution in placental mammals, in particular since ZBED6 is widely expressed during development. For instance, it would be interesting to perform ChIP-seq experiments using brain tissue from humans and chimpanzee to explore to which extent ZBED6 binding sites differ between these two species that show marked differences in cognitive functions23 but few differences at the protein level.19 A comparison between the ZBED6 binding sites established using mouse C2C12 cells and the homologous sites in the human genome indicated that about 10% of these are conserved in the human genome; the IGF2 site is one of them. This is consistent with the general observation of a dynamic evolution of transcription factor binding sites.24–26 However, ChIP-seq data from multiple species are required to better assess to which extent the localization of ZBED6 bindings sites are evolutionarily conserved.
We assume that loss-of-function mutations for ZBED6 have deleterious effects. This assumption is based on the high sequence conservation among placental mammals (the DNA binding domains are essentially 100% identical between species), the many putative downstream targets with important biological functions, and the marked phenotypic effects observed in pigs lacking only one of the ZBED6 binding sites. It is also possible that ZBED6 will show haploinsufficiency, so that loss-of-function mutations will be associated with rare dominant disorders in humans. Finally, mutations destroying or creating ZBED6 binding sites may have profound phenotypic effects, like the IGF2 mutation in pigs, and may therefore affect disease associations in humans.
The IGF2 locus in pigs represents one of the few examples where the causative mutation for a QTL has been identified as a single nucleotide substitution and where the mechanism of action for the mutation has been established. This achievement has revealed a new mechanism regulating muscle growth and heart development in placental mammals and led to the discovery of ZBED6, a previously unknown protein that apparently has a crucial function in placental mammals. We foresee that in the not too distant future the first examples of associations between ZBED6 and human disorders will be revealed.
Acknowledgements
We thank Shumaila Sayyab for help with bioinformatic analysis. The work on the ZBED6 transcription factor is funded by the Swedish Research Council and the Foundation for Strategic Research.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/13343
Supplementary Material
References
- 1.Jeon JT, Carlborg Ö, Törnsten A, Giuffra E, Amarger V, Chardon P, et al. A paternally expressed QTL affecting skeletal and cardiac muscle mass in pigs maps to the IGF2 locus. Nat Genet. 1999;21:13–15. doi: 10.1038/5938. [DOI] [PubMed] [Google Scholar]
- 2.Nezer C, Moreau L, Brouwers B, Coppieters W, Detilleux J, Hanset R, et al. An imprinted QTL with major effect on muscle mass and fat deposition maps to the IGF2 locus in pigs. Nat Genet. 1999;21:155–156. doi: 10.1038/5935. [DOI] [PubMed] [Google Scholar]
- 3.Van Laere AS, Nguyen M, Braunschweig M, Nezer C, Collette C, Moreau L, et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature. 2003;425:832–836. doi: 10.1038/nature02064. [DOI] [PubMed] [Google Scholar]
- 4.Ojeda A, Huang LS, Ren J, Angiolillo A, Cho IC, Soto H, et al. Selection in the making: a worldwide survey of haplotypic diversity around a causative mutation in porcine IGF2. Genetics. 2008;178:1639–1652. doi: 10.1534/genetics.107.084269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markljung E, Jiang L, Jaffe JD, Mikkelsen TS, Wallerman O, Larhammar M, et al. ZBED6, a novel transcription factor derived from a domesticated DNA transposon regulates IGF2 expression and muscle growth. PloS Biol. 2009;7:e1000256. doi: 10.1371/journal.pbio.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 7.Butter F, Kappei D, Buchholz F, Vermeulen M, Mann M. A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep. 2010;11:305–311. doi: 10.1038/embor.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind L, The BED finger. a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci. 2000;25:421–423. doi: 10.1016/s0968-0004(00)01620-0. [DOI] [PubMed] [Google Scholar]
- 9.Calvi BR, Hong TJ, Findley SD, Gelbart WM. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator and Tam3. Cell. 1991;66:465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- 10.International Human Genome Sequencing Consortium, author. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 11.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 12.Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci. 2005;30:87–96. doi: 10.1016/j.tibs.2004.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 14.Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, et al. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci USA. 2008;105:6632–6637. doi: 10.1073/pnas.0800970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali SA, Huang MB, Campbell PE, Roth WW, Campbell T, Khan M, et al. Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proc Natl Acad Sci USA. 2010;107:4165–4169. doi: 10.1073/pnas.1000620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsukage A, Hirose F, Yoo MA, Yamaguchi M. The DRE/DREF transcriptional regulatory system: a master key for cell proliferation. Biochim Biophys Acta. 2008;1779:81–89. doi: 10.1016/j.bbagrm.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 20.Haygood R, Babbitt CC, Fedrigo O, Wray GA. Contrasts between adaptive coding and noncoding changes during human evolution. Proc Natl Acad Sci USA. 2010;107:7853–7857. doi: 10.1073/pnas.0911249107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 22.Wright D, Boije H, Meadows JRS, Bed'hom B, Gourichon D, Vieaud A, et al. Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. PLoS Genet. 2009;5:e1000512. doi: 10.1371/journal.pgen.1000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fisher SE, Marcus GF. The eloquent ape: genes, brains and the evolution of language. Nat Rev Genet. 2006;7:9–20. doi: 10.1038/nrg1747. [DOI] [PubMed] [Google Scholar]
- 24.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meireles-Filho AC, Stark A. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr Opin Genet Dev. 2009;19:565–570. doi: 10.1016/j.gde.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grützner F, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.