Abstract
Bifidobacteria constitute an important component of the microbiota of the gastrointestinal tract of humans and other mammals. Various bifidobacterial strains are commercially exploited because of their perceived beneficial role in the maintenance of gut homeostasis. We have determined the response of B. breve UCC2003, a Gram-positive bacterium originally isolated from the nursling stool of a breast-fed infant, to several stresses (heat, osmotic, solvent) using transcriptomics, classical techniques and in silico analysis, as well as the transcriptional response of B. breve UCC2003 to oxidative stresses caused by the exposure to diamide, peroxide and environmental oxygen. Integration of these results allowed the formulation of a model for an interacting regulatory network for stress response in B. breve UCC2003, where HspR controls SOS response and the ClgR regulon, which in turn regulates and is regulated by HrcA. This model of an interacting regulatory network is believed to represent the paradigm for stress adaptation in bifidobacteria.
Key words: bifidobacterium, stress response, transcriptomics
Bifidobacteria represent one of the most common inhabitants of the animal gastrointestinal tract (GIT). From a phylogenetic perspective the genus Bifidobacterium belongs to the subclass Actinobacteridae of the phylum Actinobacteria of high GC-content, Gram-positive bacteria.1 In the human GIT their presence has been associated with beneficial effects, which have led to the widespread use of many bifidobacterial strains as components of health-promoting or probiotic foods.2
From production of the probiotic to storage and digestion, bifidobacteria, which are strict anaerobes, encounter a plethora of environmental stresses. In particular, exposure to oxygen or other oxygen-derived radicals, organic acids, osmotic stress, heat and cold have a major negative impact on bifidobacterial viability. To cope with these life-threatening challenges they can employ a variety of coping and survival strategies.
Like other bacteria, bifidobacteria are capable of synthesizing a particular set of proteins protecting the cell from damage caused by the accumulation of unfolded and/or misfolded proteins. Several of these protective proteins act as molecular chaperones, such as GroEL (Hsp60), DnaK (Hsp70) and ClpB (Hsp100), or as proteases, such as Lon, ClpP and ClpC, and play key roles in several posttranslational events to prevent protein denaturation, aggregation and misfolding.3,4
Because of their strict anaerobic life style, bifidobacteria are exquisitely sensitive to oxidative stress. Both proteins and DNA suffer from oxidative damage, and in order to alleviate this damage, bifidobacteria, like other bacteria, can upregulate systems that reduce reactive oxygen species, or reverse or remove the damage. To reduce organic peroxides it can upregulate peroxiredoxins, such as that specified by the AhpC/F system, which has also been shown to function as a chaperone in H. pylori.5 Additionally, it can protect DNA by upregulating the expression of the so-called DPS proteins, which sequester iron and protect DNA from damage by forming a crystalline structure around the DNA.6,7
In high GC-content, Gram-positive bacteria, regulation of the AhpC/F system is likely to occur by OxyR and a CRP/FNR type regulator.8,9 Members of these families are commonly associated with the response to changes in intracellular oxygen levels, redox-state or oxidative stresses.10,11 Bioinformatic-based analysis of the B. breve UCC2003 genome sequence has revealed the presence of just a single regulator-encoding gene annotated as a putative member of the Crp/FNR family. Furthermore, the UCC2003 genome contains putative Crp/FNR type motifs not only in the promoter regions of ahpC/F, but also in the promoter region of hrcA. Preliminary microarray analysis shows the upregulation of ahpC/F as well as hrcA under oxidative stress caused by the exposure of B. breve UCC2003 to diamide, peroxide and environmental oxygen. Additionally, other overlaps in upregulated genes between the heat shock response and oxidative response were detected such as clpB and clgR (Table 1).
Table 1.
Stress induced genes of B. breve UCC2003
| Locus | Product | Description | 42°C | 44°C | 47°C | 50°C | EtOH | Osmo | Diamide | H2O2 | Shaking |
| Bbr_0038 | AhpC | Alkyl hydroperoxide reductase | −2.0 | −3.2 | 0.3 | −10.2 | 3.9 | −1.3 | −1.1 | −1.1 | 4.1 |
| Bbr_0039 | AhpF | Flavoenzyme | −1.8 | −2.3 | −1.2 | −3.2 | 3.6 | 1.2 | 2.9 | 1.7 | 7.0 |
| Bbr_0076 | Hsp20 | Chaperone | 19.4 | 26.7 | 51.8 | 119.3 | 1.8 | 5.4 | 66.1 | 8.0 | 1.8 |
| Bbr_0124 | DnaK | Chaperone protein | 3.5 | 7.0 | 10.2 | 29.3 | 3.6 | 16.7 | 20.3 | 2.7 | 1.3 |
| Bbr_0125 | GrpE | GrpE protein | 3.2 | 6.7 | 7.8 | 22.8 | 3.6 | 13.1 | 15.5 | 2.2 | 1.2 |
| Bbr_0126 | DnaJ1 | Chaperone protein | 3.6 | 8.2 | 10.2 | 24.9 | 1.6 | 16.2 | 22.5 | 1.7 | 1.3 |
| Bbr_0127 | HspR | Regulator | 2.6 | 6.1 | 8.8 | 23.8 | 1.9 | 15.4 | 9.0 | 1.8 | −1.1 |
| Bbr_0216 | Modification methylase | −1.3 | 1.3 | 5.3 | 3.1 | −1.1 | 1.9 | 1.0 | 3.7 | 2.4 | |
| Bbr_0496 | MutY | DNA glycosylase | 1.1 | 2.3 | 2.5 | −1.7 | 3.9 | 1.7 | 1.9 | −2.4 | 2.1 |
| Bbr_0585 | ImpB | DNA repair | 1.3 | 4.3 | 33.0 | 18.3 | 1.9 | 4.7 | 1.2 | 6.3 | 2.3 |
| Bbr_0794 | ClpP1 | Protease | 1.0 | 1.2 | 1.0 | −1.3 | 3.4 | −2.3 | 1.0 | −1.1 | 1.1 |
| Bbr_0795 | ClpP2 | Protease | −1.1 | 1.2 | −1.4 | −3.5 | 3.3 | −1.5 | 0.9 | 1.3 | 1.1 |
| Bbr_1004 | HrcA | Regulator | 2.0 | 3.0 | 11.2 | 3.5 | 5.7 | 6.9 | 3.7 | 2.0 | 1.5 |
| Bbr_1005 | DnaJ2 | Chaperone protein | 1.4 | 2.4 | 2.0 | −4.4 | 3.8 | 1.5 | 1.8 | 1.9 | 0.9 |
| Bbr_1013 | RuvA | Holliday junction DNA helicase | 1.1 | 2.0 | 5.7 | 6.7 | 2.6 | 1.5 | 1.6 | 1.8 | 1.1 |
| Bbr_1014 | RuvB | Holliday junction DNA helicase | 1.0 | 1.9 | 2.2 | −2.4 | 2.4 | 1.0 | −2.2 | 2.2 | 1.4 |
| Bbr_1046 | Nfo | Endonuclease IV | 1.1 | 1.7 | 8.9 | 18.1 | 5.0 | 1.9 | 2.4 | 1.3 | 1.8 |
| Bbr_1171 | HrdB | principal sigma factor | 1.1 | 1.7 | 5.8 | 13.1 | 1.7 | 1.3 | 1.0 | −2.4 | −1.6 |
| Bbr_1180 | RecX | Recombination regulator | 1.2 | 2.9 | 17.7 | 10.9 | 3.5 | 2.6 | 5.7 | 3.6 | 2.1 |
| Bbr_1181 | RecA | Recombinase | 1.4 | 3.3 | 10.2 | 12.0 | 4.3 | 2.4 | 8.6 | 5.8 | 2.1 |
| Bbr_1182 | - | - | 1.0 | 1.7 | 9.1 | 66.9 | 3.6 | 1.2 | 4.7 | 2.9 | 1.1 |
| Bbr_1183 | ClgR | Regulator | 1.6 | 3.6 | 57.2 | 416.3 | 7.1 | 3.3 | 28.9 | 7.2 | 2.2 |
| Bbr_1271 | LexA | Regulator | 1.1 | 1.9 | 2.8 | 7.0 | 3.4 | 6.8 | 1.7 | 6.2 | 3.6 |
| Bbr_1356 | ClpC | Protease | −1.1 | 1.2 | −1.2 | −2.7 | 2.4 | −1.6 | −1.8 | 1.0 | 1.1 |
| Bbr_1364 | GroEL | Chaperone protein | 2.1 | 3.6 | 3.8 | 1.1 | 3.8 | 1.1 | 1.5 | 2.1 | 1.4 |
| Bbr_1668 | GroES | Chaperone protein | 1.5 | 2.3 | 3.3 | 3.3 | 3.3 | 1.8 | 4.1 | 3.3 | 1.2 |
| Bbr_1781 | ClpB | Chaperone protein | 4.2 | 12.2 | 27.3 | 94.6 | 4.4 | 8.0 | 36.0 | 13.1 | 2.2 |
The respective stresses are given, 42°–50°, 1 hour exposure to the temperature given; EtOH, 1 hour exposure to 8% ethanol; Osmo, 1 hour exposure to 0.5 M NaCl; Diamide, 1 hour exposure to 8 mM diamide; H2O2, 1 hour exposure to 1.2 mM H2O2; Shaking, 1 hour exposure to environmental oxygen by shaking at 200 rpm. Micro array fold upregulation is given as compared to an unstressed B. breve UCC2003 culture.
To alleviate DNA damage inflicted by these environmental stresses the SOS system is activated.12 Under normal conditions, expression of genes involved in DNA damage repair is very low as they are under tight negative control. Repression of these genes is managed by the LexA regulator, which binds to the SOS box in the promoter region of these genes.13 Activation of the SOS genes occurs when DNA is damaged, which can happen as a result of heat or oxidative stress. This DNA damage results in single stranded DNA (ssDNA) accumulation at the site of (stalled) replication, which in turn activates RecA. LexA autocleavage is promoted by this activated form of RecA and derepression of the SOS genes takes place as a consequence.14
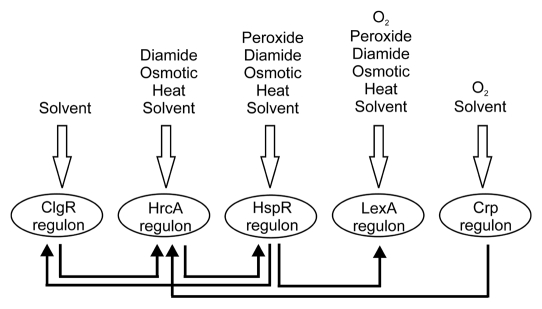
In B. breve UCC2003, SOS response occurs when the cells are subjected to both heat shock and oxidative stresses (Table 1).15 Induction of RecA expression is probably also under the control of HspR under heat shock conditions because of the proximity of recA and recX to the HspR-controlled clgR and Bbr_1182 genes, which form a transcriptional unit. Induction of recA expression by HspR could consequently activate LexA autoproteolysis and result in upregulation of the SOS genes (Fig. 1) in addition to other targets of HspR.
Figure 1.
Simplified representation of the stress gene regulatory network of B. breve UCC2003. Stress conditions that (partly) activate the regulons are indicated above; lines indicate a (putative) interaction between the regulons.
Stress responses in bifidobacteria are thus governed by a complex interactive network of regulators (ClgR, HspR, HrcA, LexA and others). A model for the stress regulatory network of B. breve UCC2003 was constructed using the current knowledge obtained from the research described in the manuscript and other available literature data.15 A model including the oxidative stress conditions is given in Figure 1. Although simplified, this model shows the complexity of stress gene regulation in B. breve UCC2003. Connections between the various regulons occur at different levels, indicative of complex interactions. The high level of conservation of the regulons and binding sites between various sequenced bifidobacterial genomes suggest that this model generally applies to all representatives of the Bifidobacterium genus.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/11477
References
- 1.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leahy SC, Higgins DG, Fitzgerald GF, van Sinderen D. Getting better with bifidobacteria. J Appl Microbiol. 2005;98:1303–1315. doi: 10.1111/j.1365-2672.2005.02600.x. [DOI] [PubMed] [Google Scholar]
- 3.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- 4.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 5.Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci USA. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kauko A, Haataja S, Pulliainen AT, Finne J, Papageorgiou AC. Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation. J Mol Biol. 2004;338:547–558. doi: 10.1016/j.jmb.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Wolf SG, Frenkiel D, Arad T, Finkel SE, Kolter R, Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 8.Hahn JS, Oh SY, Roe JH. Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2) J Bacteriol. 2002;184:5214–5222. doi: 10.1128/JB.184.19.5214-5222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rickman L, Scott C, Hunt DM, Hutchinson T, Menendez MC, Whalan R, et al. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56:1274–1286. doi: 10.1111/j.1365-2958.2005.04609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korner H, Sofia HJ, Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 11.Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 12.Erill I, Campoy S, Barbe J. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev. 2007;31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 13.Cheo DL, Bayles KW, Yasbin RE. Cloning and characterization of DNA damage-inducible promoter regions from Bacillus subtilis. J Bacteriol. 1991;173:1696–1703. doi: 10.1128/jb.173.5.1696-1703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 15.Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J Bacteriol. 2009;191:7039–7049. doi: 10.1128/JB.00897-09. [DOI] [PMC free article] [PubMed] [Google Scholar]