Abstract
Aquaporin-1 (AQP1) water channel protein expression is increased by hypertonic stress. The contribution of changes in protein stability to hypertonic induction of AQP1 have not been described. Incubation of BALB/c fibroblasts spontaneously expressing AQP1 with proteasome inhibitors increased AQP1 expression, suggesting basal proteasome-dependent degradation of the protein. Degradation by the proteasome is thought to be triggered by polyubiquitination of a target protein. To determine whether AQP1 is ubiquitinated, immunoprecipitation with anti-AQP1 antibodies was performed, and the resultant samples were probed by protein immunoblot for the presence of ubiquitin. Immunoblots demonstrated ubiquitination of AQP1 under control conditions that increased after treatment with proteasome inhibitors (MG132, lactacystin). Exposure of cells to hypertonic medium for as little as 4 h decreased ubiquitination of AQP1, an effect that persisted through 24 h in hypertonic medium. Using metabolic labeling with [35S]methionine, the half-life of AQP1 protein under isotonic conditions was found to be <4 h. AQP1 protein half-life was markedly increased by exposure of cells to hypertonic medium. These observations provide evidence that aquaporins are a target for ubiquitination and proteasome-dependent degradation. Additionally, these studies demonstrate that reduced protein ubiquitination and increased protein stability lead to increased levels of AQP1 expression during hypertonic stress.
Aquaporin-1 (AQP1) is a water channel protein expressed in many tissues, whose regulation is known to be complex (1). Age- and tissue-specific patterns of AQP1 expression have been shown for kidney, lung, brain, and eye (2, 3). Corticosteroids induce AQP1 in rat lung in vivo (3) and an erythroleukemia cell line in vitro (4); the AQP1 proximal promoter contains classical glucorticoid response elements to which activity has been localized. AQP1 expression is induced by hypertonic stress in kidney cells lines (5, 6), as well as in BALB/c fibroblasts in culture and in rat in vivo (L.K., unpublished observations). A potential role for posttranslational events in the regulation of AQP1 expression has not been described.
It is increasingly apparent that posttranslational events play an active role in determining the level of protein expression in a cell. Two major protein degradative pathways are known to operate in mammalian cells, the 26S proteasome (7), and cysteine proteases, including cytoplasmic calcium-dependent calpains (8) and lysosomal acidic cathepsins (9). The 76-aa protein ubiquitin plays a key role in proteasome-mediated protein degradation, because polyubiquitination of a target protein is a necessary signal for processing by the proteasome (7). A limited number of membrane transport proteins have been demonstrated to be ubiquitinated (10). Ubiquitination has not been shown for any member of the aquaporin family of proteins.
The studies reported here examine the role of ubiquitination and protein degradation in hypertonic induction of AQP1 expression. We demonstrate that AQP1 is ubiquitinated and degraded by the proteasome. Additionally, we observed that hypertonic stress decreases AQP1 ubiquitination and increases AQP1 protein stability, thereby contributing to overall protein induction. These findings provide an example of ubiquitination of an aquaporin water channel protein. Additionally, these studies demonstrate a mechanism for induction of a membrane protein in response to hypertonic stress.
Materials and Methods
Materials.
Polyclonal, affinity-purified rabbit antibody to AQP1 reactive with the 4-kDa carboxyl-terminal domain has been described (11). Monoclonal anti-ubiquitin antibody was purchased from Santa Cruz Biotechnology. Horseradish peroxidase-coupled secondary antibodies were from Amersham Pharmacia. ALLM, calpeptin, MG132, and lactacystin (solubilized in dimethyl sulfoxide), and ubiquitin aldehyde (solubilized in water) were purchased from Calbiochem-Novabiochem. Cycloheximide (solubilized in ethanol), chloroquine, and N-ethylmaleimide (NEM) (solubilized in dimethyl sulfoxide) were purchased from Sigma. Electrophoresis agents were from Bio-Rad, and the bicinchoninic acid (BCA) protein assay kit was from Pierce.
Cell Culture and Harvest.
BALB/c fibroblasts, which spontaneously express AQP1, were cultured in MEM supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, 40 units/ml penicillin, and 40 μg/ml streptomycin (Life Technologies, Grand Island, NY) at 37°C in 5% CO2. Where indicated, medium was made hypertonic by addition of 200 milliosmol (mosmol) sorbitol. For protein degradation studies, inhibitors were added to isotonic medium at the specified concentration for the duration of the experiment. At the time of harvest, cells were washed with ice-cold PBS, scraped, pelleted (10,000 × g, 5 min, 4°C), and frozen on dry ice. Cell pellets were resuspended in ice-cold homogenization buffer [7.5 mM sodium phosphate buffer/1 mM NaN3/1 mM EDTA/0.25 M sucrose/Complete Protease Inhibitor mixture tablet (Roche Diagnostics)], and subjected to an additional cycle of freeze-thawing. To limit deubiquitination during processing, 1 μg/ml ubiquitin aldehyde (12) and 5 mM NEM were included in the homogenization buffer. Cell lysate protein concentrations were determined by the BCA assay, using BSA as standard.
SDS/PAGE and Western Blotting.
Cell lysates or immunoprecipitated proteins were subjected to SDS/PAGE using 12% acrylamide gels (anti-AQP1 immunoblots) or 8% acrylamide gels (anti-ubiquitin immunoblots) and transferred to poly(vinylidene difluoride) membrane for protein immunoblotting as described (13). Blots were visualized by enhanced chemiluminescence (Amersham Pharmacia) and autoradiography (Kodak). Relative band intensities were determined by densitometry using MACBAS bioimaging analyzer (version 2.5; Fuji Photo Film).
Immunoprecipitation.
At the indicated times, cells were washed once with ice-cold PBS then resuspended in solubilization buffer [20 mM Tris, pH 8.0/200 mM NaCl/1% (vol/vol) Triton X-100/1% (wt/vol) deoxycholate/0.1% (wt/vol) SDS/5 mM EDTA/5 mM EGTA/Complete Protease Inhibitor mixture tablet]. For ubiquitination studies, NEM (5 mM; lysate preparation and wash) and 1 μg/ml ubiquitin aldehyde (lysate preparation only) were added to the solubilization buffer to limit deubiquitination. Insoluble material was removed by centrifugation (16,000 × g; 5 min; 4°C). The lysate was precleared by incubation with 50% protein A-Sepharose (Sigma) slurry, centrifuged (3,000 × g; 2 min; 4°C), and then incubated with 1 μg of rabbit IgG (Sigma) or polyclonal AQP1 at 4°C for a minimum of 1 h. Protein A-Sepharose, previously blocked with 10% (wt/vol) BSA, was added, and the lysate was incubated at 4°C overnight. The immunoprecipitated samples were recovered after centrifugation (3,000 × g; 2 min; 4°C), washed four times for 5 min each with solubilization buffer, then washed once for 5 min with salt-free wash buffer [20 mM Tris, pH 8.0/1% (vol/vol) Triton X-100/5 mM EDTA/5 mM EGTA]. Immunoprecipitated proteins were eluted from the Sepharose by incubation in elution buffer [10 mM Tris, pH 6.8/3% (wt/vol) SDS/10% (vol/vol) glycerol/0.01% (wt/vol) bromophenol blue/0.5 M DTT] for 30 min at 37°C. The eluted proteins were subjected to SDS/PAGE as described above.
Metabolic Labeling.
Studies of AQP1 protein stability were conducted in subconfluent monolayers that were serum- and methionine-starved for 1 h at 37°C, then labeled in glutamine- and methionine-free DMEM supplemented with 4.0 mM l-glutamine and 0.4 mCi/ml l-[35S]methionine (Amersham Pharmacia) for 1 h. Cells were then washed and incubated in MEM supplemented with 2 mM l-glutamine. For studies of protein stability in hypertonic conditions, 200 mosmol of sorbitol was added to medium beginning 4 h before the labeling procedure and remained to the end of the procedure. Samples were harvested for immunoprecipitation of AQP1 at different intervals after labeling. After SDS/PAGE (12%), gels were dried and exposed to radiographic film for 2 days.
Statistics.
Densitometric analysis of protein immunoblots and autoradiography from metabolic labeling studies are expressed as mean ± SEM for each group. Unpaired t tests were performed to assess the effect of different interventions.
Results
AQP1 Protein Expression Is Increased by Proteasome Inhibitors.
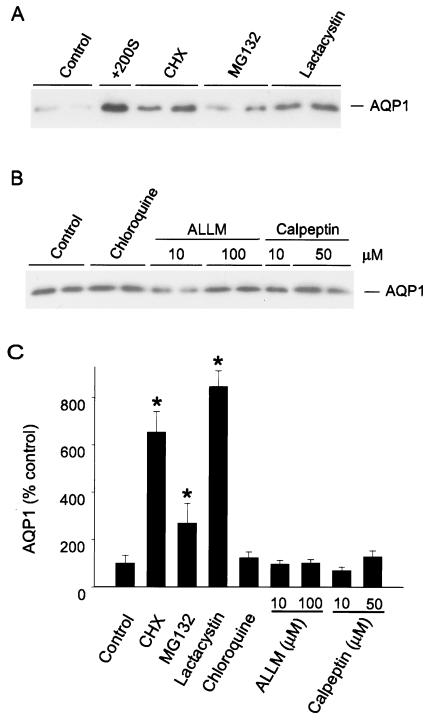
We examined a role for protein degradation in AQP1 expression using immunoblots of total cell lysates probed with anti-AQP1 antibodies. As seen in Fig. 1A, low levels of spontaneous AQP1 expression were evident in BALB/c fibroblasts at baseline. Incubation in hypertonic medium for 8 h markedly increased expression, consistent with our earlier work. Exposure of cells to the translation elongation inhibitor cycloheximide (8 h) increased AQP1 protein expression by 6-fold, a counterintuitive result suggesting that, in addition to blocking translation, cycloheximide may interfere with some aspect of protein degradation (Fig. 1 A and C). Fibroblasts then were treated with the well-characterized proteasome inhibitors MG132 or lactacystin (14) for 8 h, and immunoblots of cell lysates were probed for AQP1 expression. AQP1 protein increased 3-fold (MG132) to 8-fold (lactacystin) after proteasome inhibition (Fig. 1 A and C). In contrast, incubation of cells with a lysosomal inhibitor (chloroquine) or calpain inhibitors (ALLM, calpeptin) for 8 h had no significant effect on AQP1 protein expression (Fig. 1 B and C). These data suggest that proteasomes, but not other degradative pathways, participate in regulating basal AQP1 expression.
Figure 1.
AQP1 expression in the presence of protein processing inhibitors. (A) BALB/c fibroblasts were treated with the indicated reagent for 8 h, then harvested for protein immunoblot with affinity-purified anti-AQP1 antibody (anti-AQP1). +200S, 200 mosmol sorbitol; CHX, cycloheximide (10 μM); MG132 (10 μM) and lactacystin (10 μM), proteasome inhibitors). Autoradiographic exposure was brief (2 s) so that induced signal would be in linear range for densitometry. (B) Protein immunoblots of fibroblasts incubated with a lysosome inhibitor (chloroquine; 100 μM) or calpain inhibitors (ALLM, calpeptin) at the designated concentrations for 8 h; probed with anti-AQP1. Control signal appears increased compared with A because of longer exposure of autoradiograph (10 s). (C) Protein immunoblots from cells treated as in A and B (n ≥ 4 per group) were analyzed by densitometry and are represented as percentage of control expression (mean ± SEM; *, P < 0.05 vs. control).
AQP1 Is Ubiquitinated.
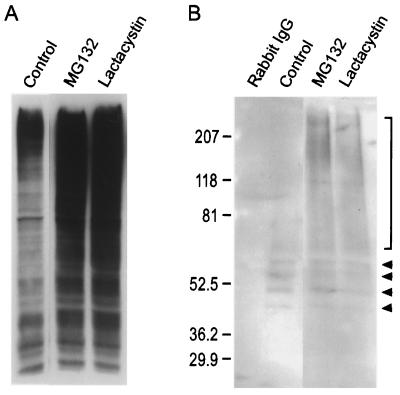
We sought evidence that AQP1 can be ubiquitinated, because signaling for proteasome-mediated protein degradation generally is initiated by covalent attachment of multiple ubiquitin moieties to the target protein (7). The reactivity of anti-ubiquitin antibodies in our system was determined with protein immunoblots of total cell lysates. These revealed a diffuse banding pattern under control conditions (Fig. 2A), as reported (15). Incubation of cells with proteasome inhibitors markedly increased the diffuse ubiquitin labeling (Fig. 2A), consistent with the expected action of these agents to allow accumulation of ubiquitinated proteins (14). To look for ubiquitination of AQP1, we performed immunoprecipitations with anti-AQP1 antibodies and probed for coimmunoprecipitation of ubiquitin (Fig. 2B). No labeling of ubiquitin was detected after immunoprecipitation with rabbit IgG. After immunoprecipitations with anti-AQP1 antibodies, a discrete ladder of bands was detected, consistent with polyubiquitination. The lowest molecular mass band migrated at approximately 44 kDa, consistent with addition of 2 ubiquitin molecules (approximately 7.5–8 kDa per molecule) to the 28-kDa AQP1 monomer. A series of higher molecular mass bands increased by 7.5–8 kDa, suggesting serial addition of ubiquitin molecules (Fig. 2B; arrowheads). After incubation of cells with MG132 or lactacystin and anti-AQP1 immunoprecipitations, discrete bands of appropriate molecular mass representing ubiquitinated AQP1 were again evident (Fig. 2B). Additionally, a higher molecular mass “smear” of ubiquitinated bands also was observed (bracket in Fig. 2B), as described (15, 16). These studies demonstrate that, under basal conditions, AQP1 is ubiquitinated.
Figure 2.
Ubiquitination of AQP1. (A) Fibroblasts were incubated in control medium or in the presence of proteasome inhibitors (MG132, lactacystin) for 8 h, and protein immunoblots of total cell lysates were probed with anti-ubiquitin antibodies. (B) Fibroblasts were incubated in control medium or with proteasome inhibitors, and AQP1 was immunoprecipitated. Rabbit IgG was used as the immunoprecipitation control. Immunoblots were probed with anti-ubiquitin antibodies. Arrowheads mark the position of discrete bands from approximately 44 to 65 kDa, consistent with addition of different numbers of ubiquitin moieties (approximately 7.5–8 kDa per ubiquitin) to the 28-kDa AQP1 monomer, beginning with an ubiquitin dimer at the lowest arrowhead. The bracket denotes higher molecular mass ubiquitinated products.
Hypertonic Stress Reduces Ubiquitination of AQP1.
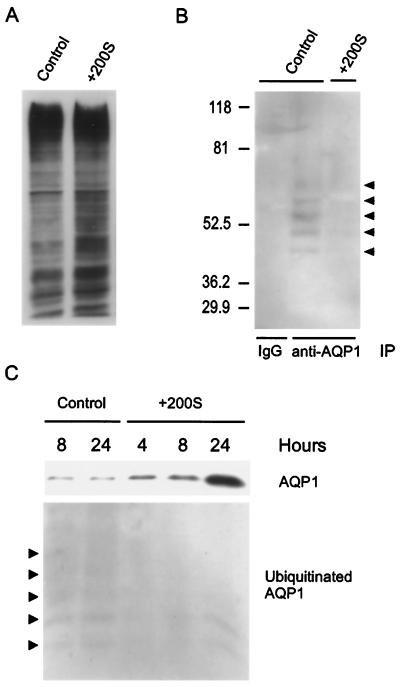
We examined the ubiquitination of AQP1 under isotonic and hypertonic conditions. First, immunoblots of total cell lysates from cells grown in isotonic or hypertonic conditions (8 h) were probed with anti-ubiquitin antibodies and revealed that total cellular ubiquitination increased slightly in response to hypertonic stress (Fig. 3A). To evaluate whether ubiquitination of AQP1 was altered by hypertonic stress, cells were incubated in isotonic or hypertonic medium for 8 h, and anti-AQP1 immunoprecipitates were probed for coimmunoprecipitation of ubiquitin (Fig. 3B). As above, no ubiquitinated AQP1 was detected after immunoprecipitation with rabbit IgG, and discrete bands representing ubiquitinated AQP1 were identified in anti-AQP1 immunoprecipitates under isotonic conditions (arrowheads, Fig. 3B). In contrast, labeling of ubiquitinated AQP1 was markedly reduced after incubation of cells in hypertonic medium (Fig. 3B). The decrease in ubiquitinated AQP1 was evident within 4 h after exposure to hypertonic medium and persisted through 24 h of hypertonic stress (Fig. 3C), although the amount of ubiquitinated AQP1 had partially returned toward baseline levels at that point. Immunoblots of these samples examined for AQP1 expression revealed that the reduction in ubiquitinated AQP1 occurred in parallel with an increase in total AQP1 protein expression (Fig. 3C).
Figure 3.
Effects of hypertonic stress on ubiquitination of AQP1. (A) Fibroblasts were incubated in isotonic or hypertonic medium for 8 h, and protein immunoblots of total cell lysates were probed with anti-ubiquitin antibodies. (B) Fibroblasts were incubated in isotonic (control) or hypertonic medium (+200S) for 8 h, and immunoprecipitation with protein A-Sepharose was performed by using rabbit (IgG) or anti-AQP1 antibodies (anti-AQP1). Protein immunoblots of the immunoprecipitates were probed with anti-ubiquitin antibodies. Arrowheads mark the position of discrete bands of ubiquitinated AQP1. (C) Cells were incubated in isotonic (control) or hypertonic medium (+200S) for the designated times, AQP1 was immunoprecipitated as described, and immunoblots were probed with anti-AQP1 (Upper) or anti-ubiquitin antibodies (Lower). Arrowheads mark discrete bands labeled by anti-ubiquitin antibodies.
Hypertonic Stress Increases AQP1 Protein Half-Life.
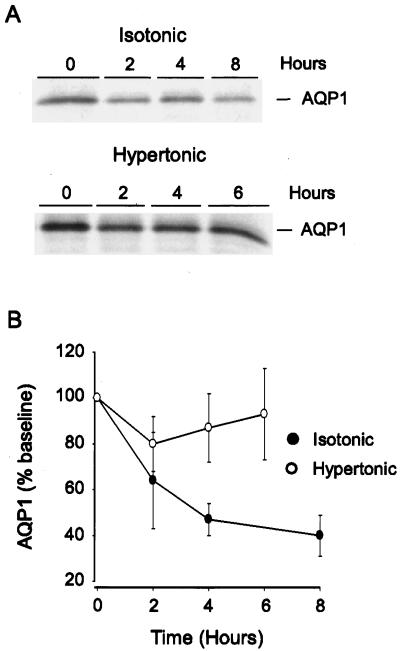
If the decrease in ubiquitination of AQP1 that occurred with hypertonic stress contributes to accumulation of the protein, the protein half-life should be altered. To explore this, fibroblasts were incubated in isotonic or hypertonic medium, then metabolically labeled with [35S]methionine as described. Anti-AQP1 immunoprecipitations were performed at different intervals after metabolic labeling, and dried SDS-PAGE gels of the immunoprecipitates were examined for expression of AQP1 by autoradiography (Fig. 4A). Under isotonic conditions, [35S]methionine-labeled AQP1 had decreased to <50% of the baseline value within 4 h (Fig. 4B; n > 4 per group). After incubation of cells in hypertonic medium for 4 h, AQP1 protein stability was markedly increased, with >80% of the labeled protein still remaining 6 h after metabolic labeling.
Figure 4.
AQP1 protein stability in isotonic and hypertonic conditions. (A) Cells were incubated in isotonic or hypertonic medium for 4 h, then metabolic labeling with [35S]methionine was performed as described. Samples were harvested at the designated times for immunoprecipitation of AQP1 as described. Samples were resolved on SDS/PAGE gels, and dried gels were exposed to x-ray film. (B) Samples treated as in A were analyzed by densitometry and are expressed as percentage of time 0 (n > 4 per group).
Discussion
Cellular responses to osmotic stress continue to be the focus of intense investigation, due in large part to the significant role they play routinely in the physiology of the kidney, and intermittently in the brain and airways of the lung. Despite an overall decrease in total DNA synthesis, RNA transcription, and protein synthesis (17), a limited number of genes are up-regulated in response to hypertonic stress (18, 19), although the number may be as high as 3% of the genome in Saccharomyces (20). Transcriptional activation of genes whose products participate in transport or synthesis of compatible osmolytes (e.g., betaine, sodium myo-inositol) in response to hyperosmolar stress is well established (21, 22). Signaling through mitogen-activated protein kinases has been implicated in hypertonic induction of the betaine transporter and sodium myo-inositol cotransporter (23, 24), AQP5 (25), and AQP1 (L.K., unpublished observations). Our findings for AQP1 in this report provide evidence that changes in ubiquitination and protein stability in response to hypertonic stress can contribute to induction of a membrane protein.
The ubiquitin-proteasome pathway plays an important role in a broad range of cellular events (7); however, little data are available on the potential role of this pathway in osmotic stress. Dictyostelium responds to hyperosmotic conditions by increasing both total ubiquitination and protein degradation of cellular proteins (26). Woo et al. (27) demonstrated that, in hypertonic (but not isotonic) conditions, proteasome inhibitors interfere with nuclear translocation of the tonicity-responsive enhancer binding protein, an osmoregulatory transcription factor, although the target for presumed ubiquitination and proteasome-dependent degradation is not known.
Processing by the ubiquitin-proteasome pathway has been demonstrated for a substantial number of cytosolic and membrane receptor proteins; however, few mammalian membrane transport proteins are known to be ubiquitinated (10). The cystic fibrosis transmembrane conductance regulator (CFTR) is ubiquitinated and rapidly degraded by the proteasome (28, 29), a process that may occur cotranslationally (30). The α- and γ-subunits of the epithelial Na channel (ENaC) are ubiquitinated, then degraded by either the proteasome or the lysosome (31). Specific mutations in ENaC subunits interfere with normal ubiquitination and protein degradation and may contribute to the pathogenesis of Liddle's syndrome (32). Ubiquitination of α-subunit isoforms of the Na, K-ATPase also has been shown, although the degradation pathway has not been described (33).
We demonstrate that AQP1 is ubiquitinated with a half-life of <4 h in basal conditions. Within 4 h of exposure to hypertonic medium, AQP1 ubiquitination decreased and AQP1 protein stability markedly increased. The decrease in ubiquitinated AQP1 in hypertonic medium was still evident after 24 h. This decrease was relatively selective, because ubiquitination of total cell lysates increased slightly in hypertonic conditions, as was described in Dictyostelium (26). The reduction in ubiquitination and increase in protein stability, previously undescribed in hypertonic stress, functions to facilitate protein induction at a time when the general pressure on the cell is to reduce protein synthesis (17).
The epithelium of the proximal tubule and descending thin limb, and endothelium of the descending vasa recta of the kidney are sites of AQP1 expression where tissue osmolality is normally high (34). Endothelial cells in the subepithelial vascular plexus around the airways also abundantly express AQP1 (3). The extent to which interstitial osmolality at that site can be elevated has not been determined; however, increased osmolality in the surface liquid of adjacent airways can trigger an increase in microvascular blood flow and fluid transudation, presumably as a means of replenishing airway water (35, 36). As was shown for AQP5 (25), AQP1 is induced in vivo in rat when the animals become hyperosmolar (L.K., unpublished observation). Each of these observations suggests that hypertonic induction of AQP1 is physiologically relevant, although the precise role for AQP1 in the cellular response to hypertonic stress is the subject of ongoing investigation. The studies reported here provide a mechanism for up-regulation of AQP1 that will likely extend to other members of the aquaporin family, as well as to other pathophysiologic conditions in which aquaporin expression is altered.
Acknowledgments
We thank Drs. Antony Rosen and Cecile Pickart for helpful discussions. We also thank Drs. Mark Knepper and Joseph Handler for critical review of the manuscript. This work was supported by National Institutes of Health Grants HL33991, HL48268, EY11239 (to P.A.), and HL03797 (to L.S.K.).
Abbreviations
- AQP1
aquaporin-1
- NEM
N-ethylmaleimide
- mosmol
milliosmol
References
- 1.King L S, Yasui M, Agre P. Mol Med Today. 2000;6:60–65. doi: 10.1016/s1357-4310(99)01636-6. [DOI] [PubMed] [Google Scholar]
- 2.Bondy C, Chin E, Smith B L, Preston G M, Agre P. Proc Natl Acad Sci USA. 1993;90:4500–4504. doi: 10.1073/pnas.90.10.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King L S, Nielsen S, Agre P. J Clin Invest. 1996;97:2183–2191. doi: 10.1172/JCI118659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moon C, King L S, Agre P. Am J Physiol. 1997;273:C1562–C1570. doi: 10.1152/ajpcell.1997.273.5.C1562. [DOI] [PubMed] [Google Scholar]
- 5.Jenq W, Mathieson I M, Ihara W, Ramirez G. Biochem Biophys Res Commun. 1998;245:804–809. doi: 10.1006/bbrc.1998.8518. [DOI] [PubMed] [Google Scholar]
- 6.Jenq W, Cooper D R, Bittle P, Ramirez G. Biochem Biophys Res Commun. 1999;256:240–248. doi: 10.1006/bbrc.1999.0306. [DOI] [PubMed] [Google Scholar]
- 7.Ciechanover A, Orian A, Schwartz A L. J Cell Biochem Suppl. 2000;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Molinari M, Carafoli E. J Membr Biol. 1997;156:1–8. doi: 10.1007/s002329900181. [DOI] [PubMed] [Google Scholar]
- 9.McGrath M E. Annu Rev Biophys Biomol Struct. 1999;28:181–204. doi: 10.1146/annurev.biophys.28.1.181. [DOI] [PubMed] [Google Scholar]
- 10.Hicke L. Trends Cell Biol. 1999;9:107–112. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 11.Smith B L, Agre P. J Biol Chem. 1991;266:6407–6415. [PubMed] [Google Scholar]
- 12.Hershko A, Rose I A. Proc Natl Acad Sci USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis J Q, Bennett V. J Biol Chem. 1984;259:1874–1881. [PubMed] [Google Scholar]
- 14.Lee D H, Goldberg A L. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 15.Mimnaugh E G, Bonvini P, Neckers L. Electrophoresis. 1999;20:418–428. doi: 10.1002/(SICI)1522-2683(19990201)20:2<418::AID-ELPS418>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.Hicke L, Riezman H. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 17.Cohen D M, Wasserman J C, Gullans S R. Am J Physiol. 1991;261:C594–C601. doi: 10.1152/ajpcell.1991.261.4.C594. [DOI] [PubMed] [Google Scholar]
- 18.Burg M B, Kwon E D, Kultz D. FASEB J. 1996;10:1598–1606. doi: 10.1096/fasebj.10.14.9002551. [DOI] [PubMed] [Google Scholar]
- 19.Blomberg A. Electrophoresis. 1997;18:1429–1440. doi: 10.1002/elps.1150180818. [DOI] [PubMed] [Google Scholar]
- 20.Rep M, Krantz M, Thevelein J M, Hohmann S. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- 21.Uchida S, Yamauchi A, Preston A S, Kwon H M, Handler J S. J Clin Invest. 1993;91:1604–1607. doi: 10.1172/JCI116367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamauchi A, Uchida S, Preston A S, Kwon H M, Handler J S. Am J Physiol. 1993;264:F20–F23. doi: 10.1152/ajprenal.1993.264.1.F20. [DOI] [PubMed] [Google Scholar]
- 23.Sheikh-Hamad D, Di Mari J, Suki W N, Safirstein R, Watts B A, Rouse D. J Biol Chem. 1998;273:1832–1837. doi: 10.1074/jbc.273.3.1832. [DOI] [PubMed] [Google Scholar]
- 24.Denkert C, Warskulat U, Hensel F, Haussinger D. Arch Biochem Biophys. 1998;354:172–180. doi: 10.1006/abbi.1998.0661. [DOI] [PubMed] [Google Scholar]
- 25.Hoffert J, Leitch V, Agre P, King L S. J Biol Chem. 2000;275:9070–9077. doi: 10.1074/jbc.275.12.9070. [DOI] [PubMed] [Google Scholar]
- 26.Zischka H, Oehme F, Pintsch T, Ott A, Keller H, Kellerman J, Schuster S C. EMBO J. 1999;18:4241–4249. doi: 10.1093/emboj/18.15.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woo S K, Maouyo D, Handler J S, Kwon H M. Am J Physiol. 2000;278:C323–C330. doi: 10.1152/ajpcell.2000.278.2.C323. [DOI] [PubMed] [Google Scholar]
- 28.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 29.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 30.Sato S, Ward C L, Kopito R R. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- 31.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staub O, Hugues A, Plant P, Ishikawa T, Kanelis V, Saleki R, Horisberger J-D, Schild L, Rotin D. Kidney Int. 2000;57:809–815. doi: 10.1046/j.1523-1755.2000.00919.x. [DOI] [PubMed] [Google Scholar]
- 33.Coppi M V, Guidotti G. FEBS Lett. 1997;405:281–284. doi: 10.1016/s0014-5793(97)00182-8. [DOI] [PubMed] [Google Scholar]
- 34.Knepper M A, Wade J B, Terris J. Kidney Int. 1996;49:1712–1717. doi: 10.1038/ki.1996.253. [DOI] [PubMed] [Google Scholar]
- 35.Prazma J, Coleman C C, Schockley W W, Boucher R. J Appl Physiol. 1994;76:2275–2280. doi: 10.1152/jappl.1994.76.6.2275. [DOI] [PubMed] [Google Scholar]
- 36.Matsui H, Davis C W, Tarran R, Boucher R C. J Clin Invest. 2000;105:1419–1427. doi: 10.1172/JCI4546. [DOI] [PMC free article] [PubMed] [Google Scholar]