Abstract
A diverse population of bacteria, archaea and fungi, collectively known as the microbiota, abounds within the gastrointestinal tract of the mammalian host. This microbial population makes many important contributions to host physiology through inter-kingdom signalling and by providing nutrients that have both local and systemic effects. In a healthy state the overall host-microbial interaction is symbiotic; however, a growing number of diseases have been associated with a dysregulated microbiota. To avoid these consequences, the host exerts substantial effort to maintain proper regulation of the microbiota with respect to localization and composition. Although important to maintaining microbial balance, the host immune response can also be the cause of a disrupted microbiota, contributing to disease severity. Here, we discuss the role of the host in both maintaining and disrupting a balanced gastrointestinal microbiota.
Key words: microbiota, gut mucosal defense, microbial detection, dysbiosis, microbial regulation
Introduction
In return for a hospitable environment and provision of nutrients, the gastrointestinal microbiota provides the host with many major advantages. The capacity of the microbiota in mediating a diverse set of beneficial roles for the host is vast,1 with metabolic activity equal to that of the liver. Roles include the synthesis of vitamins, the degradation of xenobiotics, the metabolism of bile and host hormones,2 immune development and competitive exclusion of pathogens.3,4 The realization that the microbiota contribute substantially to metabolic activities of the host and directly influence mammalian phenotype has resulted in terms such as ‘ecological development’ and the definition of mammals as ‘superorganisms’.5 The effects of the microbiota can be observed systemically. By comparing the plasma metabolic profiles of germ-free and conventional mice, it was found that numerous circulating metabolites were only present when the mice were colonized and metabolites that were common to germ-free and conventional mice differed significantly in their abundance.6 Furthermore, shifts in metabolic profiles have also been correlated to the composition of enteric bacteria.7
Over the past decade imbalances and changes in the composition of the gastrointestinal microbiota have been linked to numerous diseases including allergy, cancer, diabetes, obesity, inflammatory bowel diseases and enteric infection.8–11 The effort to “mine” the microbiota for therapeutic targets has created the need to understand the ecology and function of the gut microbiota. While recent studies have revealed new insights into the membership of the microbial community and how the microbiota regulates host physiology, little attention has been focused on characterizing how the host controls and regulates the microbiota and its composition. Although poorly understood, it is known that the host exerts substantial effort to regulate this population through the secretion of adaptive and innate immune proteins into the intestinal lumen including antimicrobial peptides, intelectin, resistin and IgA.12 In addition, the host genotype is one of the most important determinants in shaping the composition of the microbiota.13
When the host fails to appropriately regulate the gastrointestinal microbiota, the host can become increasingly susceptible to opportunistic and pathogenic infections, often leading to sepsis. Furthermore, recent studies have shown that disruptions in the microbiota that correlate to disease are sometimes the result of the host immune response, as in the case of Crohn disease (CD)14 and in Salmonella induced gastroenteritis.4 There is evidence that numerous components of the adaptive and innate immune system are important in this regulation of the microbiota, however, it is not clear if certain components are more important than others. While the focus of this review will be the host factors that regulate changes in the microbiota, it must be acknowledged that the ability of the host to do so is dependent on microbial activation.
Role of the Microbiota in Shaping Host Response
The microbiota plays a pivotal role in the organization of the immune system from birth. In recent years, the use of germ-free mice has provided critical evidence towards the essential role of the enteric bacteria on immune function. For instance, the four major structures that make up the gut associated lymphoid tissue including, mesenteric lymph nodes (MLN), peyers patches, cryptopatches and isolated lymphoid follicles, are all poorly developed in germ free mice.15 However, colonization with a single bacterial species has been sufficient to reverse these defects.15,16
Recently, there has been a great deal of evidence presented, which suggests that the gut microbiota are important in shaping and directing the host immune response. Various groups have demonstrated a role for the microbiota in intestinal epithelial homeostasis, angiogenesis and the development of the gut's innate and adaptive immune systems.15,17–20 Microbial colonization after birth is essential for the generation of intestinal T helper (Th) 17 cells as well as the production of IL–17 by these cells.21 They also showed that Th17 differentiation was particularly directed by a segmented filamentous bacteria (SFB).22 A second study found that treatment of mice with SFB or Clostridia promote the development of intraepithelial lymphocytes in small and large intestines.23 Finally, germ-free or mono-colonized mice do not develop oral tolerance to ovalbumin compared to wild type controls, which suggests a breakdown in the induction of antigen specific regulatory T cells (Treg).24 The use of probiotics to protect against enterocolitis, diabetes, allergies and IBD further substantiate the importance of continual regulation by the gut microbiota.25 In fact, a recent paper showed that mice treated with Lactobacillus paracasei NCC2461 resulted in increased Th1 responses.26 These are only a few of the studies that describe the regulation of the immune system by the gut microbiota, but they provide evidence that our immune responses are dependent on the microbiota within our guts.24 The microbiota are not only important in influencing the development of the immune system and intestinal structure. Continuous stimulation of the mucosal immune system is also required to maintain host mucosal defences. These will be discussed below, particularly with reference to mucus and antimicrobial secretion.
Defining a Balanced Microbiota
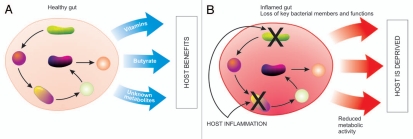
Before delving into how the host maintains a ‘balanced’ microbiota, we must first indicate our limited understanding of what a balanced microbiota is. Studies have characterized the colonization of the gut microbiota after birth and have observed differences in microbial communities depending on various factors such as, environment, mode of birth, diet and administration of antibiotics.27,28 No matter the diet or geographical location, the human gut is consistently colonized by the same bacterial phyla and only a single division of archaea. This limited phyla diversity indicates that only certain divisions have evolved close associations with the human gut.29 However, there is large inter-individual variation at the species level.30,31 Composition of the microbiota is also affected by many factors, including diet, parents, siblings, sex partners and interactions within the microbial community.32 Recent estimates based on 16S ribosomal RNA (rRNA) libraries from human fecal samples suggest that 800–1,000 different bacterial species and more than 7,000 strains inhabit the gastrointestinal tract.33 The application of 2nd generation sequencing approaches, such as 454 pyrosequencing of 16S rRNA have revealed as many as 3,300 unique operational taxonomic units (OTU) in a single individual.34 Despite this inter-individual variation, efforts at defining a core microbiota have been met with limited success. Although no OTU (defined at 98% identity) could be found in a set of 17 individuals, a group of 66 OTUs was defined as the microbial core based on presence in at least 50% of individuals. While these OTUs represented only 2.1% of all OTUs found, they represented 35.8% of the sequences in the study.31 This core microbiota has representation from three main phyla (Firmicutes, Bacteroidetes and Actinobacteria) and is dominated by the families Ruminococcaceae, Lachnospiraceae, Eubacteriaceae and Bacteroidaceae.31 Core members of the microbiota may be used as indicator species for the overall health of the population. In cases such as CD, there is a disappearance of core members of the microbiota that coincide with reduced production of butyrate as well as increased concentrations of bile acids.35 When inflammation disrupts the core microbiota the host is deprived of important functions that the microbiota provides (depicted in Fig. 1).
Figure 1.
Disrupting microbial function. (A) In a healthy gut bacteria work synergistically, with each bacteria being an integral link in the production of metabolites that are ultimately used by the host, such as vitamins and fatty acids. (B) Host inflammation targets certain groups of bacteria resulting in a loss of core members and core functions, therefore reduced metabolic activity and a deprived host.
One of the defining features of a balanced microbiota is that it is serving its key functional roles upon which the host has become dependent through co-evolution. It may therefore be of value to examine the microbiota functionally rather than based on community composition. Studying how host factors affect the functions of the microbiota through metagenomics, metabolomics or metaproteomics, rather than through a microbiomic approach may give greater insight. This is highlighted by the fact that a population of individuals with large variations in the dominant phyla had very little variation in microbial gene function,30 as shown by metagenomic analysis. Therefore, even with variations at a species level the same set of core functions could be performed. Deviations in the core microbiome at a functional level have been associated with obesity,30 and likely have implications for other diseases. Metabolomic studies taking advantage of techniques including gas chromatography-mass spectrometry (GC-MS), nuclear magnetic resonance (NMR) 1H Spectroscopy and Ion cyclotron resonance Fourier transform mass spectrometry (ICR-FT/MS) have revealed the complexity of the metabolic contributions of the microbiota.6,36 By combining microbiomic and metabolomic studies, certain populations within the microbiota have been tied to specific metabolic changes.7,35
Evidence for a Role for the Host
While dietary intake of the host is a major determining factor of microbial composition, it would appear that bacterial species that colonize are especially suited to the physiological characteristics of the host.37,38 One of the most compelling demonstrations of the host's role in shaping the composition of the microbiota was through the reciprocal transplantation of gut microbiota between mice and zebrafish.37 It was found that when the microbiota from a mouse, which is dominated by Bacteroidetes and Firmicutes, was introduced to a germ-free zebrafish, the relative abundances of community members shifted to resemble the normal microbial composition of the zebrafish, which is dominated by Proteobacteria.37 The reverse transplantation from the zebrafish to the mouse demonstrated the same phenomenon.
While there is variation between animal species, there is also variation between individuals. The microbial composition of each individual is unique, varying in composition just like that of blood chemistries.39 However, it is relatively stable over time indicating that the unique physiology of each host is important in dictating microbial composition. The fact that the microbiota of related individuals are more similar than that of unrelated individuals sharing the same diet and environment indicates the importance of host genotype.13 A more recent study using clone libraries and pyrosequencing looking at obese and lean twins and their mothers, revealed that related individuals had more species in common and a more similar bacterial community structure than unrelated individuals, regardless of obesity status.30 While twin studies reveal the significance of host genotype in determining microbial composition, the study of discordant twins (where one is healthy and the other sick) indicate that there is opportunity to regulate the microbiota in composition and function through changes in host physiology as seen in CD and obesity.30,35,40
Another aspect that demonstrates the importance of the host in regulating the microbiota is the return of the microbiota after disruption. Following disruption of the human microbiota with antibiotics, the microbiota returns relatively quickly to a population very similar to that before disruption.34 Although certain bacterial species may have been disrupted for longer periods of time,41 the overall composition was regained. The microbiota also recovers after infection with enteric pathogens such as Citrobacter rodentium or Salmonella enterica, where the population saw reduction in number and dramatic shifts in membership.42,43 It is essential to the health of the host that the microbiota are re-established after disruption, as important functions, including butyrate production, are greatly inhibited when the microbiota are altered.44 All of these pieces of evidence indicate that each individual establishes a unique equilibrium with the microbiota; however, the host factors regulating which bacterial species colonize are poorly understood.
Innate Immunity
Innate immunity plays a crucial role in defence against bacterial and viral infections.45,46 This is especially true for mucosal surfaces where the vast majority of pathogens initiate infection.47 In addition, the innate immune system plays an important role in regulating the millions of commensal bacteria that reside at the mucosal surface. This is evidenced by the abundance of innate immune defence proteins found in the feces of healthy patients in a proteomic study, indicating the extensive effort of the host in regulating the intestinal microbiota.12 The innate immune system in the gut includes mechanical barriers, chemical barriers and an induced cellular innate immune response. These components can also be divided into two highly interconnected parts, involving mechanisms for detecting the microbiota and the responses that act upon the microbiota.
Mechanical Barrier
The mucus layer that coats the length of the gastrointestinal tract is one of the most important mechanisms through which the host prevents continuous mucosal stimulation by enteric bacteria. There are two distinct mucus layers, one that is firmly adhered to the surface epithelium and is devoid of bacteria, and a second loosely adherent layer, which is heavily colonized by bacteria.48,49 The mucus layers are composed of a combination of different mucins that vary in structure and glycolsylation patterns. Mucin 2 (MUC2), a major mucin of the colonic firm and loose mucus layers, seems to be essential to the maintenance of mucosal homeostasis. For example, MUC2−/− mice develop colitis, which is dependent on direct bacterial contact with the epithelial surface.48
While the firmly adherent mucus layer provides an important barrier, the loosely adherent mucus layer is an important nutrient source for colonizing bacteria. The composition of the mucus layer is also a mechanism through which the host can maintain the microbial composition. Whole genome sequencing of representative organisms of the main bacterial families present in the human gastrointestinal tract has revealed a diverse set of glycosidases, particularly in members of the genus Bacteroides, that allow them to utilize the mucus layer as an energy source.50 When Bacteroides thetaiotaomicron is not able to obtain sufficient carbohydrate from the diet, whether it is due to limitations of the diet or competition with other bacteria, it signals the host to produce fucosylated glycans that it, but not all other members of the gut microbiota, can utilize.50 One might expect that variations in the glycosylation pattern impact the composition of the microbiota based on their glycosidase repertoire, however, this has not been tested.
Importantly, bacteria and their metabolic products can contribute to the regulation of mucin gene expression and secretion by the host. Lactobacillus casei GG increased mucin gene, MUC2, expression in intestinal epithelial cell monolayers as compared to untreated cells,51 while other bacteria have been shown to increase the expression of both MUC2 and MUC3 by goblet cells.52,53 Bacteria can also alter the pattern of the glycoconjugate repertoire expressed by absorptive enterocytes.54 Before weaning rodents express mostly sialylated surface oligosaccharides.55 An increase in fucosylation after weaning was in large part due to effects of changes in the bacterial population that occur during development, including an increase in Bacterioides spp. representation.54
Chemical Barriers
In the last several years there has been a great deal of attention given to antimicrobial proteins and peptides in protection against enteric infections. However, since these factors, including antimicrobial peptides and proteins, lysozyme, secretory phospholipase A2, C-type lectins, α-defensins and cryptdin-related sequence peptides, do not discriminate commensals from pathogens, they are also important in controlling the growth of commensal bacteria. Although Paneth cells are the main producers of antimicrobial peptides, other cell types such as enterocytes, intraepithelial lyphocytes, macrophages and neutrophils have also been reported to secrete these defense peptides.56–59 Each of these antimicrobial factors has distinct roles in maintaining the microbiota to ensure optimal health. The secretion of lysozyme results in the elimination of Gram-positive and Gram-negative bacteria by hydrolyzing the 1,4 β-glycosidic linkages that make up peptidoglycan,60 while secretory phosplipase A2 can non-specifically damage bacterial cell surfaces.61 RegIIIγ, a C-type lectin, which is expressed by Paneth cells as well as enterocytes in the small intestine, also targets Gram-positive bacteria by binding to peptidoglycan.59 In addition, RegIIIγ (human ortholog HIP/PAP) is important for controlling the population of some Gram-positive opportunistic pathogens, as depletion in its production and secretion facilitates the growth of vancomycin resistant Enterococcus.62 Defensins, which include a large family of antimicrobial peptides, are cationic membrane disrupting molecules that can target both Gram-positive and Gram-negative bacteria, as well as some fungi, viruses and protozoa and hence are essential for the control of pathogenic microbes as well as the enteric microbial populations.63,64 One study showed that a-defensin production was reduced in the ileum of CD patients, and furthermore, transgenic mice expressing Human Defensin (HD)5 showed alterations in their commensal bacterial populations compared to wild type controls.65 A very recent study found that while α defensins do not affect total microbial numbers they do affect microbial composition.66 They found that mice unable to produce active α-defensins (matrix metalloproteinase 7 knockout) had an increased ratio of Firmicutes to Bacteroidetes, whereas transgenic mice expressing HD5 demonstrated a reduced ratio of Firmicutes to Bacteroidetes as well as a disappearance of SFB compared to wildtype controls.66 Relatively little is known about the cathelicidins, which are similar to defensins and are capable of disrupting Gram-positive and Gram-negative bacteria as well as some fungi.67,68 Although the mechanism of action on bacterial growth and survival of most antimicrobial peptides is known, only a few studies have begun to identify how expression affects composition of the microbiota. The expression and secretion of antimicrobial peptides is more complex than simply sensing the microbiota, as the microbiota can alter host antimicrobial peptide production for their benefit. For example, while B. thetaiotaomicron (Gram-negative) stimulates expression of genes responsible for antimicrobial activity targeting Gram-positive bacteria, Bifidobacterium longum (Gram-positive) suppresses the expression of these same genes.69
The majority of antimicrobial activity is localized to the mucus layer providing an antibacterial barrier to prevent bacterial attachment and invasion.70 Localization within the mucus layer indicates that the host regulates microbial populations within the mucus layer rather than in the luminal contents. Caution should therefore be used when interpreting microbial abundance and composition based on fecal samples. While bacteria found in the mucosal layer of the tract will also be found in the feces, assessing the effects of the host on microbial composition in the feces may result in important changes being missed. For example, increased bacterial abundance observed in the ileum of nucleotide-binding oligomerization domain (NOD)2 deficient mice, discussed below, was not seen in fecal samples.71
Innate Immune Induction
Another component of the innate immune system that plays a key role in recognizing and controlling microbes is the family of pattern recognition receptors (PRR). These pattern recognition receptors include the Toll-like receptors (TLR) as well as NOD-containing proteins. In the gut it has been determined that there is expression of TLR-2, TLR-4, TLR-5, TLR-9 and NOD1 and NOD2.72–74 These receptors recognize conserved microbe-associated molecular patterns (MAMP) including peptidoglycan, lipopolysaccharide (LPS), flagella, bacterial DNA, diaminopimelic acid containing peptides (in peptidoglycans of Gram-negative bacteria and some Gram positive bacilli) and muramyldipeptide (common to all bacterial peptidoglycans), respectively.
New evidence reveals the role of specific receptors in regulating the microbiota. For example, mice deficient in NOD2, an intracellular receptor that recognizes Gram-positive and Gram-negative bacteria, have reduced bactericidal activity and an expansion in mucosal microbiota colonization in the terminal ileum.71 As well as being important for the regulation of the microbial load, NOD2 also appears to be important for host protection against some pathogenic bacteria, such as Helicobacter hepaticus, a common mouse pathogen.71 Mutations in NOD2 have been implicated in diseases involving disrupted microbiota. In fact, the first susceptibility gene implicated in CD was the CARD15 gene, encoding human NOD2, a mutation that is prevalent in patients with CD.75 Much more research is available on the role of the TLRs. One clinical study reported that there was an increase in expression of TLR2, TLR4 and CD14 in the terminal ileum, cecum and rectum of IBD patients.76 Although this enhanced expression may be the result of microbial dysbiosis and a disruption in immune regulation, it is not unfair to also suggest that the increase in TLR expression may be the cause of IBD in these patients. For instance, it is possible that this increase in expression resulted in increased activation of the PRRs by commensal microbes and hence an inflammatory state that initiated the pathogenesis of IBD. Additionally, a very recent study using MyD88 deficient mice found that these mice showed colonization in the spleen and liver by commensal bacteria.77 This suggests that some level of TLR recognition and signalling is necessary to maintain the commensal bacteria populations and ensure proper compartmentalization. By selectively re-establishing MyD88 expression in paneth cells, Vaishnava et al.78 demonstrated the importance of MyD88 signaling in paneth cells for limiting bacterial penetration of host tissues. Finally, there has been a significant amount of research on TLR5 deficient mice. TLR5 recognizes flagellin and is located only on the basolateral surface of the epithelial cells of the gut, hence it only reacts to flagellin on bacteria that have been able to cross the epithelial barrier and may be harmful to the host.79 Approximately 40% of TLR5−/− mice were found to develop spontaneous colitis, which was correlated with increased levels of total bacteria in the cecum and colon of these mice, and also increased translocation of commensal bacteria to the liver and spleen. When TLR5−/− mice were crossed with IL-10 deficient mice (IL-10−/− mice also develop spontaneous colitis) it was found that 100% of the mice developed colitis. This implied that TLR5 activation and resulting IL-10 production is necessary to control the commensal bacteria populations. Furthermore, the TLR5−/− mice were found to have an increase in expression of TLR4, CD14 and LBP, therefore increasing the inflammatory response to commensal microbes whereas TLR5/TLR4 double knockout mice did not develop colitis.80 These studies suggest a complex mechanism of regulation of the commensal flora. In humans a cohort of Jewish subjects were found to have reduced TLR5 expression, but none of these subjects developed CD.81 The combination of these studies suggests that, although a decrease in TLR5 expression may not be detrimental to the regulation of commensal bacteria, a complete loss of TLR5 expression can result in the inability to manage the commensal population.
The innate mechanisms described above have only begun to be examined in this role and much more research is required. Some of the key innate and adaptive components important in regulating the microbiota that have been described are presented in Table 1. The direction of the adaptive immune response is influenced by the innate immune system and this increases the many ways in which the immune system is able to regulate the microbiota. In the absence of an adaptive immune system the innate immune response compensates to maintain microbial regulation. One suggestion of this was that the intestines of RAG1−/− mice showed increased expression of 87 innate immune genes.82 Conversely, the innate immune response was necessary to prevent the establishment of specific immunity (IgG and IgM) to the microbiota, as commensal specific immunoglobulins were observed in specific pathogen free (SPF) Myd88−/− and Ticam1−/− mice.77 Development of commensal specific IgG appears to be a broad phenomenon of deficiencies in innate immunity as Nos2−/− and Cybb−/− mice also showed serum antibodies specific to commensal bacteria.77 It is also apparent that the innate immune system is necessary to maintain the microbiota within intestinal sites as translocation to the spleen after the administration of E. coli K12 was only observed in Myd88−/− and Ticam−/− mice.77
Table 1.
Examples of changes in host physiology resulting in changes in the gut microbiota
| Component | Function | Findings | Ref |
| MUC2 | Physical barrier | MUC2 deficient mice have bacteria in direct contact with the mucosa leading to inflammation | 48 |
| RegIIIγ | Antimicrobial activity | Reduced expression results in the expansion of opportunistic Gram-positive pathogens | 62 |
| IgA | Antimicrobial activity | Commensal specific IgG develops in absence of IgA expression | 90 |
| MyD88 | Microbial detection | MyD88−/− mice have commensal bacteria in liver and spleen | 77 |
| NOD2 | Microbial detection | NOD2−/− mice have reduced bactericidal activity and an expansion in mucosal microbial colonization | 70 |
| MHC | Antigen presentation | Bacterial composition and activity varies according to MHC haplotype | 87 |
MUC, mucin; Ig, immunoglobulin; NOD, nucleotide-binding oligomerization domain; MHC, major histocompatibility complex.
The Adaptive Immune Response
In the absence of an adaptive immune system the innate immune response is more pronounced, demonstrating the significance of the adaptive immune response in regulating the intestinal microbiota.59,83 The adaptive immune response in the gut consists of IgA, γβT cells, CD8+ T cells and CD4+ T cells. CD4+ T cells can be further differentiated into Treg cells and Th17 cells. Together these cell types, and the cytokines they produce, are important in regulating the inflammatory response in the gut and in controlling commensal microbes, both in abundance and activity. There is a great deal of evidence that describes how the adaptive response is capable of controlling inflammation and avoiding excessive immune responses in the gut.84,85
Induction of an adaptive immune response relies on the assistance of antigen presenting cells including macrophages and dendritic cells (DC). A study by Macpherson and Uhr showed that macrophages killed any commensal microbes that are taken up within 4 hours, while DCs could retain small numbers of live commensal bacteria for days.86 This study found that the DC then travelled to the MLN where they induced local commensal specific IgA production (reviewed in ref. 84). It would also appear that antigen presentation is important in the regulation of the microbial population, as changes in microbial composition in inbred mouse strains varied based on major histocompatability complex (MHC) haplotype.87 The nature of the change in response to MHC haplotype at the phylogenetic level is yet unclear as changes in the microbiota were assessed based on bacterial cellular fatty acid profiles.87 The MLN play an important role in compartmentalization of the adaptive immune response since in the absence of the MLN, the DCs are able to induce a systemic and potentially dangerous response to the gut microbiota. The compartmentalization of the adaptive response to commensal pathogens is also assured by the high predominance of IgA in the gut mucosa. In fact, more than 80% of all plasma cells are located in the gut lamina propria and they produce more IgA than the other Ig isotypes together.88
Humoral Immunity
IgA is produced by B cells in the lamina propria, is transcytosed across enterocytes into the lumen and is maintained within the mucus layer. The importance of IgA responses in controlling gut commensals is evidenced in many studies. Secretory IgA (s-IgA) is non-specific, and has been shown to coat endogenous bacteria.89 It was found that the IgA produced was mainly from B1 cells in the peritoneum and was independent of T-cell help.90 IgA is important for limiting access of bacteria to the intestinal surface,91 as a study of mice unable to produce IgA showed an abnormal increase in the anaerobic bacteria population, particularly SFB, in the intestinal lumen.92,93 Reconstitution of the mice with IgA normalized the microbial population.92,93 It would appear that non-specific IgA is sufficient to limit the penetration of bacteria through the intestinal epithelium as demonstrated by the protective effect of non-specific neonatal IgA.94 However, specific IgA also appears to have a role in inducing changes in bacterial physiology. Specific IgA has been shown to cause bacteria to change the expression of surface epitopes and metabolic activity, specifically reducing the microbial ability to deal with oxidative bursts.95 The changes in surface epitope expression by B. thetaiotaomicron results in a bacterium that induces less inflammation, therefore helping maintain the balanced mucosal inflammation.95 A commensal specific Ig response is not found in germ free mice, whereas a commensal specific IgA response can be found in the intestine but not the serum of SPF mice.90 However, when IgA was absent a commensal specific IgG response in the serum of SPF mice was noted.90 This suggested that antigen specific Ig also played an important role in sequestering commensal microbes to the gut mucosa.
Cell Mediated Immunity
Although the IgA response is paramount to controlling gut commensals, the T-cell response is also important. The cell mediated response in the gut mucosa is more of a regulatory response directed in order to avoid chronic inflammation and autoimmunity. One study found that patients with CD had increased levels of IL-12 and interferon-γ suggesting that a dysregulated T-cell response directed against the intestinal microbiota may be a cause of the disease.88 The regulatory role of IL-10 has been suggested to be important in inhibiting effector T-cell responses and avoiding chronic inflammation. IL-10 is a cytokine that is produced mainly by CD4+ Treg cells, and a recent report has identified IL-10 as a susceptibility locus for human ulcerative colitis (UC).96 In fact IL-10−/− mice have been shown by many groups to develop spontaneous colitis.97,98 Furthermore, treatment with exogenous IL-10 in mice with T-cell induced colitis reverses the outcome. IL-10 is also capable of decreasing the antigen presenting capacity of macrophages and DCs, which may assist in inhibiting robust responses to commensal microbes.99 Although IL-10 is important as an anti-inflammatory cytokine, there is no evidence thus far that suggests if and how it may be acting directly to control the gut microbiota.
Tregs are characterized by constitutively high levels of the transcription factor Foxp3, and are important in suppressing excessive immune responses that may be deleterious to the host. Since Tregs are the class of T cells that produce the highest levels of IL-10, studies were conducted on the effect of Treg cells in the development and prevention of IBD. It was found that depletion of Treg cells resulted in a moderate form of colitis in Helicobacter hepaticus induced model of inflammation, while Treg cell reconstitution of mice with bacterial induced colitis could inhibit colitis development.100 Treg cells were also capable of producing transforming growth factor (TGF)β, which is one of the cytokines needed for the differentiation of Th17 cells.
Th17 cells are a subset of activated CD4+ T cells and are characterized by the production of IL-17, IL-21 and IL-22. Th17 cell differentiation is controlled by both TGFβ and IL-6 and their survival and expansion is controlled by IL-23.101–104 The cytokines produced by Th17 cells have recently been a focus of many studies and have been described to be important in the control of IBD by regulating the microbiota. For instance, IL-17 production has been shown to regulate granulopoiesis, neutrophil recruitment and the induction of antimicrobial peptides, which are all important in controlling the gut microbiota to maintain a homeostatic balance. A recent study found that IL-22 expression in the gut is protective against colitis.105 A second study confirmed this finding and showed that IL-22 was produced by CD4+ T cells and NK cells.106 In fact, overexpression of IL-22 leads to the induction of the mucin genes, MUC1, MUC3 and MUC13, which resulted in enhanced mucus production and decreased the translocation of commensal bacteria across the epithelial barrier.105 IL-22 was also capable of inducing defensin expression in colonocytes leading to further protection against microbiota-induced inflammation. Overall, the role of Th17 cells in controlling gut microbiota is becoming more evident and further research will determine how to capitalize on this knowledge.
Some cell types, while not important to regulating the microbiota during the healthy state, are important for controlling the microbiota in cases of mucosal injury. For example, γδ intraepithelial T lymphocytes play a critical role in coordinating the regulation of cytoprotective, immunomodulatory and antimicrobial factors upon mucosal damage with dextran sodium sulfate (DSS). Using TCRδ knockout mice it was found that γδ T cells are essential for controlling mucosal penetration of commensal bacteria upon DSS induced damage.107
Although the innate immune system is largely able to sequester the commensal microbes, the cooperation of the adaptive immune response is imperative to maintain this control. In many instances there is a feedback loop that occurs between the two arms of the immune response to ensure that proper management of gut commensals is maintained and, if necessary, immune mechanisms are activated. An example of this is outlined above where the production of IL-22 by CD4+ T cells of the adaptive immune system can initiate upregulation of mucin genes and hence the innate immune response in an effort to control the microbiota. Instances where components of the immune response are insufficient, the host loses control of the microbiota as depicted in Figure 2. For instance, secretion of cytokines by epithelial cells and macrophages such as IL-6 and TGFβ ensure the differentiation of a Treg response, and the products of this response, such as IL-22, work to increase mucus production and defensin secretion. A second example was evident in the high levels of IL-10 produced by CD4+ T regs, which assisted in decreasing antigen presenting cell capability, hence reducing the potential of an effector T-cell response by the adaptive arm of the immune system. There is a great deal of further research required to understand how other components of both innate and adaptive immunity may be important in regulating the microbiota.
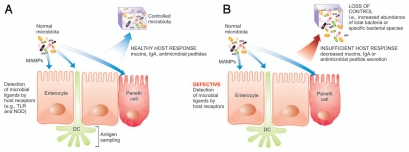
Figure 2.
Sensing and controlling the microbiota. (A) The appropriate detection and antigen sampling of the microbiota through many mechanisms results in a healthy host response leading to control of the microbiota. (B) Defective detection of the microbiota and an insufficient host response leads to a loss of control of the microbiota, including the expansion of the total bacteria or specific bacterial populations. DC, dendritic cell; MAMP, microbe associated molecular pattern; IgA, immunoglobulin A; TLR, toll-like receptor; NOD, nucleotide-binding oligomerization domain.
Disrupting the Balance
If the delicate balance between the host immune system and the microbiota is altered, the host can disrupt the beneficial functions of the microbiota. This is seen in cases of intestinal inflammation such as CD where a disrupted microbiota has been correlated to reduced levels of butyrate108 as well as disrupted fatty acid, bile acid and amino acid metabolism.35 The intestinal microbiota changes substantially in response to intestinal inflammation, whether induced chemically, by a bacterial pathogen or the result of an inflammatory bowel disease.42 The specifics of these changes, however, are not consistent between each type of inflammation. For example, changes in the microbiota in CD and UC are distinctly different, to the extent that microbial composition can be used as a diagnostic marker to differentiate active CD and UC.14 There are, however, some changes in the microbial community that are shared in CD and UC patients including reduced diversity (particularly Firmicutes),109–114 presence of bacteria that are not normally considered to be commensal members of the microbiota,110 and increased concentrations of E. coli.40,115–118 In CD, there is a particular disappearance of members of the family Ruminococcaceae, specifically the core member Faecalibacterium prausnitzii.40,119 The fact that the disappearance of F. prausnitzii is a result of host inflammation is demonstrated by the re-emergence of this population upon treatment with infliximab (TNF blocker) or high dose cortisol treatment.14 It is expected that the differences in the microbiota of CD and UC patients is a consequence of differences in the immune response in these diseases. The nature of the immune response differs between UC and CD, as UC is associated with a Th2 response, whereas CD is typified by a Th1/Th17 response.120 Because F. prausnitzii levels are normal in UC patients, this change in flora may be dependent on the nature of the T-helper cell response. A chronic Salmonella infection that results in persistent intestinal fibrosis, with a Th1/Th17 immune response that mirrors that seen in CD,121 presents an excellent model to study the role of Th response in changing the microbiota. Other aspects of immune function have also been shown to be different in CD patients. Both reduced defensin production and the development of an IgG response to a variety of bacterial and yeast components are characteristics of CD but not UC.122,123 Reduced abundance of Treg cells, particularly CD4+ CD25+ Foxp3+, has also been observed in CD and is known to mediate the inflammatory response in the gut.124 Defining which of these aspects is responsible for the disappearance of F. prausnitzii may give new insight into the disease etiology and therapy.
While the disappearance of F. prausnitzii is a result of the host immune response, it is likely that this disruption contributes to disease progression. F. prausnitzii has been shown to display therapeutic properties in a mouse model of IBD119 and Faecalibacterium is a major butyrate producer in the gut.125 Therefore, a disappearance of this core member of the microbiota31 likely has consequences for gut energy balance and homeostasis and preventing this disappearance may be a good therapeutic target.
Another example of host-induced modifications in microbiota that lead to increased severity of disease is the mouse model of Salmonella-induced gastroenteritis. Although this model requires antibiotics, the inflammation that results in further changes in the microbiota are required for the expansion of the Salmonella population.4 This was clearly demonstrated by investigating the colonization patterns of a Salmonella enterica mutant unable to induce inflammation. This bacterium demonstrated an explosive increase in colonization when co-colonized with its isogenic wildtype strain that caused inflammation or in mouse models of spontaneous colitis, including IL10−/− and VILLIN-HACL4-CD8 mice.4 Changes in microbial population preceded diarrhea indicating that host-induced changes in the microbiota were independent of diarrhea.43 Inflammation in response to Salmonella infection also resulted in the expansion of the population of Clostridium perfringens which itself is a known inducer of intestinal damage.43 The components of the inflammatory response required for the disruption of the microbiota and expansion of Salmonella are not yet known. This virulence strategy of Salmonella enterica may be a therapeutic target for cases of Salmonella induced gastroenteritis.
The host has developed numerous mechanisms to be tolerant to the microbiota by developing mechanisms to regulate inflammation, including the Tregs and IL-10 discussed above to prevent host-induced disruptions of the microbiota. Inflammation also results in changes in bacterial localization as detected by fluorescence in situ hybridization (FISH) in inflammation induced by DSS and in IL-10 knockout mice as during inflammation the population in the intestinal crypts are amplified.113 Why some bacteria disappear in one case and not another (while others thrive in an inflammatory state) might be dependent on whether the host response involves the production of reactive oxygen species (ROS) and nitric oxide, increased or reduced release of antimicrobial factors, changes in mucus production and secretion and infiltration of neutrophils along with antimicrobial molecules into the intestinal lumen.126
Future Prospects and Challenges
It is essential that we regulate our microbiota to maintain health. In rats a diet deficient in vitamin A resulted in changes in the expression of innate immunity-related genes including mucin dynamics, characterized by decreased MUC2 expression and increased MUC3 expression, as well as reduced defensin 6 expression. These changes in the mucosal immune response resulted in an overall increase in bacterial load and a relative decrease of Lactobacillus spp. as well as a concurrent increase in the E. coli population.127 It is clear that regulation of the microbiota is important to surgical outcome. When there is trauma the fluctuations in the gut ecosystem can result in sepsis as the gut is a major origin of sepsis.128 Understanding what host factors cause disruption in the gut microbiota may lead to insights into how to modulate the gut microbiome with pre, pro and synbiotics to improve outcome of surgical patients. We must also recognize that when we disrupt the microbiota through the use of antibiotics, we must consider how this will affect the host's ability to regulate the remaining microbial population.
There are a number of challenges that will have to be overcome in understanding the ecology of the gastrointestinal microbiota. While inferences can be made between host-microbial interactions across animal species because the same main phyla, Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria are present,38 this can present challenges. For example, SFB have recently been shown to be critically important to the regulation of the Th17 response in the mouse gut, and would be considered a normal colonizer.22 While this important role of SFB in mice is a great advance in the understanding of the host-microbial interaction, SFB are not normal colonizers of the human gastrointestinal tract. It is likely that there are other microbes serving equivalent roles, however, it will be an added challenge to identify them.
Another major challenge in the study of the host-microbial relationship is the continuous dialogue and interdependency between host and microbe. How a microbe interacts with the host and in turn how the host responds to that microbe is dependent on co-colonizers.50 While gnotobiotic models give some insights into the host-microbe relationship, the development of immune structures within the gut and a balanced inflammatory response are regulated by diverse colonizers, therefore how the host responds to colonization by a single organism can be misleading. Determining the sequence of events leading to the imbalance of the microbiota and inflammation will be a difficult task. It is possible that the imbalanced microbiota is the result of a mutation or a mutation could lead to inflammation thus resulting in an imbalanced microbiota.
It is clear that the host plays an important role in shaping the microbiota in health and disease, however, our understanding of the mechanisms involved in maintaining a balanced microbiota and host factors that are involved in disrupting the microbiota are still rudimentary. Future advances will depend on the utilization of knockout models, to characterize the microbiota in an in depth manner and to dissect the localization of different bacteria. A concerted effort joining the forces of microbial ecologists with expertise in immunology will have to investigate the timing of microbial disruption in comparison to immunological changes. A greater understanding of the mechanisms through which the host regulates the microbiota in both health and disease will give new insights into the ecology of the microbiota as well as strategies to regulate the microbiota to maintain or regain a healthy state.
Acknowledgements
We would like to thank Dr. Jennifer L. Bishop, Dr. Matthew A. Croxen and Shannon L. Russell for helpful discussions and critical revision of this manuscript. Work in our laboratory is supported by operating grants from the Canadian Institutes of Health Research (CIHR), the Crohn and Colitis Foundation of Canada (CCFC), and the Howard Hughes Medical Institute (HHMI). Navkiran Gill. is the recipient of a Postdoctoral Trainee Award from the Michael Smith Foundation for Health Research (MSFHR). B. Brett Finlay is a CIHR Distinguished Investigator, an HHMI International Research Scholar, and the University of British Columbia Peter Wall Distinguished Professor.
Abbreviations
- Ig
immunoglobulin
- CD
Crohn disease
- Th
T helper
- SFB
segmented filamentous bacteria
- rRNA
ribosomal ribonucleic acid
- OTU
operational taxonomic unit
- IBD
inflammatory bowel disease
- s-IgA
secretory IgA
- DSS
dextran sodium sulfate
- UC
ulcerative colitis
- IL
interleukin
- MAMP
microbe-associated molecular pattern
- TLR
Toll-like receptor
- NOD
nucleotide-binding oligomerization domain
- MLN
mesenteric lymph node
- MHC
major histocompatibility complex
- Treg
regulatory T cell
- PRR
pattern recognition receptor
- MUC
mucin
- LPS
lipopolysaccharide
- DC
dendritic cell
- SPF
specific pathogen free
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/12520
References
- 1.Marchesi J, Shanahan F. The normal intestinal microbiota. Curr Opin Infect Dis. 2007;20:508–513. doi: 10.1097/QCO.0b013e3282a56a99. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Bry L, Falk PG, Gordon JI. Host-microbial symbiosis in the mammalian intestine: exploring an internal ecosystem. Bioessays. 1998;20:336–343. doi: 10.1002/(SICI)1521-1878(199804)20:4<336::AID-BIES10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Hudault S, Guignot J, Servin AL. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut. 2001;49:47–55. doi: 10.1136/gut.49.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1110. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekirov I, Tam NM, Jogova M, Robertson ML, Li YL, Lupp C, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 12.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 13.Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, Arjan J, de Visser GM, de Vos WM. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis. 2001;13:129–134. [Google Scholar]
- 14.Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn's disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflam Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 15.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class-II molecules and fucosyl asialo Gm1 glycolipids on the small-intestinal epithelial-cells in the ex-germ-free mouse. Microbiol Immunol. 1995;39:555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 17.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 18.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451–1545. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cebra JJ. Influences of microbiota on intestinal immune system development. Amer J Clin Nutr. 1999;69:1046–1051. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov II, de Frutos RL, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009 doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rask C, Evertsson S, Telemo E, Wold AE. A full flora, but not monocolonization by Escherichia coli or lactobacilli, supports tolerogenic processing of a fed antigen. Scand J Immunol. 2005;61:529–535. doi: 10.1111/j.1365-3083.2005.01598.x. [DOI] [PubMed] [Google Scholar]
- 25.Ruemmele FM, Bier D, Marteau P, Rechkemmer G, Bourdet-Sicard R, Walker WA, et al. Clinical evidence for immunomodulatory effects of probiotic bacteria. J Pediat Gastroenterol Nutr. 2009;48:126–141. doi: 10.1097/MPG.0b013e31817d80ca. [DOI] [PubMed] [Google Scholar]
- 26.Vidal K, Benyacoub J, Moser M, Sanchez-Garcia J, Serrant P, Segura-Roggero I, et al. Effect of Lactobacillus paracasei NCC2461 on antigen-specific T-cell mediated immune responses in aged mice. Rejuv Res. 2008;11:957–964. doi: 10.1089/rej.2008.0780. [DOI] [PubMed] [Google Scholar]
- 27.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 28.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program. 2008;62:13–29. doi: 10.1159/000146245. [DOI] [PubMed] [Google Scholar]
- 29.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 32.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cel. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 33.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 34.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by Deep 16S rRNA sequencing. PLoS Biol. 2008;6:2383–2400. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4:6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claus SP, Tsang TM, Wang YL, Cloarec O, Skordi E, Martin FP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tannock GW. The search for disease-associated compositional shifts in bowel bacterial communities of humans. Trends Microbiol. 2008;16:488–495. doi: 10.1016/j.tim.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflam Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 41.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 42.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bender A, Breves G, Stein J, Leonhard-Marek S, Schroder B, Winckler C. Colonic fermentation as affected by antibiotics and acidic pH: Application of an in vitro model. Zeitschrift Fur Gastroenterologie. 2001;39:911. doi: 10.1055/s-2001-18537. [DOI] [PubMed] [Google Scholar]
- 45.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol. 1999;17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 46.Biron CA, Brossay L. NK cells and NKT cells in innate defense against viral infections. Curr Opin Immunol. 2001;13:458–464. doi: 10.1016/s0952-7915(00)00241-7. [DOI] [PubMed] [Google Scholar]
- 47.Freihorst J, Ogra PL. Mucosal immunity and viral infections. Ann Med. 2001;33:172–177. doi: 10.3109/07853890109002074. [DOI] [PubMed] [Google Scholar]
- 48.Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Nat Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol. 2001;280:922–929. doi: 10.1152/ajpgi.2001.280.5.G922. [DOI] [PubMed] [Google Scholar]
- 50.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Nat Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mattar AF, Teitelbaum DH, Drongowski RA, Yongyi F, Harmon CM, Coran AG. Probiotics upregulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediat Surg Internat. 2002;18:586–590. doi: 10.1007/s00383-002-0855-7. [DOI] [PubMed] [Google Scholar]
- 52.Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut. 2003;52:827–833. doi: 10.1136/gut.52.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mack DR, Michail S, Wei S, McDougall L, Hollingsworth MA. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol-Gastrointest Liver Physiol. 1999;276:941–950. doi: 10.1152/ajpgi.1999.276.4.G941. [DOI] [PubMed] [Google Scholar]
- 54.Hooper L, Xu J, Falk P, Midtvedt T, Gordon J. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Nat Acad Sci USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torrespinedo R, Mahmood A. Postnatal changes in biosynthesis of microvillus membrane glycans of rat small-intestine 1. Evidence of a developmental shift from terminal sialylation to fucosylation. Biochem Bioph Res Co. 1984;125:546–553. doi: 10.1016/0006-291x(84)90574-6. [DOI] [PubMed] [Google Scholar]
- 56.Ouellette AJ, Greco RM, James M, Frederick D, Naftilan J, Fallon JT. Developmental regulation of cryptdin, a corticostatin defensin precursor messenger-RNA in mouse sall intestinal crypt epithelium. J Cell Biol. 1989;108:1687–1695. doi: 10.1083/jcb.108.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harwig SSL, Tan L, Qu XD, Cho Y, Eisenhauer PB, Lehrer RI. Bactericidal properties of murine intestinal phospholipase A2. J Clin Invest. 1995;95:603–610. doi: 10.1172/JCI117704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hornef MW, Putsep K, Karlsson J, Refai E, Andersson M. Increased diversity of intestinal antimicrobial peptides by covalent dimer formation. Nature Immunol. 2004;5:836–843. doi: 10.1038/ni1094. [DOI] [PubMed] [Google Scholar]
- 59.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganz T. Antimicrobial polypeptides. J Leukocyte Biol. 2004;75:34–38. doi: 10.1189/jlb.0403150. [DOI] [PubMed] [Google Scholar]
- 61.Vadas P, Browning J, Edelson J, Pruzanski W. Extracellular phospholipase-A2 expression and inflammation—the relationship with associated disease states. J Lipid Mediators. 1993;8:1–30. [PubMed] [Google Scholar]
- 62.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–808. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nature Immunol. 2005;6:551–557. doi: 10.1038/ni1206. [DOI] [PubMed] [Google Scholar]
- 64.Hecht G. Innate mechanisms of epithelial host defense: spotlight on intestine. Am J Physiol-Cell Physiol. 1999;277:351–358. doi: 10.1152/ajpcell.1999.277.3.C351. [DOI] [PubMed] [Google Scholar]
- 65.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Nat Acad Sci USA. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hase K, Eckmann L, Leopard JD, Varki N, Kagnoff MF. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect Immun. 2002;70:953–963. doi: 10.1128/iai.70.2.953-963.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bals R, Wilson JM. Cathelicidins—a family of multifunctional antimicrobial peptides. Cellular Molec Life Sci. 2003;60:711–720. doi: 10.1007/s00018-003-2186-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. 2008;57:764–771. doi: 10.1136/gut.2007.141481. [DOI] [PubMed] [Google Scholar]
- 71.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Nat Acad Sci USA. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol. 2005;174:4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 73.Sanderson IR, Walker WA. TLRs in the Gut I. The role of TLRs/Nods in intestinal development and homeostasis. Am J Physiol-Gastrointest Liver Physiol. 2007;292:6–10. doi: 10.1152/ajpgi.00275.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fukata M, Abreu MT. Role of Toll-like receptors in gastrointestinal malignancies. Oncogene. 2008;27:234–243. doi: 10.1038/sj.onc.1210908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berrebi D, Maudinas R, Hugot JP, Chamaillard M, Chareyre F, De Lagausie P, et al. Card15 gene overexpression in mononuclear and epithelial cells of the inflamed Crohn's disease colon. Gut. 2003;52:840–846. doi: 10.1136/gut.52.6.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Honda K, Takeda K. Regulatory mechanisms of immune responses to intestinal bacteria. Mucosal Immunol. 2009;2:187–196. doi: 10.1038/mi.2009.8. [DOI] [PubMed] [Google Scholar]
- 77.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MAE, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Nat Acad Sci USA. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: Bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 80.Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gewirtz AT, Vijay-Kumar M, Brant SR, Duerr RH, Nicolae DL, Cho JH. Dominant-negative TLR5 polymorphism reduces adaptive immune response to flagellin and negatively associates with Crohn's disease. Am J Physiol-Gastrointest Liver Physiol. 2006;290:1157–1163. doi: 10.1152/ajpgi.00544.2005. [DOI] [PubMed] [Google Scholar]
- 82.Jima DD, Shah RN, Orcutt TM, Joshi D, Law JM, Litman GW, et al. Enhanced transcription of complement and coagulation genes in the absence of adaptive immunity. Mol Immunol. 2009;46:1505–1516. doi: 10.1016/j.molimm.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keilbaugh SA, Shin ME, Banchereau RF, McVay LD, Boyko N, Artis D, et al. Activation of RegIIIbeta/gamma and interferon gamma expression in the intestinal tract of SCID mice: an innate response to bacterial colonisation of the gut. Gut. 2005;54:623–629. doi: 10.1136/gut.2004.056028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Ann Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 85.Kelsall BL. Innate and adaptive mechanisms to control pathological intestinal inflammation. J Pathol. 2008;214:242–259. doi: 10.1002/path.2286. [DOI] [PubMed] [Google Scholar]
- 86.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 87.Toivanen P, Vaahtovuo J, Eerola E. Influence of major histocompatibility complex on bacterial composition of fecal flora. Infect Immun. 2001;69:2372–2377. doi: 10.1128/IAI.69.4.2372-2377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suzuki K, Ha SA, Tsuji M, Fagarasan S. Intestinal IgA synthesis: A primitive form of adaptive immunity that regulates microbial communities in the gut. Seminars Immunol. 2007;19:127–135. doi: 10.1016/j.smim.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 89.Mestecky J, Russell MW. Specific antibody activity, glycan heterogeneity and polyreactivity contribute to the protective activity of S-IgA at mucosal surfaces. Immunol Lett. 2009;124:57–62. doi: 10.1016/j.imlet.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 91.Brandtzaeg P. Induction of secretory immunity and memory at mucosal surfaces. Vaccine. 2007;25:5467–5484. doi: 10.1016/j.vaccine.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 92.Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424–1427. doi: 10.1126/science.1077336. [DOI] [PubMed] [Google Scholar]
- 93.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Nat Acad Sci USA. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harris NL, Spoerri I, Schopfer JF, Nembrini C, Merky P, Massacand J, et al. Mechanisms of neonatal mucosal antibody protection. J Immunol. 2006;177:6256–6262. doi: 10.4049/jimmunol.177.9.6256. [DOI] [PubMed] [Google Scholar]
- 95.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 96.Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nature Gen. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 97.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 98.Spencer SD, Di Marco F, Hooley J, Pitts-Meek S, Bauer M, Ryan AM, et al. The orphan receptor CRF2-4 is an essential subunit of the interleukin 10 receptor. J Exp Med. 1998;187:571–578. doi: 10.1084/jem.187.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moore KW, Malefyt RD, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Ann Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 100.Maloy KJ, Salaun L, Cahill R, Dougan G, Saunders NJ, Powrie F. CD4+CD25+ T-R cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med. 2003;197:111–119. doi: 10.1084/jem.20021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biologic Chem. 2003;278:1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- 102.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 103.Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu JF, et al. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 104.Korn T, Oukka M, Kuchroo V, Bettelli E. Th17 cells: Effector T cells with inflammatory properties. Seminars Immunol. 2007;19:362–371. doi: 10.1016/j.smim.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sugimoto K, Ogawa A, Mizoguchi E, Shimomura Y, Andoh A, Bhan AK, et al. IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J Clin Invest. 2008;118:534–544. doi: 10.1172/JCI33194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ismail AS, Behrendt CL, Hooper LV. Reciprocal interactions between commensal bacteria and gamma delta intraepithelial lymphocytes during mucosal injury. J Immunol. 2009;182:3047–3054. doi: 10.4049/jimmunol.0802705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Prot Res. 2007;6:546–551. doi: 10.1021/pr060470d. [DOI] [PubMed] [Google Scholar]
- 109.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflam Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 111.Scanlan PD, Shanahan F, O'Mahony C, Marchesi JR. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol. 2006;44:3980–3988. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dicksved J, Halfvarson J, Rosenquist M, Jarnerot G, Tysk C, Apajalahti J, et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2008;2:716–727. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]
- 115.Giaffer MH, Holdsworth CD, Duerden BI. Virulence properties of Escherichia coli strains isolated from patients with inflammatory bowel-disease. Gut. 1992;33:646–650. doi: 10.1136/gut.33.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+ D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterol. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 118.Willing B, Halfvarson J, Dicksved J, Rosenqist M, Järnerot G, Engstrand L, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 119.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Nat Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fuss IJ. Is the Th1/Th2 paradigm of immune regulation applicable to IBD? Inflamm Bowel Dis. 2008;14:110–112. doi: 10.1002/ibd.20683. [DOI] [PubMed] [Google Scholar]
- 121.Grassl GA, Valdez Y, Bergstrom KSB, Vallance BA, Finlay BB. Chronic enteric Salmonella infection in mice leads to severe and persistent intestinal fibrosis. Gastroenterol. 2008;134:768–780. doi: 10.1053/j.gastro.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 122.Mokrowiecka A, Daniel P, Slomka M, Majak P, Malecka-Panas E. Clinical utility of serological markers in inflammatory bowel disease. Hepato-Gastroenterol. 2009;56:162–166. [PubMed] [Google Scholar]
- 123.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, Petras RE, et al. Reduced paneth cell alpha-defensins and antimicrobial activity in ileal Crohn's disease. Inflamm Bowel Dis. 2006;12:20. [Google Scholar]
- 124.Saruta M, Yu QT, Fleshner PR, Mantel PY, Schmidt-Weber CB, Banham AH, et al. Characterization of FOXP3+CD4+ regulatory T cells in Crohn's disease. Clin Immunol. 2007;125:281–290. doi: 10.1016/j.clim.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 125.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 126.Sansonetti PJ. Host-bacteria homeostasis in the healthy and inflamed gut. Curr Opin Gastroenterol. 2008;24:435–439. doi: 10.1097/MOG.0b013e32830007f7. [DOI] [PubMed] [Google Scholar]
- 127.Amit-Romach E, Uni Z, Cheled S, Berkovich Z, Reifen R. Bacterial population and innate immunity-related genes in rat gastrointestinal tract are altered by vitamin A-deficient diet. J Nutr Biochem. 2009;20:70–77. doi: 10.1016/j.jnutbio.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 128.Kinross J, von Roon AC, Penney N, Holmes E, Silk D, Nicholson JK, et al. The gut microbiota as a target for improved surgical outcome and improved patient care. Curr Pharmaceut Design. 2009;15:1537–1545. doi: 10.2174/138161209788168119. [DOI] [PubMed] [Google Scholar]