Abstract
We recently showed that disruption of damX causes bile sensitivity in Salmonella enterica. The damX gene is part of an operon that contains genes with heterogeneous functions: DNA adenine methylation, biosynthesis of aromatic compounds, carbohydrate metabolism, and tRNA charging. The damX gene encodes a protein with a predicted size of 46 kDa. In Salmonella, DamX is found in the inner membrane of both dividing and non-dividing cells. The DamX protein contains a peptidoglycan-binding SPOR domain, and accumulates in the E. coli septal ring. E. coli mutants lacking DamX are bile-sensitive like their Salmonella counterparts.
Key words: DamX, bile resistance, inner membrane, peptidoglycan, SPOR domain
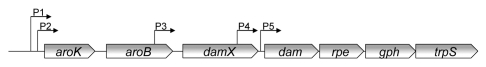
The damX gene maps immediately upstream of dam, the gene for DNA adenine methyltransferase, in the chromosomes of Escherichia coli and Salmonella enterica. Initially, damX was described as an unidentified reading frame located at centisome 74.3 on the E. coli chromosome; for this reason, it is also known as urf74.3.1 damX is the third gene in an operon that contains at least six additional genes with heterogeneous functions (Fig. 1): aroK, encoding shikimic acid kinase;2,3 aroB, encoding 3-hydroquinate synthase;4 dam;5,6 rpe, encoding ribulose-5-phosphate 3-epimerase;7 gph, encoding 2-phosphoglycolate phosphatase;8 and trpS, encoding tryptophanyl-tRNA synthetase.9 Until recently, damX was the only gene in the operon whose function remained unknown. Below we summarize recent studies on DamX structure and function.
Figure 1.
Diagram of the damX-containing operon, indicating the ORFs and promoters described in E. coli.
Structure of DamX
DamX is highly conserved in E. coli and Salmonella (≈77% identity). DamX is an inner membrane protein with a single transmembrane segment, an N-terminal cytoplasmic domain of around 100 amino acid residues and a relatively large, unstructured C-terminal periplasmic domain of about 300 amino acids. The predicted molecular weight of DamX is around 46 kDa. However, for reasons still unknown, the protein runs more slowly than expected in SDS polyacrylamide gels.1,10,11 In both E. coli and Salmonella, DamX harbors a conserved peptidoglycan-binding domain at its C-terminus, known as the SPOR domain.12 This domain is present in some septal ring components, and is thought to be important for septal ring localization (see below).
Expression of the damX Gene
In E. coli, transcription of damX is driven by at least three promoters, named P1, P2 and P3,13 (Fig. 1). All three promoters resemble a classical σ70-dependent promoter. P1 and P2 are located upstream of aroK, while P3 is inside the aroB coding sequence.13,14 P2 is the strongest promoter, followed by P1 and P3. It has been proposed that P1 and P2 ensure basal levels of expression of the entire operon, while P3 may modulate the expression level of downstream genes.13 Two additional promoters (P4 and P5) have been identified in the E. coli damX-containing operon, but neither of them contributes to damX transcription. P4 is inside the damX coding sequence, while P5 is located in the intergenic region between damX and dam.13,15,16 P1 and P2 are highly conserved in Salmonella, while P3, P4 and P5 are not. However, additional promoters may be present in Salmonella.11
Phenotypes of damX Mutants
Several phenotypes have been associated with lack and overexpression of DamX. However, some of the phenotypes resemble those found in strains lacking the downstream gene dam, and may be due to a polar effect on dam expression and/or to disruption of the P4 and P5 promoters (see below).
Cell division.
In 1995, it was shown that overproduction of DamX in E. coli produces cell filamentation, suggesting that DamX could be somehow involved in cell division.1 Almost fifteen years later, two independent studies have shown that the E. coli DamX protein contains a peptidoglycan-binding domain at its C terminus, known as the SPOR domain.10,17 The same domain is also present in S. enterica DamX, according to the Pfam database.12 The SPOR domain makes DamX accumulate in the septal ring during cell division, probably by direct binding to septal peptidoglycan.10,17 Lack of DamX does not produce any observable division defect. However, a damX mutation alters the division defects observed in mutants with alterations in other components of the septal ring:
DedD is a component of the septal ring that also contains a SPOR domain at its C terminus. Cells of a dedD mutant are elongated and form small chains of cells (“chaining phenotype”).10,17 If damX is mutated in a dedD background, both the elongation and the chaining phenotype are enhanced.10,17
ftsN is an essential component of the septal ring that also contains a SPOR domain at its C terminus. An FtsN null mutation is lethal in E. coli and Salmonella, but some viable alleles have been constructed. One such allele is ftsNslm117, which contains a transposon insertion at codon 119 in the ftsN coding sequence.17 The ftsNslm117 allele promotes mild elongation and chaining. As in the case of dedD, lack of DamX aggravates both phenotypes.
An intriguing phenotype of damX mutants is rescue of a mutant containing an ftsQ thermosensitive allele.10 FtsQ is an essential component of the septal ring, but thermosensitive alleles have been obtained. After a shift to 42°C, strains with an ftsQ1(Ts) allele undergo cell filamentation and a strong decrease in plating efficiency. Introduction of a damX mutation suppresses both phenotypes. However, an ftsQ null mutation is still lethal in a damX background. These results suggest that DamX might antagonize FtsQ function. A direct interaction between FtsQ and DamX has been reported.10
DamX shows septal localization in around 80% of dividing E. coli cells, suggesting that DamX may be recruited to the septal ring early during septal ring assembly.10 This possibility is supported by the following evidence: the components of the septal ring are thought to be recruited in a defined order, reflected by a remarkably linear set of dependencies. Certain components do not localize to the septal ring in the absence of other specific components. The first step in the assembly of the septal ring is polymerization of FtsZ at the inner face of the cytoplasmic membrane, to form the Z ring.18,19 The Z ring then serves as a scaffold for the assembly of the remaining components. FtsZ is the only protein required for recruitment of DamX to the septal ring. In strains lacking functional components located downstream in the recruitment cascade, DamX still localizes to the septal ring.10
Bile resistance.
Bile is a complex fluid containing bile salts, cholesterol, bilirubin and other organic molecules.20 Bile is stored and concentrated in the gall bladder. During digestion, bile is secreted into the duodenum. Bile salts are the main component of bile. The most abundant bile salts in humans are cholate and deoxycholate. Bile salts cause emulsification and solubilization of lipids, and their detergent activity endows bile with strong antimicrobial capacity. Enteric bacteria are intrinsically resistant to both bile and individual bile salts.21 However, specific mutations can render E. coli and Salmonella sensitive to the antimicrobial effects of bile salts. In Salmonella enterica serovar Typhimurium, null damX alleles cause sensitivity to deoxycholate.11 Similar results have been reported in E. coli.10 Interestingly, a similar phenotype is observed in mutants lacking the downstream gene, dam.22 However, several lines of evidence suggest that bile sensitivity in damX mutants is not due to a polar effect on dam expression:
Complementation of a damX mutation with a functional version of DamX restores bile resistance to wild type levels in S. enterica and E. coli.10,11
Sensitivity to bile in S. enterica dam mutants is suppressed by inactivation of either the MutHLS system or the AsmA protein.22,23 In S. enterica damX mutants, however, bile sensitivity is suppressed by inactivation of AsmA, but not by inactivation of the MutHLS system, suggesting that the causes of bile sensitivity in dam and damX mutants are different.11
Bile exerts its primary effects on cell membranes.24 Most bile-sensitive mutants have defects in proteins involved in the maintenance of envelope integrity.24 This may explain the bile sensitive phenotype of damX mutants.
Other phenotypes.
Overexpression of damX alters biofilm formation in E. coli rendering a “filamentous biofilm”, usually associated with elongated cells.25 Thus, altered biofilm formation may be a side effect of cellular filamentation when damX is overexpressed. Filamentous biofilms are also observed when certain cell division proteins are overproduced.25
Two additional phenotypes have been associated with damX mutations: (i) constitutive induction of the E. coli SOS response;26 and (ii) deficient invasion by Salmonella typhi.27 In both cases, however, the damX mutation had been generated by transposon insertion, potentially polar on dam expression. In both E. coli and Salmonella, lack of Dam methylase has pleiotropic consequences including constitutive induction of the SOS response.28,29 O'Reilly and Kreuzer (2004) considered that polarity on dam might indeed explain constitutive SOS induction. Furthermore, the same authors observed that insertions in dam produced stronger SOS induction than insertions in damX. The invasion defect observed in the S. typhi damX mutant might likewise be due to a polar effect on dam expression. Salmonella dam mutants show reduced invasion of epithelial cells,30 which correlates with reduced expression of genes necessary for invasion.31,32 Thus, if the insertion in damX had a polar effect on dam expression, invasion would be reduced.
Cytokinesis and envelope integrity: a dual role for DamX?
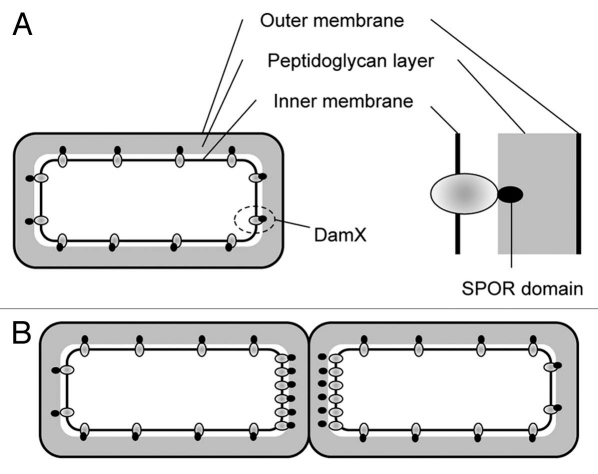
Two functions have been attributed to DamX: participation in cytokinesis as a component of the septal ring and contribution to bile resistance. Many bile-sensitive mutants described in E. coli and Salmonella have defects in envelope integrity. However, none of the bile-sensitive mutants so far identified has defects in septal ring components. DamX is an inner membrane protein, and its presence in both dividing and non-dividing cells11 suggests that, besides its role in cytokinesis, DamX may contribute to the maintenance of envelope integrity (Fig. 2).
Figure 2.
Model of DamX localization in growing and non-growing cells. (A) In non-growing cells, DamX may be distributed evenly in the inner membrane. (B) During cell division, DamX accumulates in the septum, perhaps reflecting the affinity of the SPOR domain for septal peptidoglycan.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/12079
References
- 1.Lyngstadaas A, Løbner-Olesen A, Boye E. Characterization of three genes in the dam-containing operon of Escherichia coli. Mol Gen Genet. 1995;247:546–554. doi: 10.1007/BF00290345. [DOI] [PubMed] [Google Scholar]
- 2.Løbner-Olesen A, Marinus MG. Identification of the gene (aroK) encoding shikimic acid kinase I of Escherichia coli. J Bacteriol. 1992;174:525–529. doi: 10.1128/jb.174.2.525-529.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whipp MJ, Pittard AJ. A reassessment of the relationship between aroK- and aroL-encoded shikimate kinase enzymes of Escherichia coli. J Bacteriol. 1995;177:1627–1629. doi: 10.1128/jb.177.6.1627-1629.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millar G, Coggins JR. The complete amino acid sequence of 3-dehydroquinate synthase of Escherichia coli K12. FEBS Lett. 1986;200:11–17. doi: 10.1016/0014-5793(86)80501-4. [DOI] [PubMed] [Google Scholar]
- 5.Lacks S, Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977;114:153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- 6.Hattman S, Brooks JE, Masurekar M. Sequence specificity of the P1 modification methylase (M.eco P1) and the DNA methylase (M.eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978;126:367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- 7.Lyngstadaas A, Sprenger GA, Boye E. Impaired growth of an Escherichia coli rpe mutant lacking ribulose-5-phosphate epimerase activity. Biochim Biophys Acta. 1998;1381:319–330. doi: 10.1016/s0304-4165(98)00046-4. [DOI] [PubMed] [Google Scholar]
- 8.Lyngstadaas A, Løbner-Olesen A, Grelland E, Boye E. The gene for 2-phosphoglycolate phosphatase (gph) in Escherichia coli is located in the same operon as dam and at least five other diverse genes. Biochim Biophys Acta. 1999;1472:376–384. doi: 10.1016/s0304-4165(99)00146-4. [DOI] [PubMed] [Google Scholar]
- 9.Hall CV, Yanofsky C. Cloning and characterization of the gene for Escherichia coli tryptophanyl-transfer ribonucleic acid synthetase. J Bacteriol. 1981;148:941–949. doi: 10.1128/jb.148.3.941-949.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arends SJ, Williams K, Scott RJ, Rolong S, Popham DL, Weiss DS. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD and RlpA. J Bacteriol. 2010;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Garrido J, Cheng N, Garcia-Quintanilla F, Garcia-del Portillo F, Casadesus J. Identification of the Salmonella enterica damX gene product, an inner membrane protein involved in bile resistance. J Bacteriol. 2010;192:893–895. doi: 10.1128/JB.01220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H, et al. The pfam protein families database. Nucleic Acids Res. 2008;36:281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Løbner-Olesen A, Boye E, Marinus MG. Expression of the Escherichia coli dam gene. Mol Microbiol. 1992;6:1841–1851. doi: 10.1111/j.1365-2958.1992.tb01356.x. [DOI] [PubMed] [Google Scholar]
- 14.Jonczyk P, Hines R, Smith DW. The Escherichia coli dam gene is expressed as a distal gene of a new operon. Mol Gen Genet. 1989;217:85–96. doi: 10.1007/BF00330946. [DOI] [PubMed] [Google Scholar]
- 15.Arraj JA, Wu TH, Marinus MG. Expression of a DNA methylation (dam) gene in Escherichia coli K-12. Current Microbiology. 1990;20:133–136. [Google Scholar]
- 16.Wu TH, Grelland E, Boye E, Marinus MG. Identification of a weak promoter for the dam gene of Escherichia coli. Biochim Biophys Acta. 1992;1131:47–52. doi: 10.1016/0167-4781(92)90097-j. [DOI] [PubMed] [Google Scholar]
- 17.Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weiss DS. Bacterial cell division and the septal ring. Mol Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- 19.Adams DW, Errington J. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman AF. Bile secretion and enterohepatic circulation of bile acids. In: Feldman M, Scharschmidt BF, Sleisenger MH, editors. Sleisenger and fordtran's gastrointestinal disease. 6th ed. Philadelphia: W.B. Saunders & Co; 1998. [Google Scholar]
- 21.Gunn JS. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2000;2:907–913. doi: 10.1016/s1286-4579(00)00392-0. [DOI] [PubMed] [Google Scholar]
- 22.Prieto AI, Ramos-Morales F, Casadesus J. Bile-induced DNA damage in Salmonella enterica. Genetics. 2004;168:1787–1794. doi: 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto AI, Hernandez SB, Cota I, Pucciarelli MG, Orlov Y, Ramos-Morales F, et al. Roles of the outer membrane protein AsmA of Salmonella enterica in the control of marRAB expression and invasion of epithelial cells. J Bacteriol. 2009;191:3615–3622. doi: 10.1128/JB.01592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Tenorio E, Saeki T, Fujita K, Kitakawa M, Baba T, Mori H, et al. Systematic characterization of Escherichia coli genes/ORFs affecting biofilm formation. FEMS Microbiol Lett. 2003;225:107–114. doi: 10.1016/S0378-1097(03)00507-X. [DOI] [PubMed] [Google Scholar]
- 26.O'Reilly EK, Kreuzer KN. Isolation of SOS constitutive mutants of Escherichia coli. J Bacteriol. 2004;186:7149–7160. doi: 10.1128/JB.186.21.7149-7160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leclerc GJ, Tartera C, Metcalf ES. Environmental regulation of Salmonella typhi invasion-defective mutants. Infect Immun. 1998;66:682–691. doi: 10.1128/iai.66.2.682-691.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson KR, Wertman KF, Mount DW, Marinus MG. Viability of Escherichia coli K-12 DNA adenine methylase (dam) mutants requires increased expression of specific genes in the SOS regulon. Mol Gen Genet. 1985;201:14–19. doi: 10.1007/BF00397979. [DOI] [PubMed] [Google Scholar]
- 29.Torreblanca J, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium and a novel dam-regulated locus. Genetics. 1996;144:15–26. doi: 10.1093/genetics/144.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Del Portillo F, Pucciarelli MG, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion and M cell cytotoxicity. Proc Natl Acad Sci USA. 1999;96:11578–11583. doi: 10.1073/pnas.96.20.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balbontin R, Rowley G, Pucciarelli MG, Lopez-Garrido J, Wormstone Y, Lucchini S, et al. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:8160–8168. doi: 10.1128/JB.00847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Garrido J, Casadesus J. Regulation of Salmonella enterica pathogenicity island 1 (SPI-1) by DNA adenine methylation. Genetics. 2010;184:637–649. doi: 10.1534/genetics.109.108985. [DOI] [PMC free article] [PubMed] [Google Scholar]