Abstract
The study of probiotics and prebiotics is an expanding field of interest and scientific research that has resulted in insights related to the host immune response. Recent advances have naturally led to key questions. What are the specific probiotic components that mediate immunomodulation? Can we extrapolate the results of in vitro studies in animal and human trials? Which biomarkers and immune parameters should be measured in probiotic and prebiotic intervention studies? These questions were part of a discussion entitled “How Can Probiotics and Prebiotics Impact Mucosal Immunity” at the 2009 Annual Meeting of the International Scientific Association for Probiotics and Prebiotics (ISAPP). This review highlights recent knowledge about the modulation of mucosal immunity by probiotics and prebiotics, as well as considerations for measuring their effects on mucosal immunity. A list of biomarkers and immune parameters to be measured in human clinical trials is included.
Key words: probiotic, prebiotic, immunomodulation, immune, human trials, biomarkers, gastrointestinal tract, microbiota
Introduction
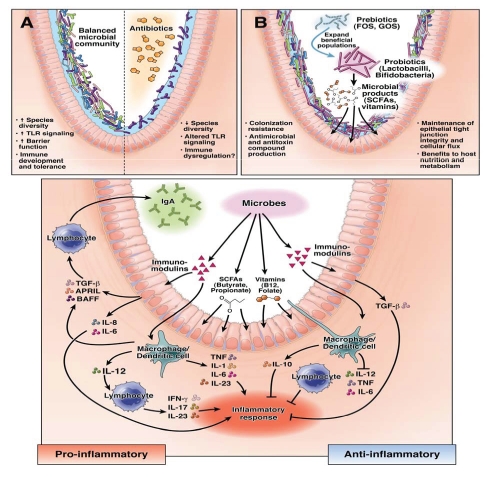
The immune system provides the first line of defense against pathogens and ingested toxins. A growing body of evidence suggests that host-microbial interactions may result in dysregulated mucosal immune responses, causing chronic inflammation such as Crohn disease or ulcerative colitis. A modest stimulation of the immune system by commensal bacteria may prevent infections. Immunomodulation is interpreted more broadly and includes antibodies, complement and cytokines, effects on gut barrier function and induction of antimicrobial compounds by the host. Microbes in the gastrointestinal tract (GIT) can exert numerous effects on different cells of the mucosal immune system and, in turn, induce the production of cytokines, which prime additional immune cells (Fig. 1). Depending on the immune stimulus, Toll-like receptors (TLRs) on the surfaces of immune cells are differentially stimulated and allow the immune system to discriminate between pathogens and the gut microbiota.1 The soluble cytoplasmic NOD-like receptors, NLRs, also mediate communication between the GIT and gut microbiota. The NLRs and TLRs act synergistically, resulting in the induction of immune cascades such as the NFκB pathway, which ultimately leads to the induction of chemokines and cytokines.2
Figure 1.
Microbial manipulation strategies and effects on intestinal biology. Reprinted from reference 72 with permission from Elsevier.
Probiotics, “live microorganisms which when administered in adequate amounts confer a health benefit on the host”3 and prebiotics have been used more or less successfully to improve the host immune response in different conditions. The most recent definition for prebiotics was defined at the 2008 ISAPP meeting, which states, “A dietary prebiotic is a selectively fermented ingredient that results in specific changes, in the consumption and/or activity of the gastrointestinal microbiota, thus conferring benefit(s) upon host health.” Without a doubt, our understanding of the mechanisms of action of probiotics and prebiotics has been facilitated by recent advances in genomics, transcriptomics, metabolomics and advances in studies of immune function. Studies of interest include investigations of interactions of probiotics and prebiotics with the host at the mucosal interface, including the GIT. In addition, bacterial components may be responsible for immunomodulation and by using transcriptomics transcriptional effects on both the host and probiotics may be determined.
Researchers are capable of studying immunological effects on the host after exposure to intact bacteria, cell surface-associated factors, metabolites and secreted proteins. These effects are dependent on the bacterial strain and ultimately the effector molecules produced by probiotics or beneficial microbes in general. Challenges include the determination of biomarkers and evaluation of the bases for human trials. What are the biomarkers and what do we consider positive results? What is the relevance of results obtained from in vitro studies, and how do the laboratory data compare to in vivo studies? What is the relevance of immunomodulation for clinical trials, and how can laboratory studies be linked to a measurable health benefit? The first part of this review discusses the effects of probiotics, their effector molecules and prebiotic compounds on mucosal immunity, while the second part addresses the extrapolation of the immunomodulatory effects of probiotics and prebiotics in vitro to in vivo models such as interventional studies in humans. Additional questions that require discussion and further research are outlined in Table 1. This review highlights the discussions and research reviewed by the discussion group: “How Can Probiotics and Prebiotics Impact Mucosal Immunity” from the 2009 Annual Meeting of the International Scientific Association for Probiotics and Prebiotics (ISAPP).
Table 1.
Key questions require further study and discussion
|
Current Knowledge About Probiotics and Prebiotics and Their Effects on Mucosal Immunity
Probiotics and probiotics-derived effector molecules; gut barrier function and immune defense.
Gut barrier function is vital for maintenance of gut health, with barrier dysfunction contributing to intestinal diseases including Crohn disease and irritable bowel syndrome (IBS). Probiotics and their effector molecules can influence the gut barrier by numerous methods including modulation of mucus production, reduction of bacterial adhesion, enhancement of tight junctions, enhancement of cell survival and induction of defensins or IgA. These effects can be accomplished by indirect influences on the permeability of tight junctions4 and direct alterations of the tight junction by modulation of tight junction proteins and protein distribution in the membrane.5–8 Moreover, activation of TLR2 [which responds to the presence of gram-positive cell wall components such as lipoteichoic acid (LTA) and peptidoglycan] by the gut microbiota is necessary for maintenance of gut homeostasis and protection from injury.9
A study by Mennigen et al.,8 demonstrated that the probiotic VSL#3 mixture protected the epithelial barrier in a murine model of colitis by preventing apoptosis and maintaining tight junction protein expression. Previous work demonstrated that conditioned media from Bifidobacterium infantis prevented a reduction in transepithelial resistance of intestinal epithelial cells and reduced ileal and colonic permeability in IL-10-deficient mice.10 Another study demonstrated that prior exposure of an intestinal epithelial cell line to viable Lactobacillus acidophilus, but not heat-inactivated L. acidophilus, limited the adverse effects induced by an Escherichia coli strain, such as a decline in transepithelial resistance, increased epithelial permeability and physiological dysfunction. Additionally, viable L. acidophilus reduced phosphorylation of tight junction proteins induced by E. coli.6 Two proteins, p40 and p75, were purified from the culture supernatant of Lactobacillus rhamnosus LGG, and both proteins prevented TNF-induced apoptosis and intestinal barrier disruption in colonic epithelial cells.11
The effects of probiotics and their effector molecules on barrier function in clinical trials are sparse. In one study, the efficacy of a placebo, VSL#3 and probiotic-derived sonicates, was studied for modulation of intestinal permeability in patients in the intensive care unit with multiple organ dysfunction syndrome.12 Patients that received VSL#3 exhibited a larger increase in serum IgA and IgG concentrations than the patients who received placebo or sonicates. However, no significant difference in intestinal permeability was observed between the patients who received VSL#3, probiotic sonicates and placebo.12 In another study, a short-term improvement in mucosal barrier function in patients with IBS was observed after administration of a probiotic fermented milk (Streptococcus thermophilus, Lactobacillus bulgaricus, L. acidophilus and Bifidobacterium longum) compared to a milk beverage containing no bacteria.13 These studies emphasize the importance of strain selection and viability. In addition, as noted in other studies with prebiotics and probiotics, effects on gut barrier function varied between strains and effector molecules.6,10,14 It is clear that probiotic immunomodulatory components consist of a varied array of effector molecules (Table 2), including surface layer proteins, cell wall polysaccharides, adhesins, teichoic acids and heat shock proteins. Other immunomodulatory components have been understudied or remain to be characterized. While in vitro work to identify important candidate effector molecules (Table 2) continues, the effects on mucosal immunity in human intervention studies still needs to be determined.
Table 2.
Potential effector molecules derived from probiotics
| Bacteria | Molecule | Effect | References |
| Lactobacillus reuteri | Secreted factors by L. reuteri biofilms | Suppression of human TNF production by LPS-activated monocytoid cells | 54 |
| L. reuteri | Secretion of reuterin (antimicrobial glycerol derivative) by L. reuteri biofilms | Secretion of reuterin by biofilms demonstrated | 54 |
| Intestinal bacteria | Butyrate (a short chain fatty acid) | Increased intestinal barrier function in Caco2 cell lines | 55 |
| Lactobacillus acidophilus NCFM | S layer protein | DC-SIGN ligand involved in the modulation of DCs and T cells functions | 56 |
| Faecalibacterium prausnitzii | F. prausnitzii supernatant | Reduced the severity of TNBS colitis and tended to correct the dysbiosis associated with TNBS colitis | 57 |
| Lactobacillus casei Shirota | Cell wall polysaccharide moiety | Inhibit macrophage and splenocyte activity | 58 |
| Bifidobacteria infantis | B. infantis conditioned medium | Reduced colonic permeability in mice and attenuated inflammation in IL-10-deficient mice | 10 |
| Lactobacillus helveticus | S layer protein | Inhibition of Escherichia coli 0157:H7 adherence to intestinal cell lines | 59 |
| Lactobacillus rhamnosus GG | Modified teichoic acids | Increased sensitivity to gastric juice and human beta-defensin-2 but no difference in immunomodulation | 60 |
| L. rhamnosus GG | P40 and P75 secreted proteins | Prevention of cytokine-induced apoptosis in human and mouse intestinal epithelial cells | 11 |
| Lactobacillus salivarius UCC118 | Secretion of bacteriocin Abp118 | Protected mice against infection with the pathogen Listeria monocytogenes | 61 |
| Escherichia coli Nissle 1917 | Flagellin | Induction of beta-defensin-2 in intestinal cell lines | 62 |
| Lactobacillus johnsonii La1 | GroEL | Bind to components of the gastrointestinal mucosa and stimulates interleukin-8 secretion in macrophages and HT29 cells in a CD14-dependent mechanism | 63 |
| Lactobacillus delbrueckii ssp. bulgaricus OLL1073R | Extracellular polysaccharides (EPS) | Stimulation of mouse splenocytes and significant increase in interferon-gamma production | 64 |
| Lactobacillus plantarum NCIMB8826 | Modified teichoic acids | Protective against colitis | 23 |
| L. johnsonii NCC533 | Adhesin; elongation factor Tu (EF-Tu) | Mucin binding, proinflammatory response in the presence of soluble CD14 | 65 |
Live bacteria, dead bacteria or bacterial supernatants.
Effects on host gene expression and mucosal immune responses. Studies have been performed using live, dead and bacterial supernatants to assess the mucosal immune response. In particular, probiotic Lactobacillus plantarum WCFS1 has been studied using in situ-based -omic models in the GIT. Gene expression of beneficial bacteria15 and the mammalian host16,17 has been studied in parallel. Gene expression profiles in ceca of germ-free mice fed a standard low fat or western diet were investigated to determine the effects of host diet on bacterial (probiotic) gene expression in the GIT.15 The western diet was high in simple sugars and fat, and the standard diet (low fat diet) was high in complex plant polysaccharides.15 In addition to transcriptomics studies, colonization levels of L. plantarum were measured in mice fed both diet types, and was determined to be ten times higher in mice receiving the standard feed compared to the western diet. Gene expression was compared to a reference group of differentially expressed genes, namely mid-logarithmic L. plantarum grown in vitro in MRS, chemically defined medium and chow medium. The main results from this study indicated that bacterial genes involved in carbohydrate metabolism and cell surface functions were upregulated in mice fed the standard diet compared to the culture media. In addition, it was determined that the western diet resulted in a nutritionally restricted, growth-limiting environment for L. plantarum in the distal gut.15 In mice fed both diets, gene sets encoding cell surface-related functions were differentially expressed, which included genes involved in D-alanylation of LTA, a probiotic effector molecule (Table 2). The authors suggested that the probiotic bacteria modified its gene expression to reduce levels of LTA on the cell surface, as a means to restrict exposure to components of the host immune system in the GIT of mice fed both diet types.
The growth phase and viability of the probiotic cultures is also a factor that varies between reported experiments. For example, in a study by van Baarlen and co-workers,17 differences in gene expression in the duodenum of healthy humans was observed depending on the growth phase and viability of the probiotic L. plantarum WCFS1. Individuals were administered logarithmic, stationary or dead cells and samples were taken from the duodenum for analysis after 6 hours. Results from this small randomized, double-blind, placebo-controlled crossover study identified gene expression patterns at the mucosal surface and within cellular pathways that were related to immune tolerance. While all three conditions resulted in the induction of genes involved in the immune response, differences in the type of induction and number of genes induced were observed between logarithmic and stationary phase cells. Logarithmic phase cells induced a response targeted towards metabolic functions such as nucleic acid metabolism and cytoplasmic organization. However, stationary phase bacteria resulted in induction of genes such as the NFκB and JUN transcription factors involved in the establishment of immune tolerance.17 Interestingly, the dead cells (heat-killed stationary phase) induced the highest fold changes, and the mucosal responses to dead and stationary phase bacteria were comparable to each other but different than the responses to logarithmic phase bacteria. These studies demonstrated the importance of reference sets of gene expression data used for comparisons in vivo, in addition to diet, growth media and growth phase of probiotics. Factors such as the stability of each probiotic strain, expression and stability of effector molecules, growth phase of the probiotic strain and site of action of probiotic-derived effector molecules should be considered for in vivo studies. Finally, can immunomodulation in the laboratory be connected with health benefits in humans and animals?
Prebiotics and mucosal immunity.
Prebiotic benefits include modulation of the gut microbiota, increased mineral adsorption, modulation of lipid metabolism and inhibition of pathogens.18 While studies of prebiotics have documented their effects on mucosal immunity, the majority of these reports included animal models, with relatively few human studies published to date. Some published studies have included other factors such as antioxidants and vitamins, which may also contribute to mucosal immunity. In studies with synbiotics, it is difficult to distinguish the effects of prebiotics from that of probiotics.
A few studies examined immunomodulation by dietary prebiotics (reviewed in ref. 19). Prebiotic supplementation increased fecal secretory IgA and postnatal immune development in infants.20 In disease states, moderate effects were seen in patients with Crohn disease,21 but results were encouraging enough to warrant further exploration. Because prebiotic compounds per definition modulate the composition of the gut microbiota, it is difficult to know whether these effects are direct or are the consequences of a shift of certain bacterial groups. Clinical trials have included an analysis of microbial composition and associated effects on short chain fatty acids (SCFA). Products of bacteria that have been reported to show anti-inflammatory properties include SCFAs (Table 2).22 Other bacterial metabolites that could be responsible for the modulation of the immune system, remain to be characterized.
Additional animal and human studies are needed, as there is no clear consensus on the effects of prebiotics on mucosal immunity. Points to consider include the nature of prebiotics and consequent indirect interactions such as system-wide changes of gut microbiota and the subsequent effects on mucosal immunity and/or health benefits and direct interactions of prebiotics with the mucosa. The challenge is determining these effects on health and designing better trials including the use of biomarkers (discussed below).
Can We Extrapolate Immunomodulatory Effects of Probiotics and Prebiotics In Vitro to Humans?
Factors influencing the immune response in vivo and in human clinical trials.
Extrapolation of immunomodulatory effects found in the laboratory and in animal studies with outcomes in human trials presents a difficult challenge. Immunomodulatory effects conferred by L. plantarum WCFS1 in vitro,23 in animal models,16,23 but also in humans24,17 highlights the difficulties of comparing similar effects by a single strain in different contexts. A few examples show correlations between in vitro and in vivo immunomodulatory properties of lactic acid bacteria.23,25,26 The study by Foligne et al.25 for instance, demonstrated the possibility of translating results from an in vitro assay with peripheral blood mononuclear cells (PBMCs) obtained from different donors, to a murine model of colitis using the IL-10/IL-12 ratio. Generally, the discrepancies between in vitro and in vivo results observed in published trials can be partly explained by the host contribution (genetic factors, different baseline immune functions between individuals, microbiome diversity, differences in the body sites targeted, intra-person variation) as well as environmental factors (diet, stress, etc.) partially controlled by each individual. Different factors can affect outcomes in studies reporting immunomodulatory effects of probiotic or prebiotic interventions in vivo.
Variation between individuals and effects on host response. Genetic factors. Genetic variability strongly influences the immune response of the host. Specific polymorphisms in genes encoding antigens (human leukocytes27) or proteins involved in the immune response can influence quantities of cytokines28 in a healthy population or disease state. A growing body of evidence indicates that individuals respond to bacterial signals (TLR ligands) differently due to single nucleotide polymorphisms (SNPs) within TLR genes.29 A frameshift mutation in NOD2 is a strong risk factor in patients with inflammatory bowel disease (Crohn disease), and this evidence supports the importance of SNPs and inter-individual differences in TLR signaling.30,31
Microbiome. Preliminary observations that all vertebrates have a microbiome that co-evolved with the host, resulted in the hypothesis that the adaptive immune system (memory-based) evolved in the vertebrate lineage because of the intimate co-existence with complex communities of beneficial microbes.32 Whether this is true or not still needs to be demonstrated, but it certainly highlights the complex links between our microbiome and our immune system. Because we are in the early stages of characterizing the phylogenetic or functional core microbiomes in human and animal models, it is difficult to fully understand relationships between the host microbiota and immune system. However, the relative paucity of microbes such as Faecalibacterium prausnitzii in inflammatory bowel disease (Crohn disease) has been demonstrated by different studies.33 Baseline immune functions and microbiomes may differ and depend on each other so that aggregate responses and disease susceptibilities in individuals may represent a combination of microbial composition, microbial functions and host genotypes.
Influence of environmental factors controlled by the host such as diet. The gut microbiota is a dynamic environment, and changes induced by the diet, the uptake of antibiotics or even physiologic stresses may induce rapid changes in microbial composition and functions,34 with a concomitant impact on the host immune response. Rapid changes in microbial composition have been well documented with a high fat diet.35 These results should encourage investigators to control diets of subjects and support the use of standardized diets when measuring the host immune response. Dietary components may have a direct effect on the response of the immune system and vitamins, for example, may have an effect on cytokine production.36 To make the story more complex, effects of dietary components may be partly determined by genetic polymorphisms of cytokines such as TNF.37
Consideration of body site contribution. Small intestine versus colon. Immune parameters measured in different body sites will certainly be different and may have an impact on results and outcomes of studies with probiotics and prebiotics. In the gut, the colon has been more extensively investigated than the small intestine or other mucosal sites of the GIT such as mouth or stomach,38 and certainly merits further investigation. As the composition of the mucus layer and mucus barrier is different at these sites, the contribution of the mucus layer and its interaction with probiotics and prebiotics or other members of the gut microbiota should be considered and investigated further.
Which biomarkers and immune parameters shall we use in probiotic/prebiotic interventions in humans?
No universal biomarkers have been identified to assess how probiotics and prebiotics impact mucosal immunity. However, specific biomarkers can be defined for certain populations and disease states, and are summarized in Table 3. Measuring these parameters is still challenging in healthy people, and limited by the ability to analyze markers only in blood, saliva, fecal samples or urine. In an experimental setting with disease patients, it is usually possible to get biopsies from the gut. New techniques and potential biomarkers are still emerging and may offer promising alternatives in the future. For instance, recent discoveries on the role of C-reactive protein (CRP) isoforms and their role in inflammation could facilitate an improved understanding of the importance of different isoforms.39 Metabolites that distinguish patients with IBD, IBS or other gut diseases have emerged.39,40 Choices of immune markers will certainly depend on the population studies and diseases studied in probiotic and prebiotic interventions.
Table 3.
Possible immune biomarkers and parameters for assessment of mucosal and systemic immunity in vivo in response to probiotic or prebiotic interventions
| Marker | Advantages | Drawbacks | References |
| Cytokines (TNF, IL6, IL10, IL12) FoxP3 | Easy to measure | Level in blood may not reflect levels in other body sites | 66 |
| CRP and acute phase reactants | Easy to measure | Measure other outcome | 39 |
| Different conformational change | |||
| Antibody (IgA) | Easy to measure | Not helpful without immune challenge | 67 |
| Gene expression profile in tissues and peripheral blood (targeted vs. global) | Extensive survey of gene activated | Difficulties for obtaining tissues in healthy humans | 17, 24 |
| Proteomics and metabolomics in urine and blood and fecal water | Extensive survey of metabolites produced | Difficult to analyze | 44, 68, 69 |
| Lack of references | |||
| Calprotectin | Easy to measure in stool | Standardization necessary | 70, 71, 67 |
Consideration of interventional trials with probiotics and prebiotics.
Choices of animal models. Different animal models have been used to study the effects of probiotics and prebiotics on mucosal immunity. Effects in mouse models of IBD have included IL-10 knockout mice, trinitrobenzene sulfonic acid (TNBS)- or dextran sodium sulphate (DSS)-induced colitis. Based on the characterization of the microbiota in different knockout mice and wild-type mice using genotyping and 16S rRNA gene analyses, large differences in the composition of the microbiota can be observed in different animal models,41 as well as between animal models obtained from different vendors.42 Differences in microbial composition between C57BL/6 mice obtained from the Jackson laboratory and Taconic laboratory were investigated recently using a high density microarray (PhyloChip). These mice differed with respect to the relative proportions of Th17 cells that contribute to inflammation and may be mediators in immune protection and immunopathology. Comparative analyses of 766 bacterial taxa detected in both groups of mice demonstrated large differences in the relative abundance of more than half of the taxa, with two taxa [Lactobacillus murinus and segmented filamentous bacterium (SFB)] being more than 25-fold more abundant in Taconic mice. The Taconic mice in this study had elevated proportions of Th17 cells.43 Most importantly, it was demonstrated that colonization of mice housed in the Jackson laboratory with one SFB species (Arthromitus spp.), was sufficient to induce the appearance of Th17 cells derived from a CD4+ T helper cell lineage.43 Furthermore, this colonization was correlated with enhanced expression of genes associated with inflammation and antimicrobial defenses, resulting in enhanced resistance to the murine intestinal pathogen Citrobacter rodentium. Thus, characterizing the microbiomes in animals at sites where immune parameters will be measured, before intervention, may provide further insights into the possible roles of certain microbes and outcomes with probiotics or prebiotics. Murine models with “humanized” microbiomes offer promising systems for evaluation of microbial effects on host immune responses. Thus, humanized microbiome murine models (for example with a baby microbiota)44 have similarities with that of formula-fed neonates.45 However, the relevance of these humanized rodent models to human diseases remains to be seen.
Response to vaccine challenges. Vaccine challenges are particularly useful to assess the efficacy of probiotics that may enhance the immune status in a healthy population. Probiotics enhance the immunogenicity of several vaccines including rotavirus, influenza, poliovirus, hepatitis B and pneumococcus.46–48 Measurement of antibody responses to vaccines is straightforward, and such human studies may provide excellent opportunities to evaluate the immunologic efficacy of probiotics.
Non-responders. Separating responders from non-responders may facilitate interpretations of effects of the probiotics or prebiotics on the immune system. Only a few in vivo trials have been performed.49
Influence of age, gender, diet. Because immune parameters differ according to age, gender, health status, activity and dietary habits, human studies should use crossover placebo-controlled design while trying to match and compare different groups.
Core or functional microbiomes. In the future, integrated approaches using microbiology, genomics, transcriptomics and proteomics tools would certainly highlight the mechanisms and the microbes that may influence the immune system.50 It is still debatable whether a core microbiome exists at the phylogenetic level51 or functional level.52 However, recent studies such as those by Gordon and colleagues suggest the existence of a functional core at the level of shared gene families, which suggests that different species could fulfill similar functional roles in the GIT. As pointed out by Shop et al.53 however, an interesting follow-up question regarding the outcome of this study is whether the lack of shared phylotypes is still as pronounced if rare populations are taken into account.
Summary and Conclusions
Research scientists face numerous challenges including the demonstration and articulation of specific beneficial effects and associated mechanisms by probiotics and prebiotics. A clear definition of human health, if one exists, is a central consideration. What is considered “healthy” as a baseline condition may vary depending on age, gender, ethnicity, diet and numerous environmental factors. Although we have some answers with certain parameters (leukocytes, cytokines), baseline parameters including quantitative “normal” ranges of immune biomarkers remain to be defined. Non-invasive techniques that evaluate blood, fecal water or urine metabolites need to be further developed. Many studies are needed to understand various metabolites and how they may provide measurable standards for interventional studies. Additionally, the involvement of neuro-immune interactions in the GIT is an understudied area of research with respect to probiotics and prebiotics.
The difficulty to establish health claims such as “strengthen the immune response” for probiotics has been illustrated recently in Europe by the absence of health claims related to the stimulation of the immune system authorized for probiotic foods by the European Food Safety Authority (http://www.isapp.net/docs/ISAPP_responds_to_EFSA_oct09.pdf). A growing body of evidence has documented the beneficial effects of probiotics and prebiotics in disease treatment and management studies. However, long-term studies are still needed to identify their prophylactic effects towards inflammatory disorders in different populations.
Before using probiotics in interventional studies, many factors remain to be considered. For example, viable cells are generally more effective at stimulating adaptive immunity, and the method of cell killing should be considered if nonviable cells are used. If cell supernatants are used, the active component(s) should be purified and the stability and physiologic effects of these compounds must be considered. Doses of bacteria and growth phase at time of harvest are additional considerations in tandem with traditional methods of determining strain robustness or functional effects. On the host side, genetic influences, site of action, the mucus barrier, route of administration, diet and microbial composition of the host contribute to results of interventional trials published to date. In future trials, individual factors that potentially affect the efficacy of probiotics and prebiotics must be addressed. Continuing advancement in technologies, knowledge of the immune system, gut microbiota and improved biomarkers are essential to making human interventional studies with probiotics and prebiotics successful.
Acknowledgements
The authors would like to thank all participants of the discussion group and in particular the investigators who shared their presentations from the meeting: Peter Bron, Michael Hsieh, Flavia Indrio, Bob Rastall, Karen Madsen and Fang Yan. S.O.F. would like to thank Todd R. Klaenhammer. The work in the J.V. laboratory is supported by the National Institutes of Diabetes, Digestive and Kidney Disease (NIDDK) (R01 DK56338 and P30 DK065075) and National Center for Complementary and Alternative Medicine (NCCAM) (R01 AT004326).
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/12924
Conflicts of Interest
J.V. receives research support from Biogaia AB and serves as a scientific advisor for Danone.
References
- 1.Lebeer S, Vanderleyden J, De Keersmaecker SC. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol. 2010;8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 2.Netea MG, van de Veerdonk FL, Kullberg BJ, Van der Meer JW, Joosten LA. The role of NLRs and TLRs in the activation of the inflammasome. Expert Opin Biol Ther. 2008;8:1867–1872. doi: 10.1517/14712590802494212. [DOI] [PubMed] [Google Scholar]
- 3.FAO/WHO, author. Guidelines for the Evaluation of Probiotics in Food. London Ontario, Canada: 2002. [Google Scholar]
- 4.Garcia-Lafuente A, Antolin M, Guarner F, Crespo E, Malagelada JR. Modulation of colonic barrier function by the composition of the commensal flora in the rat. Gut. 2001;48:503–507. doi: 10.1136/gut.48.4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madsen K, Cornish A, Soper P, McKaigney C, Jijon H, Yachimec C, et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. doi: 10.1053/gast.2001.27224. [DOI] [PubMed] [Google Scholar]
- 6.Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC) Gut. 2003;52:988–997. doi: 10.1136/gut.52.7.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol. 2007;9:804–816. doi: 10.1111/j.1462-5822.2006.00836.x. [DOI] [PubMed] [Google Scholar]
- 8.Mennigen R, Nolte K, Rijcken E, Utech M, Loeffler B, Senninger N, et al. Probiotic mixture VSL#3 protects the epithelial barrier by maintaining tight junction protein expression and preventing apoptosis in a murine model of colitis. Am J Physiol Gastrointest Liver Physiol. 2009;296:1140–1149. doi: 10.1152/ajpgi.90534.2008. [DOI] [PubMed] [Google Scholar]
- 9.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, et al. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol. 2008;295:1025–1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 11.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alberda C, Gramlich L, Meddings J, Field C, McCargar L, Kutsogiannis D, et al. Effects of probiotic therapy in critically ill patients: a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr. 2007;85:816–823. doi: 10.1093/ajcn/85.3.816. [DOI] [PubMed] [Google Scholar]
- 13.Zeng J, Li YQ, Zuo XL, Zhen YB, Yang J, Liu CH. Clinical trial: effect of active lactic acid bacteria on mucosal barrier function in patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2008;28:994–1002. doi: 10.1111/j.1365-2036.2008.03818.x. [DOI] [PubMed] [Google Scholar]
- 14.Nissen L, Chingwaru W, Sgorbati B, Biavati B, Cencic A. Gut health promoting activity of new putative probiotic/protective Lactobacillus spp. strains: a functional study in the small intestinal cell model. Int J Food Microbiol. 2009;135:288–294. doi: 10.1016/j.ijfoodmicro.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 15.Marco ML, Peters TH, Bongers RS, Molenaar D, van Hemert S, Sonnenburg JL, et al. Lifestyle of Lactobacillus plantarum in the mouse caecum. Environ Microbiol. 2009;11:2747–2757. doi: 10.1111/j.1462-2920.2009.02001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marco ML, Bongers RS, de Vos WM, Kleerebezem M. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl Environ Microbiol. 2007;73:124–132. doi: 10.1128/AEM.01475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Baarlen P, Troost FJ, van Hemert S, van der Meer C, de Vos WM, de Groot PJ, et al. Differential NFkappaB pathways induction by Lactobacillus plantarum in the duodenum of healthy humans correlating with immune tolerance. Proc Natl Acad Sci USA. 2009;106:2371–2376. doi: 10.1073/pnas.0809919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steed H, Macfarlane S. Mechanisms of Prebiotic Impact on Health. In: DC, Rastall RA, editors. Prebiotics and Probiotics Science and Technology. New York: Springer; 2009. [Google Scholar]
- 19.Seifert S, Watzl B. Inulin and oligofructose: review of experimental data on immune modulation. J Nutr. 2007;137:2563–2567. doi: 10.1093/jn/137.11.2563S. [DOI] [PubMed] [Google Scholar]
- 20.Bakker-Zierikzee AM, Tol EA, Kroes H, Alles MS, Kok FJ, Bindels JG. Faecal SIgA secretion in infants fed on pre- or probiotic infant formula. Pediatr Allergy Immunol. 2006;17:134–140. doi: 10.1111/j.1399-3038.2005.00370.x. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay JO, Whelan K, Stagg AJ, Gobin P, Al-Hassi HO, Rayment N, et al. Clinical, microbiological, and immunological effects of fructo-oligosaccharide in patients with Crohn's disease. Gut. 2006;55:348–355. doi: 10.1136/gut.2005.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, et al. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci USA. 2005;102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Troost FJ, van Baarlen P, Lindsey P, Kodde A, de Vos WM, Kleerebezem M, et al. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genom. 2008;9:374. doi: 10.1186/1471-2164-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foligne B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S, et al. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol. 2007;13:236–243. doi: 10.3748/wjg.v13.i2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena JA, Rogers AB, Ge Z, Ng V, Li SY, Fox JG, et al. Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect Immun. 2005;73:912–920. doi: 10.1128/IAI.73.2.912-920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howell WM, Calder PC, Grimble RF. Gene polymorphisms, inflammatory diseases and cancer. Proc Nutr Soc. 2002;61:447–456. doi: 10.1079/pns2002186. [DOI] [PubMed] [Google Scholar]
- 28.Yaqoob P, Newsholme EA, Calder PC. Comparison of cytokine production in cultures of whole human blood and purified mononuclear cells. Cytokine. 1999;11:600–605. doi: 10.1006/cyto.1998.0471. [DOI] [PubMed] [Google Scholar]
- 29.Schroder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 30.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 31.Hampe J, Cuthbert A, Croucher PJ, Mirza MM, Mascheretti S, Fisher S, et al. Association between insertion mutation in NOD2 gene and Crohn's disease in German and British populations. Lancet. 2001;357:1925–1928. doi: 10.1016/S0140-6736(00)05063-7. [DOI] [PubMed] [Google Scholar]
- 32.McFall-Ngai M. Adaptive immunity: care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 33.Cucchiara S, Iebba V, Conte MP, Schippa S. The microbiota in inflammatory bowel disease in different age groups. Dig Dis. 2009;27:252–258. doi: 10.1159/000228558. [DOI] [PubMed] [Google Scholar]
- 34.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scalabrino G. Vitamin-regulated cytokines and growth factors in the CNS and elsewhere. J Neurochem. 2009;111:1309–1326. doi: 10.1111/j.1471-4159.2009.06417.x. [DOI] [PubMed] [Google Scholar]
- 37.Grimble RF, Howell WM, O'Reilly G, Turner SJ, Markovic O, Hirrell S, et al. The ability of fish oil to suppress tumor necrosis factor alpha production by peripheral blood mononuclear cells in healthy men is associated with polymorphisms in genes that influence tumor necrosis factor alpha production. Am J Clin Nutr. 2002;76:454–459. doi: 10.1093/ajcn/76.2.454. [DOI] [PubMed] [Google Scholar]
- 38.Lam EK, Tai EK, Koo MW, Wong HP, Wu WK, Yu L, et al. Enhancement of gastric mucosal integrity by Lactobacillus rhamnosus GG. Life Sci. 2007;80:2128–2136. doi: 10.1016/j.lfs.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Eisenhardt SU, Thiele JR, Bannasch H, Stark GB, Peter K. C-reactive protein: how conformational changes influence inflammatory properties. Cell Cycle. 2009;8:3885–3892. doi: 10.4161/cc.8.23.10068. [DOI] [PubMed] [Google Scholar]
- 40.Kajander K, Myllyluoma E, Kyronpalo S, Rasmussen M, Sipponen P, Mattila I, et al. Elevated pro-inflammatory and lipotoxic mucosal lipids characterise irritable bowel syndrome. World J Gastroenterol. 2009;15:6068–6074. doi: 10.3748/wjg.15.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pena JA, Li SY, Wilson PH, Thibodeau SA, Szary AJ, Versalovic J. Genotypic and phenotypic studies of murine intestinal lactobacilli: species differences in mice with and without colitis. Appl Environ Microbiol. 2004;70:558–568. doi: 10.1128/AEM.70.1.558-568.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. doi: 10.1038/msb4100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69:1035–1045. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 46.Soh SE, Ong DQ, Gerez I, Zhang X, Chollate P, Shek LP, et al. Effect of probiotic supplementation in the first 6 months of life on specific antibody responses to infant Hepatitis B vaccination. Vaccine. 2010;28:2577–2579. doi: 10.1016/j.vaccine.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 47.Isolauri E, Joensuu J, Suomalainen H, Luomala M, Vesikari T. Improved immunogenicity of oral D × RRV reassortant rotavirus vaccine by Lactobacillus casei GG. Vaccine. 1995;13:310–312. doi: 10.1016/0264-410x(95)93319-5. [DOI] [PubMed] [Google Scholar]
- 48.de Vrese M, Rautenberg P, Laue C, Koopmans M, Herremans T, Schrezenmeir J. Probiotic bacteria stimulate virus-specific neutralizing antibodies following a booster polio vaccination. Eur J Nutr. 2005;44:406–413. doi: 10.1007/s00394-004-0541-8. [DOI] [PubMed] [Google Scholar]
- 49.Reiff C, Delday M, Rucklidge G, Reid M, Duncan G, Wohlgemuth S, et al. Balancing inflammatory, lipid and xenobiotic signaling pathways by VSL#3, a biotherapeutic agent, in the treatment of inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:1721–1736. doi: 10.1002/ibd.20999. [DOI] [PubMed] [Google Scholar]
- 50.Reiff C, Kelly D. Inflammatory bowel disease, gut bacteria and probiotic therapy. Int J Med Microbiol. 2010;300:25–33. doi: 10.1016/j.ijmm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 51.Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 52.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tschop MH, Hugenholtz P, Karp CL. Getting to the core of the gut microbiome. Nat Biotechnol. 2009;27:344–346. doi: 10.1038/nbt0409-344. [DOI] [PubMed] [Google Scholar]
- 54.Jones SE, Versalovic J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009;9:35. doi: 10.1186/1471-2180-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci USA. 2008;105:19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasuda E, Serata M, Sako T. Suppressive effect on activation of macrophages by Lactobacillus casei strain Shirota genes determining the synthesis of cell wall-associated polysaccharides. Appl Environ Microbiol. 2008;74:4746–4755. doi: 10.1128/AEM.00412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson-Henry KC, Hagen KE, Gordonpour M, Tompkins TA, Sherman PM. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol. 2007;9:356–367. doi: 10.1111/j.1462-5822.2006.00791.x. [DOI] [PubMed] [Google Scholar]
- 60.Perea Velez M, Verhoeven TL, Draing C, Von Aulock S, Pfitzenmaier M, Geyer A, et al. Functional analysis of D-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2007;73:3595–3604. doi: 10.1128/AEM.02083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corr SC, Li Y, Riedel CU, O'Toole PW, Hill C, Gahan CG. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlee M, Wehkamp J, Altenhoefer A, Oelschlaeger TA, Stange EF, Fellermann K. Induction of human beta-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect Immun. 2007;75:2399–2407. doi: 10.1128/IAI.01563-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, Corthesy-Theulaz IE. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect Immun. 2006;74:425–434. doi: 10.1128/IAI.74.1.425-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Makino S, Ikegami S, Kano H, Sashihara T, Sugano H, Horiuchi H, et al. Immunomodulatory effects of polysaccharides produced by Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J Dairy Sci. 2006;89:2873–2881. doi: 10.3168/jds.S0022-0302(06)72560-7. [DOI] [PubMed] [Google Scholar]
- 65.Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthesy-Theulaz IE. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immun. 2004;72:2160–2169. doi: 10.1128/IAI.72.4.2160-2169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwon HK, Lee CG, So JS, Chae CS, Hwang JS, Sahoo A, et al. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc Natl Acad Sci USA. 2010;107:2159–2164. doi: 10.1073/pnas.0904055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohan R, Koebnick C, Schildt J, Mueller M, Radke M, Blaut M. Effects of Bifidobacterium lactis Bb12 supplementation on body weight, fecal pH, acetate, lactate, calprotectin and IgA in preterm infants. Pediatr Res. 2008;64:418–422. doi: 10.1203/PDR.0b013e318181b7fa. [DOI] [PubMed] [Google Scholar]
- 68.Williams HR, Cox IJ, Walker DG, North BV, Patel VM, Marshall SE, et al. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol. 2009;104:1435–1444. doi: 10.1038/ajg.2009.175. [DOI] [PubMed] [Google Scholar]
- 69.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Konikoff MR, Denson LA. Role of fecal calprotectin as a biomarker of intestinal inflammation in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:524–534. doi: 10.1097/00054725-200606000-00013. [DOI] [PubMed] [Google Scholar]
- 71.Rouge C, Piloquet H, Butel MJ, Berger B, Rochat F, Ferraris L, et al. Oral supplementation with probiotics in very-low-birth-weight preterm infants: a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr. 2009;89:1828–1835. doi: 10.3945/ajcn.2008.26919. [DOI] [PubMed] [Google Scholar]
- 72.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–2031. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]